Abstract
We first determined the complete mitochondrial genome of the White-spotted guitarfish Rhynchobatus australiae (Rajiformes, Rhinobatidae). The complete mitogenome was 16,804 bp, with a base composition of 32.3% A, 27.5% T, 26.9% C and 13.3% G, containing 13 protein-coding genes, two rRNAs, 22 tRNAs and a control region (D-loop). It had 35 bp short intergenic spaces and 39 bp overlaps between genes. The gene order and composition of R. australiae was similar to most other fishes. The codon usage followed the typical vertebrate mitochondrial pattern (ATG or GTG for start codon and TAA or T for stop codon). The phylogenetic result showed that R. australiae was clustered with the Rhinobatos.
Key words: Rajiformes, Rhynchobatus australiae, Rhinobatidae
The White-spotted guitarfish Rhynchobatus australiae (Rajiformes, Rhinobatidae) was a locally common species found in the Western Pacific from the Gulf of Thailand and the Philippines to Queensland in Australia, mainly inhabiting inshore waters (Compagno & Last 1999; White et al. 2006). The molecular and genetic research of this species were rare. In this study, we first determined the complete mitochondrial genome of R. Australiae with GenBank accession no. KU746824 and analyzed the phylogenetic relationship in Rajiformes.
One specimen of R. australiae, which was captured from the Gulf of Thailand, was landed on a pier in Songkhla, Thailand. Tissue of R. Australiae was preserved in Wenzhou Medical University by 95% alcohol with voucher THSK20121613. The whole specimen of R. Australiae could not be preserved for its great size. The experimental protocol and data analysis methods followed Chen et al. (2015). We constructed the phylogenetic tree using the Bayesian method. Thirteen species of Rajiformes (including R. Australiae) with the complete mitogenomes available in the Genbank were selected. The outgroup was Dasyatis akajei (Myliobatoformes).
The complete mitogenome of R. Australiae was 16,804 bp, containing 13 protein-coding genes, two rRNAs, 22 tRNAs and a control region identical to other skates (Chen et al. 2015). The mitogenome of R. Australiae had 35 bp short intergenic spaces located in 13 gene junctions and 39 bp overlaps located in six gene junctions. The nucleotide composition of the genome was strongly A–T skewed, with A + T content of 59.8% for the entire sequence (32.3% A, 27.5% T, 26.9% C and 13.3% G). Except for ND6 gene and eight tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser1(UGA), Glu and Pro), all other genes were encoded on the H-strand. Twelve out of 13 mitochondrial protein-coding genes used ATG as the start codon (COI gene started with GTG). Two types of stop condons were used, except for COII and ND4 ending with the incomplete stop condon T, the rest protein-coding genes all ending with TAA. The 22 tRNA genes were ranged from 66 bp (tRNA-Ser2) to 75 bp (tRNA-Leu1), and all tRNAs could be folded into the typical cloverleaf secondary structures except the tRNA-Ser2, which replaced the dihydrouridine arm by a simple loop. Both 12S rRNA (967 bp) and 16S rRNA (1, 690 bp) genes were between tRNA-Phe and tRNA-Leu1 genes, separated by tRNA-Val gene. The origin of light-strand replication (39 bp) was located between tRNA-Asn and tRNA-Cys genes. The control region was 1106 bp in length, rich in A + T (64.8%) and poor in G (13.3%).
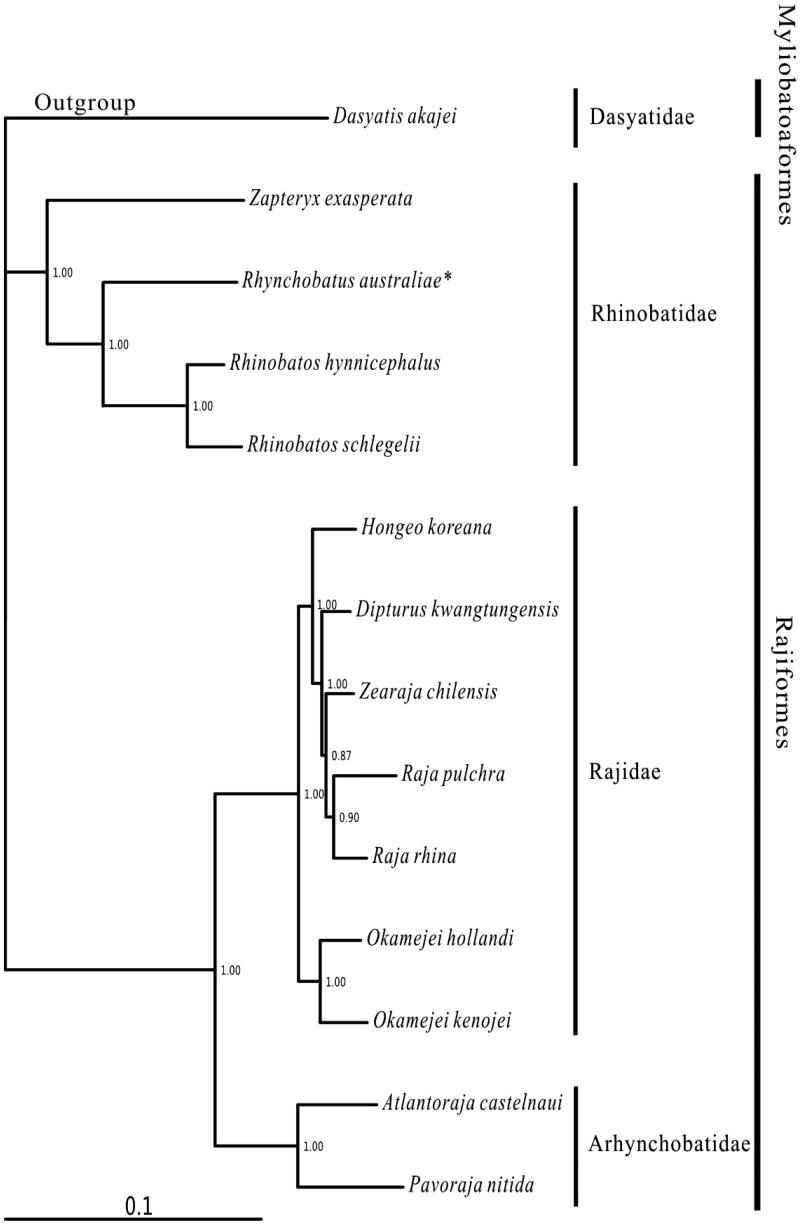
All 13 mitogenomes available in Rhinobatidae, representing three families, were used to analyze the phylogenic position of R. australiae in this study. These three families formed a (Rhinobatidae + (Rajidae + Arhynchobatidae)) relationship (Figure 1), which was consistent to the morphological result (McEachran & Dunn 1998). Rhynchobatus australiae was sister to the Rhinobatos clade, then this clade was clustered to Zapteryx exasperata, which showed Rhynchobatus were closer with Rhinobatos than Zapteryx in phylogeny.
Figure 1.
Phylogenetic position of Rhynchobatus australiae. Dasyatis akajei (NC_021132.1) was selected as the outgroup. The 13 species of Rajiformes were Zapteryx exasperata (NC_024937.1), Rhynchobatus australiae (KU746824), Rhinobatos hynnicephalus (NC_022841.1), R. schlegelii (NC_023951.1), Hongeo koreana (NC_021963.1), Dipturus kwangtungensis (NC_023505.2), Zearaja chilensis (KJ913073.2), Raja pulchra (NC_025498.1), Raja rhina (KC914434.1), Okamejei hollandi (KP756687.1), O. kenojei (NC_007173.1), Atlantoraja castelnaui (NC_025942.1) and Pavoraja nitida (NC_024599.1).
Acknowledgments
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This study was supported by Ministry of Science and Technology of Zhejiang Province (2013F50015).
References
- Chen X, Ai W, Shi X, Gao T.. 2015. Mitochondrial genome of the ringstraked guitarfish Rhinobatos hynnicephalus (Elasmobranchii: Rajiformes). Mitochondrial DNA. 26:653–654. [DOI] [PubMed] [Google Scholar]
- Compagno LJV, Last PR.. 1999. Rhinidae (=Rhynchobatidae). Wedgefishes In: Carpenter KE, Niem V, editors. FAO identification guide for fishery purposes. The Living Marine Resources of the Western Central Pacific. Rome: FAO; p. 1418–1422. [Google Scholar]
- McEachran JD, Dunn KA.. 1998. Phylogenetic analysis of skates, a morphologically conservative clade of elasmobranchs (Chondrichthyes: Rajidae). Copeia. 1998:271–290. [Google Scholar]
- White WT, Last PR, Stevens JD, Yearsley GK, Fahmi D.. 2006. Economically important sharks and rays of Indonesia. [Hiu dan pari yang bernilai ekonomis penting di Indonesia]. Canberra, Australia: Australian Centre for International Agricultural Research; p. 208. [Google Scholar]