Abstract
SARS-CoV-2 is a recently identified coronavirus accountable for the current pandemic disease known as COVID-19. Different patterns of disease progression infer a diverse host immune response, with interferon (IFN) being pivotal. IFN-I and III are produced and released by virus-infected cells during the interplay with SARS-CoV-2, thus establishing an antiviral state in target cells. However, the efficacy of IFN and its role in the possible outcomes of the disease are not yet defined, as it is influenced both by factors inherent to the virus and to the host. The virus exhibits multiple strategies to counteract the innate immune response, including those shared by SARS-CoV and MERS-CoV and other novel ones. Inborn errors in the host may affect IFN-related effector proteins or decrease its levels in plasma upon neutralization by preexistent autoantibodies. This battle between the IFN response triggered upon SARS-CoV-2 infection, its magnitude and timing, and the efficacy of its antiviral tools in dispute against the viral evasion strategies together with the genetic factors of the host, generate a scenario whose fate contributes to defining the severity of COVID-19.
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; HCoV, human coronaviruses; MERS-CoV, Middle East respiratory syndrome coronavirus; ORFs, open reading frames; S, spike; E, envelope; M, membrane; N, nucleocapsid; ACE2, angiotensin-converting enzyme 2; SARS, severe acute respiratory syndrome; IFN, interferon; IFNAR, type I IFN receptor; IFNLR, IFN-λ receptor; ISGs, interferon-stimulated genes; PRRs, pattern recognition receptors; PAMPs, pathogen-associated molecular patterns; RIG-I, retinoic acid-inducible gene 1; MDA5, melanoma differentiation-associated gene 5; LGP2, laboratory of genetics and physiology-2; MAVS, mitochondrial antiviral signaling protein; TLRs, Toll-like receptors; IRF, IFN regulatory factor; TRAF3, tumor necrosis factor receptor-associated factor 3; TANK, TRAF family member-associated NF-κB activator; TBK1, TANK-binding kinase 1; IκB, inhibitor of nuclear factor κB; IKKε, IκB kinase-ε; NF-κB, transcription factors nuclear factor-κB; Tyk2, Jak tyrosine kinases tyrosine kinase 2; JAK1, Janus kinase 1; STATs, signal transducer and activator of transcription; ISGF3, IFN-stimulated growth factor 3; ISRE, IFN-stimulated response element; AAV, adeno-associated virus; SOCS, suppressor of cytokine signaling proteins; HIF1α, hypoxia-inducible factor-1α; IGFBP3, insulin-like growth factor-binding protein 3; KLF-5, Krüpple-like factor 5; dsRNA, double stranded RNA; TBK1, TANK binding kinase 1; NPC, nuclear pore complex; KPNA2, karyopherin-α2; IFITM, interferon-induced transmembrane proteins; TMPRSS2, transmembrane serine protease 2; scRNA-seq, single-cell RNA-sequencing; MOI, multiplicity of infection
Keywords: COVID-19, Human coronavirus, Interferon, Immune evasion, Inborn errors
1. Introduction: Type I and III interferons as the first line of defense against viruses
Among the cytokines initially involved in the innate immune response, interferon (IFN) I and III encompass the first line of defense against viruses [1]. Their production and release at the cellular level occur after the detection of viral infections by innate immune sensors. Common facts for IFN-I and III include their induction by a viral infection, activation of downstream signaling pathways, and transcriptional events. However, there are functional differences between them including the cellular receptor that, while IFN-I (IFN-α, IFN-β, IFN-ε, IFN-κ, IFN-ω) binds to the type I IFN receptor (IFNAR) widely expressed in all nucleated cells, IFN-III binds to the IFN-λ receptor (IFNLR), which cellular expression is restricted in the epithelial cells of the respiratory, gastrointestinal, and urogenital systems, keratinocytes, hepatocytes, endothelial cells, and some subsets of innate immune cells (such as neutrophils, macrophages, and dendritic cells). IFNAR is formed by two subunits: IFNAR1 and IFNAR2, while IFNLR consists of subunits IFNLR1 and IL-10Rβ (shared by the IL-10 receptor) [1].
A sophisticated antiviral defense status is triggered after IFNs engage with their cellular receptors. Such protection involved the transcription of hundreds of interferon-stimulated genes (ISGs), able to synthesize multiple proteins committed to “interfere” with every step of viral replication [2]. Comparatively, a faster ISGs induction and decline is promoted by IFN-I. Nevertheless, current data suggests that both IFNs can differentially engage their receptors to produce different signaling outcomes [3,4].
Type I and type III IFNs not only create an intracellular antiviral state but also activate the adaptive immune responses against viruses. The latter claims a fine-tuning regulation to avoid undesirable consequences. Particularly, when uncontrolled response, IFN-I signaling can be deleterious through its systemic, pro-inflammatory effects [5]. Whether the IFN response has a protective or pathogenic effect against SARS and MERS seems to be dependent on the context in which IFN signaling is induced.
Interestingly, many viruses have evolved to avoid, subvert, or directly interfere with the IFN function of their hosts. Such viral immune evasion includes viral proteins that work at different stages of the IFNs-pathway. These strategies are deployed by viruses that cause both acute and persistent infections. Among the former, SARS-CoV and MERS-CoV are typical examples [6,7].
The objective of this review will be focused on offering an update description based on the knowledge regarding the mechanisms involved in the production, magnitude and timing, signaling, and genesis of type I and III interferon response against SARS-CoV-2 infection. The still-controversial arguments that link these cytokines to the immune damage of COVID-19 patients will be included. Finally, the focus will be aimed at the pivotal counterattack strategies to interferon-mediated antiviral actions, where we include the mechanisms of evasion and the antiviral effectors occurring in the virus, and the events of congenital errors observed in patients seriously ill with COVID-19.
2. Brief description of SARS-CoV-2
As a novel betacoronavirus, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) shares its genomic organization and exhibits high genome sequence identity against the other two human coronaviruses (HCoV), causing severe respiratory diseases: almost 80% with SARS-CoV and 50% with Middle East respiratory syndrome coronavirus (MERS-CoV). The single-stranded RNA genome contains six functional open reading frames (ORFs) ordered from 5′ to 3′: replicase (ORF1a/ORF1b), spike (S), envelope (E), membrane (M), and nucleocapsid (N), and also including thee seven putative ORFs encoding accessory proteins intermingled between the structural genes. The vast majority of the SARS-CoV-2 encoded proteins show similar length to the homologous ones in SARS-CoV, sharing more than 90% amino acid identity in four structural genes, except for the S gene. The largest gene coding for the replicase covers two-thirds of the 5′ genome and encodes a large polyprotein (pp1ab), which renders 16 nonstructural proteins after its proteolytical cleavage. These nonstructural proteins (nsp) are involved in transcription and virus replication and exhibit greater than 85% amino acid sequence identity with SARS-CoV [8]. SARS-CoV-2 interacts with the angiotensin-converting enzyme 2 (ACE2) receptor, highly expressed in pneumocytes type II, and targets the lower part of the respiratory tract causing severe acute respiratory syndrome (SARS).
3. Type I and III interferon-mediated immune response against viruses
3.1. Type I and III IFN synthesis and signaling cascades
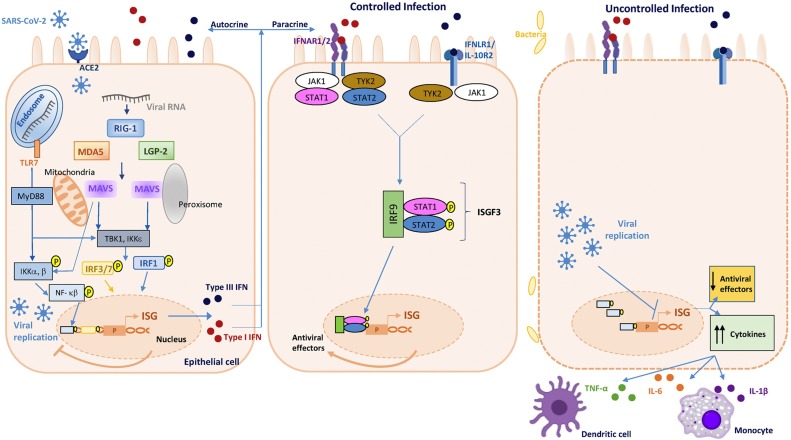
As depicted in Fig. 1 , a rapid IFN-I production is induced into the cell once the pattern recognition receptors (PRRs) sense pathogen-associated molecular patterns (PAMPs) such as viral nucleic acids (RNA from SARS-CoV-2), triggering a range of signaling cascades and subsequent gene activation [9]. As mentioned before, IFN-I is produced and released acting in both autocrine and paracrine ways, and generates an antiviral state supported by the rapid expression of ISGs [2]. Concomitantly, this antiviral defense also implies different immune cells recruited and activated by IFN-I induced proinflammatory cytokines to promote virus clearance and activation of a well-controlled and sustained adaptive immune response [10,11].
Fig. 1.
Type I and III interferons response following SARS-CoV-2 sensing during controlled (left and central cell) and uncontrolled (right cell) infection. Schematic representing the canonical pathway of pathogen recognition receptor (PRR) sensing of intracellular pathogens. Pathogen-associated molecular patterns (PAMPs) -such as viral RNA- are recognized by pathogen recognition receptors –PRRs- (RIG-1, MDA5, and LGP2) lead to activation and phosphorylation of IRF3/7 and IRF1, their nuclear traslocation, and drive the expression of both types I and III interferons. Both interferons are translated and secreted from the infected cell in an autocrine or paracrine manner. Upon engagement of IFN-I to the IFNAR and of IFN-III to the IFNLR receptor, signal transductions start, leading to the expression of interferon-stimulated genes (ISG). These opposite scenarios compare the IFN-I/III optimal response controlling SARS-CoV-2 infection (left and central cell), and the counteracted and/or deficient IFN-I/III response (right cell) associated with high virus replication, immune-mediated complications (including down-regulation of ISGs, together with exacerbated NF-κB activation), and lung immunopathology (“cytokine storm”, accumulation of monocytes, vascular damage, and abnormal T cell response).
The IFN-production process starts with an early event in the cytoplasm when cellular PRRs -including retinoic acid-inducible gene 1 (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and the still enigmatic laboratory of genetics and physiology-2 (LGP2) along with the cooperation of MDA5, sense viral RNA. Upon PAMP (viral RNA)-PPRs (RIG-I/MDA-/LGP-2) interaction, sensors are activated and interact with the mitochondrial antiviral signaling protein (MAVS), a downstream adaptor, which promote its activation [9,12].
Besides its cytosolic sensing, viral RNA is detected when located in the endosomal membrane by the Toll-like receptors (TLRs). Specifically, TLR3 detect double-stranded RNA, while single-stranded RNA is recognized by TLR7 -particularly involved in SARS-CoV and MERS-CoV- and TLR8. Innate sensing signaling cascade continues with the downstream adaptor protein molecules for TLRs, MyD88 (for TLR4, TLR7, TLR8) and TRIF (for TLR3, TLR4), important for protection against human coronavirus infections [13].
The downstream central event that stimulates type I and III interferons is the phosphorylation of the IFN gene “master regulators”, IFN regulatory factor (IRF)3 and IRF7 through tumor necrosis factor receptor-associated factor 3 (TRAF3), TRAF family member-associated NF-κB activator (TANK)-binding kinase 1 (TBK1), and the inhibitor of nuclear factor κB (IκB) kinase-ε (IKKε) [[14], [15], [16]]. Upon phosphorylation, IRF3 and/or IRF7 dimerize and translocate into the nucleus, where they induce the expression (and release) of IFN-I as well as a subset of early ISGs [17]. In parallel, the transcription nuclear factor-κB (NF-κB) stimulates the production of pro-inflammatory cytokines (e.g., IL-1, IL-6, TNF-α).
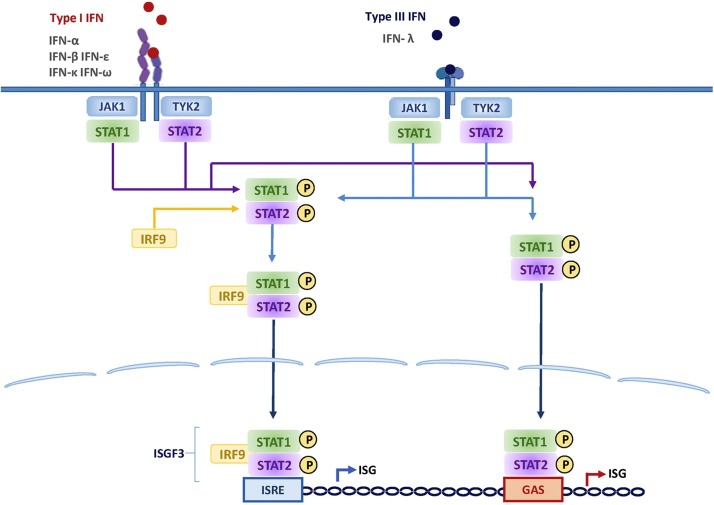
Once released, IFN-I binds to the interferon-α and β receptor (IFNAR, composed of the IFNAR1 and IFNAR2 subunits), leading to the activation of Jak tyrosine kinases, tyrosine kinase 2 (Tyk2) and Janus kinase 1 (JAK1), already pre-associated and able to phosphorylate specific tyrosine residues of the IFN receptor chains. In this condition, these domains of the IFNAR serve as docking sites for the recruitment of signal transducer and activator of transcription (STATs) [18]. Once the STAT1 and STAT2 are phosphorylated and activated, they are heterodimerized, and associated with the DNA binding protein IRF9 to form a complex known as IFN-stimulated growth factor 3 (ISGF3). ISGF3 translocates to the nucleus and binds DNA containing an IFN-stimulated response element (ISRE) to activate gene transcription of ISGs, thus inducing the expression of numerous ISG products that establish the antiviral state at the site of the viral infection [19,20] (Fig. 1, Fig. 2 ). Nevertheless, it has been reported that different states of ISGF3 are able to control the switch from homeostatic to enhanced ISGs expression in macrophages used in a murine model. Hence, IFNs switch cells from a resting state to induced gene expression by alternating subunits of the transcription factor ISGF3. Whereas the preformed STAT2-IRF9 complexes control the basal expression of ISGs, type I IFNs (and IFN-II or IFN-γ) cause the promoter binding of a complete ISGF3 complex containing STAT1, STAT2, and IRF9 [21]. Likewise, viral RNA may also be recognized by its double-stranded RNA elements using the cellular protein kinase receptor (PKR). This enzyme blocks protein synthesis in infected cells, and since it is an ISG, its expression is also under IFN-I influence [22].
Fig. 2.
IFN signaling through the Jak-STAT pathway. Type I (left panel) and type III (right panel) IFNs bind to distinct receptors but activate the same downstream signaling events, and induce almost identical sets of genes mainly through the activation of IFN-stimulated gene factor 3 (ISG3) and STAT1 homodimers.
3.2. Uncontrolled interferon-mediated response during SARS-Cov-2 infection
Beyond their antiviral activity, IFNs are also responsible for activities related to the regulation of metabolism and cell death, as well as the recruitment and activation of immunocompetent cells. In this context, during human coronavirus infections, an elevated virus replication may eventually lead to widespread inflammation and exacerbated cytokines release, also known as a “cytokine storm” as an uncontrolled response that propitiates inflammatory diseases [23]. Host-related factors such as the timing in the IFN-response may enhance such deleterious outcomes. For instance, when the IFN-I response is delayed, a failure to obstruct rapid replication of MERS infection in mice was observed. Subsequently, increased infiltration and activation of pulmonary mononuclear cells and neutrophils were present, as well as enhanced proinflammatory cytokine expression favoring severe pneumonia [23,24]. Among COVID-19 patients, it is critical to understand the regulation of the IFN-I response upon infection, with a clear negative impact of a delayed IFN-I response on viral control and disease severity. Hadjadj et al. have recently studied COVID-19 patients at an earlier disease stage who exhibited high viremia accompanied by exacerbated IFN-I response. Among them, they found upregulated IFNAR signaling, down-regulation of ISGs, together with exacerbated NF-κB activation. Their immune phenotypic profiling in peripheral blood mononuclear cells revealed disproportionate cytokine release, and hyperinflammation directly related to clinical severity [25] (Fig. 1).
However, the factors related to the SARS-CoV-2 infection itself and to those dependent on the host capable of controlling the temporality of the IFN-I response remain obscure. In this sense, it is of particular relevance to clarify the mechanisms that regulate the on/off of the IFN-I response as well as its autocrine and paracrine effects.
Upon viral infection, IFN-III is the most abundant in the mucosal, respiratory, gastrointestinal, and urogenital tracts. They are secreted by epithelial cells, macrophages, CD8 + T lymphocytes, NK cells, Treg lymphocytes, dendritic cells, and hepatocytes [1]. At least, during the respiratory infection caused by type A influenza virus, it seems to be produced earlier [26]. Regarding SARS-CoV-2, upon infection of intestinal epithelial cells, using both colon-derived cell lines and primary colon organoids, the IFN-III response appears to be more efficient than IFN-I at controlling viral replication [27,28].
The PRRs contributed to the induction of IFN-III expression overlapping with those eliciting IFN-α and IFN-β expressions. However, the selective IFN-III expression, but not IFN-I, is mediated by Ku70 (a cytosolic DNA sensor) [29]. Differences between IFN-I and IFN-III in the gene promoter support the higher dependence of IFN-III on NF-κB than IFN-I expression [30]. Likewise, IFN-III production involves MAVS signaling from the peroxisome -highly abundant in epithelial cells-, in contrast to the mitochondrial location for IFN-β expression [31].
Similar to IFN-I, IFN-III activates the JAK–STAT signaling pathway upon IFNLR engagement (Fig. 1, Fig. 2). The IFNLR is expressed on cells of epithelial origin, including respiratory epithelial cells [32]. IFN-III is present in the lower, but not upper, airways of patients with COVID-19. Critically ill patients with COVID-19 showed increased amounts of IFN-III mRNA in nasopharyngeal and bronchoalveolar samples but suppressed IFN-I in peripheral blood [22,29]. Among these patients, different reports offer evidence of the growing interest in the use of IFN-III as a therapeutic alternative based on its antiviral properties [28,33,34]. However, some pathophysiological roles of IFN-III induced by RNA viruses have generated controversies. Dendritic cells in the lung may cause an IFN-III mediated barrier damage that compromises the tissue integrity thus predisposing to lethal bacterial superinfection [32]. Likewise, Major et al. reported that IFN-III impairs the differentiation of alveolar epithelial progenitor cells into secretory and ciliated cell subtypes, and further identified that such impairment on epithelial proliferation is dependent on the expression of IFNLR. Furthermore, IFN-III caused expansion and restructuring of the nasal microbiota, and impaired the epithelial barrier function, which allowed bacteria to invade and colonize the tissue [35]. These findings provide a strong rationale for rethinking the still controversial IFN-III use in clinical practice, including COVID-19 patients.
4. HCoV infections and IFN-mediated response
4.1. Host and viral factors influencing the magnitude and dynamics of IFN-I response
The innate immune response to viruses shows widespread heterogeneity at the level of single cells. According to the cell type and its microenvironment, different dynamics of IFN-I response have revealed that it is induced only in a fraction of virus-infected cells and, subsequently, a portion of cells exposed to IFN upregulates ISGs. Likewise, using single-cell analysis, the amplitude and kinetics of the response appear to be heterogeneous for a given cell type. Such heterogeneity in the IFN-I response is also sustained by dissimilar cellular metabolism, translational and transcriptional states at the activation time that influences the level of sensors, adaptors, and receptors. The dissimilar timing of the IFN response upon viral replication is crucial to define its efficacy in controlling viral spread [[36], [37], [38], [39], [40]]. The preferential distribution of IFNLR on epithelial cells suggests functional differences between IFN-I and IFN-III at epithelial surfaces, despite their redundant functions. Comparatively, faster antiviral protection is generated by IFN-I signaling upon a rapid robust induction of ISGs, thereby leading to differential antiviral environments [[36], [37], [38]].
Among the viral factors influencing the IFN-I response, viral fitness also appears to shape the dynamics of the host antiviral response at the whole population level [1]. Even more, the capability of the virus to modulate IFN signaling is an important hallmark for virulence. For instance, a stronger IFN-I response was detected in cells infected with HCoV-229E rather than those infected with SARS-CoV or MERS-CoV, thus, thus allowing inferring IFN synthesis suppression [6], or a delayed response in the lung with IFN-I detectable after the peak of the viral replication achieved [41]. Using models of the human airway, it was reported that SARS-CoV and MERS infections show a peak of its replication preceding the delayed ISGs response. In this regard, the virus-induced epigenetic modulation is pivotal on the transcriptional activity of STAT1 and PKR pathways, thus deferring the induction of ISGs expression. Induction of IFN-β and ISGs in SARS-CoV and MERS-CoV infection appeared to occur later compared with type A influenza virus infection [42].
4.2. Timing of the IFN-I response for the SARS-CoV-2 infection
This is still a controversial issue. On the one hand, as early as 3 days after infection and later beyond clearance of the virus, the immune response analysis of COVID-19 patients reported a lack of IFN-I/III response in serum despite robust cytokine and chemokine production; pro-inflammatory cytokines and chemokines were strongly elevated without detectable levels of type I and III IFNs. Comparing transcriptional profiles obtained from post-mortem lung samples from COVID-19-positive patients with biopsied healthy lung tissue from uninfected individuals demonstrated ∼2000 differentially expressed genes significantly induced in response to SARS-CoV-2 including a subset of ISGs with no IFN-I or IFN-III. Peripherally derived macrophages predominated in the lungs of severe cases which correlates with high levels of chemokines, including CCL2, CCL8, and CCL11, and a significant increase in circulating IL-6, IL1RA, CCL2, CCL8 CXCL2, CXCL8, CXCL9, and CXCL16 levels was detected in serum samples but consistently tested negative for IFN-β and the IFN-λ family of IFNs. However, this response is detected at low multiplicity of infection (MOI), when the virus is not a strong inducer of the IFN-I and -III system, as opposed to conditions where MOI is high [43]. Other studies suggest that rather than its complete absence, IFN response may be delayed at an earlier stage of infection. Therefore, in contrast with early reports, accumulating data obtained from patients with severe COVID-19 offers evidence of a strong IFN-I response, after the initial delayed, or even suppressed, interferon response. Such vigorous IFN-I response could exacerbate hyperinflammation in the progression to severe COVID-19 through diverse mechanisms. Moreover, a study based on four COVID-19 patients revealed that the levels of IFN-α (but not IFN-β) and ISGs were associated with viral load levels and disease severity [44]. Despite such elevated IFN-I levels found in severe infections, the viral load level remained almost unchanged (Fig. 1). Further understanding of the roles of IFN-I at different stages of infection and in patients with mild versus severe COVID-19 will provide insights for the therapeutic use of interferon administration or, JAK inhibitors [45]. Thus, both viral and host factors can affect the timing of IFN response and allow to distinguish two alternative scenarios: at the early stages of infection, when the viral load is low and IFN is early produced, viral replication can be limited soon. In contrast, when the IFN response is delayed and/or deficient by limiting initial viral replication, inflammation and lung injury may occur (Fig. 1). It is plausible among older hosts with a compromised IFN induction or when the initial viral load is high, and the viral population has a higher chance to counteract the IFN response using its escape strategy, thus delaying its induction [46]. Surprisingly, high levels of IFN-α and ISGs correlated with disease severity and viral RNA copies among SARS and MERS patients. Elevated levels of IFN-induced chemokines were sustained even after the resolution of acute illness in patients who exhibited severe hypoxemia [47,48].
4.3. Animal models for assessing IFN-response in HCoV infections
Indeed, the viral pathogenesis studies in animal models do not fully reproduce human diseases associated with SARS-CoV-2, SARS-CoV, and MERS-CoV infection considering that there are host-specific factors in determining IFN response and disease outcomes that limit such extrapolation. For instance, whereas a subset of ISGs is shared among mammalian species, many other ISGs are species-specific [49].
Particularly for SARS-CoV-2 infection, non-human primates do not fully replicate COVID-19 [50]. Likewise, mice are scarcely susceptible to SARS-CoV-2 infection and, on its lungs, the IFN-I response can restrict viral replication. Nevertheless, the development of a mouse model of SARS-CoV-2 based on adeno-associated virus (AAV)-mediated expression of ACE2 was able to support viral replication and exhibit pathological findings observed in COVID-19 patients. It includes the IFN-I inability to control SARS-CoV-2 replication in vivo but its pivotal role as a driver of pathological responses [51]. In contrast to mice, Syrian hamsters are highly permissive to SARS-CoV-2 infection and virus-induced lung pathology characterized by a robust inflammatory response, neutrophil infiltration, and edema, developing bronchopneumonia. Such immune-mediated damage highlights the role of STAT2 as a double-edged sword, limiting systemic virus dissemination on the one hand, yet promoting severe lung injury on the other [52,53].
Several animal species are suitable disease models of SARS-CoV, but most laboratory animals are refractory or only partially susceptible to MERS-CoV infection [54]. Using the mice model, the IFN-I and III signaling during SARS-CoV infection is influenced by the genetic background of the animal. In C57BL/6 or 129 (type I, type II, or type III interferons knockout) mice, the IFN signaling contributes to protection by enhancing viral clearance but does not contribute to pathogenesis. However, even developing only mild SARS, the viral pathogenesis is regulated by a STAT1 dependent mechanism. In contrast, BALB/c mice develop lethal SARS-CoV disease, IFN-I induction appears delayed compared to viral replication, and the subsequent infiltration of inflammatory monocytes and macrophages with lung tissues was detrimental. IFN-I remains detectable until after virus titer peak, but early IFN-I administration ameliorates immunopathology, highlighting the relevance of early IFN-I mediated antiviral activities. Such abnormal IFN response timing during SARS-CoV infection in mice does not correlate with the human response kinetics [23,55]. On the other hand, during MERS-CoV infection in mice, the IFN-I response is induced without any delay, resulting in a protective event from disease and fatal outcomes [56]. These studies emphasize the pivotal role of IFN-I schedule of action concerning virus infection for determining its outcome such as an early IFN-I induction (or exogenous administration when IFN-I expression is delayed or reduced) confers protection, but a delayed IFN-I response fails to control the virus and can also promote immunopathology (Fig. 1).
5. Modulation of innate antiviral response by HCoV
5.1. SARS-CoV and MERS strategies to counteract antiviral IFN-response
Despite the strong antiviral cellular state achieved when IFN-I and III induce hundreds of antiviral effectors or ISGs, HCoV infections remain pathogenic. This is at least partially due to several viral strategies to counteract the IFN response on common pathways, more specifically the mechanisms used by DNA and RNA viruses to block the IFN-III mediated responses [57,58]. Clinical studies showed that coronaviruses evade innate immunity during the first 10 days of infection, which corresponds to a period of widespread inflammation and steadily increasing viral load [59].
As mentioned above, the IFN response involves a multistep process that starts at the early PAMPs sensing, followed by its production and release, the JAK-STAT signaling after IFNAR engagement, ending with the ISG-mediated antiviral activities, and its feedback control. Such a cascade of events may be interrupted at different points by the direct action of at least 10 SARS-CoV structural and nonstructural proteins [8].
At the early sensing phase, HCoV encoded proteins are committed to escape innate response by PRRs, particularly those related to viral RNA recognition. SARS-CoV and other HCoV such as SARS-CoV-2, replicate in the interior of double-membrane vesicles to prevent RIG-I activation by dsRNA replication intermediates [60]. SARS-CoV can evade RIG-I and MDA5 recognition of the viral RNA by using its nonstructural protein 14 (nsp14). This protein has a guanine-N7-methyltransferase activity that can mimic the cap structure on viral RNA, thus appearing as “host mRNA-like” [61]. Besides, the nsp16 from SARS-CoV and MERS-CoV, using its 2′-O-methyltransferase activity, may also modify the cap structure thus allowing the viruses to be recognized by MDA5 [62].
Moreover, IFN-I and IFN-III production may be inhibited by HCoV [43]. To this aim, SARS-CoV uses its membrane (M) protein to sequester innate sensors and signaling molecules into cytoplasmic compartments, thus impeding the formation of TRAF3/TANK/TBK1/IKKε complex [63]. Furthermore, SARS-CoV interferes with IRF3 function using different viral proteins and strategies. The nsp3 (papain-like protease) of SARS, when anchored in the membrane, can physically interact and inhibit STING/TBK1/IKKε and disrupt the phosphorylation and dimerization of IRF3, which are activated by STING and TBK1 [64]. Likewise, IRF-3 phosphorylation and nuclear translocation are inhibited in cells expressing ORF 3b, ORF 6, or N protein, while NF-κB is inhibited in cells expressing N protein. ORF6 is the only one able to inhibit STAT1 translocation [65], but none of these proteins impair STAT1 phosphorylation, as nsp1does [66]. Thus, the synthesis of interferons is inhibited by the four viral proteins, but interferon signaling is only inhibited by nsp1, ORF 3b, and ORF 6 proteins that block transcription of ISGs [67,68]. Similarly, the M, ORF 4a, 4b, and 5 proteins of MERS-CoV inhibit nuclear translocation of IRF3 [69].
Finally, HCoV can counteract directly the antiviral actions of ISGs-derived effector proteins such as the 2′,5′-oligoadenylate synthetase (OAS)-ribonuclease L (RNase L) complex. For instance, MERS-CoV accessory protein ORF4b has 2′-5′-phosphodiesterase activity and can destroy neosynthesized oligoadenylates thus avoiding the activation of RNase L [70], while ORF4a is a dsRNA binding protein that is probably sequestering it, thus acting as a stress response antagonist. Since dsRNA is sensed and provokes the PKR-mediated phosphorylation and inhibition of eIF2α, leading to the arrest of cellular and viral translation and the formation of stress granules, ORF4a rescues translation inhibition and prevents activation of the stress response pathway [71,72]. The PKR-induced translational arrest shuts down the negative feedback on the IFN-I response, which can thus result in a prolonged and/or amplified IFN-I response [73]. Because PKR is an ISG, the translational arrest is, in turn, potentiated by the IFN-I response. This underlines a contradictory situation in which translation arrest prevents viral replication but also sets a threshold of viral detection to commensurate the host transcriptional antiviral response to the level of infection [73]. Whether the PKR pathway is modulated by SARS-CoV2 is unknown.
Finally, SARS-CoV may enhance the host feedback control of the IFN pathway by perturbing cellular ISGs that repress the IFN-I response, such as the suppressor of cytokine signaling proteins (SOCS1 and SOCS3). Particularly in plasmacytoid dendritic cells, SOCS1 and SOCS3 strongly suppressed TLR7-mediated IFN-I production by binding the IRF7, a pivotal transcription factor of the TLR7 pathway, through the SH2 domain to promote its proteasomal degradation. The S protein of SARS-CoV could induce phenotypic conversion of human B cells, either from peripheral blood or B lymphoma cells, to macrophage-like cells expressing the marker Mac-1. Moreover, the S protein enhanced the expression of SOCS3 among several other proteins such as CD86, hypoxia-inducible factor-1α (HIF1α), C/EBPβ, insulin-like growth factor-binding protein 3 (IGFBP3), Krüpple-like factor 5 (KLF-5), and CD54 [74,75].
5.2. SARS-CoV-2 strategies to counteract IFN-response
Remarkably, SARS-CoV-2 has its own anti-IFN proteins (Fig. 3 ) [25,43,76]. A virus-cell protein interaction map identified several innate immune signaling proteins as partners of viral proteins in cells [77]. One of the first proteins produced during viral infection, nsp1, binds to the host ribosome and blocks the mRNA entry channel, thus inhibiting cellular translation but not the viral translation; such viral escape is mediated by the cis-acting RNA hairpin SL1 in the 5′UTR of SARS-CoV-2 [78]. Other viral proteins such as nsp8, nsp9, and nsp16 are produced early during viral replication, before the generation of double-stranded RNA (dsRNA). Such consecutive events could interfere with the recognition of dsRNA by cellular sensors and the virus could replicate its genome. Nsp16 suppresses mRNA splicing by binding to the mRNA recognition domains of the U1 and U2 splicing RNAs. Finally, nsp8 and nsp9 bind to the 7SL RNA in the Signal Recognition Particle and interfere with protein trafficking to the cell membrane upon infection [79].
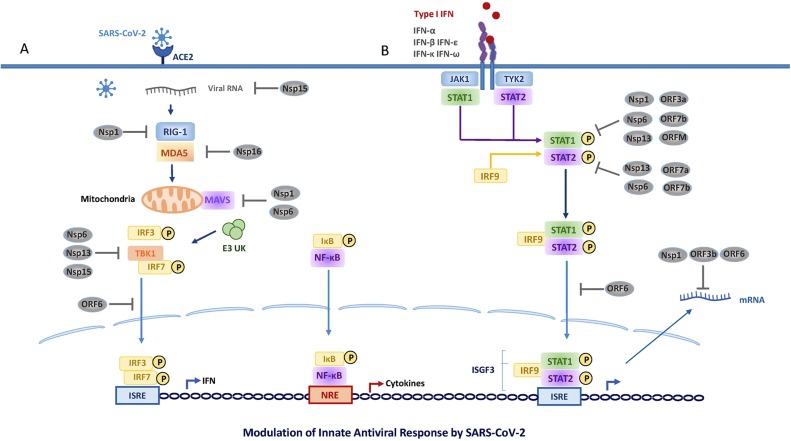
Fig. 3.
SARS-CoV-2 proteins already reported to interfere with IFN-I pathways are indicated. The left panel (A) shows viral interference mechanisms during IFN-I production; the right panel (B) depicts those strategies that enable the virus to counteract the IFN-I signaling.
The high identity (greater than 90%) found in the amino acid sequence between SARS-CoV-2 and SARS-CoV nsp16 suggests that evasion of viral RNA recognition by its 5′cap modification may also be exhibited by SARS-CoV-2 [80]. Even the potent interferon-antagonizing and protease activities of SARS-CoV nsp3 are mirrored by the homologous of SARS-CoV-2, despite sharing high amino acid sequence similarity. Other viral proteins with lower sequence identity, such as ORF3b and ORF6, have their capacity altered to antagonize-IFNs by increasing (truncated ORF3b) or abolishing (missing two amino acids ORF6b) [65,80,81].
Among SARS-CoV-2 proteins able to antagonize IFN-I production, nsp6 (membrane protein) binds TANK binding kinase 1 (TBK1) to suppress IRF3 phosphorylation, both nsp13, and nsp15 bind and block TBK1 phosphorylation, and ORF6 by localizing at the nuclear pore complex (NPC), binds importin karyopherin-α2 (KPNA2) thus targeting the nuclear import pathway and inhibiting the IRF3 nuclear translocation [[82], [83], [84], [85]]. Other strategies to counteract IFN induction include the nsp15 that can cleave the 5′-polyU of the negative-sense viral RNA [25,86], as well as ORF8 and nucleocapsid proteins that showed strong inhibition on IFN-β and NF-κB-responsive promoter by inhibiting ISRE [87].
The ISGs transcription may furtherly be altered by several SARS-CoV-2 viral proteins able to block IFNAR and IFNLR signaling. This pathway is affected through blocking STAT1/STAT2 phosphorylation or nuclear translocation using at least three different approaches, such as (1) nsp1, nsp6, nsp13, ORF3a, M, and ORF7b suppress STAT1 phosphorylation; (2) nsp6, nsp13, ORF7a, and ORF7b inhibit STAT2 phosphorylation; and (3) ORF6 blocks STAT1 nuclear translocation [83]. Remarkably, SARS-CoV-2 nsp1 and nsp6 suppress IFN-I signaling more efficiently than SARS-CoV and MERS-CoV, while ORF9b of both SARS-CoV viruses associate with Tom70 to promote the interaction with MAVS [77,88].
The protein synthesis is disrupted by SARS-CoV-2 at different steps including host mRNA splicing, protein translation, and membrane protein trafficking. Like to SARS-CoV and other HCoV, SARS-CoV-2, nsp1 and nsp11 inhibit host mRNA translation, including that of IFN-I, by binding the 40S 18S ribosomal subunits, respectively. Particularly, the nsp1 interference shutdowns the host protein production. Thus, the major parts of the innate immune system that depend on the translation of antiviral defense factors such as IFN-β or RIG-I are disarmed. Hence, this viral protein almost completely prevents translation not only of IFNs and other proinflammatory cytokines but also IFN-stimulated antiviral ISGs [89]. To produce its own viral proteins, the SARS-CoV-2 uses structural features present in the 5′UTR of all SARS-CoV mRNAs, to circumvent such Nsp1-mediated block of translation [90]
6. Host genetic factors that impair type I IFNs antiviral activities in severe COVID-19
6.1. Inborn errors of TLR3- and IRF7-dependent type I interferon response
The antiviral efficacy of IFN-I may also be affected by host-related factors. Arunachalam et al. using single-cell transcriptomics analysis in the myeloid cells of patients with severe COVID-19 revealed the absence of IFN-I, low HLA-DR, and transient expression of IFN-stimulated genes [91]. Such findings resemble severe immunodeficiencies with dysfunction in the IFN-I pathway associated with gene encoding members of the cascade. Therefore, the presence of rare inborn errors among 8 out of 13 loci that govern TLR3- and IRF7-dependent type I IFN immunity was characterized among life-threatening COVID-19 patients in a recent multicenter study that analyzed exome or genome sequences in more than 600 patients with this clinical condition. The viral load level measured in immune cells from these patients was higher than those observed in cells from healthy donors [92]. At least 3.5% of patients of various ages (17–77 years), ancestries, and both genders, with life-threatening COVID-19 pneumonia had known (autosomal recessive IRF7 and IFNAR1 deficiencies or autosomal dominant TLR3, TICAM1, TBK1, and IRF3 deficiencies) or new (autosomal dominant UNC93B1, IRF7, IFNAR1, and IFNAR2 deficiencies) genetic defects at eight of the 13 candidate loci involved in the TLR3- and IRF7-dependent induction and amplification of type I interferon. Plasmacytoid dendritic cells from IRF7-deficient patients produced no type I IFN on infection with SARS-CoV-2. These findings, in line with their roles reported previously in pulmonary immunity to influenza virus, define the essential role of both the double-stranded RNA sensor TLR3 and type I IFN cell-intrinsic immunity in the control of SARS-CoV-2 infection in the lungs.
6.2. Presence of neutralizing auto-antibodies against type I interferon
Importantly, patients treated with IFN-α or IFN-β have revealed the presence of neutralizing IgG autoantibodies against such cytokines, thus diminishing their plasma concentration [93]. This endogenous condition was recently analyzed in a multicenter study involving almost 1000 patients with life-threatening COVID-19 pneumonia from more than 15 countries. It has been shown that almost 14% of them had IgG autoantibodies against at least one type of IFN-I, except IFN-β, exhibiting a strong excess of male and slightly older patients (> 90%). The serum level of the neutralizing IgG autoantibodies against IFN-α2 and/or IFN-ω during the acute phase of infection was higher than those of IgM and IgA auto-antibodies against IFN-ω and IFN- α2, respectively. Either way, the patients’ blood carried sufficiently large amounts of auto-antibodies to neutralize the corresponding type I IFNs and block their antiviral activity in vitro against SARS-CoV-2. Moreover, all the patients who were tested for auto-antibodies against IFN-α2 also had auto-antibodies against all 13 IFN-α subtypes (IFN-α1, -α2, -α4, -α5, -α6, -α7, -α8, -α10, -α13, -α14, -α16, -α17, and -α21). Remarkably, these antibodies were not induced by SARS-CoV-2 but were detected before the infection occurred. Among the life-threatening COVID-19 patients, the presence of auto-auto-antibodies against other cytokines (IFN-γ, GM- CSF, IL-6, IL-10, IL-12p70, IL-22, IL-17A, IL-17 F, and/or TNF- β) were also found but only a minority of them were neutralizing ones. Despite SARS-CoV-2 is a poor IFN-I inducer, these autoantibodies were clinically non-evident until the patients developed life-threatening COVID-19 pneumonia, suggesting that the small amounts of IFN induced by the virus are important for protection against severe diseases. Even though scarce IFN-I levels may enhance its induction, these autoantibodies were preexistent and were not triggered by SARS-CoV-2 infection, and considering its high prevalence in men, it could be X-linked supporting at least in part, the observed sex bias for the worst course of COVID-19. In contrast, the prevalence of autoantibodies against IFN-I in the general population was estimated at 0.33% [94].
6.3. Host-genetic variants in IFN-related genes associated with severe COVID-19
Using Mendelian randomization in a genome-wide association study, a significant host-genetic association with severe COVID-19 was defined among more than 2200 critically ill patients. Genetic variants for two IFN-related genes that act early in disease (IFNAR2 and OAS genes) were characterized among life-threatening patients. These include a cluster of genes that encode antiviral restriction enzyme activators (2′,5′-oligoadenylate synthetase OAS1, OAS2, and OAS3), TYK2 (encoding a tyrosine kinase), DPP9 (dipeptidyl peptidase gene), and IFNAR2 (interferon receptor gene). The low expression of IFNAR2 and high expression of TYK2 appear as causative of severe COVID-19. Moreover, in lung tissue from these critically ill patients, a high expression of the monocyte/macrophage chemotactic receptor CCR2 was revealed by a transcriptome-wide association study [95].
7. Are there beneficial IFN-mediated activities in SARS-CoV-2 replication?
Recent advances in systematic screening strategies have revealed the existence of a small subset of ISGs that paradoxically exhibit activities that favor viral replication [96,97]. These ISGs act directly facilitating viral entry [97] or downregulating IFN signaling for feedback control, as mentioned above. Instead, ISGs with antiviral activities such as interferon-induced transmembrane proteins (IFITM2 and IFITM3) able to block the entry of several enveloped viruses [109], can be hijacked by human coronavirus OC43 (HCoV-OC43) to favor infection in human cells [98].
SARS-CoV-2 uses ACE2, transmembrane serine protease 2 (TMPRSS2), and neuropilin-1 to enter cells [99,100]. Analysis of human, nonhuman primate, and mouse single-cell RNA-sequencing (scRNA-seq) data sets, generated from healthy or infected individuals, revealed that the expression of ACE2 is primarily limited to type II pneumocytes in the lung, absorptive enterocytes within the gut, and goblet secretory cells of the nasal mucosa [101]. Interestingly, this meta-analysis identified an association between ACE2 expression and canonical ISGs or components of the IFN-α signaling pathway in different tissues. Independent analysis of publicly available data sets concluded that the ACE2 expression pattern is similar to ISGs [102]. In vitro validations were performed by treating primary human upper airway cells with numerous inflammatory cytokines. IFNα2, and to some extent, IFNγ led to a greater and more significant ACE2 up-regulation compared with all other cytokines tested [101]. Substantial upregulation of ACE2 was also observed in primary skin and primary bronchial cells treated with either IFN-I or IFN-II. Moreover, ACE2 expression was also upregulated upon ex vivo type A influenza virus infection in human lung explants isolated following surgical resection [101]. Because the majority of cells robustly upregulating ACE2 were epithelial, this observation potentially explains why previous analyses aimed at defining canonical ISGs within immune populations did not identify ACE2 as an induced gene [96]. Finally, STAT1, STAT3, IRF8, and IRF1 binding sites were identified within −1500 to +500 bp of the ACE2 transcription start site [101].
However, a recent report demonstrates the existence of a novel, transcriptionally independent truncated isoform of ACE2, named deltaACE2 (dACE2). This dACE2 isoform, but not ACE2, is inducible (ISG) by IFNs (types I, II, and III), and viruses that induce IFN responses. Moreover, its expression is induced in vitro by SARS-CoV-2 infection. Nevertheless, the dACE2 isoform is nonfunctional as a SARS-CoV-2 receptor and carboxypeptidase. It will be important to examine a potential cross-talk between the ISG-type induction of dACE2 and its role in the regulation of ACE2 protein levels, especially in COVID-19 conditions [103,104]. Despite the need for direct evidence that IFNs upregulate ACE2 in target cells in vivo, altogether these studies suggest that ACE2 could be an ISG that enhances SARS-CoV-2 internalization in human epithelial cells [99,101]. Elucidating the tissue and cell-type specificity of ISGs and their mechanisms of action is essential for understanding the potential dual role of IFNs during human SARS-CoV-2 infection, as well as to guide the use of IFNs in clinical trials.
8. Conclusions
Given the complexity of IFN responses in SARS-CoV-2 infection, further insight into their impact on the disease is of extreme importance for the continued development of drugs targeting the IFN pathways. Research is needed to establish whether IFN-III and IFN-I have similar effects or whether one is more beneficial or detrimental than the other. It should be categorically established whether type I IFN responses are enlarged in the lungs of COVID-19 patients in contrast to the inhibited type I IFN responses observed in blood. Further research will be required to define whether suppression of blood type I IFN in critically ill COVID-19 patients occurs due to the ability of SARS-CoV-2 protein to obstruct IFN signaling. Alternatively, low type I IFN may be the consequence rather than the cause of generalized immunosuppression induced by high viral titers.
Finally, type I and III interferons, with different shades, share antiviral and immunomodulatory activities. However, during COVID-19, the IFN response is complex, and it is still undefined whether IFN-I and III have similar effects or one of them is more beneficial than the other. Their location, timing, and duration of exposure are pivotal parameters to define the infection outcome, which are influenced by inherited host factors and viral mechanisms able to evade antiviral activities. SARS-CoV-2 proteins antagonize IFN-I at multiple steps in the IFN-I production/signaling pathways, that collectively determine the whole counteracting force against IFN-I restriction in infected hosts. This virus employs strategies that were presented previously by SARS-CoV, but whose inhibitory capacity may vary among different coronaviruses, probably influencing the outcome of viral replication, transmission, and pathogenesis. Along with high levels of viral replication, the IFN response may be impaired causing generalized immunosuppression. Thus, further research is deserved to evaluate the lung pathogenicity of IFNs in SARS-CoV-2 infection as well as to assess whether IFNs or IFN blockade are potential therapeutic choices.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Jorge Quarleri: Conceptualization, Methodology, Writing - review & editing. M. Victoria Delpino: Conceptualization, Methodology, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Biographies

Jorge Quarleri obtained his Biochemist degree at University of Buenos Aires (UBA) in 1989. He completed his postgraduate Degree in Microbiology at the University of Buenos Aires and obtained his PhD in Virology in 1999 with a project focused on the Genomic characterization of hepatitis C virus. Currently, his positions are, Adjunct Professor in Microbiology at School of Medical Sciences (UBA), Senior Researcher at the National Scientific and Technical Research Council (CONICET). He works at the Institute of Biomedical Research in Retroviruses and Aids (INBIRS). His main research fields include molecular pathogenesis of HIV at the central nervous system, viral mechanisms of HIV persistence, and role of coinfections (Hepatitis B and C viruses, Trypanosoma cruzi) in the HIV pathogenesis.

María Victoria Delpino is investigator for National Scientific and Technical Research Council (CONICET) in the Institute of Immunology, Genetics and Metabolism (INIGEM). María Victoria Delpino received her Biologist degree by the University of Luján and her PhD in University of Buenos Aires (UBA). Her laboratory has been involved in investigate different aspects of Brucella infection including the pathogenic aspect of osteoarticular and liver damage and how adrenal steroids modulate immune responses.
References
- 1.Mesev E.V., LeDesma R.A., Ploss A. Decoding type I and III interferon signaling during viral infection. Nat. Microbiol. 2019;4(6):914–924. doi: 10.1038/s41564-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crosse K.M., Monson E.A., Beard M.R., Helbig K.J. Interferon-stimulated genes as enhancers of antiviral innate immune signaling. J. Innate Immun. 2018;10(2):85–93. doi: 10.1159/000484258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazear H.M., Schoggins J.W., Diamond M.S. Shared and distinct functions of type I and type III interferons. Immunity. 2019;50(4):907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forero A., Ozarkar S., Li H., Lee C.H., Hemann E.A., Nadjsombati M.S., Hendricks M.R., So L., Green R., Roy C.N., Sarkar S.N., von Moltke J., Anderson S.K., Gale M., Jr., Savan R. Differential activation of the transcription factor IRF1 underlies the distinct immune responses elicited by type I and type III interferons. Immunity. 2019;51(3):451–464. doi: 10.1016/j.immuni.2019.07.007. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14(1):36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human Coronaviruses: A Review of Virus-Host Interactions. Diseases. 2016;4(3) doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2(3):264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutta S., Das N., Mukherjee P. Picking up a fight: fine tuning mitochondrial innate immune defenses against RNA viruses. Front. Microbiol. 2020;11:1990. doi: 10.3389/fmicb.2020.01990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crouse J., Kalinke U., Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol. 2015;15(4):231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- 11.Makris S., Paulsen M., Johansson C. Type I interferons as regulators of lung inflammation. Front. Immunol. 2017;8:259. doi: 10.3389/fimmu.2017.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duic I., Tadakuma H., Harada Y., Yamaue R., Deguchi K., Suzuki Y., Yoshimura S.H., Kato H., Takeyasu K., Fujita T. Viral RNA recognition by LGP2 and MDA5, and activation of signaling through step-by-step conformational changes. Nucleic Acids Res. 2020;48(20):11664–11674. doi: 10.1093/nar/gkaa935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S., Channappanavar R., Kanneganti T.D. Coronaviruses: innate immunity, inflammasome activation, inflammatory cell death, and cytokines. Trends Immunol. 2020;41(12):1083–1099. doi: 10.1016/j.it.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma S., tenOever B.R., Grandvaux N., Zhou G.P., Lin R., Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300(5622):1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 15.Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 2007;282(21):15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 16.Zevini A., Olagnier D., Hiscott J. Crosstalk between cytoplasmic RIG-I and STING sensing pathways. Trends Immunol. 2017;38(3):194–205. doi: 10.1016/j.it.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oshiumi H., Kouwaki T., Seya T. Accessory factors of cytoplasmic viral RNA sensors required for antiviral innate immune response. Front. Immunol. 2016;7:200. doi: 10.3389/fimmu.2016.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schindler C., Levy D.E., Decker T. JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 2007;282(28):20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 19.Stark G.R., Cheon H., Wang Y. Responses to cytokines and interferons that depend upon JAKs and STATs. Cold Spring Harb. Perspect. Biol. 2018;10(1) doi: 10.1101/cshperspect.a028555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raftery N., Stevenson N.J. Advances in anti-viral immune defence: revealing the importance of the IFN JAK/STAT pathway. Cell. Mol. Life Sci. 2017;74(14):2525–2535. doi: 10.1007/s00018-017-2520-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Platanitis E., Demiroz D., Schneller A., Fischer K., Capelle C., Hartl M., Gossenreiter T., Muller M., Novatchkova M., Decker T. A molecular switch from STAT2-IRF9 to ISGF3 underlies interferon-induced gene transcription. Nat. Commun. 2019;10(1):2921. doi: 10.1038/s41467-019-10970-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J., Qian C., Cao X. Post-translational modification control of innate immunity. Immunity. 2016;45(1):15–30. doi: 10.1016/j.immuni.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 23.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pene F., Marin N., Roche N., Szwebel T.A., Merkling S.H., Treluyer J.M., Veyer D., Mouthon L., Blanc C., Tharaux P.L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kerneis S., Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galani I.E., Triantafyllia V., Eleminiadou E.E., Koltsida O., Stavropoulos A., Manioudaki M., Thanos D., Doyle S.E., Kotenko S.V., Thanopoulou K., Andreakos E. Interferon-lambda mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity. 2017;46(5):875–890. doi: 10.1016/j.immuni.2017.04.025. e6. [DOI] [PubMed] [Google Scholar]
- 27.Stanifer M.L., Kee C., Cortese M., Zumaran C.M., Triana S., Mukenhirn M., Kraeusslich H.G., Alexandrov T., Bartenschlager R., Boulant S. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020;32(1) doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreakos E., Tsiodras S. COVID-19: lambda interferon against viral load and hyperinflammation. EMBO Mol. Med. 2020;12(6) doi: 10.15252/emmm.202012465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., Brann T.W., Zhou M., Yang J., Oguariri R.M., Lidie K.B., Imamichi H., Huang D.W., Lempicki R.A., Baseler M.W., Veenstra T.D., Young H.A., Lane H.C., Imamichi T. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J. Immunol. 2011;186(8):4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iversen M.B., Ank N., Melchjorsen J., Paludan S.R. Expression of type III interferon (IFN) in the vaginal mucosa is mediated primarily by dendritic cells and displays stronger dependence on NF-kappaB than type I IFNs. J. Virol. 2010;84(9):4579–4586. doi: 10.1128/JVI.02591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazear H.M., Nice T.J., Diamond M.S. Interferon-lambda: Immune Functions at Barrier Surfaces and Beyond. Immunity. 2015;43(1):15–28. doi: 10.1016/j.immuni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broggi A., Ghosh S., Sposito B., Spreafico R., Balzarini F., Lo Cascio A., Clementi N., De Santis M., Mancini N., Granucci F., Zanoni I. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369(6504):706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prokunina-Olsson L., Alphonse N., Dickenson R.E., Durbin J.E., Glenn J.S., Hartmann R., Kotenko S.V., Lazear H.M., O’Brien T.R., Odendall C., Onabajo O.O., Piontkivska H., Santer D.M., Reich N.C., Wack A., Zanoni I. COVID-19 and emerging viral infections: the case for interferon lambda. J. Exp. Med. 2020;217(5) doi: 10.1084/jem.20200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nile S.H., Nile A., Qiu J., Li L., Jia X., Kai G. COVID-19: Pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev. 2020;53:66–70. doi: 10.1016/j.cytogfr.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Major J., Crotta S., Llorian M., McCabe T.M., Gad H.H., Priestnall S.L., Hartmann R., Wack A. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 2020;369(6504):712–717. doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wimmers F., Subedi N., van Buuringen N., Heister D., Vivie J., Beeren-Reinieren I., Woestenenk R., Dolstra H., Piruska A., Jacobs J.F.M., van Oudenaarden A., Figdor C.G., Huck W.T.S., de Vries I.J.M., Tel J. Single-cell analysis reveals that stochasticity and paracrine signaling control interferon-alpha production by plasmacytoid dendritic cells. Nat. Commun. 2018;9(1):3317. doi: 10.1038/s41467-018-05784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pervolaraki K., Rastgou Talemi S., Albrecht D., Bormann F., Bamford C., Mendoza J.L., Garcia K.C., McLauchlan J., Hofer T., Stanifer M.L., Boulant S. Differential induction of interferon stimulated genes between type I and type III interferons is independent of interferon receptor abundance. PLoS Pathog. 2018;14(11) doi: 10.1371/journal.ppat.1007420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talemi S.R., Hofer T. Antiviral interferon response at single-cell resolution. Immunol. Rev. 2018;285(1):72–80. doi: 10.1111/imr.12699. [DOI] [PubMed] [Google Scholar]
- 39.Stanifer M.L., Pervolaraki K., Boulant S. Differential regulation of type I and type III interferon signaling. Int. J. Mol. Sci. 2019;20(6) doi: 10.3390/ijms20061445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krey K., Babnis A.W., Pichlmair A. System-based approaches to delineate the antiviral innate immune landscape. Viruses. 2020;12(10) doi: 10.3390/v12101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshikawa T., Hill T.E., Yoshikawa N., Popov V.L., Galindo C.L., Garner H.R., Peters C.J., Tseng C.T. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS One. 2010;5(1):e8729. doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menachery V.D., Eisfeld A.J., Schafer A., Josset L., Sims A.C., Proll S., Fan S., Li C., Neumann G., Tilton S.C., Chang J., Gralinski L.E., Long C., Green R., Williams C.M., Weiss J., Matzke M.M., Webb-Robertson B.J., Schepmoes A.A., Shukla A.K., Metz T.O., Smith R.D., Waters K.M., Katze M.G., Kawaoka Y., Baric R.S. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio. 2014;5(3):e01174–14. doi: 10.1128/mBio.01174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A. B.R. tenOever, imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–1045. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei L., Ming S., Zou B., Wu Y., Hong Z., Li Z., Zheng X., Huang M., Luo L., Liang J., Wen X., Chen T., Liang Q., Kuang L., Shan H., Huang X. Viral invasion and type I interferon response characterize the immunophenotypes during Covid-19 infection. Cell. 2020 [Google Scholar]
- 45.Lee J.S., Shin E.C. The type I interferon response in COVID-19: implications for treatment. Nat. Rev. Immunol. 2020;20(10):585–586. doi: 10.1038/s41577-020-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park A., Iwasaki A. Type I. And type I. Interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27(6):870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim E.S., Choe P.G., Park W.B., Oh H.S., Kim E.J., Nam E.Y., Na S.H., Kim M., Song K.H., Bang J.H., Park S.W., Kim H.B., Kim N.J., Oh M.D. Clinical progression and cytokine profiles of middle east respiratory syndrome coronavirus infection. J. Korean Med. Sci. 2016;31(11):1717–1725. doi: 10.3346/jkms.2016.31.11.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiel V., Weber F. Interferon and cytokine responses to SARS-coronavirus infection. Cytokine Growth Factor Rev. 2008;19(2):121–132. doi: 10.1016/j.cytogfr.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lukhele S., Boukhaled G.M., Brooks D.G. Type I interferon signaling, regulation and gene stimulation in chronic virus infection. Semin. Immunol. 2019;43 doi: 10.1016/j.smim.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A., Schipper D., van Run P., Leijten L., Sikkema R., Verschoor E., Verstrepen B., Bogers W., Langermans J., Drosten C., Fentener van Vlissingen M., Fouchier R., de Swart R., Koopmans M., Haagmans B.L. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368(6494):1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Israelow B., Song E., Mao T., Lu P., Meir A., Liu F., Alfajaro M.M., Wei J., Dong H., Homer R.J., Ring A., Wilen C.B., Iwasaki A. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. bioRxiv. 2020 doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boudewijns R., Thibaut H.J., Kaptein S.J.F., Li R., Vergote V., Seldeslachts L., Van Weyenbergh J., De Keyzer C., Bervoets L., Sharma S., Liesenborghs L., Ma J., Jansen S., Van Looveren D., Vercruysse T., Wang X., Jochmans D., Martens E., Roose K., De Vlieger D., Schepens B., Van Buyten T., Jacobs S., Liu Y., Marti-Carreras J., Vanmechelen B., Wawina-Bokalanga T., Delang L., Rocha-Pereira J., Coelmont L., Chiu W., Leyssen P., Heylen E., Schols D., Wang L., Close L., Matthijnssens J., Van Ranst M., Compernolle V., Schramm G., Van Laere K., Saelens X., Callewaert N., Opdenakker G., Maes P., Weynand B., Cawthorne C., Vande Velde G., Wang Z., Neyts J., Dallmeier K. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 2020;11(1):5838. doi: 10.1038/s41467-020-19684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., Yamada S., Fan S., Chiba S., Kuroda M., Guan L., Takada K., Armbrust T., Balogh A., Furusawa Y., Okuda M., Ueki H., Yasuhara A., Sakai-Tagawa Y., Lopes T.J.S., Kiso M., Yamayoshi S., Kinoshita N., Ohmagari N., Hattori S.I., Takeda M., Mitsuya H., Krammer F., Suzuki T., Kawaoka Y. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. U S A. 2020;117(28):16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1) doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smits S.L., de Lang A., van den Brand J.M., Leijten L.M., van I.W.F., Eijkemans M.J., van Amerongen G., Kuiken T., Andeweg A.C., Osterhaus A.D., Haagmans B.L. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog. 2010;6(2) doi: 10.1371/journal.ppat.1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao J., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J., Gale M.J., Jr., Baric R.S., Enjuanes L., Gallagher T., McCray P.B., Jr., Perlman S. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. U S A. 2014;111(13):4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou J.H., Wang Y.N., Chang Q.Y., Ma P., Hu Y., Cao X. Type III interferons in viral infection and antiviral immunity. Cell. Physiol. Biochem. 2018;51(1):173–185. doi: 10.1159/000495172. [DOI] [PubMed] [Google Scholar]
- 58.Grandvaux N., tenOever B.R., Servant M.J., Hiscott J. The interferon antiviral response: from viral invasion to evasion. Curr. Opin. Infect. Dis. 2002;15(3):259–267. doi: 10.1097/00001432-200206000-00008. [DOI] [PubMed] [Google Scholar]
- 59.Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., Law K.I., Tang B.S., Hon T.Y., Chan C.S., Chan K.H., Ng J.S., Zheng B.J., Ng W.L., Lai R.W., Guan Y., Yuen K.Y., Group H.U.S.S. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan L., Zhang Y., Ge J., Zheng L., Gao Y., Wang T., Jia Z., Wang H., Huang Y., Li M., Wang Q., Rao Z., Lou Z. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat. Commun. 2020;11(1):5874. doi: 10.1038/s41467-020-19770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y., Tao J., Sun Y., Wu A., Su C., Gao G., Cai H., Qiu S., Wu Y., Ahola T., Guo D. Structure-function analysis of severe acute respiratory syndrome coronavirus RNA cap guanine-N7-methyltransferase. J. Virol. 2013;87(11):6296–6305. doi: 10.1128/JVI.00061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H., Thiel V., Sen G.C., Fensterl V., Klimstra W.B., Pierson T.C., Buller R.M., Gale M., Jr., Shi P.Y., Diamond M.S. 2’-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468(7322):452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siu K.L., Chan C.P., Kok K.H., Chiu-Yat Woo P., Jin D.Y. Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell. Mol. Immunol. 2014;11(2):141–149. doi: 10.1038/cmi.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5(5):369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81(18):9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81(21):11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81(2):548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.T., Baker S.C., Li K. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282(44):32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Y., Zhang L., Geng H., Deng Y., Huang B., Guo Y., Zhao Z., Tan W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4(12):951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thornbrough J.M., Jha B.K., Yount B., Goldstein S.A., Li Y., Elliott R., Sims A.C., Baric R.S., Silverman R.H., Weiss S.R. Middle east respiratory syndrome coronavirus NS4b protein inhibits host RNase l activation. mBio. 2016;7(2) doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Comar C.E., Goldstein S.A., Li Y., Yount B., Baric R.S., Weiss S.R. Antagonism of dsRNA-Induced innate immune pathways by NS4a and NS4b accessory proteins during MERS coronavirus infection. mBio. 2019;10(2) doi: 10.1128/mBio.00319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rabouw H.H., Langereis M.A., Knaap R.C., Dalebout T.J., Canton J., Sola I., Enjuanes L., Bredenbeek P.J., Kikkert M., de Groot R.J., van Kuppeveld F.J. Middle east respiratory coronavirus accessory protein 4a inhibits PKR-Mediated antiviral stress responses. PLoS Pathog. 2016;12(10) doi: 10.1371/journal.ppat.1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dalet A., Arguello R.J., Combes A., Spinelli L., Jaeger S., Fallet M., Vu Manh T.P., Mendes A., Perego J., Reverendo M., Camosseto V., Dalod M., Weil T., Santos M.A., Gatti E., Pierre P. Protein synthesis inhibition and GADD34 control IFN-beta heterogeneous expression in response to dsRNA. EMBO J. 2017;36(6):761–782. doi: 10.15252/embj.201695000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang S., Liu K., Cheng A., Wang M., Cui M., Huang J., Zhu D., Chen S., Liu M., Zhao X., Wu Y., Yang Q., Zhang S., Ou X., Mao S., Gao Q., Yu Y., Tian B., Liu Y., Zhang L., Yin Z., Jing B., Chen X., Jia R. SOCS proteins participate in the regulation of innate immune response caused by viruses. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.558341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patra T., Meyer K., Geerling L., Isbell T.S., Hoft D.F., Brien J., Pinto A.K., Ray R.B., Ray R. SARS-CoV-2 spike protein promotes IL-6 trans-signaling by activation of angiotensin II receptor signaling in epithelial cells. PLoS Pathog. 2020;16(12) doi: 10.1371/journal.ppat.1009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tidu A., Janvier A., Schaeffer L., Sosnowski P., Kuhn L., Hammann P., Westhof E., Eriani G., Martin F. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA. 2020 doi: 10.1261/rna.078121.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., Thai J., Miller Z.D., Lin A.E., Schmidt M.M., Stewart D.G., Goldfarb D., De Lorenzo G., Rihn S.J., Voorhees R.M., Botten J.W., Majumdar D., Guttman M. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020 doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lokugamage K.G., Hage A., de Vries M., Valero-Jimenez A.M., Schindewolf C., Dittmann M., Rajsbaum R., Menachery V.D. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020;94(23) doi: 10.1128/JVI.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., Sauter D., Gifford R.J., Consortium U.-C., Nakagawa S., Sato K. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32(12) doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuen C.K., Lam J.Y., Wong W.M., Mak L.F., Wang X., Chu H., Cai J.P., Jin D.Y., To K.K., Chan J.F., Yuen K.Y., Kok K.H. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020;9(1):1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., Menachery V.D., Rajsbaum R., Shi P.Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33(1) doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cai Z., Zhang M.X., Tang Z., Zhang Q., Ye J., Xiong T.C., Zhang Z.D., Zhong B. USP22 promotes IRF3 nuclear translocation and antiviral responses by deubiquitinating the importin protein KPNA2. J. Exp. Med. 2020;217(5) doi: 10.1084/jem.20191174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., Danziger O., White K.M., Rathnasinghe R., Uccellini M., Gao S., Aydillo T., Mena I., Yin X., Martin-Sancho L., Krogan N.J., Chanda S.K., Schotsaert M., Wozniak R.W., Ren Y., Rosenberg B.R., Fontoura B.M.A., Garcia-Sastre A. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U S A. 2020;117(45):28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sa Ribero M., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16(7) doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li J.Y., Liao C.H., Wang Q., Tan Y.J., Luo R., Qiu Y., Ge X.Y., ORF6 The. ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shi C.S., Qi H.Y., Boularan C., Huang N.N., Abu-Asab M., Shelhamer J.H., Kehrl J.H. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193(6):3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., Straub J.H., Sturzel C.M., Frohlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K.M.J., Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369(6508):1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tanaka T., Kamitani W., DeDiego M.L., Enjuanes L., Matsuura Y. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 2012;86(20):11128–11137. doi: 10.1128/JVI.01700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., Wagh D., Coller J., Pellegrini K.L., Kazmin D., Alaaeddine G., Leung W.S., Chan J.M.C., Chik T.S.H., Choi C.Y.C., Huerta C., Paine McCullough M., Lv H., Anderson E., Edupuganti S., Upadhyay A.A., Bosinger S.E., Maecker H.T., Khatri P., Rouphael N., Peiris M., Pulendran B. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369(6508):1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., Rosain J., Bilguvar K., Ye J., Bolze A., Bigio B., Yang R., Arias A.A., Zhou Q., Zhang Y., Onodi F., Korniotis S., Karpf L., Philippot Q., Chbihi M., Bonnet-Madin L., Dorgham K., Smith N., Schneider W.M., Razooky B.S., Hoffmann H.H., Michailidis E., Moens L., Han J.E., Lorenzo L., Bizien L., Meade P., Neehus A.L., Ugurbil A.C., Corneau A., Kerner G., Zhang P., Rapaport F., Seeleuthner Y., Manry J., Masson C., Schmitt Y., Schluter A., Le Voyer T., Khan T., Li J., Fellay J., Roussel L., Shahrooei M., Alosaimi M.F., Mansouri D., Al-Saud H., Al-Mulla F., Almourfi F., Al-Muhsen S.Z., Alsohime F., Al Turki S., Hasanato R., van de Beek D., Biondi A., Bettini L.R., D’Angio M., Bonfanti P., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Oler A.J., Tompkins M.F., Alba C., Vandernoot I., Goffard J.C., Smits G., Migeotte I., Haerynck F., Soler-Palacin P., Martin-Nalda A., Colobran R., Morange P.E., Keles S., Colkesen F., Ozcelik T., Yasar K.K., Senoglu S., Karabela S.N., Rodriguez-Gallego C., Novelli G., Hraiech S., Tandjaoui-Lambiotte Y., Duval X., Laouenan C., Clinicians C.-S., Clinicians C., Imagine C.G., French C.C.S.G., Co V.C.C., Amsterdam U.M.C.C.-B., Effort C.H.G., Group N.-U.T.C.I., Snow A.L., Dalgard C.L., Milner J.D., Vinh D.C., Mogensen T.H., Marr N., Spaan A.N., Boisson B., Boisson-Dupuis S., Bustamante J., Puel A., Ciancanelli M.J., Meyts I., Maniatis T., Soumelis V., Amara A., Nussenzweig M., Garcia-Sastre A., Krammer F., Pujol A., Duffy D., Lifton R.P., Zhang S.Y., Gorochov G., Beziat V., Jouanguy E., Sancho-Shimizu V., Rice C.M., Abel L., Notarangelo L.D., Cobat A., Su H.C., Casanova J.L. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370(6515) doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vallbracht A., Treuner J., Flehmig B., Joester K.E., Niethammer D. Interferon-neutralizing antibodies in a patient treated with human fibroblast interferon. Nature. 1981;289(5797):496–497. doi: 10.1038/289496a0. [DOI] [PubMed] [Google Scholar]
- 94.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Beziat V., Manry J., Shaw E., Haljasmagi L., Peterson P., Lorenzo L., Bizien L., Trouillet-Assant S., Dobbs K., de Jesus A.A., Belot A., Kallaste A., Catherinot E., Tandjaoui-Lambiotte Y., Le Pen J., Kerner G., Bigio B., Seeleuthner Y., Yang R., Bolze A., Spaan A.N., Delmonte O.M., Abers M.S., Aiuti A., Casari G., Lampasona V., Piemonti L., Ciceri F., Bilguvar K., Lifton R.P., Vasse M., Smadja D.M., Migaud M., Hadjadj J., Terrier B., Duffy D., Quintana-Murci L., van de Beek D., Roussel L., Vinh D.C., Tangye S.G., Haerynck F., Dalmau D., Martinez-Picado J., Brodin P., Nussenzweig M.C., Boisson-Dupuis S., Rodriguez-Gallego C., Vogt G., Mogensen T.H., Oler A.J., Gu J., Burbelo P.D., Cohen J.I., Biondi A., Bettini L.R., D’Angio M., Bonfanti P., Rossignol P., Mayaux J., Rieux-Laucat F., Husebye E.S., Fusco F., Ursini M.V., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Castagnoli R., Montagna D., Licari A., Marseglia G.L., Duval X., Ghosn J., Lab H., NIAID-USUHS G., Clinicians C., Clinicians C.-S., Imagine C.G., French C.C.S.G., Milieu Interieur C., Co V.C.C., Amsterdam U.M.C.C.-B., Effort C.H.G., Tsang J.S., Goldbach-Mansky R., Kisand K., Lionakis M.S., Puel A., Zhang S.Y., Holland S.M., Gorochov G., Jouanguy E., Rice C.M., Cobat A., Notarangelo L.D., Abel L., Su H.C., Casanova J.L. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370(6515) doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., Furniss J., Richmond A., Gountouna E., Wrobel N., Harrison D., Wang B., Wu Y., Meynert A., Griffiths F., Oosthuyzen W., Kousathanas A., Moutsianas L., Yang Z., Zhai R., Zheng C., Grimes G., Beale R., Millar J., Shih B., Keating S., Zechner M., Haley C., Porteous D.J., Hayward C., Yang J., Knight J., Summers C., Shankar-Hari M., Klenerman P., Turtle L., Ho A., Moore S.C., Hinds C., Horby P., Nichol A., Maslove D., Ling L., McAuley D., Montgomery H., Walsh T., Pereira A., Renieri A., Gen O.I., Investigators I., Initiative C.-H.G., I. and Me, B. Investigators, Gen C.I., Shen X., Ponting C.P., Fawkes A., Tenesa A., Caulfield M., Scott R., Rowan K., Murphy L., Openshaw P.J.M., Semple M.G., Law A., Vitart V., Wilson J.F., Baillie J.K. Genetic mechanisms of critical illness in Covid-19. Nature. 2020 doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 96.Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mar K.B., Rinkenberger N.R., Boys I.N., Eitson J.L., McDougal M.B., Richardson R.B., Schoggins J.W. LY6E mediates an evolutionarily conserved enhancement of virus infection by targeting a late entry step. Nat. Commun. 2018;9(1):3603. doi: 10.1038/s41467-018-06000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao X., Guo F., Liu F., Cuconati A., Chang J., Block T.M., Guo J.T. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc Natl Acad Sci U S A. 2014;111(18):6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Anton-Plagaro C., Shoemark D.K., Simon-Gracia L., Bauer M., Hollandi R., Greber U.F., Horvath P., Sessions R.B., Helenius A., Hiscox J.A., Teesalu T., Matthews D.A., Davidson A.D., Collins B.M., Cullen P.J., Yamauchi Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370(6518):861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., 2nd, Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.E., Barbry P., Leslie A., Kiem H.P., Flynn J.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J., H.C.A.L.B.N.E.a. lung-network@humancellatlas.org, H.C.A.L.B. Network SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016–1035. doi: 10.1016/j.cell.2020.04.035. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhuang M.W., Cheng Y., Zhang J., Jiang X.M., Wang L., Deng J., Wang P.H. Increasing host cellular receptor-angiotensin-converting enzyme 2 expression by coronavirus may facilitate 2019-nCoV (or SARS-CoV-2) infection. J. Med. Virol. 2020 doi: 10.1002/jmv.26139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Onabajo O.O., Banday A.R., Stanifer M.L., Yan W., Obajemu A., Santer D.M., Florez-Vargas O., Piontkivska H., Vargas J.M., Ring T.J., Kee C., Doldan P., Tyrrell D.L., Mendoza J.L., Boulant S., Prokunina-Olsson L. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 2020 doi: 10.1038/s41588-020-00731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Blume C., Jackson C.L., Spalluto C.M., Legebeke J., Nazlamova L., Conforti F., Perotin J.M., Frank M., Butler J., Crispin M., Coles J., Thompson J., Ridley R.A., Dean L.S.N., Loxham M., Reikine S., Azim A., Tariq K., Johnston D.A., Skipp P.J., Djukanovic R., Baralle D., McCormick C.J., Davies D.E., Lucas J.S., Wheway G., Mennella V. A novel ACE2 isoform is expressed in human respiratory epithelia and is upregulated in response to interferons and RNA respiratory virus infection. Nat. Genet. 2021 doi: 10.1038/s41588-020-00759-x. [DOI] [PubMed] [Google Scholar]