ABSTRACT
The third outbreak of coronavirus (CoV) infection (after SARS-CoV and MERS-CoV) caused by a novel CoV (SARS-CoV-2) of the genus Beta-coronavirus has become a global pandemic. CoVs are enveloped viruses whose proteins include spike (S), membrane (M), and envelope (E) which are embedded in the viral envelope. The glycosylated S protein, which forms homo-trimeric spikes on the surface of the viral particle, mediates viral entry into host cells. SARS-CoV-2, like SARS-CoV, uses the Angiotensin-Converting Enzyme 2 (ACE2) cell surface protein for cellular entry. An attractive anti-viral approach is targeting virus entry into cells, for which three strategies are suggested: 1) direct targeting of the viral glycoprotein; 2) targeting the viral receptor on the cell surface; and 3) using soluble (s) ACE2 that binds to S protein thereby neutralizing the virus. In this article, the advantages and disadvantages of these strategies are explained. Moreover, we propose that fusion of the sACE2 to anti-CD16 to produce a bi-specific molecule could be a promising anti-viral strategy.
KEYWORDS: COVID-19, sars-CoV-2, sACE2, cd16, sACE2-Anti-CD16
Introduction
The Coronavirus (CoVs) family was discovered in the 1960 s and was classified as the family of Coronaviridae, which is the largest family of Nidovirales. The Coronaviridae family consists of two main subfamilies, including Orthocoronavirinae and Torovirinae. Orthocoronavirinae family is comprised of four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus.1
CoVs are commonly found in mammals and birds. Alpha and Beta-coronaviruses circulate in mammals, including bats. Gamma viruses mainly infect avian and some mammalian species, while Deltacorona viruses infect birds and mammals. Before 2002, CoVs were considered as nuisances but never as serious viruses.1,2
The world experienced the first outbreak of coronavirus (CoV) infection in 2002–2003 by Severe Acute Respiratory Syndrome (SARS) and the second, in 2011 by Middle East Respiratory Syndrome (MERS). In both cases, the causative agents, SARS-CoV and MERS-CoV were newly identified as coronavirus in the Betacoronavirus genus with zoonotic origin.3–5
Since December 2019, some hospitals in Wuhan, China have been suffering from pneumonia of unknown causes that later was identified as a novel CoV (nCoV) in the Betacoronavirus genus and was formerly named as 2019-nCoV (Coronavirus disease 2019) but recently named as SARS-CoV-2. This virus caused the third outbreak of coronavirus infection that became a pandemic.5 Fever, cough, fatigue, and pneumonia are the main clinical symptoms of SARS-CoV-2 infection.
Coronaviruses, including the newly discovered SARS-CoV-2, are spherical single-stranded RNA viruses, characterized by spike proteins, projecting from the virion surface. The spherical morphology of the viral particle together with the spike projections led to the name of coronavirus from the Latin word corona that means crown, due to the appearance of the virus as a royal crown under the electron microscope.6,7
Host cells entrance pathway of the SARS-CoV-2
Coronaviruses are lipid bilayer (from host cell membrane) enveloped viruses shaped by structural proteins, including spike (S), membrane (M), and envelope (E) proteins, which are all embedded in the viral envelope. The S protein is heavily glycosylated and forms homotrimeric spikes on the surface of the viral particle and mediates viral entry into host cells.8 The tissue tropism of CoVs is determined by the S protein interaction with the receptors on host cells. Several cellular molecules have been described as the receptors for CoVs. Angiotensin-converting enzyme 2 (ACE2) has been identified as the main receptor of SARS‐CoV-S and SARS-CoV-2-S proteins.
ACE2 is one of the main enzymes in the renin–angiotensin system (RAS) that regulates blood pressure, fluids and electrolyte balance, and is involved in systemic vascular resistance.9–13
Activation of local pulmonary RAS in lungs may affect the pathogenesis of lung injury through several mechanisms, such as increase of vascular permeability and alterations of alveolar epithelial cells.14,15 Pulmonary RAS activation involves renin enzyme that is the primary enzyme for the activation of RAS cascade. Angiotensinogen, as a globular protein, is cleaved by renin to produce angiotensin I (Ang I, a decapeptide hormone). ACE converts Ang I to active Ang II (an octapeptide hormone). Ang II uses vasoactive effects via binding to angiotensin II type I (AT1) and type II (AT2) receptors.
ACE2 is a homologue of ACE and has a key role in balancing responses that is initiated by ACE.11–13 ACE2 hydrolyzes Ang I to produce Ang-(1–9) and Ang II to generate Ang-(1–7) that binds to the G-protein coupled receptor MAS16,17 in order to antagonize several Ang II-mediated effects. Overall, ACE2 functions as a counter-regulatory enzyme by decreasing local Ang II concentrations. Recently, it has been shown that human recombinant soluble ACE2 (hrs ACE2) inhibits the growth of SARS-CoV-2 in Vero cell line.18 This inhibition is due to competitive interaction of SARS-CoV-2 with ACE2 and hrsACE2.
High levels of Ang II can increase the vascular permeability and pulmonary edema.19–21 In mice models of acute respiratory distress syndrome, mice with knockdown ACE2 expression showed more severe disease and symptoms, but over-expression of ACE2 had protective effects.22 Viral replication and viral spike protein have been shown to decrease the expression of ACE2 but not ACE in mice models infected with SARS-CoV.23 Also, SARS-CoV can induce rapid downregulation of ACE2 from the cell surface24,25 and release catalytically active ACE2 ectodomains.26–28 Current results propose that the physiological balance between Ang II/Ang (1–7) and ACE/ACE2 has been possibly interrupted by SARS-CoV infection. This virus-mediated effect may have a pathogenic role in lung injury.29,30 Moreover, inoculation of SARS-CoV spike protein into mice induced a significant increase of Ang II protein in lung tissue and aggravated acid-induced acute lung injury.23 Data from these earlier animal studies may suggest a possible mechanism of SARS-CoV infection and can cause severe lung failure that is probably mediated through high protein levels of Ang II that result from the inhibition of ACE2 by the viral spike protein. It has been suggested that the pathogenic mechanism of the disease might be shared between SARS-CoV-2 and SARS-CoV, when their spike proteins interact with ACE2.31 Accordingly, the balance of ACE/ACE2 function and compensation of ACE2 may improve the virus-induced severe lung injury.32
Analysis of the receptor-binding motif (RBM) of S protein, a part of the receptor-binding domain (RBD) that binds to ACE2, revealed that amino acid residues essential for binding of SARS-CoV-S to ACE2 are conserved in the SARS-CoV-2-S protein. However, most of these residues are absent from S protein of SARS-CoV-related virus from bats and were not found to be used by ACE2 for entry.33–36 It has been shown that anti-human ACE2 serum can block the entry of virus through SARS-S and SARS-CoV-2-S proteins, but not that of MERS-S virus protein into BHK-21 cells. Furthermore, SARS-CoV-2 could infect the ACE2 transfected BHK-21 cells with high efficiency but did not infect parental BHK-21 cells, indicating that SARS-CoV-2-S, like SARS-S, used ACE2 for entry. A recent study showed that SARS-CoV-2-S protein has a higher affinity to ACE2 than SARS‐S protein. It has been demonstrated that binding of SARS-S to ACE2 induced ACE2 shedding from the cells with higher efficiency than NL63-S protein. Indeed, the pathogenicity of NL63 virus (HCoV-NL63) is milder than SARS virus that is related to the lower affinity of NL63 for human ACE2.24,37,38
Neutralization of the virus before entry
The first approach to combat SARS-CoV-2 is virus neutralization before entering human cells. Several strategies can be used for neutralizing virus, including:
1) Targeting viral glycoprotein directly by neutralizing antibodies. The genome sequence of SARS-CoV-2 virus is available for gene synthesis, enabling the use of S protein to immunize mice or rabbits. Screening immunized animals for neutralizing antibodies using phage or yeast display libraries that express antibody fragments could be used to identify the antibody candidates for viral neutralization.39,40 The challenge is that antibody candidates need to be validated in cell culture and animal models to confirm their ability to neutralize SARS-CoV-2 and prevent infection. A cocktail of different antibodies might be required to ensure full protection for patients. Due to the high rate of mutations in RNA viruses, several virus mutants should be examined in the population to test if sufficient breadth of coverage is obtained with the neutralizing antibody. Despite all these efforts, this method cannot guarantee that neutralizing antibodies can be resistant to other mutants of the virus may have in the future.
2) Targeting the viral receptor on the human cell surface is another strategy. The co-incidence of SARS-CoV and SARS-CoV-2 using ACE2 as the receptor opens up the possibility exploiting the extensive studies on the entry of SARS-CoV and applying it to SARS-CoV-2. Several possible blocking methods could be considered, which have been shown to be effective in preventing infection in SARS-CoV models. The advantage of this technique is that the ACE2 protein of the host cells will not change, so, the possibility of virus escape from binding to therapeutic agents is low. The first method is using soluble reagents that bind to human ACE2. For example, small or large molecule inhibitors could be proper reagents that prevent the binding of virus to human ACE218 or small RBD of SARS-CoV-S protein that has been demonstrated to be the key domain for binding to ACE2 during the entry process. Although the administration of this domain has been shown to block the entry of SARS-CoV in cell culture, it is unclear if the equivalent RBD of SARS-CoV-2 has the same blocking effect as SARS-CoV and SARS-CoV-2 may not share the same binding site on ACE2. Second: administration of antibodies such as single-chain variable fragments (ScFv) that bind to ACE2 protein may prevent SARS-CoV-2 infection.
3) Using soluble ACE2 (sACE2), which may bind to the S protein of SARS-CoV-2 and neutralize the virus. A recent study has shown the positive effects of this approach. This study has proven that soluble recombinant human ACE2 can inhibit the entry of the SARS-CoV-2 to human cells.18 Soluble ACE2 was noted to block the SARS virus from infecting cells in culture. The binding affinity of sACE2 to the SARS-S protein has been reported to be 1.70 nM that is comparable to the affinities of monoclonal antibodies (mAbs). As mentioned above, SARS-CoV-2 has more affinity for ACE2 compared to SARS-CoV. Thus, it could be a promising strategy for treatment or prevention of COVID-19 disease.41,42
Moreover, it might be possible to construct a chimeric immunoadhesin molecule through binding of sACE2 to human immunoglobulin G (IgG) Fc fragment. Several studies have demonstrated that the ACE2 amino acids 18–615 are sufficient for SARS-S protein binding, which also covers the peptidase domain necessary for ACE2 enzymatic function.33 Although the structure of the virus S protein and its binding to the ACE2 is largely unknown, the same ACE2 protein domains may be employed by the SARS-CoV-2 virus as used by SARS-CoV to bind and infect the human cells.
The main advantage of the Fc fragment of the chimeric molecule is endowing a longer half-life on the sACE2. Recombinant ACE2 half-life was extended from less than 2 h to over 1 week in mice model when formatted as recombinant ACE2-Fc.43
A key difference of the ACE2-Fc compared to neutralizing antibodies is that the effector functions of the Fc fragment may recruit macrophages, natural killer, and dendritic cells through the CD16 receptor against the particles of the virus or infected cells. This may facilitate faster activation of immune response and eliminate the virus. There are evidences that illustrate Fc engaging antibodies were more potent in eliminating SARS via activation of phagocytic cells than antibodies that neutralize virus alone.44 On the other hand, Fc domain of the chimeric protein may direct SARS-CoV-2 toward Fc receptor (CD16) positive cells, which has been shown in vitro for neutralizing antibodies in MERS.45 The clinical significance of this phenomenon is unclear in in vivo situation. However, clinical trials may reveal the side effects of ACE2-Fc treatment.
The sACE2-anti-CD16 VHH bi-specific molecule
Human receptors for IgG (FcγR) are divided into three classes, including CD64 (FcγRI), CD32 (FcγRII) and CD16 (FcγRIII). CD16 (FcγRIII) and CD64 (FcγRI) are activatory and some CD32 (FcγRII) isoforms are inhibitory receptors. CD16 is a transmembrane isoform with low affinity for IgG and is expressed on NK cells, a small subpopulation of T lymphocytes, as well as monocytes and macrophages. It is an activating receptor involved in antibody-dependent cell-mediated cytotoxicity (ADCC), phagocytosis, endocytosis, and cytokine release. The Fc domain of IgG can bind not only to activatory but also to inhibitory Fc receptors (FcγRIIB) that are expressed on B cells and myeloid cells.46,47 However, due to the size of the Fc fragment and its low affinity for CD16, chimeric ACE2-Fc molecule might not be effective against the virus.
Therefore, a chimeric molecule consisting of single-domain antibodies (sdAbs) with the variable domain of the camelid heavy-chain antibodies (also named VHH or nanobodies), might be a proper strategy for inhibition and treatment of COVID-19 disease.48 These small antibody domains are endowed with a large number of properties making them very attractive for antibody engineering. Despite the reduced size of their antigen-binding surface, VHH domains exhibit affinities in the range of those of conventional mAbs.49,50 The single-domain nature of VHH permits the amplification and subsequent cloning of the corresponding genes, without requiring the use of artificial linker peptide (as for ScFv) or of bi-cistronic constructs (as for Fab fragments). This feature allows direct cloning of large VHH repertoires from immunized animals, without disruption of CH2-CH3/CH2-CH3 pairing when generating Fc fragment. The VHH format is likely responsible for the high production when these domains or VHH-based fusion molecules are expressed. Moreover, VHH fragments show exquisite refolding capabilities and physical stability.51 Finally, genes encoding VHH show a large degree of homology with the IGVH3 family of human IGVH genes,52 which might confer a low antigenicity in humans. Altogether, these data show that VHH might be an excellent candidate to engineer multi-specific or multi-functional proteins for immunotherapy. Notably, sdAbs directed against CD16 could be linked to sACE2 to generate bi-specific molecules suitable for bridging effector killer cells and target cells.
The sACE2-anti-CD16 VHH bi-specific molecule (Figure 1) may have several advantages compared to ACE2-Fc, including binding to CD16 with high affinity and binding to activating receptors. The small size of this molecule allows rapid permeation into different tissues, and can be produced in large quantities in prokaryotic and eukaryotic cell lines.53–55
Figure 1.
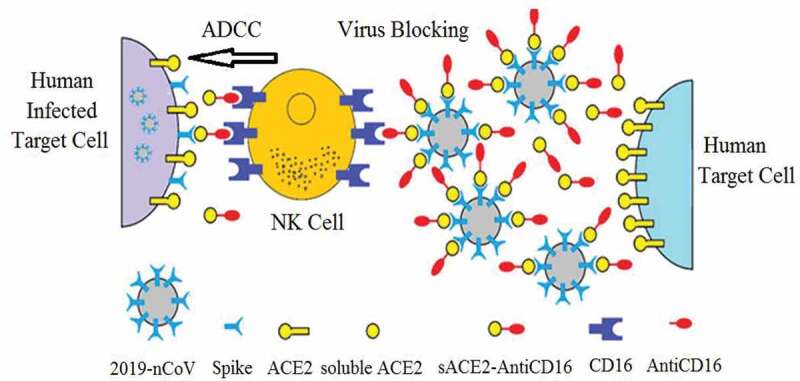
The spike protein of the coronavirus binds to ACE2 on target cells (include lung and gastrointestinal tissues in the human body) leading to cell entry. The sACE2-anti-CD16 VHH bi-specific molecule not only blocks SARS-CoV-2 from infecting cells but also mediates ADCC by NK cells. Anti-CD16 VHH fused to soluble ACE2 would prolonged the construct circulation half-life in the body
ace
Therefore, the sACE2-anti-CD16 VHH bi-specific molecule (Figure 1) may block the virus S protein or leaves the ACE2 free to perform its physiologic function in lung.
Bridging the NK cells to SARS-CoV-2 may help to infect them by the virus
NK cells are the first arm of the cellular immune response against virus infection and tumor cells. But how they are protected from SARS-CoV-2 and other viruses?
Although the immune system of COVID-19 patients produces specific antibodies step by step against the virus, natural antibodies also target virus S protein, but these antibodies have two disadvantages: first, they have low affinity for the Fc receptors and cannot induce appropriate ADCC. Second, they can activate the complement system and cause severe leukocyte invasion and inflammation that potentially results in tissue damage.
On the other hand, it has been noted that plasma from infected patients recovered from the disease contains high levels of opsonizing antibodies against the virus and induces the virus neutralization via ADCC, before exacerbating the inflammation.56 These experiments are ongoing for SARS-CoV-2 infected patients, and initial results appear promising.56 These antibodies have several functions, including opsonization process, ADCC, complement activation and recruitment of NK, MQ, and other immune effector cells. The sACE2-anti-CD16 VHH bi-specific molecule can block S protein and provides proper balances between ACE2/ACE and angiotensin II/I, followed by removing edema and pulmonary permeability. It does not activate complement system and has high affinity for FcγRIII (CD16) that results in activation of ADCC and rapidly prevents the proliferation of virus-infected cells.
Conclusion
To help alleviate virus symptoms and alleviate SARS-CoV-2 disease, it is necessary to treat subjects as quickly as possible, especially before irreversible lung damage, during and after SARS-CoV-2 vaccine development. This could be done through using sACE2-anti-CD16 VHH bi-specific molecule, which function through three mechanisms, including treatment of ACE2 deficiency and lung injury, virus neutralization, and recruitment of immune effector cells. As mentioned before, the affinity of S protein for ACE2 in SARS-CoV-2 is even more than of SARS-CoV. Therefore, virus replication in infected cells is dangerous as the virus transfers from cell to cell, using S protein on infected cells to bind to ACE2 on healthy cells, which results in rapid lung infection and can progress to death. It has been described that the pathogenicity of SARS-CoV compared to the milder coronavirus NL63 is due to the lower affinity of NL63 for human ACE2 versus SARS-CoV.57 Therefore, it is predicted if SARS-CoV-2 escapes from ACE2 neutralization via decreasing affinity, it will mutate into a less pathogenic virus, as occurred in reemergent SARS-CoV in 2003–2004 that had lower affinity for ACE2 and resulted in less severe infection without secondary transmission.58 Thus, SARS-CoV-2 could be presented with an evolutionary trap when faced with potential sACE2-anti-CD16 VHH therapy, leading to milder clinical symptoms.
Author contributions
AS planned, structured, wrote, and revised the manuscript. MHF contributed to the writing and revision of the manuscript.
References
- 1.Woo PCY, Lau SKP, Lam CSF, Lau CCY, Tsang AKL, Lau JHN, Bai R, Teng JLL, Tsang CCC, Wang M, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus.. J Virol. 2012;86(7):3995–4008. doi: 10.1128/Jvi.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woo PC, Huang Y, Lau SK, Yuen KY.. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804–20. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. The New England Journal of Medicine. 2003;348(20):1967–76. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA.. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. The New England Journal of Medicine. 2012;367(19):1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. China Novel Coronavirus I, Research T. A Novel Coronavirus from Patients with Pneumonia in China, 2019. The New England Journal of Medicine. 2020;382(8):727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barcena M, Oostergetel GT, Bartelink W, Faas FG, Verkleij A, Rottier PJ, Koster AJ, Bosch BJ. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(2):582–87. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neuman BW, Adair BD, Yoshioka C, Quispe JD, Orca G, Kuhn P, Milligan RA, Yeager M, Buchmeier MJ. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J Virol. 2006;80(16):7918–28. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77(16):8801–11. doi: 10.1128/jvi.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res. 2000;87(5):E1–9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 10.Imai Y, Kuba K, Ohto-Nakanishi T, Penninger JM. Angiotensin-converting enzyme 2 (ACE2) in disease pathogenesis. Circ J. 2010;74(3):405–10. doi: 10.1253/circj.cj-10-0045. [DOI] [PubMed] [Google Scholar]
- 11.Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G, Turner AJ. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. The Journal of Biological Chemistry. 2000;275(43):33238–43. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 12.Paul M, Poyan Mehr A, Kreutz R. Physiology of local renin-angiotensin systems. Physiol Rev. 2006;86(3):747–803. doi: 10.1152/physrev.00036.2005. [DOI] [PubMed] [Google Scholar]
- 13.Zimmerman BG, Dunham EW. Tissue renin-angiotensin system: a site of drug action? Annu Rev Pharmacol Toxicol. 1997;37:53–69. doi: 10.1146/annurev.pharmtox.37.1.53. [DOI] [PubMed] [Google Scholar]
- 14.Kuba K, Imai Y, Penninger JM. Angiotensin-converting enzyme 2 in lung diseases. Curr Opin Pharmacol. 2006;6(3):271–76. doi: 10.1016/j.coph.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Specks U, Martin WJ 2nd, Rohrbach MS. Bronchoalveolar lavage fluid angiotensin-converting enzyme in interstitial lung diseases. Am Rev Respir Dis. 1990;141(1):117–23. doi: 10.1164/ajrccm/141.1.117. [DOI] [PubMed] [Google Scholar]
- 16.Reudelhuber TL. The renin-angiotensin system: peptides and enzymes beyond angiotensin II. Curr Opin Nephrol Hypertens. 2005;14(2):155–59. doi: 10.1097/00041552-200503000-00011. [DOI] [PubMed] [Google Scholar]
- 17.Santos RA, Simoes E Silva AC, Maric C, Silva DM, Machado RP, de Buhr I, Heringer-Walther S, Pinheiro SV, Lopes MT, Bader M, et al. Angiotensin-(1-7) is an endogenous ligand for the G protein-coupled receptor Mas. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(14):8258–63. doi: 10.1073/pnas.1432869100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monteil V, Kwon H, Prado P, Hagelkruys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado Del Pozo C, Prosper F, et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell. 2020. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fyhrquist F, Saijonmaa O. Renin-angiotensin system revisited. J Intern Med. 2008;264(3):224–36. doi: 10.1111/j.1365-2796.2008.01981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall RP. The pulmonary renin-angiotensin system. Curr Pharm Des. 2003;9(9):715–22. doi: 10.2174/1381612033455431. [DOI] [PubMed] [Google Scholar]
- 21.Marshall RP, Gohlke P, Chambers RC, Howell DC, Bottoms SE, Unger T, McAnulty RJ, Laurent GJ. Angiotensin II and the fibroproliferative response to acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2004;286(1):L156–64. doi: 10.1152/ajplung.00313.2002. [DOI] [PubMed] [Google Scholar]
- 22.Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, Yang P, Sarao R, Wada T, Leong-Poi H, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436(7047):112–16. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11(8):875–79. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glowacka I, Bertram S, Herzog P, Pfefferle S, Steffen I, Muench MO, Simmons G, Hofmann H, Kuri T, Weber F, et al. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J Virol. 2010;84(2):1198–205. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S, Guo F, Liu K, Wang H, Rao S, Yang P, Jiang C. Endocytosis of the receptor-binding domain of SARS-CoV spike protein together with virus receptor ACE2. Virus Res. 2008;136(1–2):8–15. doi: 10.1016/j.virusres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haga S, Yamamoto N, Nakai-Murakami C, Osawa Y, Tokunaga K, Sata T, Yamamoto N, Sasazuki T, Ishizaka Y. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(22):7809–14. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia HP, Look DC, Tan P, Shi L, Hickey M, Gakhar L, Chappell MC, Wohlford-Lenane C, McCray PB Jr.. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am J Physiol Lung Cell Mol Physiol. 2009;297(1):L84–96. doi: 10.1152/ajplung.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert DW, Yarski M, Warner FJ, Thornhill P, Parkin ET, Smith AI, Hooper NM, Turner AJ. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2). The Journal of Biological Chemistry. 2005;280(34):30113–19. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imai Y, Kuba K, Penninger JM. The discovery of angiotensin-converting enzyme 2 and its role in acute lung injury in mice. Exp Physiol. 2008;93(5):543–48. doi: 10.1113/expphysiol.2007.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto K, Ohishi M, Katsuya T, Ito N, Ikushima M, Kaibe M, Tatara Y, Shiota A, Sugano S, Takeda S, et al. Deletion of angiotensin-converting enzyme 2 accelerates pressure overload-induced cardiac dysfunction by increasing local angiotensin II. Hypertension. 2006;47(4):718–26. doi: 10.1161/01.HYP.0000205833.89478.5b. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H, Zhu J, Zhang Q, Wu J, Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. Journal of Medical Virology. 2020. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaparianos A, Argyropoulou E. Local renin-angiotensin II systems, angiotensin-converting enzyme and its homologue ACE2: their potential role in the pathogenesis of chronic obstructive pulmonary diseases, pulmonary hypertension and acute respiratory distress syndrome. Curr Med Chem. 2011;18(23):3506–15. doi: 10.2174/092986711796642562. [DOI] [PubMed] [Google Scholar]
- 33.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–68. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 34.Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–38. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann M, Muller MA, Drexler JF, Glende J, Erdt M, Gutzkow T, Losemann C, Binger T, Deng H, Schwegmann-Wessels C, et al. Differential sensitivity of bat cells to infection by enveloped RNA viruses: coronaviruses, paramyxoviruses, filoviruses, and influenza viruses. PLoS One. 2013;8(8):e72942. doi: 10.1371/journal.pone.0072942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menachery VD, Dinnon KH 3rd, Yount BL Jr., McAnarney ET, Gralinski LE, Hale A, Graham RL, Scobey T, Anthony SJ, Wang L, et al. Trypsin Treatment Unlocks Barrier for Zoonotic Bat Coronavirus Infection. J Virol. 2020;94:5. doi: 10.1128/JVI.01774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–54. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–63. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin YW, Chang KH, Hong GW, Yeo SG, Jee Y, Kim JH, Oh MD, Cho DH, Kim SH. Selection of Vaccinia Virus-Neutralizing Antibody from a Phage-Display Human-Antibody Library. J Microbiol Biotechnol. 2019;29(4):651–57. doi: 10.4014/jmb.1812.12024. [DOI] [PubMed] [Google Scholar]
- 40.Keck ZY, Wang Y, Lau P, Foung SKH. Isolation of HCV Neutralizing Antibodies by Yeast Display. Methods Mol Biol. 2019;1911:395–419. doi: 10.1007/978-1-4939-8976-8_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–54. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sui J, Li W, Murakami A, Tamin A, Matthews LJ, Wong SK, Moore MJ, Tallarico AS, Olurinde M, Choe H, et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(8):2536–41. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu P, Wysocki J, Souma T, Ye M, Ramirez V, Zhou B, Wilsbacher LD, Quaggin SE, Batlle D, Jin J. Novel ACE2-Fc chimeric fusion provides long-lasting hypertension control and organ protection in mouse models of systemic renin angiotensin system activation. Kidney Int. 2018;94(1):114–25. doi: 10.1016/j.kint.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 44.Yasui F, Kohara M, Kitabatake M, Nishiwaki T, Fujii H, Tateno C, Yoneda M, Morita K, Matsushima K, Koyasu S, et al. Phagocytic cells contribute to the antibody-mediated elimination of pulmonary-infected SARS coronavirus. Virology. 2014;454-455:157–68. doi: 10.1016/j.virol.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, He L, Chen Y, Wu J, Shi Z, et al. Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry. J Virol. 2020;94:5. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siberil S, Dutertre CA, Boix C, Bonnin E, Menez R, Stura E, Jorieux S, Fridman WH, Teillaud JL. Molecular aspects of human FcgammaR interactions with IgG: functional and therapeutic consequences. Immunol Lett. 2006;106(2):111–18. doi: 10.1016/j.imlet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat Med. 2000;6(4):443–46. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 48.Muyldermans S. Single domain camel antibodies: current status. J Biotechnol. 2001;74(4):277–302. doi: 10.1016/s1389-0352(01)00021-6. [DOI] [PubMed] [Google Scholar]
- 49.Spinelli S, Frenken LG, Hermans P, Verrips T, Brown K, Tegoni M, Cambillau C. Camelid heavy-chain variable domains provide efficient combining sites to haptens. Biochemistry. 2000;39(6):1217–22. doi: 10.1021/bi991830w. [DOI] [PubMed] [Google Scholar]
- 50.Alvarez-Rueda N, Behar G, Ferre V, Pugniere M, Roquet F, Gastinel L, Jacquot C, Aubry J, Baty D, Barbet J, et al. Generation of llama single-domain antibodies against methotrexate, a prototypical hapten. Mol Immunol. 2007;44(7):1680–90. doi: 10.1016/j.molimm.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 51.Dumoulin M, Conrath K, Van Meirhaeghe A, Meersman F, Heremans K, Frenken LG, Muyldermans S, Wyns L, Matagne A. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002;11(3):500–15. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su C, Nguyen VK, Nei M. Adaptive evolution of variable region genes encoding an unusual type of immunoglobulin in camelids. Mol Biol Evol. 2002;19(3):205–15. doi: 10.1093/oxfordjournals.molbev.a004073. [DOI] [PubMed] [Google Scholar]
- 53.Pant N, Hultberg A, Zhao Y, Svensson L, Pan-Hammarstrom Q, Johansen K, Pouwels PH, Ruggeri FM, Hermans P, Frenken L, et al. Lactobacilli expressing variable domain of llama heavy-chain antibody fragments (lactobodies) confer protection against rotavirus-induced diarrhea. J Infect Dis. 2006;194(11):1580–88. doi: 10.1086/508747. [DOI] [PubMed] [Google Scholar]
- 54.Frenken LG, van der Linden RH, Hermans PW, Bos JW, Ruuls RC, de Geus B, Verrips CT. Isolation of antigen specific llama VHH antibody fragments and their high level secretion by Saccharomyces cerevisiae. J Biotechnol. 2000;78(1):11–21. doi: 10.1016/s0168-1656(99)00228-x. [DOI] [PubMed] [Google Scholar]
- 55.Bazl MR, Rasaee MJ, Foruzandeh M, Rahimpour A, Kiani J, Rahbarizadeh F, Alirezapour B, Mohammadi M. Production of chimeric recombinant single domain antibody-green fluorescent fusion protein in Chinese hamster ovary cells. Hybridoma (Larchmt). 2007;26(1):1–9. doi: 10.1089/hyb.2006.037. [DOI] [PubMed] [Google Scholar]
- 56.Patel P, Nandwani V, Vanchiere J, Conrad SA, Scott LK. Use of therapeutic plasma exchange as a rescue therapy in 2009 pH1N1 influenza A-An associated respiratory failure and hemodynamic shock. Pediatr Crit Care Me. 2011;12(2):E87–E9. doi: 10.1097/PCC.0b013e3181e2a569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kruse RL. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res. 2020;9:72. doi: 10.12688/f1000research.22211.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li W, Zhang C, Sui J, Kuhn JH, Moore MJ, Luo S, Wong SK, Huang IC, Xu K, Vasilieva N, et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. Embo J. 2005;24(8):1634–43. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]


