Abstract
In this study, the complete mitochondrial genome of Inez’s red-backed vole Caryomys inez was sequenced and analyzed as the first species in genus Caryomys. The complete mitochondrial genome of C. inez is 16,354 bp in length and shows a typical vertebrate pattern with 13 PCGs, 22 tRNAs, 2 rRNAs and 2 non-coding regions. To gain a clear phylogenetic position of C. inez, a ML phylogenetic tree was constructed based on 12 PCGs on H-strand from 23 rodent species except for ND6 gene which is on L-strand. As a result, C. inez is clustered with genera Eothenomys and Myodes, showing their close phylogenetic relationships. The present study may facilitate further investigation of the taxonomic studies and phylogenetic analyses of the genus Caryomys.
Keywords: Caryomys inez, complete mitochondrial genome, phylogeny
The Inez’s red-backed vole, Caryomys inez, is a Chinese native species and is distributed in central and northern China (Li & Xue 2008). Genus Caryomys is similar to genus Eothenomys in external morphology and was once regarded as a subgenus of Eothenomys (Allen 1940). Ma and Jiang (1996) reinstated Caryomys as a valid genus and had been accepted by systematists (Luo et al. 2000; Wang 2003; Wilson & Reeder 2005). Liu et al. (2012) confirmed this view based on morphology and two mitochondrial genes (CO1 and Cytb). In this study, we captured a specimen of C. inez in Lingchuan, Shanxi Province (N35.565°, E113.366°), China, and sequenced its complete mitochondrial genome (GenBank accession no. KU200225), and analyzed its phylogenetic relationships to close genera Eothenomys, Myodes, Microtus and Cricetulus in family Cricetidae. The voucher specimen was labelled as S0862 and deposited at Shandong University (Weihai).
The complete mitochondrial genome of C. inez is 16,354 bp in length and consists of 13 protein-coding genes (PCGs), 22 tRNAs, 2 rRNAs and 2 non-coding regions, with a base composition of 33.25% A, 26.99% T, 26.80% C and 12.96% G. Apart from ND6 which is on the L-strand, all PCGs are encoded on the H-strand.
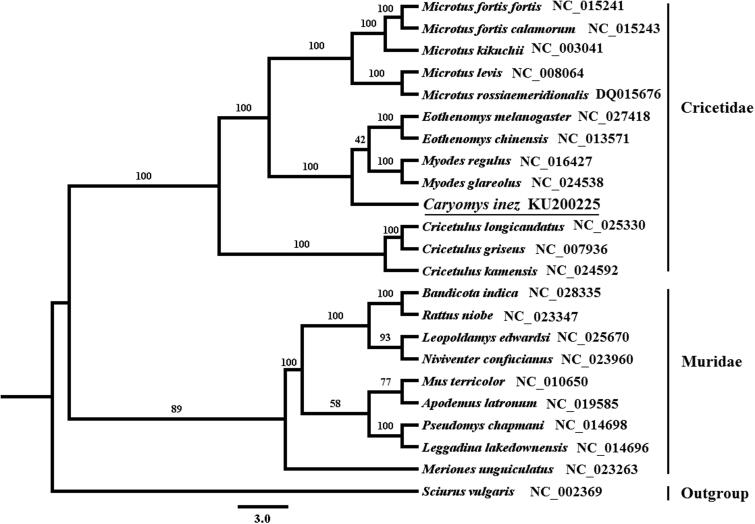
The nucleotide sequence data of 12 H-strand PCGs of C. inez and other 22 rodent species were used for phylogenetic analyses with Sciurus vulgaris as outgroup taxa. The best-fitting model (GTR + I + G) of sequence evolution was obtained by the software jModelTest 2.1.3 (Darriba et al. 2012) under the Akaike Information Criterion (Akaike 1974). The phylogenetic tree of Maximum likelihood (ML) was constructed in RAxML-VI-HPC (Stamatakis 2006) with 1000 bootstrap replicates (Figure 1), the 22 ingroup species used in ML analysis belong to two families, Cricetidae and Muridae. Our analysis shows that the Cricetidae species examined here forms a well-supported monophyletic group and the monophyly of the family Muridae is also supported with high bootstrap value. In our ML phylogenetic tree, C. inez is clustered with genera Eothenomys and Myodes (bootstrap value =100). This study firstly provided the complete mitochondrial genome of species in genus Caryomys, and we expect our data and result may facilitate further investigation of the taxonomic studies and phylogenetic analyses of the genus.
Figure 1.
Maximum likelihood (ML) phylogenetic tree based on 12 PCGs located on the H-strand under GTR + I + G model. Numbers represent node support inferred from ML bootstrap analyses.
Acknowledgements
We would like to thank the anonymous reviewers for providing valuable comments on the article.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Funding information
This research was funded by the Major International (Regional) Joint Research Project of NSFC (Grant No. 31110103910).
References
- Akaike H. 1974. A new look at the statistical model identification. Automat Control IEEE Transact. 19:716–723. [Google Scholar]
- Allen GM. 1940. The mammals of China and Mongolia. Natural history of Central Asia. Vol. XI New York: The American Museum of Natural History. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. JModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YX, Xue XX.. 2008. The microtine distribution and its environmental significance since the Middle Pleistocene in Zhangping Caves of the Qinling Mountains. Sci China Ser D Earth Sci. 51:294–299. [Google Scholar]
- Liu SY, Liu Y, Guo P, Sun ZY, Robert WM, Fan ZX, Fu JR, Zhang YP.. 2012. Phylogeny of oriental voles (Rodentia: Muridae: Arvicolinae): molecular and morphological evidence. Zool Sci. 29:610–622. [DOI] [PubMed] [Google Scholar]
- Luo ZX, Chen W, Guo B.. 2000. Fauna sinica. Mammalia. Vol. 6, Rodentia, Part III: Cricetidae. Beijing: Scientific Publisher. [Google Scholar]
- Ma Y, Jiang JQ.. 1996. The reinstatement of the status of genus Caryomys (Rodentia: Microtinae). Acta Zootaxonom Sin. 21:493–497. (in Chinese with English abstract). [Google Scholar]
- Wilson DE, Reeder DM.. 2005. Mammal species of the world: a taxonomic and geographic reference. Baltimore: The Johns Hopkins University Press. [Google Scholar]
- Stamatakis A. 2006. . RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Wang YX. 2003. A complete checklist of mammal species and subspecies in China: a taxonomic and geographical reference. Beijing: China Forestry Publishing House. [Google Scholar]