Abstract
In this study, complete chloroplast genome sequences of Rhus chinensis was characterized by de novo assembly using whole genome sequence data. The chloroplast genome of R. chinensis were 149,011bp long, which was comprised of a large single copy region of 96,882 bp, a small single copy region of 18,647bp, and a pair of inverted repeats of 16,741 bp. The genome contained 77 protein-coding genes, four rRNA genes and 30 tRNA genes. Phylogenetic tree revealed that R. chinensis was closely grouped with Spondias species, S. tuberosa and S. bahiensis, belonging to the Anacardiaceae family.
Keywords: Rhus chinensis, chloroplast, genome sequence
The Rhus genus in the Anacardiaceae family consists of 250 species that are mainly distributed in temperate and tropical regions (Rayne & Mazza 2007). Rhus chinensis has been used as a traditional medicine in Asia for treatment of cold, fever and malaria (Rayne & Mazza 2007). In particular, extract of the galls, induced by infection with Schlechtendalia chinensis (Bell), is known to be effective on bacterial control since galls are rich in gallotannin (50–70%) along with phenolic compounds such as gallic acid (Djakpo & Yao 2010). Previous R. chinensis studies have mainly focused on its biological activities, however, there is no report regarding genome of this plant species until now. In this study, we characterized a chloroplast genome sequence of R. chinensis and analyzed its phylogenetic relationship with other species. The genome information can be further applied to genetic and molecular study of this plant species.
Chloroplast genome of the R. chinensis was characterized based on the previous studies (Kim et al. 2015a,b). In brief, we isolated total genomic DNA from leaf tissues of R. chinensis which were collected from Yanggu, Gangwon Province (38°17′33.2″N, 128°08′37.1″E) in South Korea (Seoul National University voucher No. IM151120-1) and performed whole genome sequencing using an Illumina MiSeq platform (Illumina, San Diego, CA). High quality paired end reads of about 0.8 Gb were assembled using CLC genome assembler 4.6 (v. beta 4.6, CLC Inc., Aarhus, Denmark). The contigs representing chloroplast sequence were retrieved, ordered and combined into a single sequence by comparing with chloroplast genome of Azadirachta indica (NC_023792.1).
Complete chloroplast genome of R. chinensis (KX447140) was 149,011 bp in length. The genome consisted of four distinct parts such as a large single copy region of 96,882 bp, a small single copy region of 18,647bp, and a pair of inverted repeats of 16,741 bp. In the genome, 77 protein-coding genes, four rRNA genes, and 30 tRNA genes were identified through DOGMA annotation (http://dogma.ccbb.utexas.edu/) and BLAST searches.
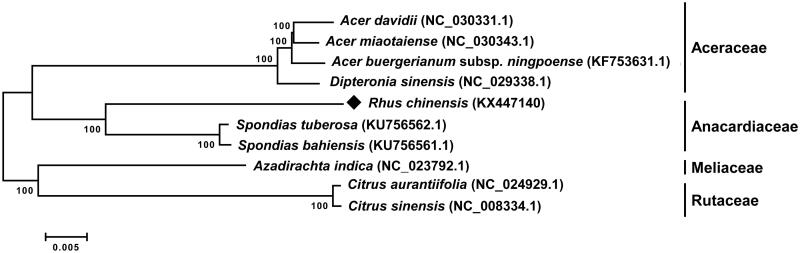
Complete chloroplast genome sequence of R. chinensis was subjected to phylogenetic analysis with nine species belonging to the Sapindales order by neighbour-joining method with 1000 bootstrap values in the MEGA 6.0 (MEGA Inc., Englewood, NJ) (Figure 1) (Tamura et al. 2013). The phylogenetic analysis revealed that R. chinensis was closely located with two Spondias species, S. tuberosa (KU756562.1) and S. bahiensis (KU756561.1), belonging to the Anacardiaceae family.
Figure 1.
Phylogenetic tree showing relationship between Rhus chinensis and nine species belonging to the Sapindales order. The phylogenetic tree was constructed using complete chloroplast genome sequences of the ten species and analyzed neighbour-joining method with 1000 bootstrap values in MEGA 6.0. The numbers in the nodes of phylogenetic tree indicated the bootstrap support values.
Acknowledgments
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- Djakpo O, Yao W.. 2010. . Rhus chinensis and Galla Chinensis-folklore to modern evidence: review. Phytother Res. 24:1739–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ.. 2015a. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 10:e0117159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015b. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayne S, Mazza G.. 2007. Biological activities of extracts from sumac (Rhus spp.): a review. Plant Foods Hum Nutr. 62:165–175. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]