Abstract
Here, we present the first complete mitochondrial genome within the gastropod family Pachychilidae, using the viviparous freshwater snail Tylomelania sarasinorum. This species is a representative member of the lacustrine Tylomelania radiations of the Malili-Lakes-System (Sulawesi, Indonesia). The mitochondrial genome was 16,632 bp long and contained 13 protein-coding genes, 2 rRNA genes and 22 tRNA genes. A pronounced A + T bias was observed with an overall base composition of 29.5% A, 35.7% T, 18.3% G and 16.6% C. Tylomelania sarasinorum exhibited a novel mitochondrial gene arrangement, differing from all Caenogastropoda mitochondrial genomes published to date.
Keywords: Gastropoda, Mollusca, Pachychilidae, Tylomelania sarasinorum
Tylomelania (Caenogastropoda: Cerithioidea: Pachychilidae) is a genus of viviparous freshwater snails endemic to Sulawesi, Indonesia (von Rintelen et al. 2010). Renowned for their adaptive radiations in the ancient Malili Lakes System, Tylomelania represent a model system for freshwater invertebrate speciation (von Rintelen et al. 2010, 2014). While Caenogastropoda are the most speciose group of extant gastropods with ∼85,000 species (Colgan et al. 2007), the phylogeny of gastropods in general, and Caenogastropoda in particular, is still a matter of debate (Colgan et al. 2007; Strong et al. 2011). Here, we present the mitochondrial genome of Tylomelania sarasinorum (Kruimel 1913), which constitutes the first complete mitochondrial genome within the family Pachychilidae and the third within the superfamily Cerithioidea (Zeng et al. 2014).
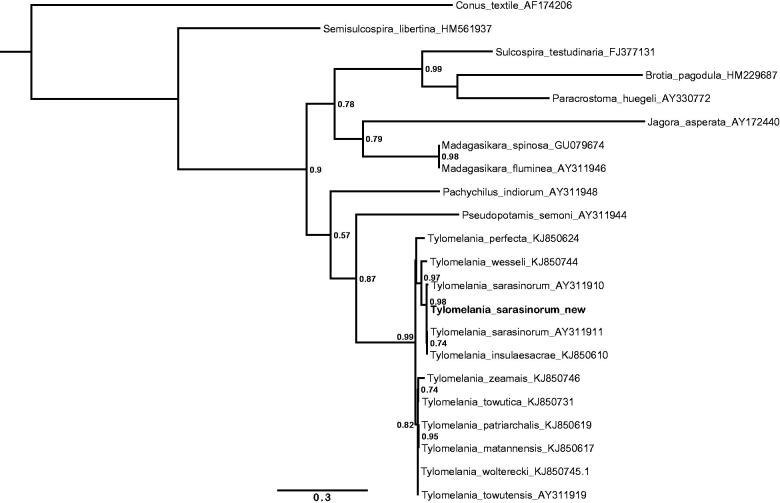
The specimen was collected at Loeha Island (Lake Towuti, South Sulawesi, 2.76075 S 121.5586 E) and is stored at the Museum für Naturkunde Berlin (accession number: ZMB Moll. 119994). DNA was extracted using CTAB extraction (Winnepenninckx et al. 1993) and shotgun sequenced (150-bp paired end) on an Illumia MiSeq® (Illumina, San Diego, CA) generating ∼50 mio reads. The mitochondrial genome was reconstructed with MITObim (Hahn et al. 2013) using the COI sequence of the sister species T. wallacei (KJ933844) as seed reference and the following parameters: “-denovo”, “-pair”, “-proofread” and “-clean”. The assembled mitogenome had a mean coverage of 495× and was manually inspected for repeats at the ends of the assembly to confirm circularity. Annotations were carried out with MITOchondrial genome annotation server (MITOS) (Bernt et al. 2012), followed by manual validation of the coding regions using the NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The annotated sequence file was submitted to NCBI (KU878411). The phylogenetic position of the new sequence is shown in Figure 1.
Figure 1.
Maximum likelihood tree illustrating the position of the new Tylomelania sarasinorum 16s rRNA gene sequence within a subset of pachychilid species. Tree topology is largely in agreement with earlier work (Koehler & Glaubrecht 2010), but fails to resolve clades down to species level in the rapidly radiating genus Tylomelania which is in line with previous results (von Rintelen et al. 2004). Sequences were aligned using MAFFT 7.271 and highly divergent or poorly aligned regions were removed with Gblocks 0.91b (Castresana 2000) allowing for gap positions and smaller blocks. Trees were calculated using PhyML 3.1 (Guindon et al. 2010) with 12 rate categories, optimized equilibrium frequencies, GTR model of sequence evolution and combined heuristics (Nearest Neighbor Interchange and Subtree Pruning and Rerafting).
The complete mitochondrial genome was 16,632 bp in length, contained 13 protein-coding genes (PCGs), 2 rRNA genes and 22 tRNA genes. As described for other gastropods, in the mitochondrial genome of T. sarasinorum an A + T bias was evident with an overall base composition of 29.5% A, 35.7% T, 18.3% G and 16.6% C. The gene arrangement was most similar to the mitogenomes of the two cerithioideans Semisulcospira libertina (Gould 1859) and Koreoleptoxis globus ovalis (Burch & Jung 1987), differing only in the position of tRNAArg and tRNAGln (Cunha et al. 2009; Zeng et al. 2014; Osca et al. 2015). The light strand was clustered in the following order: tRNACys, tRNAAla, tRNAAsn, tRNATrp, tRNAGlu, tRNATyr, tRNAArg, tRNAGln, tRNALys, COX3, tRNAMet, CYTB, ND6, tRNAPro, ND1, tRNALeu2, tRNALeu1, 16S-rRNA, tRNAVal, tRNAGly, tRNAThr, 12S-rRNA. The heavy strand genes clustered: tRNASer1, ND2, tRNAAsp, ATP8, ATP6, tRNAIle, ND3, COX1, COX2, tRNASer2, ND4L, ND4, tRNAHis, ND5, tRNAPhe. All PCGs had ATG as initiation codon. TAA was the most used termination codon with the exception of COX3, CYTB, ND2, ATP8 and ND3, which used a TAG termination codon. The 12S and 16S genes had a length of 899 and 1382 bp, respectively. Overlaps were observed between CYTB and ND6 (47 bp), 16S-rRNA and tRNAVal (18 bp), and 16S-ND4L and ND4 (7 bp), tRNALeu1 and 16S-rRNA (7 bp), ND2 and tRNAAsp(2 bp) and between tRNAThr and 12S-rRNA.
Acknowledgements
We wish to thank Björn Stelbrink and Bert Van Bocxlaer for collecting specimens in the field.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding information
This study was funded by the German Research Council (DFG) grant Ri 1738/9-1.
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF.. 2012. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet E. 7:332–336. [DOI] [PubMed] [Google Scholar]
- Burch JB, Jung Y.. 1987. A new freshwater prosobranch snail (Mesogastropoda pleuroceridae) from Korea. Walerana. 2:411–453. [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis . Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Colgan DJ, Ponder WF, Beacham E, Macaranas J.. 2007. Molecular phylogenetics of Caenogastropoda (Gastropoda: Mollusca) . Mol Phylogenet Evol. 42:717–737. [DOI] [PubMed] [Google Scholar]
- Cunha RL, Grande C, Zardoya R.. 2009. Neogastropod phylogenetic relationships based on entire mitochondrial genomes. BMC Evol. Biol. 9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B.. 2013. . Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler F, Glaubrecht M.. 2010. Uncovering an overlooked radiation: molecular phylogeny and biogeography of Madagascar’s endemic river snails (Caenogastropoda: Pachychilidae: Madagasikara gen. nov.). Biol J Linn Soc. 99:867–894. [Google Scholar]
- Kruimel JH. 1913. Verzeichnis der von Herrn E. C. Abendanon in Celebes gesammelten Süsswasser-Mollusken. Bijdragen Tot De Dierkunde. 19:215–235. [Google Scholar]
- Osca D, Templado J, Zardoya R.. 2015. Caenogastropod mitogenomics . Mol Phylogenet Evol. 93:118–128. [DOI] [PubMed] [Google Scholar]
- von Rintelen T, von Rintelen K, Glaubrecht M.. 2010. The species flocks of the viviparous freshwater gastropod Tylomelania (Mollusca: Cerithioidea: Pachychilidae) in the ancient lakes of Sulawesi, Indonesia: the role of geography, trophic morphology and color as driving forces in adaptive radiation In: Glaubrecht M, editor. Evolution in action. Berlin: Springer Berlin Heidelberg; p. 485–512. [Google Scholar]
- von Rintelen T, Stelbrink B, Marwoto RM, Glaubrecht M.. 2014. A snail perspective on the biogeography of Sulawesi, Indonesia: origin and intra-island dispersal of the viviparous freshwater gastropod Tylomelania. PLos One. 9:e98917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Rintelen T, Wilson AB, Meyer A, Glaubrecht M.. 2004. Escalation and trophic specialization drive adaptive radiation of freshwater gastropods in ancient lakes on Sulawesi, Indonesia. Proc R Soc Lond B. 271:2541–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong EE, Colgan DJ, Healy JM, Lydeard C, Ponder WF, Glaubrecht M.. 2011. Phylogeny of the gastropod superfamily Cerithioidea using morphology and molecules. Zool J Linn Soc. 162:43–89. [Google Scholar]
- Winnepenninckx B, Backeljau T, De Wachter R.. 1993. Extraction of high molecular weight DNA from molluscs . Trends Genet. 9:407. [DOI] [PubMed] [Google Scholar]
- Zeng T, Yin W, Xia R, Fu C, Jin B.. 2014. Complete mitochondrial genome of a freshwater snail, Semisulcospira libertina (Cerithioidea: Semisulcospiridae). Mitochondrial DNA. 26:897–898. [DOI] [PubMed] [Google Scholar]