Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has rapidly spread within the human population. Although SARS-CoV-2 is a novel coronavirus, most humans had been previously exposed to other antigenically distinct common seasonal human coronaviruses (hCoVs) before the coronavirus disease 2019 (COVID-19) pandemic. Here, we quantified levels of SARS-CoV-2-reactive antibodies and hCoV-reactive antibodies in serum samples collected from 431 humans before the COVID-19 pandemic. We then quantified pre-pandemic antibody levels in serum from a separate cohort of 251 individuals who became PCR-confirmed infected with SARS-CoV-2. Finally, we longitudinally measured hCoV and SARS-CoV-2 antibodies in the serum of hospitalized COVID-19 patients. Our studies indicate that most individuals possessed hCoV-reactive antibodies before the COVID-19 pandemic. We determined that ∼20% of these individuals possessed non-neutralizing antibodies that cross-reacted with SARS-CoV-2 spike and nucleocapsid proteins. These antibodies were not associated with protection against SARS-CoV-2 infections or hospitalizations, but they were boosted upon SARS-CoV-2 infection.
Keywords: antibodies, coronavirus, SARS-CoV-2, pre-existing immunity
Graphical abstract

Analysis of human serum samples before and after the onset of the COVID-19 pandemic show that antibodies against common seasonal human coronaviruses are cross-reactive against SARS-CoV-2 but do not confer cross-protection against infection or hospitalization.
Introduction
Coronaviruses commonly infect humans (Dijkman et al., 2012; Friedman et al., 2018; Gaunt et al., 2010; Killerby et al., 2018). The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged at the end of 2019 and has rapidly spread among humans, many of whom have been previously exposed to common seasonal human coronaviruses (hCoVs) (Edridge et al., 2020). Common seasonal hCoVs include the betacoronaviruses HKU1 and OC43 and the alphacoronaviruses 229E and NL63 (Pfefferle et al., 2009; Pyrc et al., 2006; Vijgen et al., 2006; Woo et al., 2005). SARS-CoV-2 belongs to the betacoronavirus genus and is more closely related to HKU1 and OC43 than to the alphacoronaviruses 229E and NL63 (Jaimes et al., 2020; Okba et al., 2020). A recent study examining electronic medical records suggested that recent hCoV infections are not associated with decreased SARS-CoV-2 infections but are associated with reducing the severity of coronavirus disease 2019 (COVID-19) (Sagar et al., 2021). It is unclear whether this apparent cross-protection is mediated by antigen-specific cellular or humoral immunity or whether it is due to short-term general cross-protection similar to what has been recently reported with rhinovirus and influenza virus infections (Wu et al., 2020). It is unknown whether prior hCoV exposures elicit antibodies that prevent or alter the outcomes of SARS-CoV-2 infections. Further, it is unknown whether differently aged individuals have distinct hCoV immune histories that can affect SARS-CoV-2 susceptibility. To address this, we completed a serological survey using serum samples collected from differently aged humans prior to the COVID-19 pandemic. We quantified levels of antibodies reactive to viral proteins from hCoVs and determined whether these antibodies were associated with SARS-CoV-2 protection. Finally, we completed a series of studies using serum collected from COVID-19 patients to determine whether antibodies reactive to hCoVs are boosted upon SARS-CoV-2 infections.
Results
Identification of SARS-CoV-2-reactive antibodies in human sera collected prior to the COVID-19 pandemic
We completed ELISAs to quantify levels of pre-pandemic SARS-CoV-2-reactive IgG antibodies in 431 human serum samples collected in 2017. We tested serum samples collected from 263 children (ages 1–17) at the Children’s Hospital of Philadelphia which had been originally collected for lead testing and 168 adults (ages 18–90) who had been recruited into the Penn Medicine Biobank. We tested Penn Medicine Biobank samples from individuals who had no medical history of cancer or organ transplantation, pregnancy during the previous 9 months, or an infectious disease within the previous 28 days prior to blood draw. With these samples, we previously found that differently aged individuals possess H3N2 influenza virus antibodies that have different specificities (Gouma et al., 2020).
We found that 4.2% of serum samples collected in 2017 contained IgG antibodies that reacted to the SARS-CoV-2 full-length spike (S) protein (Figure 1 A), 0.93% of samples contained antibodies that reacted to the receptor binding domain (RBD) of the SARS-CoV-2 S protein (Figure 1B), and 16.2% of samples contained antibodies that reacted to the SARS-CoV-2 nucleocapsid (N) protein (Figure 1C). Several pre-pandemic serum samples contained antibodies that were at similar levels as those in serum from PCR-confirmed COVID-19 recovered donors (Figures 1A–1C). We found no obvious differences in levels of SARS-CoV-2 cross-reactive antibodies among donors with different birth years (Figures S1 A–S1C). We obtained similar results when ELISAs were completed with unpurified serum antibodies and purified IgG (Figure S2 ). Most serum samples with antibodies reactive to the SARS-CoV-2 full-length S protein did not have antibodies that reacted to the SARS-CoV-2 S-RBD protein (Figure 1D), which is consistent with recent studies showing that some individuals possessed pre-pandemic antibodies against the S2 domain of the SARS-CoV-2 S protein (Nguyen-Contant et al., 2020; (Shrock et al., 2020)). There was a poor correlation between N and S antibody titers in pre-pandemic samples (Figure S3 ).
Figure 1.
Identification of pre-existing cross-reactive SARS-CoV-2 antibodies in human serum prior to the pandemic
(A–C) ELISAs were completed to quantify levels of serum antibodies binding to the SARS-CoV-2 full-length S protein (A), the S-RBD (B), and the N protein (C). Dashed line denotes lower limit of detection (LOD = 50); dotted line represents a threshold set 2-fold above LOD (>100). We tested samples collected from 431 individuals in the summer of 2017, prior to the global pandemic. We also tested samples collected from 15 individuals after confirmed SARS-CoV-2 infections as well as from recovered adults.
(D) The relationship between antibody titers in donors with detectable IgG against the S-RBD and/or full-length S is shown.
(E) SARS-CoV-2 pseudotype neutralization assays were completed with pre-pandemic serum samples with (n = 14) and without (n = 29) cross-reactive SARS-CoV-2 antibodies, as well as serum samples from individuals after confirmed SARS-CoV-2 infections (n = 15); one-way ANOVA Tukey’s multiple comparisons of log2 transformed antibody titers ∗∗∗∗p < 0.0001; dotted line denotes lower LOD (=10).
(F–H) ELISAs were completed to quantify levels of serum antibodies binding to the full-length S proteins from 229E, NL63, and OC43 with pre-pandemic serum samples with (n = 17) and without (n = 17) cross-reactive SARS-CoV-2 antibodies. Unpaired t tests of log2 transformed antibody titers ∗∗p < 0.002. Horizontal lines indicate geometric mean and error bars represent standard deviation. See also Figure S1, Figure S2, Figure S3, Figure S4, and S5.
Figure S1.
There are no obvious age-related differences in pre-pandemic SARS-CoV-2 and hCoV reactive antibodies, related to Figure 1
ELISAs were completed to measure levels of serum antibodies binding to the SARS-CoV-2 full-length spike (S) protein (A), SARS-CoV-2 receptor binding domain (S-RBD) of S (B), SARS-CoV-2 nucleocapsid (N) protein (C), 229E S protein (D), NL63 S protein (E), and OC43 S protein (F). Serum samples collected from 431 individuals in the summer of 2017 were tested. Reciprocal titer from serially-diluted serum samples (A-C) and optical densities at 450nm wavelength (OD450) of 1:500 dilution of serum (D-F) are shown. Dashed line denotes lower limit of detection (LOD = 50), dotted line represents a threshold set 2-fold above LOD (> 100).
Figure S2.
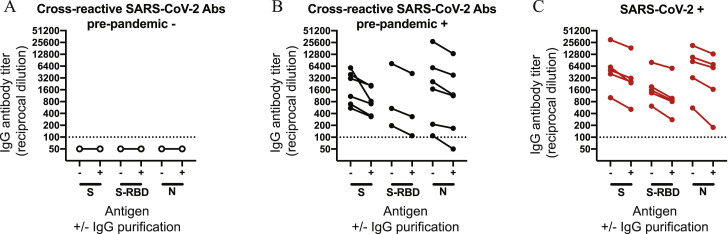
Comparison of ELISA data using unpurified and purified serum IgG antibodies, related to Figure 1 and 2
IgG was purified from sera samples from individuals without (A; n = 5) and with (B; n = 11) pre-pandemic cross-reactive antibodies. IgG was also purified from serum samples from individuals who had recovered from a confirmed SARS-CoV-2 infection (C; n = 5). ELISAs were completed to quantify levels of serum antibodies binding to SARS-CoV-2 full length S, S-RBD, and N protein with and without IgG magnetic bead purification. The dotted line represents a threshold set 2-fold above the limit of detection (> 100).
Figure S3.
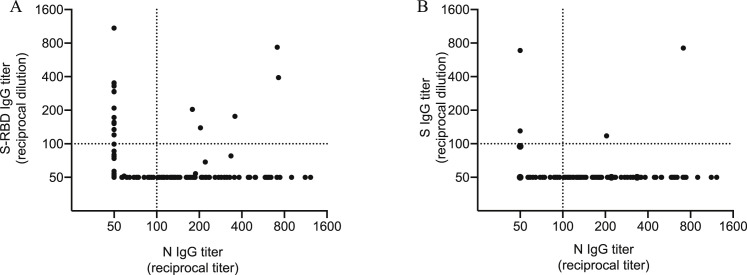
Correlation between N, S, and S-RBD antibody titers in pre-pandemic samples, related to Figure 1
Shown are the relationships between serum IgG antibody titers against the SARS-CoV-2 N protein and S-RBD (A) or full length S (B) from 431 individuals whose samples were collected prior to the pandemic in the summer of 2017. Dotted line represents a threshold set 2-fold above the limit of detection (> 100).
We completed neutralization assays with a SARS-CoV-2 vesicular stomatitis virus (VSV) pseudotype platform. In contrast to serum antibodies isolated from PCR-confirmed COVID-19 recovered donors, serum antibodies from individuals collected before the pandemic had very low or undetectable levels of SARS-CoV-2 neutralizing antibodies, regardless of whether or not the sample possessed cross-reactive antibodies against SARS-CoV-2 S and N proteins (Figures 1E and S4 ). We obtained similar results when we tested pre-pandemic serum samples with a bona fide BSL3-level SARS-CoV-2 neutralization assay (Figure S5 ).
Figure S4.
SARS-CoV-2 pseudotype neutralization curves, related to Figure 1
Raw neutralization curves for data from Figure 1E are shown, including samples from individuals who did not have pre-pandemic cross reactive SARS-CoV-2 antibodies (A), individuals who possessed pre-pandemic cross reactive SARS-CoV-2 antibodies (B), and individuals following confirmed SARS-CoV-2 infection (C). Mean and error bars are shown for each replicate; dotted line denotes the cut-off for foci reduction neutralization of 50% (FRNT50).
Figure S5.
Pre-pandemic cross-reactive antibodies do not neutralize SARS-CoV-2 in bonafide BSL3-level neutralization assays, related to Figure 1
Neutralization assays with live SARS-CoV-2 were completed using 9 pre-pandemic samples with cross-reactive SARS-CoV-2 antibodies, 7 pre-pandemic samples without cross-reactive SARS-CoV-2 antibodies, and 5 samples from individuals who recovered from a PCR-confirmed SARS-CoV-2 infection. The pre-pandemic samples for these experiments were collected in 2019 and are different from those shown in Figure 1 (which were collected in 2017).
Humans with pre-pandemic SARS-CoV-2-reactive antibodies had elevated levels of antibodies against previously circulating betacoronaviruses
We completed ELISAs to quantify levels of pre-pandemic hCoV-reactive IgG antibodies in all 431 human serum samples collected in 2017. Most serum samples possessed antibodies that reacted to the S protein of 229E and NL63 (both alphacoronaviruses) as well as that of OC43 (a betacoronavirus) (Figures S1D–S1F). There were no major differences in levels of these antibodies among individuals with different birth years; however, serum from very young children possessed lower levels of antibodies reactive to the OC43, 229E, and NL63 S proteins (Figures S1D–S1F). We completed full antibody titrations to directly compare levels of hCoV antibodies in a subset of pre-pandemic samples from individuals who either did (n = 17) or did not (n = 17) possess cross-reactive SARS-CoV-2 antibodies (Figures 1F–1H). Pre-pandemic antibody levels against the 229E and NL63 alphacoronavirus S proteins were similar among individuals with and without SARS-CoV-2-reactive antibodies (Figures 1F and 1G). In contrast, antibody levels against the betacoronavirus OC43 S protein were higher in individuals with SARS-CoV-2-reactive antibodies than in individuals who did not possess pre-pandemic SARS-CoV-2-reactive antibodies (Figure 1H). These data suggest that pre-pandemic SARS-CoV-2-reactive antibodies were likely elicited by previously circulating betacoronavirus strains, such as OC43.
Pre-existing hCoV cross-reactive antibodies were not associated with protection from SARS-CoV-2 infections
It is unknown whether antibodies elicited by prior hCoV infections protect against SARS-CoV-2 infections and/or prevent severe COVID-19. To address this, we measured SARS-CoV-2 IgG antibodies in pre-pandemic serum samples from 251 individuals who subsequently went on to become PCR-confirmed infected with SARS-CoV-2 and in a control group of pre-pandemic samples from 251 matched individuals who did not become infected with SARS-CoV-2. Pre-pandemic samples were collected by the Penn Medicine BioBank from August 2013 to March 2020 and PCR-confirmed SARS-CoV-2 infections were identified by nasopharyngeal swab PCR testing results in electronic health records. We found that 2.2% of samples possessed pre-pandemic antibodies reactive to the SARS-CoV-2 full-length S protein, 0.6% of samples possessed pre-pandemic antibodies reactive to the SARS-CoV-2 S-RBD, and 23.9% of samples possessed pre-pandemic antibodies reactive to the SARS-CoV-2 N protein. Importantly, we found no differences in SARS-CoV-2-reactive antibodies in serum samples from individuals who did or did not become subsequently infected with SARS-CoV-2 (Figure 2 A; S protein, p = 0.62; S-RBD, p = 0.49; N protein, p = 0.34; see also Tables S1 and S2). We also measured antibodies reactive to the OC43 S protein and found no differences among samples from individuals who did or did not become infected with SARS-CoV-2 (Figure 2A; p = 0.90; see also Tables S1 and S2). Among those with PCR-confirmed SARS-CoV-2 infections, we found no relationship between SARS-CoV-2 and OC43 antibody titers and hospitalization or disease severity among hospitalized patients (Tables S1 and S2). We found no relationship between SARS-CoV-2 and OC43 antibody titers and the need for respiratory support and admittance into the ICU after SARS-CoV-2 infection (Tables S1 and S2).
Figure 2.
Pre-pandemic SARS-CoV-2 and OC43-reactive antibodies are not associated with protection from SARS-CoV-2 infection
(A and B) We quantified antibody levels in pre-pandemic serum samples collected from individuals who later became SARS-CoV-2 infected (cases; n = 251) and those who did not become SARS-CoV-2 infected (controls; n = 251). ELISAs were completed to quantify levels of antibodies reactive to SARS-CoV-2 proteins (S, S-RBD, and N) and the OC43 S protein. Shown are data using samples collected from the entire cohort between August 2013 and March 2020 (A) and samples from a smaller subset of individuals collected between April 2019 and March 2020 (B). Antibody titers between cases and controls were not significantly different as determined by unpaired t tests of log2 transformed antibody titers. Dashed line denotes lower limit of detection (LOD = 50), dotted line represents a threshold set 2-fold above LOD (>100). See also Tables S1 and S2.
Previous studies indicated that immunity to hCoV can be short-lived (Huang et al., 2020), and a recent study documented that antibody titers against hCoV can fluctuate over time (Edridge et al., 2020), presumably due to repetitive hCoV exposures. In our study, pre-pandemic serum samples were collected from 2013–2020, and, therefore, it is possible that antibody levels in some of the samples collected several years prior to 2020 do not accurately reflect antibody levels present during the COVID-19 pandemic. To address this, we compared SARS-CoV-2 and OC43 IgG antibody titers in the serum of individuals in our cohort who had samples collected within one year of the pandemic (between April 2019 and March 2020). Using this smaller cohort (n = 39 SARS-CoV-2 cases and n = 57 controls), we still found no differences in levels of antibodies reactive to the SARS-CoV-2 S protein, S-RBD protein, N protein, or OC43 S protein (Figure 2B). Taken together, our data suggest that a subset of humans possessed non-neutralizing cross-reactive antibodies against SARS-CoV-2 S and N proteins prior to the COVID-19 pandemic, but these antibodies were not associated with protection from SARS-CoV-2 infections or reducing hospitalizations upon SARS-CoV-2 infections.
SARS-CoV-2 boosts antibodies reactive to other human betacoronaviruses
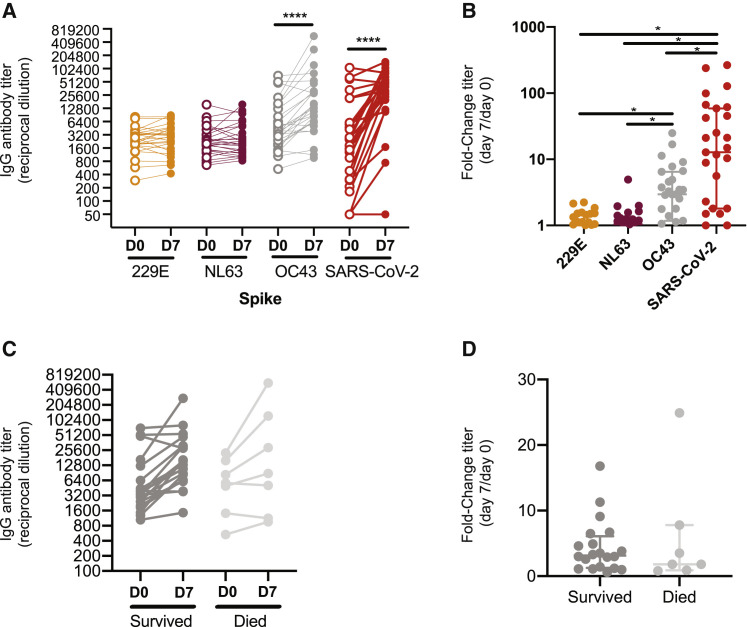
Recent studies indicate that COVID-19-recovered donors possess higher levels of antibodies against seasonal betacoronaviruses (Nguyen-Contant et al., 2020; (Shrock et al., 2020)). To determine whether antibodies against the S protein of hCoVs are boosted upon SARS-CoV-2 infection, we measured 229E, NL63, OC43, and SARS-CoV-2 S IgG antibody levels in sera collected longitudinally from 27 hospitalized COVID-19 patients. Samples from a subset of the hospitalized patients (10 of 27) were tested with an extended respiratory pathogen viral panel to confirm that they were not simultaneously co-infected with SARS-CoV-2 and a different coronavirus. Serum IgG antibodies reactive to the S protein of the 229E and NL63 alphacoronaviruses did not change over 7 days of hospitalization (Figures 3A and 3B). Conversely, serum antibodies reactive to the S protein of OC43 and SARS-CoV-2 betacoronaviruses significantly increased over the course of hospitalization (Figures 3A and 3B). We found that boosted antibodies in hospitalized patients primarily targeted the S2 domain, and not the S1 domain, of the OC43 S protein (Figures S6 A and S6B). Overall OC43 IgG antibody titers (Figure 3C) and the magnitude of OC43 S antibody boosts (Figure 3D) were not associated with outcome of disease. These data indicate that cross-reactive antibodies elicited by previous hCoV infections are not associated with protection from SARS-CoV-2 infections but are boosted after infection with SARS-CoV-2.
Figure 3.
SARS-CoV-2 infections boost antibodies that react to OC43 S protein
(A–D) We quantified antibody levels in serum collected from 27 individuals 0 and 7 days after hospitalization for COVID-19. ELISAs were completed to quantify levels of antibodies reactive to the S proteins of 229E, NL63, OC43, and SARS-CoV-2. IgG titers (A) and titer fold change (B) are shown. Levels of OC43 S-reactive antibodies (C) and fold change in OC43 S-reactive antibodies (D) were not associated with disease outcome. Paired t test of log2 transformed antibody titers, ∗∗∗∗p < 0.0001. One-way ANOVA Tukey’s multiple comparisons fold-change in antibody titers, ∗p < 0.04. Horizontal lines indicate the median and error bars show interquartile ranges. See also Figure S6.
Figure S6.
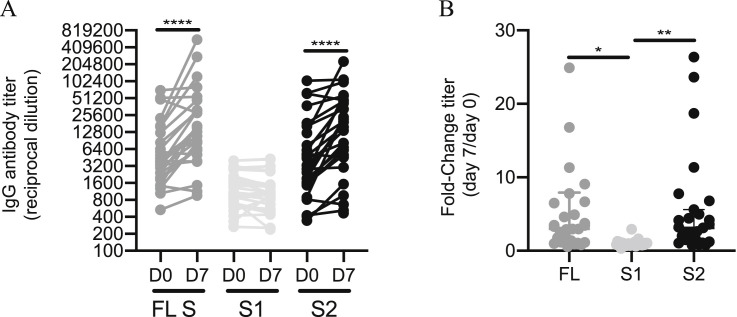
Antibodies directed to the S2 region of OC43 spike are boosted during SARS-CoV-2 infection, related to Figure 3
We quantified antibody levels in serum collected from 27 individuals 0 and 7 days after hospitalization for COVID-19. ELISAs were completed to measure levels of serum antibodies binding to the OC43 full-length spike (FL) protein and the individual S1 and S2 subunits of the OC43 spike. (A) IgG titers and (B) titer fold change are shown. Paired t test of log2 transformed antibody titers, ∗∗∗∗p < 0.0001. One-way ANOVA Tukey’s multiple comparisons fold-change in antibody titers, * p < 0.02 ∗∗p < 0.005. Horizontal lines indicate the median and error bars show interquartile range.
Discussion
Our study demonstrates that ∼20% of individuals possessed SARS-CoV-2 cross-reactive serum antibodies prior to the COVID-19 pandemic. Using samples collected in 2017, we found that pre-pandemic cross-reactive antibodies directed against the SARS-CoV-2 N protein were more prevalent than those directed against the SARS-CoV-2 S protein (16.2% seropositive versus 4.2% seropositive). We found that most individuals possessed pre-pandemic serum antibodies reactive to the S proteins of 229E, NL63, and OC43 (Figure S1); however, pre-pandemic samples with detectable levels of SARS-CoV-2 antibodies had higher levels of antibodies against the OC43 S protein (Figure 1H). Although our data suggest that prior infections with seasonal human betacoronaviruses (such as OC43) likely elicit antibodies that cross-react with SARS-CoV-2 proteins, it is unclear why only a subset of OC43 seropositive individuals possessed antibodies reactive to SARS-CoV-2 prior to the pandemic. Further studies will be needed to determine the temporal relationship between seasonal human betacoronavirus infections and the induction of SARS-CoV-2 cross-reactive antibodies. Further studies investigating the relationship of pre-pandemic antibodies against other betacoronaviruses, such as HKU1, with pre-pandemic SARS-CoV-2 cross-reactive antibodies are also needed.
Our study is consistent with a recent manuscript demonstrating a lack of SARS-CoV-2 neutralizing activity in pre-pandemic sera (Poston et al., 2020). In contrast, a different study reported that pre-pandemic serum from young children possess SARS-CoV-2 neutralizing antibodies (Ng et al., 2020). It is unclear whether these differences are due to the specific assays used in each study or other factors such as geographic differences in sampling. For example, the study by Ng et al. (2020) used a pseudotyped neutralization assay using cells that lack ACE2, which is the cellular receptor for SARS-CoV-2. Our study is unique in that we were able to directly assess whether pre-pandemic antibodies were associated with protection from SARS-CoV-2 infections and hospitalizations. Although we found no differences in pre-pandemic antibody levels against SARS-CoV-2 and OC43 among those infected and not infected with SARS-CoV-2 (Figure 2) and among SARS-CoV-2-infected individuals with different disease severities (Tables S1 and S2), larger cohorts including individuals with a large range of different clinically defined disease severities will be required to determine whether pre-pandemic levels of antibodies are associated with reducing some aspects of severe COVID-19. Additional studies need to be completed to determine whether neutralizing antibodies elicited by SARS-CoV-2 infections protect against subsequent reinfections with SARS-CoV-2.
Further studies also need to be completed to determine how immune history affects de novo immune responses after SARS-CoV-2 infection. We find that individuals infected with SARS-CoV-2 produce antibodies reactive to both the SARS-CoV-2 S protein and OC43 S protein (Figure 3). In the case of influenza viruses, sequential infections with antigenically distinct strains can elicit antibodies against conserved epitopes between the strains, and it is unclear whether these cross-reactive antibodies inhibit de novo immune responses or affect disease severity (Cobey and Hensley, 2017). Our studies suggest that SARS-CoV-2 infection boosts antibodies reactive to the S2 domain of the OC43 S protein. Further studies are needed to precisely map the footprints of these antibodies, and additional studies need to be completed to determine whether these antibodies help resolve infections or whether they enhance disease in COVID-19 patients.
Given that our data suggest that pre-pandemic non-neutralizing antibodies elicited by hCoVs do not provide SARS-CoV-2 protection, special attention should be directed toward evaluating whether T cell responses primed against hCoV infections provide partial protection against SARS-CoV-2 infections. Recent studies have clearly shown that some individuals possessed SARS-CoV-2-specific CD4+ and CD8+ T cells prior to the COVID-19 pandemic (Braun et al., 2020; Grifoni et al., 2020; Le Bert et al., 2020; Mateus et al., 2020; Sette and Crotty, 2020; Schulien et al., 2021), and it is possible that pre-existing cellular immunity might play an important protective role in the context of pandemic viruses that only share non-neutralizing antibody epitopes with previously circulating viral strains.
Limitations of the study
The data presented here show that pre-pandemic serum antibodies that cross-react with SARS-CoV-2 do not correlate with protection against SARS-CoV-2 infections and severity of COVID-19. We generated data by using pre-pandemic samples that were collected from individuals who became PCR-confirmed infected with SARS-CoV-2. We compared antibody levels in these samples to antibody levels in pre-pandemic samples from individuals who did not get infected with SARS-CoV-2. For these studies, we included samples that were collected from August 2013 to March 2020 (Figure 2A). Because immunity to hCoVs can be short-lived (Huang et al., 2020) and fluctuate over time (Edridge et al., 2020), we also directly compared antibody titers in samples that were collected within one year of the pandemic (Figure 2B). Using both datasets, we found no correlation between pre-pandemic antibody levels and SARS-CoV-2 infections and COVID-19 severity. Nonetheless, future studies need to continue exploring the temporal relationship between seasonal coronavirus infections and the induction of SARS-CoV-2 cross-reactive antibodies to determine whether transient antibody-mediated protection is possible. Future studies should also evaluate the protective potential of pre-pandemic cross-reactive mucosal antibodies. Finally, studies need to address whether pre-existing cellular immunity limits COVID-19 severity. Our study only examined serum antibodies, and it is possible that rapid engagement of memory B and T cells and long-lived plasma cells provide protection after SARS-CoV-2 exposures of humans with unique immune histories.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-human IgG-HRP | Jackson ImmunoResearch | Cat. 109-036-098, RRID: AB_2337596 |
| mAb CR3022 | Expressed for this paper | N/A |
| mAb 1E9F9 | Absolute Antibody | Ab01402-2.0 |
| anti-dsRNA J2 | Absolute Antibody | Ab01299-2.0 |
| Goat anti-mouse IgG alexa 488 | ThermoFisher Scientific | Cat. A-11029, RRID: AB_2534088 |
| Hoescht 33342 | Sigma Aldrich | B2261 |
| Bacterial and Virus Strains | ||
| SARS-CoV-2 VSV pseudotypes | Generated for this paper | N/A |
| SARS-CoV-2 (WA-1) | BEI | NR-52281 |
| Biological Samples | ||
| Pre-pandemic adult serum samples | Penn Medicine Biobank (PMBB) | N/A |
| Pre-pandemic children serum samples | Children’s Hospital of Philadelphia (CHOP) | N/A |
| COVID-19 patient serum samples | Hospital of the University of Pennsylvania (HUP | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SARS-CoV-2 spike protein | Expressed for this paper | N/A |
| SARS-CoV-2 RBD protein | Expressed for this paper | N/A |
| SARS-CoV-2 nucleocapsid protein | Sino Biological | Cat. 40588-V08B |
| OC43 spike protein | Sino Biological | Cat. 40607-V08B |
| NL63 spike protein | Sino Biological | Cat. 40604-V08B |
| 229E spike protein | Sino Biological | Cat. 40605-V08B |
| OC43 S1 subunit protein | Expressed for this paper | N/A |
| OC43 S2 subunit protein | Expressed for this paper | N/A |
| Experimental Models: Cell Lines | ||
| 293T | ATCC | Cat. CRL-3216, RRID: CVCL_0063 |
| 293F | Laboratory of Scott Hensley, University of Pennsylvania, PA | Thermo Fisher cat. R79007 |
| VeroE6/TMPRSS | Laboratory of Stefan Pohlman, German Primate Center, Leibniz Institute for Primate Research | (Hoffmann et al., 2020) |
| Vero CCL81 | ATCC | Cat. CCL-81, RRID: CVCL_0059 |
| Recombinant DNA | ||
| Plasmid: pCAGGS SARS-CoV-2 spike | Laboratory of Florian Krammer, Mt. Sinai, NY | Amanat et al., 2020 |
| Plasmid: pCAGGS SARS-CoV-2 RBD | Laboratory of Florian Krammer, Mt. Sinai, NY | Amanat et al., 2020 |
| Plasmid: pCG1 SARS- 2 S | Laboratory of Stefan Pohlman, German Primate Center, Leibniz Institute for Primate Research | (Hoffmann et al., 2020) |
| Plasmid: OC43 rS1 | Laboratory of Scott Hensley, University of Pennsylvania, PA | This paper |
| Plasmid: OC43 rS2 | Laboratory of Scott Hensley, University of Pennsylvania, PA | This paper |
| Software and Algorithms | ||
| Prism8 | GraphPad Software | www.graphpad.com/scientific-software/prism/ |
| Flouro-X | ImmunoSpot | www.immunospot.com/index-ctl |
| Deposited Data | ||
| Original data | Mendeley Data | https://doi.org/10.17632/ygv2j9psc5.1 |
Resources Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Scott E. Hensley (hensley@pennmedicine.upenn.edu).
Materials Availability
All unique reagents generated in this study will be available from the Lead Contact upon reasonable request.
Data and Code Availability
All raw data generated in this study have been deposited on Mendeley Data: https://doi.org/10.17632/ygv2j9psc5.1.
Experimental Model and Subject Details
Pre-pandemic Human Serum Samples
Serum samples shown in Figure 1 were collected before the COVID-19 pandemic between May and August of 2017 from individuals at the Children’s Hospital of Philadelphia (CHOP; n = 263, children age 0-17 years old) and through the Penn Medicine BioBank (n = 168, adults >18 years old). Samples from CHOP were leftover de-identified blood samples collected for routine lead testing.
Serum samples shown in Figure 2 were collected via the Penn Medicine BioBank prior to the pandemic (n = 502, between August 2013 and March 2020). These samples were from adults who subsequently had a reverse transcription quantitative polymerase chain reaction (RT-qPCR) confirmed SARS-CoV-2 infection using nasopharyngeal swabs (cases, n = 251), and those who had SARS-CoV-2 PCR negative results (controls, n = 251). The RT-qPCR clinical testing results were acquired from Penn Medicine electronic health records and test results between March 2020 and August 2020 were included in the analysis. The Penn Medicine BioBank is an established repository that routinely collects blood products from donors visiting the University of Pennsylvania Healthcare system upon written informed consent. All studies were approved by the University of Pennsylvania Institutional Review Board.
Human Samples Collected After SARS-CoV-2 Infection
Serum samples were obtained from recovered convalescent donors who had a history of PCR-confirmed SARS-CoV-2 infection (n = 15). These samples were used in experiments shown in Figure 1. Additionally, plasma samples were collected from patients admitted to the Hospital at the University of Pennsylvania (HUP) with PCR-confirmed SARS-CoV-2 infections (n = 27), as previously described (Mathew et al., 2020). Hospital inpatients were categorized for pneumonia severity using a WHO ordinal scale that was based on the level of oxygen support needed at day 0 and day 7. All samples were collected after obtaining informed consent and studies were approved by the University of Pennsylvania Institutional Review Board.
Cell lines
293F cells were from Thermo fisher (Thermo Fisher cat. R79007). 293T and Vero CCL81 cells were from ATCC (ATCC cat. CRL-3216, RRID:CVCL_0063 and ATCC cat. CCL-81, RRID:CVCL_0059, respectively).VeroE6/TMPRSS2 cells were a gift from Stefan Pohlman (German Primate Center, Leibniz Institute for Primate Research) as described previously (Hoffmann et al., 2020). All cell lines were cultured using manufacturer’s guidelines and used as described in Method Details below.
Method Details
Quantification of serum antibody titers
Serum antibody titers against SARS-CoV-2 and other human coronavirus (hCoV) antigens were quantified by enzyme-linked immunosorbent assays (ELISA) as previously described (Flannery et al., 2020). Plasmids encoding the full-length SARS-CoV-2 spike (S) protein and the receptor binding domain of the S (S-RBD) were provided by Florian Krammer (Icahn School of Medicine at Mt. Sinai, New York City NY) (Amanat et al., 2020). SARS-CoV-2 S-RBD and the SARS-CoV-2 S proteins were purified from 293F transfected cells by Ni-NTA resin. SARS-CoV-2 nucleocapsid (N) protein, and full-length hCoV spike antigens (OC43, 229E, and NL63) were purchased (Sino Biological, Wayne PA; cat. 40588-V08B, 40607-V08B, 40604-V08B, and 40605-V08B, respectively) and reconstituted in Dulbecco’s phosphate buffered saline (DPBS). OC43 subunit proteins were purified by Ni-NTA resin from 293F cells transfected with plasmids encoding the S1 or S2 subunits of the OC43 spike protein. ELISA plates (Thermo Fisher Scientific: cat. 14-245-153) were coated overnight at 4°C with either 2 μg/mL SARS-CoV-2 antigen, 1.5 μg/mL hCOV antigen, or DPBS to control for background. Sera was heat-inactivated in a 56°C water bath for 1 h prior to serial dilutions starting at 1:50 in dilution buffer (DPBS supplemented with 1% milk and 0.1% Tween-20). ELISA plates were blocked with 200 μL of blocking buffer (DPBS supplemented with 3% milk and 0.1% Tween-20), washed 3 times with PBS plus 2% Tween (PBS-T), and 50 μL of diluted sera was added. After 2 h of incubation, ELISA plates were washed 3 times with PBS-T and bound antibodies were detected using a 1:5000 dilution of goat anti-human IgG conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories, West Grove, PA: cat. 109-036-098). ELISA plates were developed with the addition of 50 μL SureBlue 3, 3', 5, 5'-tetramethylbenzidine substrate (SeraCare: material number 5120-0077) and the reactions were stopped by the addition of 25 μL of 250mM hydrochloric acid after 5 min. Optical densities at 450nm wavelength were obtained on a SpectraMax 190 microplate reader (Molecular Devices, San Jose, CA). Serum antibody titers were expressed as the reciprocal serum dilution at a set OD that was based off of a standard curve from the monoclonal antibody CR3022 (a gift from Ian Wilson, Scripps) starting at 0.5 μg/mL (for S-RBD and S ELISAs) or serially diluted pooled serum (for SARS-CoV-2 N ELISAs and hCoV S ELISAs). Standard curves were included on every plate to control for plate-to-plate variation. Antibody titers for each sample were measured in at least two technical replicates performed on separate days.
Purification of IgG antibodies
For some experiments, we purified IgG from sera samples before completing ELISAs. IgG was purified from sera samples using PureProteome Protein G magnetic beads (Millipore, Darmstadt, Germany: cat. LSKMAGG02) as previously described (Arevalo et al., 2020). Sera samples were diluted in PBS and incubated with 100 μL of washed magnetic beads for 1 h at room temperature with constant mixing. Unbound fractions were removed using the magnetic stand and beads were washed with PBS. Bound IgG was eluted with the addition of 100 μL of 0.2 M glycine, pH 2.5 followed by 5 min incubation at room temperature. The eluant containing purified IgG was neutralized with 10 μL of 1.0 M Tris, pH 8.8 prior to being run in ELISA.
Generation of SARS-CoV-2 pseudotypes
SARS-CoV-2 pseudotypes were generated with a previously described vesicular stomatitis virus (VSV) pseudotype platform (Anderson et al., 2020). Briefly, pseudotyped VSV virions with SARS-CoV-2 Spike were produced through transfection of 293T with 35 μg of pCG1 SARS-CoV-2 S delta18 expression plasmid encoding a codon optimized SARS-CoV-2 S gene with an 18-residue truncation in the cytoplasmic tail (kindly provided by Stefan Pohlmann) (Hoffmann et al., 2020). 30 h post transfection, the SARS-CoV-2 spike expressing cells were infected for 2-4 h with VSV-G pseudotyped VSVΔG-RFP at a multiplicity of infection (MOI) of ∼1-3. Then, the cells were washed twice with media to remove unbound virus. 28-30 h after infection, the media containing the VSVΔG-RFP SARS-CoV-2 pseudotypes were harvested and clarified by centrifugation two times at 6000xg. SARS-CoV-2 pseudotypes were aliquoted and stored at −80°C until used for antibody neutralization analysis.
Quantification of SARS-CoV-2 pseudotype neutralizing antibody titers
Serum SARS-CoV-2 neutralizing antibodies were measured as previously described (Anderson et al., 2020). Vero E6 cells stably expressing TMPRSS2 were seeded in 100 μL at 2.5x104 cells/well in a 96 well collagen coated plate. The next day, heat inactivated serum samples were serially diluted 2-fold and mixed with 50-200 focus forming units/well of VSVΔG-RFP SARS-CoV-2 pseudotype virus and 600ng/mL of 1E9F9, a mouse anti-VSV Indiana G (Absolute Antibody, Oxford, UK: cat. Ab01402-2.0). The serum-virus mixture was incubated for 1 h at 37°C before being plated on VeroE6 TMPRSS2 cells. 23-24 h post infection, the cells were washed, fixed with 4% paraformaldehyde, and visualized on an S6 FluoroSpot Analyzer (CTL, Shaker Heights OH) and individual infected foci were enumerated. The focus reduction neutralization titer 50% (FRNT50) was measured as the greatest serum dilution at which focus count was reduced by at least 50% relative to control cells that were infected with pseudotype virus in the absence of human serum. FRNT50 titers for each sample were measured in at least two technical replicates performed on separate days.
BSL-3 SARS-CoV-2 neutralization assays
Vero CCL81 (ATCC: cat. CCL-81) cells were plated in 96 well plates (100μL/well) at a density of 25,000 cells per well. The following day, in the BSL-3, 100 plaque forming units (pfu) of SARS-CoV-2 (WA-1, BEI cat. NR-52281) was diluted into 30 μl DMEM and added to each dilution of serum samples. The serum and virus were incubated together at room temperature for 1 h and transferred to the supernatant of the Vero CCL81 cells. Each sample was prepared independently in duplicate. Cells were incubated under standard cell culture conditions at 37°C and 5% CO2 for 48 h. Cells were fixed in 4% formaldehyde/PBS for 15 min at room temperature and then washed three times with PBS-T. Cells were blocked (2% BSA/PBS-T) for 60 min and incubated in primary antibody (anti-dsRNA J2, Absolute Antibody cat: Ab01299-2.0) overnight at 4C. Cells were washed 3x PBS and incubated in secondary (anti-mouse IgG alexa 488 Thermofisher cat. A-11029, and hoescht 33342, Sigma Aldrich cat. B2261) for 2 h at room temperature. Cells were washed 3x in PBST and imaged using ImageXpress Micro (Molecular Devices, San Jose, CA) using a 10X objective. Ten sites per well were captured and wells were scored for viral infection.
Quantification and Statistical Analysis
Statistical analyses were performed using Prism version 8 (GraphPad Software, San Diego CA). Reciprocal serum dilution antibody titers were log2 transformed for statistical analysis. ELISA antibody titers below the limit of detection (LOD; reciprocal titer < 50) were set to a reciprocal titer of 25. Log2 transformed antibody titers were compared with unpaired t tests and statistical significance was set to p value < 0.05. Linear regressions were also performed using log2 transform titers and untransformed data from the other variables. We compared antibody titers in pre-pandemic serum samples from individuals who did and did not have a subsequent PCR-confirmed SARS-CoV-2 infection. For these analyses we selected serum sample from individuals with RT-PCR negative results matching sex, age, and race for each SARS-CoV-2 PCR-confirmed case (RT-PCR positive) to define controls for our cohort. In instances we did not find matched controls, we randomly selected patients with RT-PCR negative test results. We also compared antibody titers in pre-pandemic serum samples among SARS-CoV-2 PCR-confirmed individuals in relationship to hospitalization or need for respiratory support due to COVID-19. Multivariate logistic regression was used to compare the antibody differences for these studies. All the models were adjusted by sex, age, race, and analyses were performed in R (R Core Team, 2016). We compared Log2 transformed antibody titers in COVID-19 hospitalized patients at day 0 and day 7. We also compared the fold change in titer by day 7. We compared the fold change in OC43 titers between patients who survived and patients who died by day 28 of hospitalization.
Acknowledgments
This work was supported by institutional funds from the University of Pennsylvania, NIH grants U19AI082630 (E.J.W. and S.E.H.), HL137006 (N.J.M.), HL137915 (N.J.M.), R21AI129531 (P.B.), and R21AI142638 (P.B.); and Peer Reviewed Medical Research Program award PR182551 (P.B.). E.M.A. and T.B.M. were supported by the NIH Training in Virology T32 Program (T32AI007324). P.H. and C.P.A. were supported by the NIH Emerging Infectious Diseases T32 Program (T32AI055400). We thank J. Lurie, J. Embiid, J. Harris, and D. Blitzer for philanthropic support. We thank all members of the Wherry Lab and the Penn COVID-19 Sample Processing Unit (Zahidul Alam, Mary M. Addison, Katelyn T. Byrne, Aditi Chandra, Hélène C. Descamps, Yaroslav Kaminskiy, Jacob T. Hamilton, Julia Han Noll, Dalia K. Omran, Eric Perkey, Elizabeth M. Prager, Dana Pueschl, Jennifer B. Shah, Jake S. Shilan, and Ashley N. Vanderbeck) for sample procurement, processing, and logistics. We thank the staff of the PMBB. We thank F. Krammer (Mt. Sinai) for sending us the SARS-CoV-2 spike RBD expression plasmids. We thank David Anderson for assistance with the graphical abstract.
Author contributions
Serological Assays, E.M.A., E.C.G., C.P.A., M.J.B., M.E.W., S.G., C.M.M., S.R.C., P.H., T.B.M., H.R., and S.C.; Data Analyses, E.M.A., E.C.G., and A.V.; Obtaining and Processing Samples, O.O., D.M., A.E.B., D.A.O., A.R.G., J.E.W., C.A., K.D., O.K., J.D., A.P., J.K., N.H., S.A.A., A.C.H., L.A.V., L.K.-C., M.B.P., and N.J.M.; Manuscript Writing, E.M.A., E.C.G., A.V., D.J.R., and S.E.H.; Supervision, S.G., J.W., M.R.B., E.J.W., N.J.M., S.C., P.B., D.J.R., and S.E.H.; Funding Acquisition, P.B., E.J.W., N.J.M., and S.E.H.
Declaration of interests
A.C.H. is a consultant for Immunai. E.J.W. has consulting agreements with and/or is on the scientific advisory board for Merck, Elstar, Janssen, Related Sciences, Synthekine, and Surface Oncology. E.J.W. is a founder of Surface Oncology and Arsenal Biosciences. E.J.W. is an inventor on a patent (U.S. patent number 10,370,446) submitted by Emory University that covers the use of PD-1 blockade to treat infections and cancer. D.J.R. is on scientific advisory boards for Alnylam, Metrea, Novartis, Pfizer, and Verve; is a consultant for and receives research support from Regeneron for work unrelated to this report; and is a founder of Vascular Strategies and Staten Biotechnologies. S.E.H. has received consultancy fee from Sanofi Pasteur, Lumen, Novavax, and Merck for work unrelated to this report.
Published: February 9, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.02.010.
Supplemental Information
References
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.M., Diorio C., Goodwin E.C., McNerney K.O., Weirick M.E., Gouma S., Bolton M.J., Arevalo C.P., Chase J., Hicks P., et al. SARS-CoV-2 antibody responses in children with MIS-C and mild and severe COVID-19. J. Pediatric Infect. Dis. Soc. 2020:piaa161. doi: 10.1093/jpids/piaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo C.P., Le Sage V., Bolton M.J., Eilola T., Jones J.E., Kormuth K.A., Nturibi E., Balmaseda A., Gordon A., Lakdawala S.S., Hensley S.E. Original antigenic sin priming of influenza virus hemagglutinin stalk antibodies. Proc. Natl. Acad. Sci. USA. 2020;117:17221–17227. doi: 10.1073/pnas.1920321117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Cobey S., Hensley S.E. Immune history and influenza virus susceptibility. Curr. Opin. Virol. 2017;22:105–111. doi: 10.1016/j.coviro.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkman R., Jebbink M.F., Gaunt E., Rossen J.W., Templeton K.E., Kuijpers T.W., van der Hoek L. The dominance of human coronavirus OC43 and NL63 infections in infants. J. Clin. Virol. 2012;53:135–139. doi: 10.1016/j.jcv.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Flannery D.D., Gouma S., Dhudasia M.B., Mukhopadhyay S., Pfeifer M.R., Woodford E.C., Gerber J.S., Arevalo C.P., Bolton M.J., Weirick M.E., et al. SARS-CoV-2 seroprevalence among parturient women in Philadelphia. Sci. Immunol. 2020;5:eabd5709. doi: 10.1126/sciimmunol.abd5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N., Alter H., Hindiyeh M., Mendelson E., Shemer Avni Y., Mandelboim M. Human Coronavirus Infections in Israel: Epidemiology, Clinical Symptoms and Summer Seasonality of HCoV-HKU1. Viruses. 2018;10:E515. doi: 10.3390/v10100515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt E.R., Hardie A., Claas E.C., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouma S., Kim K., Weirick M.E., Gumina M.E., Branche A., Topham D.J., Martin E.T., Monto A.S., Cobey S., Hensley S.E. Middle-aged individuals may be in a perpetual state of H3N2 influenza virus susceptibility. Nat. Commun. 2020;11:4566. doi: 10.1038/s41467-020-18465-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15, e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8, e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L.C., Rattigan S.M., Borgert B.A., Moreno C.A., Solomon B.D., Trimmer-Smith L., et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., André N.M., Chappie J.S., Millet J.K., Whittaker G.R. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J. Mol. Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killerby M.E., Biggs H.M., Haynes A., Dahl R.M., Mustaquim D., Gerber S.I., Watson J.T. Human coronavirus circulation in the United States 2014-2017. J. Clin. Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., et al. UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Contant P., Embong A.K., Kanagaiah P., Chaves F.A., Yang H., Branche A.R., Topham D.J., Sangster M.Y. S Protein-Reactive IgG and Memory B Cell Production after Human SARS-CoV-2 Infection Includes Broad Reactivity to the S2 Subunit. MBio. 2020;11:e01991-20. doi: 10.1128/mBio.01991-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S., Oppong S., Drexler J.F., Gloza-Rausch F., Ipsen A., Seebens A., Müller M.A., Annan A., Vallo P., Adu-Sarkodie Y., et al. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg. Infect. Dis. 2009;15:1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poston D., Weisblum Y., Wise H., Templeton K., Jenks S., Hatziioannou T., Bieniasz P. Absence of SARS-CoV-2 neutralizing activity in pre-pandemic sera from individuals with recent seasonal coronavirus infection. Clin. Infect. Dis. 2020:ciaa1803. doi: 10.1093/cid/ciaa1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Dijkman R., Deng L., Jebbink M.F., Ross H.A., Berkhout B., van der Hoek L. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J. Mol. Biol. 2006;364:964–973. doi: 10.1016/j.jmb.2006.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2016. R: A Language and Environment for Statistical Computing [Online]. Vienna, Austria. Available.https://www.R-project.org/ Accessed. [Google Scholar]
- Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L., Mizgerd J.P. Recent endemic coronavirus infection is associated with less severe COVID-19. J. Clin. Invest. 2021;131:143380. doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar, Daul F., Salvat Lago M., Decker A., et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- Sette A., Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat. Rev. Immunol. 2020;20:457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Lemey P., Maes P., Van Reeth K., Nauwynck H., Pensaert M., Van Ranst M. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006;80:7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Mihaylova V.T., Landry M.L., Foxman E.F. Interference between rhinovirus and influenza A virus: a clinical data analysis and experimental infection study. Lancet Microbe. 2020;1:e254–e262. doi: 10.1016/s2666-5247(20)30114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data generated in this study have been deposited on Mendeley Data: https://doi.org/10.17632/ygv2j9psc5.1.