ABSTRACT
The last twenty years of seminal microbiome research has uncovered microbiota’s intrinsic relationship with human health. Studies elucidating the relationship between an unbalanced microbiome and disease are currently published daily. As such, microbiome big data have become a reality that provide a mine of information for the development of new therapeutics. Machine learning (ML), a branch of artificial intelligence, offers powerful techniques for big data analysis and prediction-making, that are out of reach of human intellect alone. This review will explore how ML can be applied for the development of microbiome-targeted therapeutics. A background on ML will be given, followed by a guide on where to find reliable microbiome big data. Existing applications and opportunities will be discussed, including the use of ML to discover, design, and characterize microbiome therapeutics. The use of ML to optimize advanced processes, such as 3D printing and in silico prediction of drug-microbiome interactions, will also be highlighted. Finally, barriers to adoption of ML in academic and industrial settings will be examined, concluded by a future outlook for the field.
KEYWORD: microbiome, machine learning, artificial intelligence, drug product development, microbial therapeutics, clinical translation, pharmaceutical sciences, COVID-19, colonic drug delivery, personalized medicines
Introduction
The human microbiome describes the genomes of trillions of microorganisms that live on and within the human body. Members of the microbiota include bacteria, viruses, fungi, archaea, and protozoa; collectively they inhabit nearly every imaginable region of their host.1 Bacteria alone encode for over 100-fold more unique genes than humans.2 Subsequently, microbiota is known to exert extensive influence upon human health and metabolism. For example, the development of metabolic syndrome, Parkinson’s disease, inflammatory bowel disease, and periodontitis are strongly associated with imbalanced, also known as ‘dysbiotic’, microbiome compositions.3–6 With regards to metabolism, the microbiome wields impactful metabolic capacity. The gut microbiome plays a crucial role in the degradation of dietary fibers, proteins, polyphenols, and starches which would be otherwise indigestible by their human host.7,8 Endogenous moieties broken down by microbiota include bile acids, hormones, and intestinal mucus.9 Drugs are also highly susceptible to metabolism by microbiota. A seminal study found that of 271 commonly administered oral drugs, 176 (64.9%) of them were significantly metabolized by at least one strain of 76 human gut bacteria.10 Due to substantial differences between individuals’ microbiomes, drug metabolism by microbiota can result in significant inter-patient variability in pharmacokinetics and pharmacodynamics.11
From the beginning of the 21st century, microbiome-based research has amassed at a progressive rate. In the mid-2000s, affordable and accurate DNA sequencing methods facilitated the commencement of the Human Microbiome Project: a multi-site collaboration leading to the genomic profiling of microbiota at key body sites.12 Later, the project assessed the role of the microbiome in human health and disease.13 In parallel to this field-leading work, many other laboratories worldwide have contributed to the now extensive knowledge base concerning the microbiome. Vast quantities of data have been made globally accessible through publications and online databases, such as the NIH Human Microbiome Project Data Portal; MicrobiomeDB; and China National GeneBank.14–16 Such data are being increasingly utilized by scientists to design microbiome-targeted therapies and predict microbiome-drug interactions. Due to the scale of available data, artificial intelligence (AI) offers a means to quickly and accurately mine, process, and analyze available information. Whilst it may take a human years to identify patterns in terabytes of data, AI algorithms can efficiently perform operations within seconds to minutes, depending on computer processing power, data size, and algorithm complexity. There are four main types of AI: machine learning (ML); natural language processing; rule-based expert systems; and robotics.17 By far, ML is the most extensively used AI technique within the microbiome field.18–20 ML uses coded computer algorithms to identify patterns in data, which facilitates the classification of new information or prediction of future outcomes (Figure 1).
Figure 1.
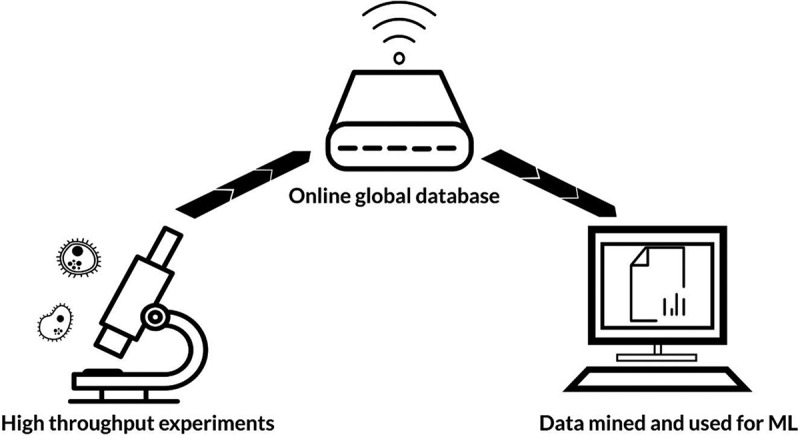
Flow of information for machine learning (ML). Vast quantities of experimental data are uploaded to online globally accessible databases. These data can then be mined and used by ML algorithms
This review will provide an accessible introduction to ML and highlight its current applications within the microbiome therapeutics field; namely, using ML as a tool for designing microbiome-targeted therapies, and predicting drug-microbiome interactions. An overview of useful databases for ML microbiome projects will also be presented. Opportunities for utilizing ML will be addressed, with a description of barriers to adoption in academic and industrial settings. Finally, an outlook for the field will be described.
Machine Learning Methods
The origins of ML date back to the 1600s, when the first mechanical calculator, and the modern binary system were created. These inventions, still integral to contemporary ML, were translated to modern ML by the mid-1950s. In 1952, IBM’s Arthur Samuel coined the phrase ‘machine learning’ upon creating a computer program that could improve its skill in the game of checkers the more it played. Later, in 1997, IBM’s Deep Blue computer rose to fame after successfully beating the chess grandmaster Garry Kasparov. Though Kasparov requested a rematch, Deep Blue was retired with its clean slate without encore. At the turn of the millennium, ML algorithms were progressively applied to tasks other than winning board games. Research on ML began hitting an exponential rate; algorithms were developed to detect cancer more accurately than radiologists, identify human faces from images, predict consumer behavior, speak like a native, and lipread, to name just a few.21 Today, ML technology has been applied to almost every sector on a global scale. Still ML capabilities are advancing at a rapid rate; new research articles are published daily that solve previously impenetrable challenges. Healthcare has especially benefitted from ML; algorithms are now available to diagnose disease, develop drugs, recommend personalized treatment plans, and predict future health outcomes. Though it will take some time for widespread clinical adoption, ML promises a future of consistent, probability, and evidence-based medicine.17
ML can be split into two parts: supervised and unsupervised learning.18 Supervised learning involves directing an algorithm to solve a pre-defined problem. For example, ‘will this newly synthesized drug be metabolized by gut microbiota?’ To generate an answer, the ML software must have access to data that have been labelled. To pertain to the previous example, the ML algorithm would be supplied with a list of drugs, and their molecular features, labeled as being susceptible or not susceptible to gut microbiota metabolism. The algorithm would then look for similarities between the newly synthesized drug, and the list of labeled drugs, by examining their molecular features. A prediction, based on probability, would then be outputted based on whether the new drug has more in common with drugs that are known to be metabolized, or those that are known to remain intact. This is known as a classification task. The ML model can then be validated by performing experiments testing microbiome-drug metabolism in a laboratory setting.
An entire dataset can be used to train an ML model, however, it is a common practice to split data into separate training and testing sets. The latter is used to test the model’s performance on new and unseen data, if obtaining new data is not readily attainable. The training set can be further split into training and validation sets, wherein the latter is used to further refine the model. Supervised algorithms can either classify new data, e.g. ‘metabolized’ or ‘not metabolized’, or they can predict a numerical value, such as ‘percentage of dose metabolized’. The output of the algorithm (classification or numerical value) depends on how the dataset is labeled, and whether a classification or regression analysis is performed. Several types of supervised ML algorithms exist, these include basic linear methods, decision trees, support vector machines, and advanced neural networks (Figure 2).
Figure 2.
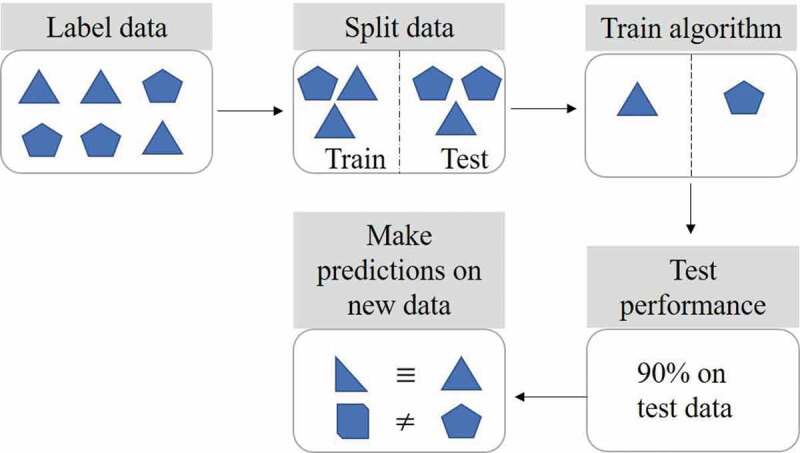
Supervised machine learning steps. Labeled data is split into training and testing sets. The algorithm is then trained to learn the differences between the labeled data. The test set is used to check and refine algorithm performance. Predictions can subsequently be made using new data previously unseen by the algorithm
Compared to supervised methods, unsupervised learning does not address any pre-defined questions.22 At all stages of data mining and ML, the chance of bias should be reduced as much as possible. One could say that introducing a question to an algorithm leads to bias, as the algorithm will look to solve that particular problem. In unsupervised learning, an ML algorithm works to identify patterns in data without any prior operator input. This can subsequently lead to elements being identified that could not be conceived by the operator. Unsupervised ML methods can produce clustering or association outputs. Clustering algorithms identify distinct groups within data; association algorithms output rules found within data. Common unsupervised ML techniques include k-means clustering, principal component analysis, and k nearest neighbors.
At the intersection between supervised and unsupervised learning is semi-supervised ML.18 Semi-supervised learning involves using a partly labeled set of data. The ML algorithm uses unsupervised learning to label the unlabeled data, by drawing conclusions from the labeled data. Following that, supervised techniques can be employed to answer defined questions about the labeled information. In practice semi-supervised learning is useful, as real-world datasets are often incompletely labeled due to poor formatting at the time of conception. Hand-labeling data can be extremely time-consuming, error-prone, and expensive, especially if datasets are particularly large and complex. The automated labeling step in semi-supervised learning provides a quick and unbiased way to format previously difficult data.
The final type of ML, reinforcement learning, is similar to supervised learning in the sense that it is goal-directed.23 Though unlike supervised learning, reinforcement learning does not require labeled data. Instead, it is supplied with a set of rules around a problem. For example, an operator may input ‘if a new formulation protects a drug from microbiome metabolism, this is a positive action. If the drug is degraded by the microbiota, this is a negative action’. Reinforcement learning solves pre-defined problems in a self-teaching, iterative manner. An algorithm will carry out actions to attempt to solve a problem; the closer an action gets to a solution, the more reward experienced in the system. If a given action results in an outcome that veers away from a solution, then the system will experience punishment. Over time, the algorithm will learn what types of actions are rewarding. In this way, a path toward problem solution is paved. In order to build an optimum path to problem solution, reinforcement learning algorithms face the dilemma of attempting new actions whilst concurrently aiming to maximize reward, this is known as the exploration vs. exploitation trade-off.24 Reinforcement learning is probably best known for its use in the mastery of games, such as the incredibly complex and ancient board game ‘Go’. In 2017, reinforcement learning reached superhuman performance within Go, without any input of data: the algorithm was its own teacher.25 Common types of reinforcement learning include temporal-difference learning, Q-learning, and state-action-reward-state-action (SARSA) learning (Table 1).24
Table 1.
Types of machine learning
| ML Type | Defined question asked? | Is data labeled? | Outputs | Example Algorithms |
|---|---|---|---|---|
| Supervised | Yes | Yes (fully) |
|
|
| Unsupervised | No | No |
|
|
| Semi-supervised | Yes | Yes (partly) |
|
Any supervised or unsupervised method |
| Reinforcement | Yes | No (rules supplied instead) | Actions moving toward a solution |
|
In all types of ML, it is important for operators to be aware of over or under fitting algorithms. Overfitting occurs when an algorithm is too sensitive to patterns in training data, leading to perfect accuracy when training, but inadequate performance during testing and validation. On the other hand, if an algorithm is not sensitive enough, and too simple, then it will not be able to model the training data and is expected to perform poorly when generalized to the testing dataset. Finding an optimal balance between algorithm underfitting and overfitting is a crucial component of ML engineering. Inadequate performance of ML algorithms is typically due to poor fitting; it can take a great deal of ML engineers’ time to tweak and perfect ML parameters to find a sweet spot (Figure 3).26
Figure 3.
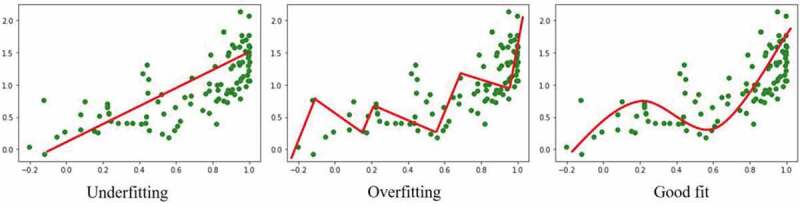
Illustration of underfitting and overfitting in simple regression machine learning. Data points are represented by green markers and model fitting by a red line
Databases For Ml Microbiome Projects
What makes a good database?
The quality of data used for ML projects is paramount. Weak, invalid data will lead to weak and invalid ML outputs: ‘garbage in equals garbage out’. Whilst large datasets are favorable for ML projects, as more information invariably leads to higher internal and external validity, big data often come with challenges.27 Firstly, it is important to validate the source of data. Within microbiome science, it is common for researchers to use individuals’ stools to sample their gut microbiomes, however it is becoming increasingly clear that stools may not be the most accurate method for profiling the gut microbiome.28 Though assaying of stools is noninvasive and cost-effective, microbiota composition changes across the intestinal tract, therefore stools cannot provide a snapshot of any one gastrointestinal niche.29,30 Many reputable databases contain stool microbiome data, and the information should not be ignored as it can provide useful insights.14,31–33 It is, however, important to be aware of sampling methodology when conducting ML analysis to allow for valid interpretation. Another detail to investigate when examining a microbiome dataset is the population sampled. Microbiome composition is well known to differ with respect to a plethora of factors, including age, sex, health, diet, ethnicity, diet, and drug use.34–39 If a ML project aims to draw universally valid interpretations, scientists must consider how globally representative the input data is.
The amount of data available in databases is another important factor. Generally, the more data available to an ML project the better, as the algorithm will naturally be exposed to more information with more variance, allowing increased prediction accuracies.40 However, the impact database size has on ML predictions does vary considerably based on the problem being analyzed, and the complexity of the algorithm. For example, if there is a very strong pattern in data, such as ‘ionized drugs are more water soluble than neutral drugs’, less data are required to teach an ML algorithm. Furthermore, some algorithms inherently require larger amounts of data, due to their intricacy and complexity. Simpler algorithms such as linear regression and basic decision trees typically need less data compared to neural networks and deep learning algorithms with multiple layers.41 Unfortunately, there is no single minimum amount of data needed for all types of ML. A decision on appropriate sample size can be reached using several methods, such as acquiring expert opinion, operator experience, and observing sample sizes in other similar published projects. For a given project, a plot of ‘dataset size vs. model prediction performance’ can be constructed using a subset of available data. This will allow reasonable projection of potential amounts of data required for a given performance score.42 Though insufficient dataset size is a common problem in the general field of ML, big data have become intrinsic to microbiome research. DNA sequencing, metabolomics, proteomics, and in silico representation of drugs produce enormous quantities of data that describe the minutiae of the microbiota-host relationship.43 Though vast quantities of data can help to build highly predictive ML models, very large datasets (reaching terabytes) do come with problems.44 For one, it is not guaranteed that big data are entirely without bias and fully globally representative. Populations less likely to be involved in microbiome research should not be disregarded; thus, it is important to always examine the source of data, no matter its scale. Next, data in repositories are commonly ‘messy’, containing missing values, unequal scaling, and background noise from poorly performed experiments or incorrect data labeling.45,46 Clearly, without rectification messy data will adversely impact the predictive power of ML algorithms. Data cleaning is a key part of ML projects. Before any algorithm training or testing begins, it is imperative that messy data are pre-processed. Contrary to common belief, machine learning engineers do not spend most of their time building, training, and refining algorithms. A 2020 study found that data pre-processing (cleaning, labeling, compiling) is on average allocated over 75% of ML project time.47 There are a myriad of techniques applied to data pre-processing, and full description of these is outside the scope of this article. A couple of existing sources summarizes this topic well.22,48 In brief, missing values can be removed or imputed with best estimates.49 Units of data features, such as drug molecular weight or LogP, will be transformed to a similar scale, to avoid giving undue weight to smaller units (e.g. 100 mg may be treated as more important than 0.1 g by an algorithm).50 Outliers in datasets may also be removed if they are judged as anomalies.
Useful databases
Projects utilizing ML within the field of microbiome therapeutics will likely require information on drugs, microbiota, and microbiota-human interactions. With a few exceptions, large databases bringing this information together do not yet exist, therefore it is necessary to mine data from multiple sources. Table 2 gives a summary of some reputable databases with potential utility for ML projects in this field.
Table 2.
Summary of key databases with potential utility in ML microbiome projects. Databases have been divided into: drugs, drug-microbiota interactions, microbiota, and microbiota-human interactions
| Database | Description | URL |
|---|---|---|
| Drugs | ||
| BindingDB | A binding affinity database focusing on drug-protein interactions. | http://www.bindingdb.org/bind/index.jsp |
| ChEMBL | A manually curated database of 2 million bioactive molecules bringing together chemical, activity, and genomic data. | https://www.ebi.ac.uk/chembl/ |
| ChemDB | A user-friendly database incorporating over 5 million small molecules annotated with important chemical features, e.g. predicted solubility. | http://cdb.ics.uci.edu/ |
| ChemSpider | A chemical structure database providing information on over 67 million entities from hundreds of sources. | http://www.chemspider.com/ |
| DrugBank | A bioinformatics and cheminformatics database providing information on over 13,500 drug entities, including biologics. | https://www.drugbank.ca/ |
| Open Babel | A wiki-led chemical toolbox suitable for use in many ML programs. | http://openbabel.org/wiki/Main_Page |
| PharmGKB | A smaller database of 708 drugs annotated with information on how genetic variation can impact clinical response. | https://www.pharmgkb.org/ |
| RDKit | A cheminformatics software for use in ML projects in Python. | https://www.rdkit.org/ |
| Super Natural | Database of over 325,000 natural products providing information on physicochemical properties and toxicity. | http://bioinf-applied.charite.de/supernatural_new/index.php?site=home |
| Drug-microbiota interactions | ||
| DrugBug | Database constructed using ML to predict the metabolism of drugs by bacteria found in the human gut.51 | http://metagenomics.iiserb.ac.in/drugbug/ |
| Microbiota | ||
| BacDive | Database providing taxonomic, physiological, and environmental data on over 80,000 strains of bacteria. | https://bacdive.dsmz.de/ |
| EnsemblBacteria | Database for bacterial and archaeal genomes. | http://bacteria.ensembl.org/index.html |
| European Nucleotide Archive | Database providing comprehensive nucleotide sequencing information for bacteria, viruses, and fungi, protozoa and archaea. | https://www.ebi.ac.uk/ena/browser/home |
| Gold | Database providing genomic information for eukaryotes, prokaryotes, and viruses. | https://gold.jgi.doe.gov/ |
| IMG/M | Metagenomic database for microbiota, annotated with functions. | https://img.jgi.doe.gov/ |
| NCBI Microbial Genomes | Comprehensive database covering information on microbial genomes, functions, and recent corresponding publications. | https://www.ncbi.nlm.nih.gov/genome/microbes/ |
| Microbiota-human interactions | ||
| Disbiome | Database covering microbiota changes in disease states. | https://disbiome.ugent.be/home |
| eHOMD | Expanded Human Oral Microbiome Database: genomic data on 775 microbial species in the upper GI and respiratory tracts. | http://www.homd.org/ |
| HMDB | The human metabolome database containing information on over 114,000 human metabolites. | https://hmdb.ca/ |
| Human Microbiome Project | Genomic characterization of microbiota at five body sites (HMP1), and information on microbiota-human interactions in disease (iHMP). | https://www.hmpdacc.org/ |
| MDB | Microbiome database from China National GeneBank, providing metadata on both host-associated (humans, rodents, pigs) and environmental microbes. | https://db.cngb.org/microbiome/ |
| MGnify | Database providing genomic, proteomic, and metabolomic data on human and environmental biomes. | https://www.ebi.ac.uk/metagenomics/ |
| MicrobiomeDB | Data mining platform for microbiome experiments. | https://microbiomedb.org/mbio/app/ |
| UniProt | Detailed database providing the proteomes of almost 200,000 microbiota and humans. | https://www.uniprot.org/ |
| Virtual Metabolic Human | Database on human and gut microbial metabolism with corresponding disease and nutritional information. | https://www.vmh.life/#home |
Applications And Opportunities
Design and discovery of microbiome therapeutics
The last few years have seen an upsurge in publications utilizing ML within microbiome research, though ML used to specifically design new therapeutics targeted to the microbiome is still in its infancy. The expansion of microbiome ML work naturally follows the curation of large accessible databases.14–16 With the elucidation of host-microbiota metabolomics, proteomics, and genomics, disease phenotypes can be identified.52–56 Whilst scouring research literature and databases for microbiome disease markers could be time-consuming for a human, ML algorithms output information much more efficiently. Once it is known what elements of the microbiome cause disease, it is possible to begin work toward preventative agents and treatments. Dysbiosis at various body sites has been associated with many diseases. For example, perturbations in the proportions of phyla Firmicutes and Bacteroidetes in the gut are strongly linked with several pathologies, including ulcerative colitis, obesity, and motor neuron disease.57–59 Etiological factors are believed to be increased intestinal permeability and inflammation, decreased short-chain fatty acid (SCFA) production, and increased levels of lipoprotein lipase.3,60 A variety of ML methods, such as support vector machines, neural networks, and logistic regression, have been utilized to identify microbiota features present in several disease states.61 Gupta et al. have developed a classification ML model that can predict individuals’ health status based on microbiome profiling.62 Elsewhere, human-bacteria protein-protein interactions have been successfully predicted.63 A study by Zeevi et al. used an ML algorithm integrating 800 people’s blood parameters, dietary habits, anthropometrics, physical activity, and gut microbiome profile to predict postprandial glycemic response.64 Such work illustrates how precision microbiome medicine can become a reality.65 The promise for ML-led microbiome precision medicine within oncology has been well described by Cammarota et al.26
Drug discovery scientists have utilized ML for years to identify novel molecules with therapeutic potential.41,66–68 Supervised, unsupervised, semi-supervised, and reinforcement learning styles have all been applied to novel drug-target predictions: methods which could be easily employed for identification of microbiome therapeutics.69–72 ML has also been applied to drug repurposing, allowing screening of thousands of marketed drugs for alternative therapeutic purposes.73 In 2018 over 1,000 marketed drugs were screened for activity against 40 gut bacteria strains: 24% of the drugs were found to inhibit growth of at least one strain, despite the majority not being marketed for antibiotic purposes.35 This highlights the potential for repurposing within microbiome medicine. Prebiotics, entities that promote the growth of beneficial microbiota, are popular and efficacious interventions within microbiome medicine.74–76 A recent study using both supervised and unsupervised ML has elucidated and quantified the importance of the structure-property relationships of beta-glucans as prebiotics.77 Probiotics, live microorganisms that when administered in adequate amounts confer a health benefit, are commonly investigated microbiome therapeutics.78 Currently identification of probiotic candidates usually occurs via a top-down approach, whereby bacteria are selected based on the observation that they are enriched in healthy individuals.65 Though this methodology has discovered useful probiotics such as Bifidobacterium, Lactobacillus, and Akkermansia muciniphila, it gives little consideration for therapeutic mechanism, dose, or heterogeneity between individuals.60,79 A bottom-up approach could exploit ML techniques to identify mechanisms of probiotic action, and search for microbiota that fulfils these actions.65 Algorithms could consider a multitude of factors before recommending a probiotic, such as efficacy, bioavailability, risk of toxicity, ease of formulation, and scalability. Deep learning techniques, which can incorporate multiple layers of ML algorithms, would be well suited to solve these kinds of complex tasks (Figure 4).41
Figure 4.
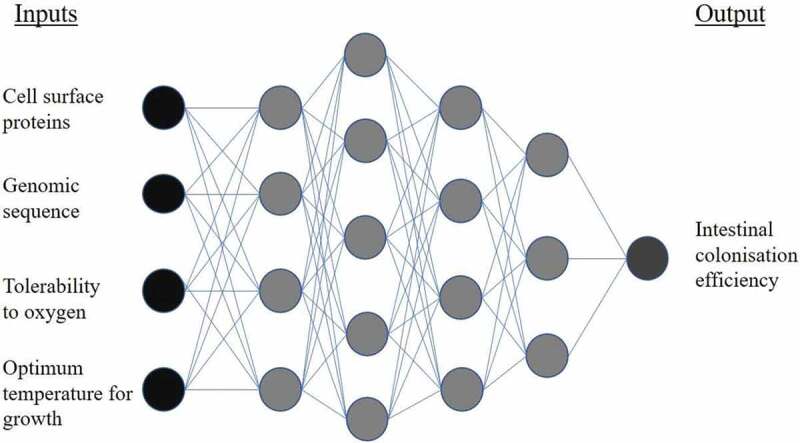
Example of a deep neural network for application in probiotic screening. Input information describing a bacterial strain is fed into hidden layers of the network. Progressing through the layers, the algorithm approaches its output: the predicted intestinal colonization efficiency of the bacteria, when administered as a probiotic
Formulation of microbiome therapeutics
Once active moieties have been selected, then pharmaceutical expertise can be used to transform a drug into a medicine. There are three main categories of microbiome therapeutics: probiotics, prebiotics, and entities that alter the microbiome microenvironment (Table 3). By far the most successful of probiotic therapies is fecal microbiota transplant (FMT) for the treatment of Clostridium difficile infection. Traditionally, FMT involves the infusion of feces from a healthy donor into the GI tract of a patient via the rectal route, to ‘reset’ their gut microbiome composition. In C. difficile infection, FMT has a 92% rate of disease resolution, far outperforming traditional antibiotics. Work is increasingly underway to investigate FMT for benefit in other diseases, and formulate fecal microbiota in oral capsules to avoid transplant of whole fecal material.4,80–82 Prebiotics are often regarded as unregulated dietary supplements, due to their natural presence in food. There have however been several clinical trials using prebiotics for amelioration of defined diseases at specified doses. For example, 20 g of specific prebiotics daily for 6 weeks has been shown to improve gut dysbiosis in human immunodeficiency virus (HIV).83,84 The final category of microbiome therapeutics, entities that alter the microbiome microenvironment, is perhaps the broadest and most novel. This category includes small molecules and biopharmaceuticals that promote the growth of beneficial microbiota or dissuade the growth of pathogens.85 Such entities include the direct delivery of beneficial bacterial metabolites to the gut, and bacteriophages designed to lyse harmful bacteria.86–89Figure 5
Table 3.
Types of microbiome therapeutics
| Category | Definition | Examples |
|---|---|---|
| Probiotics | Live microorganisms, which when administered in adequate amounts, confer a health benefit on the host.163 |
|
| Prebiotics | A substrate that is selectively utilized by host microorganisms conferring a health benefit.166 |
|
| Entities altering the microbiome microenvironment | Small molecules and biopharmaceuticals that promote the growth of beneficial microbiota or dissuade the growth of pathogens. |
|
Figure 5.
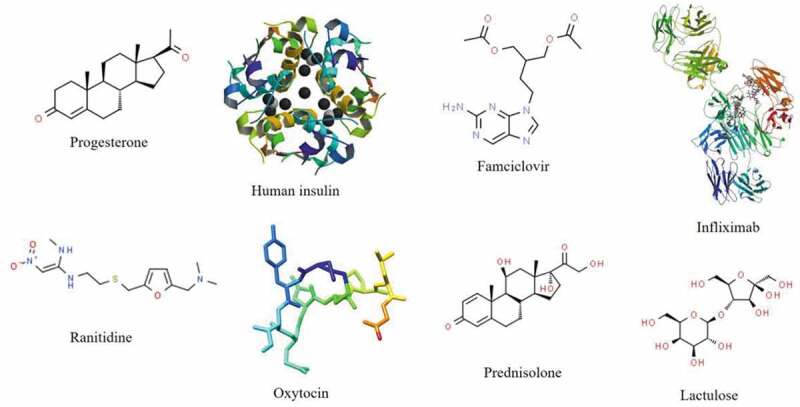
Various drugs proven to be susceptible to metabolism by gut microbiota.10,32,33,172–177
When formulating a medicine, there are hundreds if not thousands of factors to consider. These include route of administration, site of drug release, ease of manufacture, dosage form, and drug solubility, stability, and bioavailability.90 As well as being a therapeutic target, the microbiome can also be harnessed to facilitate targeted drug delivery.91,92 For example, fermentable carbohydrate coatings have been developed for accurate colonic drug delivery.93,94 Supplied with information on a drug compound, ML can quickly and accurately assess formulation options without bias.95 Due to the complex nature of formulation science, deep ML techniques are especially useful. Yang et al. have used deep ML to predict specific characteristics of formulations, such as dosage form disintegration and drug release.96 Elsewhere, deep ML has been used to predict drug solubility and oral tablet disintegration.97,98 Other types of ML have also shown utility in formulation design. Supervised ML, specifically random forest, has been used to predict the physical stability of solid dispersions over 6 months.99 Factors considered include drug molecular weight, and polymer viscosity, melting point, and topological polar surface area. The ability to predict formulation stability could save formulation scientists months of time spent iterating unstable dosage forms. Other applications include prediction of nanoparticle loading efficiency, concentration of vitamins in samples, crystallinity of drugs, and biophysical properties of monoclonal antibodies.100–103 As biopharmaceuticals, monoclonal antibodies share similarities with probiotics. Due to the inherent sensitivity of biopharmaceuticals, formulations should strike a balance between product efficacy, safety, stability, and manufacturability. Both proteins and live bacterial cells are susceptible to degradation in the acidic environment of the stomach, thus formulations for oral administration must adequately protect entities during gastrointestinal transit.104 As shown in the work by Gentiluomo et al., ML can be utilized to better understand factors that impact biotherapeutic stability.100 This is furthered echoed in work published by Pfizer, who developed an ML model to predict biophysical stability of antibodies using only the primary protein sequence.105 Perhaps in the future, the stability of probiotics in formulation could be predicted using their genomic sequence, or cell surface proteins. ML has been used to identify bacterial genes essential for life, predict antimicrobial resistance, and explore antibiotic permeation through bacterial membranes.106–108 In the case of bacterial membrane permeation, this could be applied to nanoparticles designed to enter bacterial cells for beneficial microbiome effects. Efficacy of formulations can also be predicted using ML. Luo et al. have illustrated the use of ML following a clinical trial, allowing prediction of changes to the gut microbiome after prebiotic consumption.109 This could pave the way for personalized design of formulations that maximize individual clinical outcomes. 3D printing is an additive manufacturing method with great utility in production of personalized medicines.110–120 The human microbiome is as unique as one’s fingerprint, thus 3D printing could be a useful technique for manufacture of personalized microbiome therapeutics.121–127 Such formulations could contain live microorganisms, for example oral films containing viable probiotics have been produced using inkjet printing.128 3D printing of microbiome therapeutics remains in its infancy, and so ML can be well applied to efficiently optimize printing parameters; reducing the number of empirical experiments needed.129,130 For example, ML has been used to accurately predict the printability of medicines; this could be well applied to printing of microbiome-targeted therapeutics to optimize formulation performance.131 Toxicity of formulations can also be predicted by ML. For example, the skin and genital microbiomes are sensitive to the use of certain excipients.132 ML could predict which excipients in topical formulations may exert detrimental impacts on external commensals. Clearly, there is a lot of potential, and room for innovation, within the design of microbiome therapeutics using ML.
Prediction of microbiome-drug interactions
Though not traditionally acknowledged, the metabolic capacity of the gut microbiome has long been equated to that of the liver.133 Over 180 orally administered drugs are now known to be metabolized by gut microbiota.10,11,33,134,135 Microbiota drug metabolism can lead to significant inter-individual pharmacokinetic variability.136 For example, a strain of Eggerthella lenta capable of inactivating the cardiac glycoside drug digoxin is estimated to inhabit the guts of 40% of the global population.137 Intestinal degradation of digoxin can lead to higher dose requirements of patients and make selecting a first dose difficult. Moreover, premature conversion of the Parkinson’s therapeutic levodopa by bacterial tyrosine decarboxylases in the jejunum is known to increase patients’ dose requirements.138 The number of drugs known to undergo microbiota metabolism may only represent the tip of the iceberg. Firstly, there are still many drugs which have not been tested for degradative susceptibility. Secondly, investigations have largely focused on bacterial interactions with drugs, disregarding contributions of viruses, fungi, protozoa, and archaea.139,140 Thirdly, microbiome metabolism of drugs administered by non-oral routes is under-researched. Though the human colon houses the highest density of microbiota in the body, the small intestine, skin, genital, oral, nostril, ocular, and auditory microbiomes represent unique microbial niches that could impact drugs administered by other routes.12,141,142 Microbiome-drug metabolism may be assessed using ML combined with biosensors, which can measure subtle alterations in pharmacokinetics in vivo .143,144 In addition to metabolism, the absorption of drugs across biological barriers may be impacted by perturbations in microbiome composition. For example, dysbiosis of the skin microbiome can affect barrier function, potentially altering the permeation of topical drugs.145 Moreover, dysbiosis in the colon is known to impact tight junction integrity.57 Whilst a database exists providing information on the impact of human genome variation on drug response (PharmGKB), comprehensive resources summarizing the impact of microbiome variation influence on drugs are lacking.146 The DrugBug resource curated by Sharma et al. is probably the most functional database, however it does not address pharmacokinetic concerns arising from bacterial metabolism.51
Due to the scale of work required to investigate microbiome effects on drugs by practical experimentation, ML offers a feasible way to predict interactions quickly and accurately. Though this work is by no means complete, there have been several progressive examples of ML use in recent years. In conjunction with practical high-throughput analysis of drug metabolism by gut bacteria, Zimmerman et al. used the clustering ML technique known as principal component analysis (PCA) to identify structural similarities of drugs susceptible to gut bacteria metabolism.10 Using the study’s documented 20,596 bacteria-drug interactions, PCA revealed how the presence of certain functional groups can increase the chance of metabolism. For example, drugs containing ester or amide groups are more likely to be specifically hydrolyzed by Bacteroidetes species. Other susceptible functional groups include nitro and azo moieties, which are prone to reduction by multiple anaerobic strains. This information is clearly useful for predicting untested drugs’ risk of metabolism. Another application of ML within the field of microbiome drug metabolism is work by Sharma et al.51 The group first constructed a database of substrates for human gut bacterial enzymes. Once assembled, the database included 1,609 substrates; 324,697 enzymes; and 491 bacterial genomes. PCA was then used to assess structural diversity of the included substrates to ensure a robust, representative model. Following this, random forest ML was employed to classify commercial drugs as being susceptible to biotransformation by specific gut microbiota enzymes. Upon analysis the prediction accuracy of the algorithm was judged to be >93%. This resource was subsequently made publicly available via the DrugBug online prediction platform.51 Despite being the first work of its kind, the DrugBug tool does have room for improvement. Firstly, the prediction accuracy could be improved using a larger number of drug molecules for algorithm training and testing, and by fine tweaking of algorithm parameters. Secondly, the online platform is quite slow and complicated to use. There is also no information given on the prevalence of the gut bacteria in the global population, or any clinical or pharmacokinetic implications of potential drug metabolism. DrugBug is a brilliant starting point for the ML microbiome pharmaceutics community to build on.
Whilst the microbiome can impact the actions of drugs, the same applies to the converse. A study by Maier et al. has highlighted that a significant proportion of non-antibiotic drugs impair growth of gastrointestinal microbiota.35 A couple of research groups have utilized this dataset of >1000 marketed drugs to build ML models that can predict anti-commensal activity of drugs untested by Maier et al.147,148 Such work could be highly useful in preclinical toxicity screening within the pharmaceutical industry, to predict whether new treatments could cause gut dysbiosis.
Barriers To Overcome
Currently, the use of ML to develop microbiome therapeutics lags considerably behind other areas of science.95,149,150 There are several possible reasons for this, representing challenges for the field to overcome. Firstly, data describing human-microbiome-drug interactions have only begun to reach a rapid rate of discovery in recent years, with studies such as the high-throughput screening of microbiota drug metabolism by Zimmerman et al., and work from the field-leading APC Microbiome Ireland.10,151–154 Whilst this new abundance of information is clearly positive, to facilitate ML projects data should be collated and formatted into accessible databases. Such databases could provide information on microbiota drug metabolism, formulation parameters for microbiome therapeutics, and pharmacokinetic details for microbiota-targeted medicines. With the construction of large databases containing reliable, clean data, ML work would be easier and yield stronger predictions. Clearly, collaboration and information sharing between academic institutions and commercial corporations will be necessary for the reality of global database creation. Whilst this may be difficult in the short term, the yields will far outweigh disadvantages, likely leading to the development of new therapies for patients. Where large quantities of data are available in-house, for example historical experimental data, it may be difficult to use in ML projects.155 Without the foresight of storing data for potential ML use, it is likely that substantial pre-processing steps will be required.45 Though possible, cleaning of decades of heterogeneously formatted data can take years. There is also the barrier of skillset requirements for the adoption of ML within the field. Demand for data scientists in the United Kingdom grew by 231% in the five years preceding 2019.156 In addition to ML skills, it is highly beneficial for workers to also have pharmaceutics expertise, as this will facilitate optimal data mining, cleaning, and end analysis. For example, ML scientists without a pharmaceutics background may not spot anomalies or mistakes in datasets. They may also find interpretation and application of ML algorithm outputs more challenging. Training of pharmaceutical scientists in ML skills, and training of ML scientists in pharmaceutics, is thus required. Evidently, this will require substantial investment within both academic and industry settings. Finally, it is important that settings facilitate the use of ML to obtain optimal results. More so in industry, well-defined and proven systems of work will change dramatically with the widespread adoption of ML. For example, formulation scientists may spend much less time physically compounding iterations of products due to the predictive power of ML. Implementation of ML will require restructuring of procedures, in a careful and strategic manner, with full support of team members.
Future Outlook
The explosion of big data within microbiome research in the last 20 years opens opportunities for ML like never before. Freely accessible genomic, proteomic, and metabolomic metadata facilitates commencement of ML projects with little initial investment or risk. To make the most of ML, academic and corporate institutions will need to collaborate to populate new databases with previously proprietary information, such as probiotic viability, microbiota drug metabolism, and stability studies. Whilst this will invariably raise issues around intellectual property, collaboration will offer projects not previously feasible for companies, and most importantly, will benefit patients through the development of new therapies. In order to adopt ML in a strategic, widespread, and sustainable manner, institutions will need to invest in the training of ML engineers. In the wake of a global recession instigated by the COVID-19 pandemic, institutions may find training budgets are restricted.157–159 Low- to middle-income countries will be especially affected, for example there is a projected 7.2% decrease in regional economic activity in Latin America compared to 4.7% in Europe and Central Asia.160 Though the global economic downturn will undoubtedly affect the pharmaceutical industry, it is a worthwhile investment to commit budgets to ML expertise. Once workers are trained, ML will save time and physical resources in product development. ML has the capacity to optimize and streamline the preclinical stages of microbiome therapeutics, meaning products will progress to trials faster with likely higher success rates. Major pharmaceutical companies currently showing the most activity within AI include Novartis, AstraZeneca, and Boehringer Ingelheim.155 Specialist AI companies, mostly working in the drug discovery space, include Exscientia, Atomwise, GNS Healthcare, and Berg. These specialist corporations typically partner with major pharma companies to unite expertise, a collaborative method of drug development that is likely to only increase in occurrence.161
Concluding Remarks
Clearly, opportunities for utilizing ML in development of microbiome therapeutics are manifold. Since the early 2000s big data within microbiome research have become a reality, with genomic sequencing and metabolomic mapping providing an influx of information on the host-microbiota relationship.12,13 This data has provided a deepening understanding of how the microbiome can affect health, allowing the development of precision microbiome medicine. In more recent years, the impact of microbiota metabolism on drugs has come to light, and may change pharmacokinetic modeling forever.10,162 With the amount of data currently available, it is more important than ever to utilize systematic, accurate, and unbiased tools for analysis. ML techniques offer an accessible way to interpret metadata and use it to solve problems. Whilst a human could not feasibly recognize patterns within terabytes of information, ML can do this and solve complex tasks within seconds to minutes. To obtain information for ML projects, it is important to have access to reliable databases. Though several of these exist for data on drugs, microbiota, and microbiota-host dynamics, there is a distinct lack of resources providing pharmaceutical information. Curation of such databases is important for optimal utilization of ML within the field. Several challenges face institutions looking to adopt ML. Namely, cooperation with other corporations, data management, service remodeling, and lack of skillset. Whilst the necessary initial investment may be challenging for academic and industrial settings, especially considering the current global economy, actions will be worthwhile in the long term. ML is transforming almost every sector worldwide and is here to stay.
Funding Statement
This work was supported by The Engineering and Physical Sciences Research Council under Grant [EP/S023054/1].
Disclosure statement
The authors declare no conflict of interest.
References
- 1.Rosa BA, Mihindukulasuriya K, Hallsworth-Pepin K, Wollam A, Martin J, Snowden C, et al. Improving characterization of understudied human microbiomes using targeted phylogenetics [Article]. mSystems. 2020;5:1. doi: 10.1128/mSystems.00096-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464:59-65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singer-Englar T, Barlow G, Mathur R.. Obesity, diabetes, and the gut microbiome: an updated review. Expert Rev Gastroenterol Hepatol. 2019. January;13(1):3–20. doi: 10.1080/17474124.2019.1543023. [DOI] [PubMed] [Google Scholar]
- 4.Dutta SK, Verma S, Jain V, Surapaneni BK, Vinayek R, Phillips L, et al. Parkinson’s disease: the emerging role of gut dysbiosis, antibiotics, probiotics, and fecal microbiota transplantation. J Neurogastroenterol Motil. 2019. July 1;530(1–2):363–376. doi: 10.5056/jnm19044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yadav V, Varum F, Bravo R, Furrer E, Bojic D, Basit AW. Inflammatory bowel disease: exploring gut pathophysiology for novel therapeutic targets. Transl Res. 2016. October;176:38–68. doi: 10.1016/j.trsl.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Gao L, Xu T, Huang G,Jiang S, Gu Y, Chen F. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018. May;9(5):488–500. doi: 10.1007/s13238-018-0548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ercolini D, Fogliano V. Food design to feed the human gut microbiota. J Agric Food Chem. 2018;66(15):3754–3758. doi: 10.1021/acs.jafc.8b00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ezra-Nevo G, Henriques SF, Ribeiro C. The diet-microbiome tango: how nutrients lead the gut brain axis. Curr Opin Neurobiol. 2020. [2020/06/01/];62:122–132. doi: 10.1016/j.conb.2020.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Valdes AM, Walter J, Segal E,Spector TD. Role of the gut microbiota in nutrition and health. BMJ. 2018;361:k2179. doi: 10.1136/bmj.k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R,Goodman AL. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019;570(7762):462–467. doi: 10.1038/s41586-019-1291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke G, Sandhu KV, Griffin BT,Dinan TG, Cryan JF, Hyland NP. Gut reactions: breaking down xenobiotic-microbiome interactions. Pharmacol Rev. 2019. April;71(2):198–224. doi: 10.1124/pr.118.015768. [DOI] [PubMed] [Google Scholar]
- 12.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012. 2012/06/01;486(7402):207–214.doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Proctor LM, Creasy HH, Fettweis JM, Lloyd-Price J, Mahurkar A, Zhou W, et al. The integrative human microbiome project. Nature. 2019. 2019/05/01;569(7758):641–648.doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Human Microbiome HMPNIH. Project. Maryland, USA: National Institutes of Health (NIH); 2020. [Google Scholar]
- 15.EuPathDB. MicrobiomeDB. USA: National . Institute of Allergy and Infectious Diseases (NIAD); 2020. [Google Scholar]
- 16.China National GeneBank. Microbiome Database . Shenzhen (China): China National GeneBank; 2020. [Google Scholar]
- 17.Davenport T, Kalakota R. The potential for artificial intelligence in healthcare. Future Healthc J. 2019;6(2):94–98. doi: 10.7861/futurehosp.6-2-94. doi: 10.7861/futurehosp.6-2-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camacho DM, Collins KM, Powers RK,Costello JC, Collins JJ. Next-generation machine learning for biological networks [review]. Cell. 2018;173(7):1581–1592. doi: 10.1016/j.cell.2018.05.015 [DOI] [PubMed] [Google Scholar]
- 19.Namkung J. Machine learning methods for microbiome studies. J Microbiol. 2020. March;58(3):206–216. doi: 10.1007/s12275-020-0066-8. [DOI] [PubMed] [Google Scholar]
- 20.Vujkovic-Cvijin I, Sklar J, Jiang L,Natarajan L, Knight R, Belkaid Y. Host variables confound gut microbiota studies of human disease. Nature. 2020. November;587(7834):448–454. doi: 10.1038/s41586-020-2881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Google . A history of machine learning 2020 [Accessed 20th October 2020]. Available from: https://cloud.withgoogle.com/build/data-analytics/explore-history-machine-learning/
- 22.Badillo S, Banfai B, Birzele F, Davydov II, Hutchinson L, Kam‐Thong T, Siebourg‐Polster J, Steiert B, Zhang JD. An introduction to machine learning. . Clinical Pharmacology & Therapeutics. 2020. Apr;;107(4):871–885. doi: 10.1002/cpt.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neftci EO, Averbeck BB. Reinforcement learning in artificial and biological systems. Nature Machine Intelligence. 2019/03/01 2019;1(3):133–143. doi: 10.1038/s42256-019-0025-4. [DOI] [Google Scholar]
- 24.Sledge IJ, Príncipe JC, editors. Balancing exploration and exploitation in reinforcement learning using a value of information criterion. 2017 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); 2017 5–9 March 2017. doi: 10.1109/ICASSP.2017.7952670. [DOI] [Google Scholar]
- 25.Silver D, Schrittwieser J, Simonyan K, Antonoglou I, Huang A, Guez A, et al. Mastering the game of Go without human knowledge. Nature. 2017. 2017/10/01;550(7676):354–359. doi: 10.1038/nature24270 [DOI] [PubMed] [Google Scholar]
- 26.Cammarota G, Ianiro G, Ahern A, Carbone C, Temko A, Claesson MJ, et al. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nature reviews gastroenterology & hepatology.2020; 17:635-648. [DOI] [PubMed] [Google Scholar]
- 27.Chen M, Mao S, Liu, Y. Big Data:A Survey. Mobile networks and applications. 2014;19(2):171–209. 10.1007/s11036-013-0489-0. [DOI] [Google Scholar]
- 28.Tang Q, Jin G, Wang G, Liu T, Liu X, Wang B, et al. Current sampling methods for gut microbiota: a call for more precise devices. Front Cell Infect Microbiol. 2020;10:151. doi: 10.3389/fcimb.2020.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vuik F, Dicksved J, Lam SY, Fuhler GM, van der Laan L, van de Winkel A, et al. Composition of the mucosa-associated microbiota along the entire gastrointestinal tract of human individuals. United European Gastroenterol J. 2019. August;7(7):897–907. doi: 10.1177/2050640619852255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James KR, Gomes T, Elmentaite R, Kumar N, Gulliver EL, King HW, et al. Distinct microbial and immune niches of the human colon. Nat Immunol. 2020 Mar;21(3):343–353. doi: 10.1038/s41590-020-0602-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parmanand BA, Kellingray L, Le Gall G, Basit AW, Fairweather-Tait S, Narbad A. A decrease in iron availability to human gut microbiome reduces the growth of potentially pathogenic gut bacteria; an in vitro colonic fermentation study. J Nutr Biochem. 2019. May;67:20–27. doi: 10.1016/j.jnutbio.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Yadav V, Smart AL, Tajiri S, Basit AW. Stability of peptide drugs in the colon. Eur J Pharm Sci. 2015. October;78(78):31–36. doi: 10.1016/j.ejps.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 33.Coombes Z, Yadav V, E. McCoubrey L, Freire C, W. Basit A, Conlan RS, et al. Progestogens are metabolized by the gut microbiota: implications for colonic drug delivery. Pharmaceutics. 2020;12(8). doi: 10.3390/pharmaceutics12080760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaudhari DS, Dhotre DP, Agarwal DM, Gaike AH, Bhalerao D, Jadhav P, et al. Gut, oral and skin microbiome of indian patrilineal families reveal perceptible association with age. Sci Rep. 2020. ;2020/03/30 10(1):5685. doi: 10.1038/s41598-020-62195-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018. 2018/03/01;555(7698):623–628. doi: 10.1038/nature25979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freire AC, Basit AW, Choudhary R, Piong CW, Merchant HA. Does sex matter? the influence of gender on gastrointestinal physiology and drug delivery. Int J Pharm. 2011. August 30;415(1–2):15–28. doi: 10.1016/j.ijpharm.2011.04.069. [DOI] [PubMed] [Google Scholar]
- 37.Dehingia M, Adak A, Khan MR. Ethnicity-influenced microbiota: a future healthcare perspective. Trends Microbiol. 2019/03/01/ 2019;27(3):191–193. doi: 10.1016/j.tim.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 38.So D, Whelan K, Rossi M, Morrison M, Holtmann G, Kelly J, et al. . Dietary fiber intervention on gut microbiota composition in healthy adults: A systematic review and meta-analysis. Am J Clin Nutr. 2018;05/11;107.doi: 10.1093/ajcn/nqy041. [DOI] [PubMed] [Google Scholar]
- 39.Vujkovic-Cvijin I, Sklar J, Jiang L, Natarajan L, Knight R, Belkaid Y. . Host variables confound gut microbiota studies of human disease. In: Nature. 2020; 587(7834):448-454. doi: 10.1038/s41586-020-2881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raudys SJ, Jain AK. Small sample size effects in statistical pattern recognition: recommendations for practitioners [article]. IEEE Trans Pattern Anal Mach Intell. 1991;13(3):252–264. doi: 10.1109/34.75512. [DOI] [Google Scholar]
- 41.Lecun Y, Bengio Y, Hinton G. Deep learning [Review]. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 42.Vabalas A, Gowen E, Poliakoff E, Casson AJ. . Machine learning algorithm validation with a limited sample size. PLoS One. 2019;12(3):e0224365. doi: 10.1371/journal.pone.0224365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng M, Cao L, Microbiome Big-Data NK. Mining and applications using single-cell technologies and metagenomics approaches toward precision medicine [review]. Front Genet. 2019; 10: 972. 2019-October-09. doi: 10.3389/fgene.2019.00972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox DR, Kartsonaki C, Keogh RH. Big data: some statistical issues. Stat Probab Lett. 2018. [2018/05/01/];136:111–115. doi: 10.1016/j.spl.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.García-Gil D, Luengo J, García S, Herrera F. Enabling smart data: noise filtering in big data classification. Inf Sci (Ny). 2019. . 479:135–152. 2019/04/01/. [Google Scholar]
- 46.Lazer D, Kennedy R, King G, Vespignani A. . The parable of google flu: traps in big data analysis. Science. 2014;343(6176):1203–1205. doi: 10.1126/science.1248506. [DOI] [PubMed] [Google Scholar]
- 47.Anonymous. Not so big. The Economist 2020. June 2020;13:S5–S6. [Google Scholar]
- 48.Géron A. Hands-on machine learning with scikit-learn & tensorFlow. First ed. California, USA: O’Reilly; 2017. [Google Scholar]
- 49.Morris TP, White IR, Royston P. Tuning multiple imputation by predictive mean matching and local residual draws. BMC Med Res Methodol. 2014/06/05 2014;14(1):75. doi: 10.1186/1471-2288-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koprinkova P, Petrova M. Data-scaling problems in neural-network training [Article]. Eng Appl Artif Intell. 1999;12(3):281–296. doi: 10.1016/S0952-1976(99)00008-1. [DOI] [Google Scholar]
- 51.Sharma AK, Jaiswal SK, Chaudhary N, Sharma VK. A novel approach for the prediction of species-specific biotransformation of xenobiotic/drug molecules by the human gut microbiota. Sci Rep. 2017. 2017 Aug 29;7(1):9751. doi: 10.1038/s41598-017-10203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noronha A, Modamio J, Jarosz Y, Guerard E, Sompairac N, Preciat G, et al. The virtual metabolic human database: integrating human and gut microbiome metabolism with nutrition and disease. Nucleic Acids Res. 2019. Jan 8;47(D1):D614–D624. doi: 10.1093/nar/gky992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han W, Ye Y. A repository of microbial marker genes related to human health and diseases for host phenotype prediction using microbiome data. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. 2019;24:236–247. PMID: 30864326. [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen HT, Phan NYK, Luong HH, Le TP, Tran NC. Efficient discretization approaches for machine learning techniques to improve disease classification on gut microbiome composition data [Article]. Advances in Science, Technology and Engineering Systems. 2020;5(3):547–556. doi: 10.25046/aj050368. [DOI] [Google Scholar]
- 55.Chandrasekaran SN, Ceulemans H, Boyd JD, Carpenter AE. Image-based profiling for drug discovery: due for a machine-learning upgrade? Nat rev drug discov. 2020. December 22.doi: 10.1038/s41573-020-00117-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou YH, Gallins P. A review and tutorial of machine learning methods for microbiome host trait prediction [Article]. Front Genet. 2019;10(JUN). doi: 10.3389/fgene.2019.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghyselinck J, Verstrepen L, Moens F, Van den Abbeele P, Said J, Smith B, et al. A 4-strain probiotic supplement influences gut microbiota composition and gut wall function in patients with ulcerative colitis. Int J Pharm. 2020. July;587:119648. doi: 10.1016/j.ijpharm.2020.119648. [DOI] [PubMed] [Google Scholar]
- 58.Barlow GM, Yu A, Mathur R. Role of the gut microbiome in obesity and diabetes mellitus. Nutr Clin Pract. 2015. December;30(6):787–797. doi: 10.1177/0884533615609896. [DOI] [PubMed] [Google Scholar]
- 59.Rowin J, Xia Y, Jung B, Sun J.Gut inflammation and dysbiosis in human motor neuron disease [Article]. Physiological Reports. 2017;5(18):18. doi: 10.14814/phy2.13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moens F, Van den Abbeele P, Basit AW, Dodoo C, Chatterjee R, Smith B, et al. . A four-strain probiotic exerts positive immunomodulatory effects by enhancing colonic butyrate production in vitro. Int J Pharm. 2019. 555:1–10. 2019/01/30/. doi: 10.1016/j.ijpharm.2018.11.020 [DOI] [PubMed] [Google Scholar]
- 61.Pasolli E, Truong DT, Malik F, Waldron L, Segata N. Machine learning meta-analysis of large metagenomic datasets: tools and biological insights [article]. PLoS Comput Biol. 2016;12(7):7. doi: 10.1371/journal.pcbi.1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta VK, Kim M, Bakshi U, Cunningham KY, Davis JM, Lazaridis KN, et al. A predictive index for health status using species-level gut microbiome profiling. Nat Commun. 2020;11(1):1. doi: 10.1038/s41467-020-18476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lian X, Yang S, Li H, Fu C, Zhang Z. Machine-learning-based predictor of human-bacteria protein-protein interactions by incorporating comprehensive host-network properties [article]. J Proteome Res. 2019;18(5):2195–2205. doi: 10.1021/acs.jproteome.9b00074. [DOI] [PubMed] [Google Scholar]
- 64.Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015. November 19;163(5):1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 65.Veiga P, Suez J, Derrien M, Elinav E. Moving from probiotics to precision probiotics. Nat Microbiol. 2020. May 11; 5: 878-880. doi: 10.1038/s41564-020-0721-1. [DOI] [PubMed] [Google Scholar]
- 66.Zhang L, Tan J, Han D, Zhu H. From machine learning to deep learning: progress in machine intelligence for rational drug discovery [Review]. Drug Discov Today. 2017;22(11):1680–1685. doi: 10.1016/j.drudis.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 67.Stephenson N, Shane E, Chase J, Rowland J, Ries D, Justice N, et al. Survey of machine learning techniques in drug discovery [review]. Curr Drug Metab. 2019;20(3):185–193. doi: 10.2174/1389200219666180820112457. [DOI] [PubMed] [Google Scholar]
- 68.Elbadawi M, Gaisford S, Basit AW.. Advanced machine-learning techniques in drug discovery. Drug discov today. 2020. December 5. doi: 10.1016/j.drudis.2020.12.003 [DOI] [PubMed] [Google Scholar]
- 69.Olivecrona M, Blaschke T, Engkvist O, Chen H. Molecular de-novo design through deep reinforcement learning. J Cheminform. 2017;2017/09/04 9(1):48. doi: 10.1186/s13321-017-0235-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen H, Zhang Z.A Semi-Supervised Method for Drug-Target Interaction Prediction with Consistency in Networks PLoS One. 2013;8:5. doi: 10.1371/journal.pone.0062975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bleakley K, Yamanishi Y. Supervised prediction of drug-target interactions using bipartite local models [Article]. Bioinformatics. 2009;25(18):2397–2403. doi: 10.1093/bioinformatics/btp433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taguchi YH, Iwadate M, Umeyama H. Principal component analysis-based unsupervised feature extraction applied to in silico drug discovery for posttraumatic stress disorder-mediated heart disease [Article]. BMC Bioinform. 2015;16(1):1. doi: 10.1186/s12859-015-0574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou H, Gao M, Skolnick J. Comprehensive prediction of drug-protein interactions and side effects for the human proteome. Sci Rep. 2015/06/09 2015;5(1):11090. doi: 10.1038/srep11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359(6380):1151. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- 75.Byrne CS, Chambers ES, Preston T, Tedford C, Brignardello J, Garcia-Perez I, Holmes E, Wallis GA, Morrison DJ, Frost GS, et al. Effects of Inulin propionate ester incorporated into palatable food products on appetite and resting energy expenditure: A randomised crossover study [Article]. Nutrients. 2019;11(4):4. doi: 10.3390/nu11040861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koziolek M, Alcaro S, Augustijns P, Basit AW, Grimm M, Hens B, Hoad CL, Jedamzik P, Madla CM, Maliepaard M, et al. The mechanisms of pharmacokinetic food-drug interactions – A perspective from the UNGAP group. European Journal of Pharmaceutical Sciences. 2019. 134:31–59. 2019/06/15/. doi: 10.1016/j.ejps.2019.04.003 [DOI] [PubMed] [Google Scholar]
- 77.Lam K-L, Cheng W-Y, Su Y, Li X, Wu X, Wong K-H, et al. Use of random forest analysis to quantify the importance of the structural characteristics of beta-glucans for prebiotic development. Food Hydrocolloids. 2020;108.doi: 10.1016/j.foodhyd.2020.106001. [Google Scholar]
- 78.Dodoo CC, Stapleton P, Basit AW, Gaisford S. Use of a water-based probiotic to treat common gut pathogens. Int J Pharm. 2019. February;556(556):136–141. doi: 10.1016/j.ijpharm.2018.11.075. [DOI] [PubMed] [Google Scholar]
- 79.Earley H, Lennon G, Balfe A, Coffey JC, Winter DC, O'Connell PR. The abundance of Akkermansia muciniphila and its relationship with sulphated colonic mucins in health and ulcerative colitis. Sci Rep. 2019. October 30;9(1):15683. doi: 10.1038/s41598-019-51878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fadda HM. The route to palatable fecal microbiota transplantation. AAPS PharmSciTech. 2020;21(3):3. doi: 10.1208/s12249-020-1637-z. [DOI] [PubMed] [Google Scholar]
- 81.Allegretti J, Eysenbach LM, El-Nachef N, Fischer M, Kelly C, Kassam Z. The current landscape and lessons from fecal microbiota transplantation for inflammatory bowel disease: past, present, and future [review]. Inflamm Bowel Dis. 2017;23(10):1710–1717. doi: 10.1097/MIB.0000000000001247. [DOI] [PubMed] [Google Scholar]
- 82.Allegretti JR, Fischer M, Sagi SV, Bohm ME, Fadda HM, Ranmal SR, et al. Fecal microbiota transplantation capsules with targeted colonic versus gastric delivery in recurrent clostridium difficile infection: a comparative cohort analysis of high and lose dose. Dig Dis Sci. 2019. June;64(6):1672–1678. doi: 10.1007/s10620-018-5396-6. [DOI] [PubMed] [Google Scholar]
- 83.Deusch S, Serrano-Villar S, Rojo D, Martínez-Martínez M, Bargiela R, Vázquez-Castellanos JF, et al. Effects of HIV, antiretroviral therapy and prebiotics on the active fraction of the gut microbiota [Article]. AIDS. 2018;32(10):1229–1237. doi: 10.1097/QAD.0000000000001831. [DOI] [PubMed] [Google Scholar]
- 84.Serrano-Villar S, Vázquez-Castellanos JF, Vallejo A, Latorre A, Sainz T, Ferrando-Martínez S, et al. The effects of prebiotics on microbial dysbiosis, butyrate production and immunity in HIV-infected subjects. Mucosal Immunol. 2017. 2017/09/01;10(5):1279–1293. doi: 10.1038/mi.2016.122 [DOI] [PubMed] [Google Scholar]
- 85.Daisley BA, Chanyi RM, Abdur-Rashid K, Al KF, Gibbons S, Chmiel JA, et al. Abiraterone acetate preferentially enriches for the gut commensal Akkermansia muciniphila in castrate-resistant prostate cancer patients. Nat Commun. 2020. September 24;11(1):4822. doi: 10.1038/s41467-020-18649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chambers ES, Byrne CS, Rugyendo A, Morrison DJ, Preston T, Tedford C, et al. The effects of dietary supplementation with inulin and inulin-propionate ester on hepatic steatosis in adults with non-alcoholic fatty liver disease. Diabetes Obes Metab. 2019;21(2):372–376. doi: 10.1111/dom.13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chambers ES, Viardot A, Psichas A, Morrison DJ, Murphy KG, Zac-Varghese SEK, et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut. 2015;64(11):1744–1754. doi: 10.1136/gutjnl-2014-307913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016;8(1):39. doi: 10.1186/s13073-016-0294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ghose C, Euler CW. Gram-negative bacterial lysins [Review]. Antibiotics. 2020;9(2):2. doi: 10.3390/antibiotics9020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aulton ME, Taylor K. Aulton’s pharmaceutics: the design and manufacture of medicines. Edinburgh, United Kingdom: Churchill Livingstone/Elsevier; 2018. English. [Google Scholar]
- 91.McConnell EL, Liu F, Basit AW. Colonic treatments and targets: issues and opportunities. J Drug Target. 2009. June;17(5):335–363. doi: 10.1080/10611860902839502. [DOI] [PubMed] [Google Scholar]
- 92.Bak A, Ashford M, Brayden DJ. Local delivery of macromolecules to treat diseases associated with the colon. Adv Drug Deliv Rev. 2018. November;136-137:2–27. Dec;136-137. doi: 10.1016/j.addr.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 93.Varum F, Cristina Freire A, Bravo R, Basit AW. OPTICORE, an innovative and accurate colonic targeting technology. Int J Pharm. 2020. April;583:119372. doi: 10.1016/j.ijpharm.2020.119372. [DOI] [PubMed] [Google Scholar]
- 94.Varum F, Cristina Freire A, Fadda HM, Bravo R, Basit AW. . A dual pH and microbiota-triggered coating (Phloral(TM)) for fail-safe colonic drug release. Int J Pharm. 2020. April;583:119379. doi: 10.1016/j.ijpharm.2020.119379. [DOI] [PubMed] [Google Scholar]
- 95.Damiati SA. digital pharmaceutical sciences. AAPS PharmSciTech. 2020. July 26;21(6):206. doi: 10.1208/s12249-020-01747-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang Y, Ye Z, Su Y, Zhao Q, Li X, Ouyang D. Deep learning for in vitro prediction of pharmaceutical formulations. Acta Pharm Sin B. 2019;9(1):177–185. 2019/01/01/. doi: 10.1016/j.apsb.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lusci A, Pollastri G, Baldi P. Deep architectures and deep learning in chemoinformatics: the prediction of aqueous solubility for drug-like molecules [Article]. J Chem Inf Model. 2013;53(7):1563–1575. doi: 10.1021/ci400187y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Han R, Yang Y, Li X, Ouyang D. Predicting oral disintegrating tablet formulations by neural network techniques. Asian J Pharm Sci. 2018;13(4):336–342. Jul;. doi: 10.1016/j.ajps.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han R, Xiong H, Ye Z, Yang Y, Huang T, Jing Q, et al. . Predicting physical stability of solid dispersions by machine learning techniques. Journal of Controlled Release. 2019;2019/10/01/;311–312:16–25.doi: 10.1016/j.jconrel.2019.08.030. [DOI] [PubMed] [Google Scholar]
- 100.Gentiluomo L, Roessner D, Augustijn D, Svilenov H, Kulakova A, Mahapatra S, et al. Application of interpretable artificial neural networks to early monoclonal antibodies development. European Journal of Pharmaceutics and Biopharmaceutics. 2019;2019/08/01/;141:81–89. 10.1016/j.ejpb.2019.05.017. [DOI] [PubMed] [Google Scholar]
- 101.Li Y, Abbaspour MR, Grootendorst PV, Rauth AM, Wu XY. Optimization of controlled release nanoparticle formulation of verapamil hydrochloride using artificial neural networks with genetic algorithm and response surface methodology. European Journal of Pharmaceutics and Biopharmaceutics. 2015;2015/08/01/;94:170–179. 10.1016/j.ejpb.2019.05.017. [DOI] [PubMed] [Google Scholar]
- 102.Barthus RC, Mazo LH, Poppi RJ. Simultaneous determination of vitamins C, B6 and PP in pharmaceutics using differential pulse voltammetry with a glassy carbon electrode and multivariate calibration tools. J Pharm Biomed Anal. 2005. 2005/06//;38(1):94–99. doi: 10.1016/j.jpba.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 103.Ghosh A, Louis L, Arora KK, Hancock BC, Krzyzaniak JF, Meenan P, et al. Assessment of machine learning approaches for predicting the crystallization propensity of active pharmaceutical ingredients [Article]. CrystEngComm. 2019;21(8):1215–1223.doi: 10.1039/C8CE01589A. [Google Scholar]
- 104.Dodoo CC, Wang J, Basit AW, Stapleton P, Gaisford S. Targeted delivery of probiotics to enhance gastrointestinal stability and intestinal colonisation. Int J Pharm. 2017;530(1):224–229. 2017/09/15/. doi: 10.1016/j.ijpharm.2017.07.068. [DOI] [PubMed] [Google Scholar]
- 105.King AC, Woods M, Liu W, Lu Z, Gill D, Krebs MRH. High-throughput measurement, correlation analysis, and machine-learning predictions for pH and thermal stabilities of Pfizer-generated antibodies [Article]. Protein Science. 2011;20(9):1546–1557. doi: 10.1002/pro.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Singhal A, Roy D, Mittal S, Dhar J, Singh A. A new computational approach to identify essential genes in bacterial organisms using machine learning. Advances in Intelligent Systems and Computing. 2019(p):67–79.doi: 10.1007/978-981-13-1132-1_6. [Google Scholar]
- 107.Lee MW, de Anda J, Kroll C, Bieniossek C, Bradley K, Amrein KE, et al. How do cyclic antibiotics with activity against Gram-negative bacteria permeate membranes? A machine learning informed experimental study [Article]. Biochimica Et Biophysica Acta - Biomembranes. 2020;1862:8. doi: 10.1016/j.bbamem.2020.183302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khaledi A, Weimann A, Schniederjans M, Asgari E, Kuo TH, Oliver A, et al. Predicting antimicrobial resistance in Pseudomonas aeruginosa with machine learning-enabled molecular diagnostics. EMBO Mol Med. 2020;12(3):3. doi: 10.15252/emmm.201910264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Luo YM, Liu FT, Chen MX, Tang WL, Yang YL, Tan XL, et al. A machine learning model based on initial gut microbiome data for predicting changes of bifidobacterium after prebiotics consumption [article]. Nan Fang Yi Ke Da Xue Xue Bao (Journal of Southern Medical University). 2018;38(3):251–260. doi: 10.3969/j.issn.1673-4254.2018.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goyanes A, Madla CM, Umerji A, Duran Piñeiro G, Giraldez Montero JM, Lamas Diaz MJ, et al. Automated therapy preparation of isoleucine formulations using 3D printing for the treatment of MSUD: first single-centre, prospective, crossover study in patients [Article]. Int J Pharm. 2019;567. doi: 10.1016/j.ijpharm.2019.118497. [DOI] [PubMed] [Google Scholar]
- 111.Trenfield SJ, Awad A, Madla CM, Hatton GB, Firth J, Goyanes A, et al. . Shaping the future: recent advances of 3D printing in drug delivery and healthcare [Review]. Expert Opin Drug Deliv. 2019;16(10):1081–1094. doi: 10.1080/17425247.2019.1660318. [DOI] [PubMed] [Google Scholar]
- 112.Awad A, Yao A, Trenfield SJ, Goyanes A, Gaisford S, Basit AW. . 3D printed tablets (Printlets) with braille and moon patterns for visually impaired patients [Article]. Pharmaceutics. 2020;12(2):2. doi: 10.3390/pharmaceutics12020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Elbadawi M, Ong JJ, Pollard TD, Gaisford S, Basit AW. Additive manufacturable materials for electrochemical biosensor electrodes. Adv Funct Mater. 2020. doi: 10.1002/adfm.202006407. [DOI] [Google Scholar]
- 114.Norman J, Madurawe RD, Moore CM, Khan MA, Khairuzzaman A. A new chapter in pharmaceutical manufacturing: 3D-printed drug products. Adv Drug Deliv Rev 2017 Jan 1;108:39–50. doi: 10.1016/j.addr.2016.03.001 [DOI] [PubMed] [Google Scholar]
- 115.Melocchi A, Briatico-Vangosa F, Uboldi M, Parietti F, Turchi M, von Zeppelin D, et al. Quality considerations on the pharmaceutical applications of fused deposition modeling 3D printing. Int J Pharm. 2021;592:119901. 2021 Jan 5. doi: 10.1016/j.ijpharm.2020.119901. [DOI] [PubMed] [Google Scholar]
- 116.Lim SH, Kathuria H, Tan JJY, Kang L. 3D printed drug delivery and testing systems - a passing fad or the future? Advanced Drug Delivery Reviews. 2018;132:139–168. 2018 Jul. doi: 10.1016/j.addr.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 117.Gioumouxouzis CI, Karavasili C, Fatouros DG. Recent advances in pharmaceutical dosage forms and devices using additive manufacturing technologies. Drug Discov Today. 2019. Feb;24(2):636–643. doi: 10.1016/j.drudis.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 118.Alhnan MA, Okwuosa TC, Sadia M, Wan KW, Ahmed W, Arafat B. Emergence of 3D printed dosage forms: opportunities and challenges. Pharm Res. 2016 Aug;33(8):1817–1832. doi: 10.1007/s11095-016-1933-1. [DOI] [PubMed] [Google Scholar]
- 119.Chen G, Xu Y, Chi Lip Kwok P, Kang L. Pharmaceutical applications of 3d printing. Additive Manufacturing. 2020;2020/08/01/;34:101209. 10.1016/j.addma.2020.101209. [DOI] [Google Scholar]
- 120.Awad A, Fina F, Goyanes A, Gaisford S, Basit AW. 3D printing: principles and pharmaceutical applications of selective laser sintering. Int J Pharm. 2020;586:119594. 2020 Aug 30. doi: 10.1016/j.ijpharm.2020.119594. [DOI] [PubMed] [Google Scholar]
- 121.Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJM, et al. Identifying personal microbiomes using metagenomic codes. Proceedings of the National Academy of Sciences. 2015;201423854.doi: 10.1073/pnas.1423854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Merchant HA, Liu F, Orlu Gul M, Basit AW. . Age-mediated changes in the gastrointestinal tract. Int J Pharm. 2016;512(2):382–395. 2016/10/30/. doi: 10.1016/j.ijpharm.2016.04.024. [DOI] [PubMed] [Google Scholar]
- 123.Hatton GB, Madla CM, Rabbie SC, Basit AW. All disease begins in the gut: influence of gastrointestinal disorders and surgery on oral drug performance. Int J Pharm. 2018. 2018 Sep 5;548(1):408–422. doi: 10.1016/j.ijpharm.2018.06.054. [DOI] [PubMed] [Google Scholar]
- 124.Hatton GB, Madla CM, Rabbie SC, Basit AW. Gut reaction: impact of systemic diseases on gastrointestinal physiology and drug absorption. Drug Discov Today. 2019;24(2):417–427. 2019/02/01/. doi: 10.1016/j.drudis.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 125.Capel AJ, Rimington RP, Lewis MP, Christie SDR. 3D printing for chemical, pharmaceutical and biological applications. Nature Reviews Chemistry. 2018. 2018/12/01;2(12):422–436. doi: 10.1038/s41570-018-0058-y. [DOI] [Google Scholar]
- 126.Xu X, Awad A, Robles-Martinez P, Gaisford S, Goyanes A, Basit AW. Vat photopolymerization 3D printing for advanced drug delivery and medical device applications. J Control Release 2020 Oct 5.doi: 10.1016/j.jconrel.2020.10.008. [DOI] [PubMed] [Google Scholar]
- 127.Durga Prasad Reddy R, Sharma V. Additive manufacturing in drug delivery applications: A Review. Int J Pharm. September 2020;4:119820. doi: 10.1016/j.ijpharm.2020.119820. [DOI] [PubMed] [Google Scholar]
- 128.Dodoo CC, Stapleton P, Basit AW, Gaisford S. . The potential of streptococcus salivarius oral films in the management of dental caries: an inkjet printing approach [Article]. Int J Pharm. 2020;591:591. doi: 10.1016/j.ijpharm.2020.119962. [DOI] [PubMed] [Google Scholar]
- 129.Elbadawi M, Gustaffson T, Gaisford S, Basit AW. 3D printing tablets: predicting printability and drug dissolution from rheological data. Int J Pharm. 2020;590:119868. 2020/11/30/. doi: 10.1016/j.ijpharm.2020.119868. [DOI] [PubMed] [Google Scholar]
- 130.Trenfield SJ, Tan HX, Goyanes A, Wilsdon D, Rowland M, Gaisford S, Basit AW. Non-destructive dose verification of two drugs within 3D printed polyprintlets. Int J Pharm. 2020;577:577. doi: 10.1016/j.ijpharm.2020.119066. [DOI] [PubMed] [Google Scholar]
- 131.Elbadawi M, Muniz Castro B, Gavins FKH, Ong JJ, Gaisford S, Perez G, et al. M3DISEEN: A novel machine learning approach for predicting the 3D printability of medicines. Int J Pharm 2020 Sep 20;590:119837. doi: 10.1016/j.ijpharm.2020.119837 [DOI] [PubMed] [Google Scholar]
- 132.Perez GIP, Gao Z, Jourdain R, Ramirez J, Gany F, Clavaud C, et al. Body site is a more determinant factor than human population diversity in the healthy skin microbiome [Article]. PLoS One. 2016;11(4):4. doi: 10.1371/journal.pone.0151990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Scheline RR. Metabolism of foreign compounds by gastrointestinal microorganisms. Pharmacol Rev. 1973;25:451. 10.1002/jps.2600571202. [DOI] [PubMed] [Google Scholar]
- 134.Sousa T, Paterson R, Moore V, Carlsson A, Abrahamsson B, Basit AW. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm. 2008. 2008 Nov 03;363(1–2):1–25. doi: 10.1016/j.ijpharm.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 135.Crouwel F, Buiter HJC, de Boer NK. Gut microbiota-driven drug metabolism in inflammatory bowel disease. J Crohns Colitis. 2020 Jul. 11. doi: 10.1093/ecco-jcc/jjaa143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hitchings R, Kelly KL. Predicting and understanding the human microbiome’s impact on pharmacology. Trends in Pharmacological Sciences. 2019. Jul;40(7):495–505. doi: 10.1016/j.tips.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Koppel N, Bisanz JE, Pandelia M-E, Turnbaugh PJ, Balskus EP. Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins. eLife. 2018;7:7. doi: 10.7554/eLife.33953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.van Kessel SP, Frye AK, El-Gendy AO, Castejon M, Keshavarzian A, van Dijk G, El Aidy S. Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of parkinson’s disease. Nat Commun. 2019;10(1):310. doi: 10.1038/s41467-019-08294-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Borrel G, Brugere J-F, Gribaldo S, Schmitz RA, Moissl-Eichinger C. The host-associated archaeome. Nature Reviews. Microbiology. 2020. 2020 Jul;18(11):20. doi: 10.1038/s41579-020-0407-y. [DOI] [PubMed] [Google Scholar]
- 140.Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66(6):1039. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dong Q, Brulc JM, Iovieno A, Bates B, Garoutte A, Miller D, Revanna KV, Gao X, Antonopoulos DA, Slepak VZ, et al. Diversity of bacteria at healthy human conjunctiva [Article]. Investigative Opthalmology & Visual Science. 2011;52(8):5408–5413. doi: 10.1167/iovs.10-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pollard TD, Ong JJ, Goyanes A, Orlu M, Gaisford S, Elbadawi M, Basit AW. Electrochemical biosensors: a nexus for precision medicine. Drug discovery Today. 2020; 31. 2020 Oct. doi: 10.1016/j.drudis.2020.10.021. [DOI] [PubMed] [Google Scholar]
- 144.Palmara G, Frascella F, Roppolo I, Chiappone A, Chiadò A. Functional 3D printing: approaches and bioapplications. Biosens Bioelectron 2020 Nov 24:112849.doi: 10.1016/j.bios.2020.112849. [DOI] [PubMed] [Google Scholar]
- 145.Beri K. Skin microbiome & host immunity: applications in regenerative cosmetics & transdermal drug delivery. Future Science OA. 2018;4(6):302. doi: 10.4155/fsoa-2017-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012. 2012/10/01;92(4):414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nava Lara RA, Aguilera-Mendoza L, Brizuela CA, Peña A, Del Rio G. Heterologous machine learning for the identification of antimicrobial activity in human-targeted drugs. Molecules (Basel, Switzerland). 2019. 2019 Mar 31;24(7):7. doi: 10.3390/molecules24071258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zheng S, Chang W, Liu W, Liang G, Xu Y, Lin F. Computational prediction of a new admet endpoint for small molecules: anticommensal effect on human gut microbiota. J Chem Inf Model. 2019. 2019 Mar 25;59(3):1215–1220. doi: 10.1021/acs.jcim.8b00600. [DOI] [PubMed] [Google Scholar]
- 149.Ekins S, Puhl AC, Zorn KM, Lane TR, Russo DP, Klein JJ, Hickey AJ, Clark AM. Exploiting machine learning for end-to-end drug discovery and development. Nat Mater. 2019. 2019/05/01;18(5):435–441. doi: 10.1038/s41563-019-0338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bhhatarai B, Walters WP, Hop CECA, Lanza G, Ekins S. Opportunities and challenges using artificial intelligence in ADME/Tox. Nat Mater. 2019. 2019/05/01;18(5):418–422. doi: 10.1038/s41563-019-0332-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Walsh J, Gheorghe CE, Lyte JM, van de Wouw M, Boehme M, Dinan TG, et al. 2020. Gut microbiome-mediated modulation of hepatic cytochrome P450 and P-glycoprotein: impact of butyrate and fructo-oligosaccharide-inulin. Journal of Pharmacy and Pharmacology. 2020/04/26; 72: 1072-1081. 10.1111/jphp.13276. [DOI] [PubMed] [Google Scholar]
- 152.Walsh J, Olavarria-Ramirez L, Lach G, Boehme M, Dinan TG, Cryan JF, Griffin BT, Hyland NP, Clarke G. Impact of host and environmental factors on β-glucuronidase enzymatic activity: implications for gastrointestinal serotonin. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2020. 2020/04/01;318(4):G816–G826. doi: 10.1152/ajpgi.00026.2020. [DOI] [PubMed] [Google Scholar]
- 153.Walsh J, Griffin BT, Clarke G, Hyland NP. Drug-gut microbiota interactions: implications for neuropharmacology. . British Journal of Pharmacology. 2018. Dec;;175(24):4415–4429. doi: 10.1111/bph.14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zimmermann-Kogadeeva M, Zimmermann M, Goodman AL. Insights from pharmacokinetic models of host-microbiome drug metabolism. Gut Microbes. 2019;11:3, 587-596, DOI: 10.1080/19490976.2019.1667724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schuhmacher A, Gatto A, Hinder M, Kuss M, Gassmann O. The upside of being a digital pharma player. Drug Discov Today 2020 Jun 15.doi: 10.1016/j.drudis.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 156.Royal Society Working Group. Dynamics of data science skills: how can all sectors benefit from data science talent? United Kingdom; 2019. URL: https://royalsociety.org/topics-policy/projects/dynamics-of-data-science/. [Google Scholar]
- 157.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, china [Article]. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in wuhan, china: a descriptive study [Article]. The Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A novel coronavirus from patients with pneumonia in China, 2019 [Article]. New England Journal of Medicine. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.COVID-19 to plunge global economy into worst recession since world war ii [internet]. the world bank; 2020; 8thJune 2020. Available from:https://www.worldbank.org/en/news/press-release/2020/06/08/covid-19-to-plunge-global-economy-into-worst-recession-since-world-war-ii
- 161.Mak -K-K, Pichika MR. Artificial intelligence in drug development: present status and future prospects. Drug Discovery Today. 2019. Mar;;24(3):773–780. doi: 10.1016/j.drudis.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 162.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science (New York, NY). 2019;363(6427):9931. doi: 10.1126/science.aat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Food and Agriculture Organization of the United Nations and World Health Organisation . Guidelines for the evaluation of probiotics in food. London Ontario (Canada); 2002. p. 1–11. [Google Scholar]
- 164.NuBiyota. safety and efficacy of met-3 in obese human subjects: clinicalTrials.gov; 2018 [accessed 3rd April 2020]. Available from: https://clinicaltrials.gov/ct2/show/NCT03660748?id=NCT03660748&draw=2&rank= 1
- 165.Zhao M, Shen C, Ma L. Treatment efficacy of probiotics on atopic dermatitis, zooming in on infants: a systematic review and meta-analysis. Int J Dermatol. 2018. 2018/06/01;57(6):635–641. doi: 10.1111/ijd.13873. [DOI] [PubMed] [Google Scholar]
- 166.Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, et al. Expert consensus document: the international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics [review]. Nat Rev Gastroenterol Hepatol. 2017;14(8):491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 167.Hester SN, Mastaloudis A, Gray R, Antony JM, Evans M, Wood SM. Efficacy of an anthocyanin and prebiotic blend on intestinal environment in obese male and female subjects. Journal of Nutrition and Metabolism. 2018;2018:7497260. doi: 10.1155/2018/7497260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bomhof MR, Parnell JA, Ramay HR, Crotty P, Rioux KP, Probert CS, Jayakumar S, Raman M, Reimer RA. Histological improvement of non-alcoholic steatohepatitis with a prebiotic: a pilot clinical trial [article]. Eur J Nutr. 2019;58(4):1735–1745. doi: 10.1007/s00394-018-1721-2. [DOI] [PubMed] [Google Scholar]
- 169.Chen PB, Black AS, Sobel AL, Zhao Y, Mukherjee P, Molparia B, Moore NE, Aleman Muench GR, Wu J, Chen W, et al. Directed remodeling of the mouse gut microbiome inhibits the development of atherosclerosis. Nat Biotechnol. 2020;38(11):1288–1297. 2020/06//. doi: 10.1038/s41587-020-0549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Brody LP, Sahuri-Arisoylu M, Parkinson JR, Parkes HG, So PW, Hajji N, et al. Cationic lipid-based nanoparticles mediate functional delivery of acetate to tumor cells in vivo leading to significant anticancer effects [article]. Int J Nanomedicine. 2017;12:6677–6685. doi: 10.2147/IJN.S135968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mills S, Ross RP, Hill C. Bacteriocins and bacteriophage; a narrow-minded approach to food and gut microbiology [review]. FEMS Microbiol Rev. 2017;41(1):S129–S153. doi: 10.1093/femsre/fux022. [DOI] [PubMed] [Google Scholar]
- 172.Yadav V, Gaisford S, Merchant HA, Basit AW. Colonic bacterial metabolism of corticosteroids. Int J Pharm. 2013;457(1):268–274. 2013 Nov 30. doi: 10.1016/j.ijpharm.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 173.Basit AW, Lacey LF. Colonic metabolism of ranitidine: implications for its delivery and absorption [article]. Int J Pharm. 2001;227(1–2):157–165. doi: 10.1016/S0378-5173(01)00794-3. [DOI] [PubMed] [Google Scholar]
- 174.Kim D-H. Gut microbiota-mediated drug-antibiotic interactions. Drug Metabolism and Disposition. 2015;43(10):1581. doi: 10.1124/dmd.115.063867. [DOI] [PubMed] [Google Scholar]
- 175.Basit AW, Newton JM, Lacey LF. Susceptibility of the H2-receptor antagonists cimetidine, famotidine and nizatidine, to metabolism by the gastrointestinal microflora. Int J Pharm. 2002. 2002/04/26/;237(1–2):23–33. doi: 10.1016/S0378-5173(02)00018-2. [DOI] [PubMed] [Google Scholar]
- 176.Sousa T, Yadav V, Zann V, Borde A, Abrahamsson B, Basit AW. On the colonic bacterial metabolism of azo-bonded prodrugs of 5-aminosalicylic acid. J Pharm Sci. 2014;103(10):3171–3175. 2014/10/01. doi: 10.1002/jps.24103. [DOI] [PubMed] [Google Scholar]
- 177.Yadav V, Varum F, Bravo R, Furrer E, Basit AW. Gastrointestinal stability of therapeutic anti-TNF α IgG1 monoclonal antibodies. Int J Pharm. 2016. 2016 Apr 11;502(1–2):181–187. doi: 10.1016/j.ijpharm.2016.02.014. [DOI] [PubMed] [Google Scholar]


