Abstract
We present the first mitochondrial genome of the calcified, geniculate coralline red alga Corallina officinalis (Corallinales). The circular genome consists of 26,504 bp and has a gene content consisting of 23 protein-coding genes, 26 transfer RNA genes and two ribosomal RNA genes, with an overall GC content of 30.1%.
Keywords: Calcified macroalgae, Corallina, mitochondrial genome
Corallina officinalis Linnaeus (Corallinales, Rhodophyta) is a geniculate (articulated) coralline macroalga ubiquitous in the intertidal zone of temperate coastal regions across the NE Atlantic (Brodie et al. 2013; Williamson et al. 2015). Turfing assemblages of C. officinalis provide habitat for numerous small invertebrates, shelter via their physical structure from environmental stresses associated with intertidal habitats and a substratum for the settlement of macro- and microalgae (Nelson 2009). Given these attributes, C. officinalis is considered an important autogenic ecosystem engineer (Nelson 2009). Despite this, very limited genomic data are available for this species or coralline algae in general (Kim et al. 2013). To date, a partial genome has been reported for the geniculate coralline Calliarthron tuberculosum (Chan et al. 2011), and the complete mitochondrial genome of the non-geniculate, rhodolith-forming Sporolithon durum (Sporolithales) is available (Kim et al. 2013). Further sequencing of calcified macroalgal genomes is therefore a priority in order to advance this field of research.
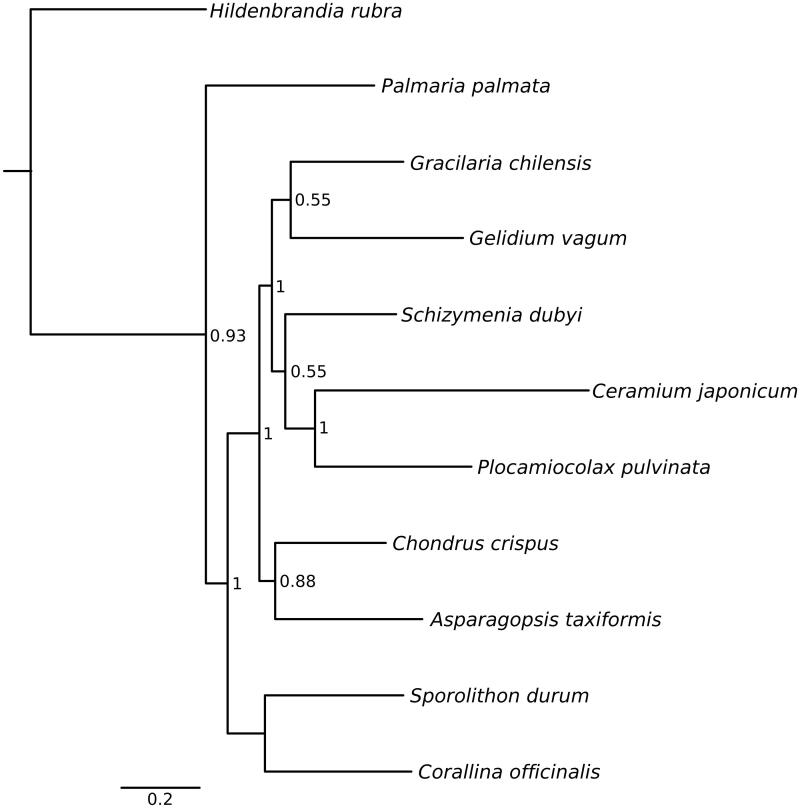
Total genomic DNA was extracted from 0.5 cm2 of a discrete C. officinalis frond (accession no. BM001215284, The Natural History Museum, London) collected from Heybrook Bay, southwest Britain (50°31′68″N, 4°11′92″W) using a modified CTAB extraction (Williamson et al. 2015). Following quantification of double-stranded DNA with a Qubit fluorometer 2.0 (Invitrogen, Waltham, MA), an indexed library was constructed using a TruSeq Nano DNA sample preparation kit (Illumina Inc., San Diego, CA) and sequenced on an Illumina MiSeq flowcell; version 3 chemistry, 2 × 300 paired end. Reads were trimmed using default settings in Geneious v.8.1.7 (http://www.geneious.com, Kearse et al. 2012) and subsequently assembled to the previously reported C. officinalis epitype cox1 sequence (Brodie et al. 2013), GenBank accession no. FM180073. Unassembled reads were iteratively mapped and reassembled to the putative cox1 sequence until the resulting contig could be circularized. Gene boundaries were annotated using MITOS (Bernt et al. 2013) and verified by visualization of open reading frames and comparison with alignments of Florideophycean mitochondrial genes. Amino acid sequences for each of 24 protein-coding genes (Yang et al. 2015) were aligned with 10 reference mt genomes (Figure 1) using Geneious. Aligned sequences were concatenated, verified by eye and subjected to phylogenetic analysis using Mr Bayes (Ronquist & Huelsenbeck 2003) using the JONES + G amino acid substitution model after Yang et al. (2015).
Figure 1.
Bayesian inference tree from analysis of concatenated protein-coding genes. Only nodes supported by pp >0.50 are annotated. GenBank accession numbers for each species: Hildenbrandia rubra NC026055, Palmaria palmata NC026056, Gracliaria chilensis NC026831, Gelidium vagum NC023077, Schizymenia dubyi KJ398163, Ceramium japonicum KJ398159, Plocamiocolax pulvinata NC014773, Chondrus crispus NC001677, Asparagopsis taxiformis NC026843, Sporolithon durum KF186230 and Corallina officinalis KU641510.
We determined the mitochondrial genome of C. officinalis to be 26,504 bp in length. The sequence is deposited in GenBank under accession no. KU641510. The mitochondrial genome codes for 51 genes: 23 protein-coding genes, 26 tRNA genes and the large and small rRNA genes. Gene order follows a similar pattern to those of other Florideophycean red macroalgae. The ATG start codon is used in all protein-coding genes except nad2 (GTG) and sdh3 (ATT). The TAA stop codon is used in all protein-coding genes except cox2, nad6 and rps12 (TAG). The overall GC content is 30.1% with a GC skew of 0.03 and AT skew of 0.06. Nucleotide composition of the entire mt genome is A= 9823 (37.1%), T = 8706 (32.8%), G = 4099 (15.5%) and C = 3876 (14.6%).
Acknowledgements
The authors would like to thank the staff at the Natural History Museum (NHM) sequencing facility.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding informtion
Funding was provided by the NHM DIF grant. This work was supported by an NHM Development Innovation Fund grant.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation . Mol Phylogenet Evol. 69:313–319. [DOI] [PubMed] [Google Scholar]
- Brodie J, Walker RH, Williamson CJ, Irvine LM.. 2013. Epitypification and redescription of Corallina officinalis L., the type of the genus, and C. elongata Ellis et Solander (Corallinales, Rhodophyta). Cryptogamie, Algol. 34:49–56. [Google Scholar]
- Chan XC, Yang EC, Banerjee T, Yoon HS, Martone PT, Estevez JM, Bhattacharya D.. 2011. . Red and green algal monophyly and extensive gene sharing found in a rich repertoire of red algal genes. Curr Biol. 21:328–333. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KM, Yang EC, Kim JH, Nelson WA, Yoon HS.. 2013. Complete mitochondrial genome of a rhodolith, Sporolithon durum (Sporolithales, Rhodophyta). Mitochondrial DNA. 26:155–156. [DOI] [PubMed] [Google Scholar]
- Nelson WA. 2009. Calcified macroalgae – critical to coastal ecosystems and vulnerable to change: a review. Marine Freshwater Res. 60:787–801. [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. . MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Williamson CJ, Walker RH, Robba L, Yesson C, Russell S, Irvine LM, Brodie J.. 2015. Toward resolution of species diversity and distribution in the calcified red algal genera Corallina and Ellisolandia (Corallinales, Rhodophyta). Phycologia. 54:2–11. [Google Scholar]
- Yang EC, Kim KM, Kim SY, Lee JM, Boo GH, Lee J-H, Nelson WA, et al. 2015. Highly conserved mitochondrial genomes among multicellular red algae of the Florideophyceae. Genome Biol E. 7:2394–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]