ABSTRACT
Blautia is a genus of anaerobic bacteria with probiotic characteristics that occur widely in the feces and intestines of mammals. Based on phenotypic and phylogenetic analyses, some species in the genera Clostridium and Ruminococcus have been reclassified as Blautia, so to date, there are 20 new species with valid published names in this genus. An extensive body of research has recently focused on the probiotic effects of this genus, such as biological transformation and its ability to regulate host health and alleviate metabolic syndrome. This article reviews the origin and biological characteristics of Blautia and the factors that affect its abundance and discusses its role in host health, thus laying a theoretical foundation for the development of new functional microorganisms with probiotic properties.
KEYWORDS: Blautia, biotransformation, probiotics, host health
Introduction
The gastrointestinal tract of mammals harbors a complex and diverse community of microorganisms, including a variety of bacteria, viruses, archaea, and eukaryotes.1 According to the results of previous 16S rRNA studies, Firmicutes is one of the core phyla of the intestinal microbiota of humans and other vertebrates, and Lachnospiraceae and Ruminococcaceae are the most abundant families of this phylum, accounting for 50% and 30% of the total intestinal microbiota, respectively.2 As a genus of the Lachnospiraceae family, Blautia has been of particular interest since its establishment for its contribution to alleviating inflammatory diseases and metabolic diseases and for its antibacterial activity against specific microorganisms.3,4 Several recent reports have indicated that the composition of and changes in the Blautia population in the intestine are related to factors such as host age, geography, diet, genotype, health, disease state, and other physiological states.5–7 This genus has also been revealed to play a certain role in biotransformation and crosstalk with other intestinal microorganisms.8,9 Although Blautia has shown a series of potential probiotic properties, there is no comprehensive understanding of this genus, probably due to the lack of a comprehensive review. The Ninth Edition of Bergey’s Manual of Systematic Bacteriology contains no description of the genus Blautia, only some introduction to the previous species belonging to the genus Clostridium or Ruminococcus and a summary of the characteristics of the type strains. With the development and popularization of high-throughput sequencing technology, increasing numbers of new species from the gut and feces are being isolated and identified, so a more comprehensive description of this new genus is urgently warranted. This article reviews the recent studies on Blautia and discusses its potential probiotic role in host health.
Morphological, physiological, and biochemical characteristics of Blautia
As opposed to traditional culture methods alone, phylogenetic analysis of 16S rRNA gene sequences in combination with culture-based analyses is an effective approach to identify microbial diversity in the host intestine and feces.10 Using phenotypic and phylogenetic analyses, Liu et al11 revealed a hitherto unknown coccus-shaped strain WAL 14507 T (=ATCC BAA-1564 T = DSM 19850 T) and established it as the new species Blautia wexlerae sp. nov. To date, 20 species with valid published names constitute the genus Blautia (Table 1), including B. coccoides, B. hansenii, and B. producta, which were originally misclassified into the genus Clostridium or Ruminococcus.11 A phylogenetic tree based on the 16S rRNA gene sequences of the representative species of Blautia is provided in Figure 1. The composition of this genus is constantly updated by adding new species and strains, but in general, the species in Blautia still form a relatively stable and coherent uniline branch.27
Table 1.
All species of Blautia reported in the literature
| Strains | Isolation source | Gram Staining | Type strain | References |
|---|---|---|---|---|
|
Blautia coccoides (Clostridium coccoides) |
mice feces | G+ | ATCC 29236 T | Kaneuchi et al.(1976)12 |
|
Blautia hansenii (Ruminococcus hansenii) |
human feces | G+ | ATCC 27752 T | Holdeman et al.(1974)13 |
|
Blautia hydrogenotrophica (Ruminococcus hydrogenotrophicus) |
human feces | G+ | DSM 10507 T | Bernalier et al.(1996)14 |
|
Blautia luti (Ruminococcus luti) |
human feces | G+ | DSM 14534 T | Simmering et al.(2002)15 |
|
Blautia producta (Ruminococcus productus) |
sputum | G+ | ATCC 27340 T | Ezaki et al.(1994)16 |
|
Blautia schinkii (Ruminococcus schinkii) |
rumen of lambs | G+ | DSMZ 105/8 | Rieu-Lesme et al. (1996)17 |
| Blautia wexlerae | children’s stool | G+ | ATCC BAA-1564 T | Liu et al.(2008)11 |
| Blautia glucerasea | dog feces | G+ | DSM 22028 T | Furuya et al.(2010)18 |
| Blautia stercoris | human feces | G+ | KCTC 5981 T | Park et al.(2012)19 |
| Blautia faecis | human feces | G+ | KCTC 5980 T | Park et al.(2013)20 |
|
Blautia obeum (Ruminococcus obeum) |
human feces | G+ | ATCC 29174 T | Moore et al.(1976)21 |
| Blautia caecimuris | mouse cecum | G+ | SJ18T | Lagkouvardos et al.(2016)22 |
| Blautia massiliensis | human fecces | G− | DSM 101187 T | Durand et al.(2017)23 |
| Blautia phocaeensis | human feces | G+ | Marseille-P3441 | Traore et al.(2017)24 |
| Blautia marasmi | Human stool | G+ | Marseille-P2377T | Pham et al.(2017)25 |
| Blautia provencensis | children’s stool | G+ | Marseille-P3502T | Pham et al.(2017)26 |
| Blautia hominis | human feces | G+ | KCTC 15618 T | Shin et al.(2018)27 |
| Blautia argi | dog feces | G+ | KCTC 15426 T | Paek et al.(2019)28 |
| Blautia brookingsii | human feces | G+ | SG772 | Ghimire et al.(2020)29 |
| Blautia faecicola | human feces | G+ | KGMB01111 T | Kim et al.(2020)30 |
Figure 1.
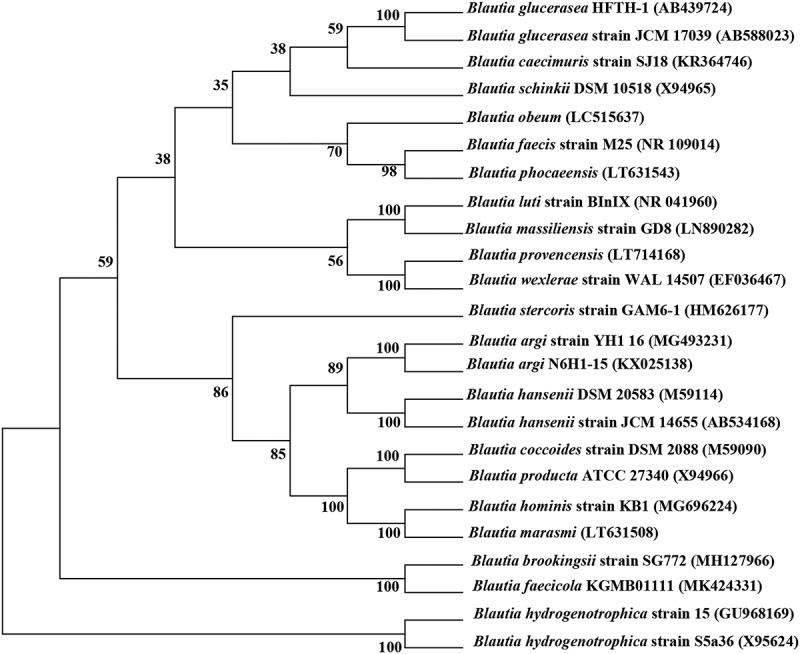
Phylogenetic consensus tree based on 16S rRNA gene sequences, reconstructed with the neighbor-joining (NJ), maximum-parsimony (MP) and maximum-likelihood (ML) algorithms. Bootstrap values calculated for 1000 subsets are shown at branch nodes
According to reports, the human intestinal microbial community can be divided into three “enterotypes,” namely, Bacteroides, Prevotella, and Ruminococcus, among which the Ruminococcus type is mostly driven by related groups of the order Clostridiales, Blautia, and unclassified Lachnospiraceae.31 Blautia is widely distributed in mammalian feces and intestines. For example, B. hydrogenotrophica and B. stercoris were first isolated from human feces;14,19 B. wexlerae and B. luti were found to be the most abundant of the Blautia spp. and are considered among the dominant species of the human intestine;32 B. coccoides was first isolated from the feces of mice fed a high-lactose diet;12 B. glucerasei was isolated from the feces of dogs;18 and some species such as B. producta and B. schinkii were even isolated from sewage and rumen.17,33 These findings indicate the importance of the survival and evolution of Blautia in the gut and other microenvironments.
Blautia species are strictly anaerobic, non-motile, 1.0–1.5 × 1.0–3.0 μm in size, usually spherical or oval, and appear in pairs or strands, with most strains being sporeless. The optimum temperature and pH for most Blautia strains are 37°C and 7.0, respectively.11 Some species such as B. producta possess both heterotrophic and autotrophic properties and can use CO, H2/CO2, and carbohydrates as energy sources.34 Carbohydrate utilization experiments have shown that all Blautia strains can use glucose, but different strains showed different abilities to use sucrose, fructose, lactose, maltose, rhamnose, and raffinose (Table 2). The final products of glucose fermentation by Blautia are acetic acid, succinic acid, lactic acid, and ethanol, and the main biochemical tests have revealed negative results for lecithin, lipase, catalase, and indole. The long-chain fatty acids produced by Blautia strains are classified into linearly saturated and monounsaturated types, with C14:0, C16:0, and C16:00 dimethyl acetal fatty acids as the main species. The GC content of Blautia DNA is 37–47 mol%, and the type species of this genus is B. coccoides.11
Table 2.
Characteristics of carbohydrates utilization in Blautia strain
| Strains | Blautia coccoides CLC-1 | Blautia hansenii ATCC 27752 T | Blautia hydrogenotrophica S5a33 | Blautia luti BInIXT | Blautia producta U-1 | Blautia schinkii strain B | Blautia wexlerae WAL 14507 T | Blautia glucerasei HFTH-1 T | Blautia stercoris GAM6-1 T | Blautia faecis M25T |
Blautia obeum ATCC 29174 T |
Blautia caecimuris | Blautia massiliensis GD9T | Blautia hominis KB1T | Blautia argi N6H1-15 T | Blautia brookingsii SG-772 | Blautia faecicola. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| End products of fermentation |
S,A | L,A | A,L | A,S | A,L, | A,S | A,S | A,F | A | L,A | A | ND | ND | A,S | A,L | ND | A |
| Glucose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - |
| Mannose | + | - | w | + | ND | + | + | - | - | - | + | ND | + | + | - | + | - |
| Arabinose | + | - | - | + | + | + | + | + | + | + | + | + | + | + | + | ND | w |
| Xylose | + | - | ND | + | + | + | + | + | - | - | + | + | + | + | - | ND | - |
| Robise | + | - | ND | + | + | + | + | ND | + | + | + | ND | ND | + | - | ND | - |
| Sucrose | + | - | - | + | + | + | D | ND | - | - | - | + | + | + | + | ND | w |
| Fructose | + | - | + | + | + | + | D | + | - | - | + | ND | ND | + | + | ND | - |
| Lactose | + | + | ND | + | + | ND | D | + | - | - | + | + | + | + | + | ND | - |
| Maltose | + | + | + | + | + | D | + | - | - | + | + | + | + | + | ND | - | |
| Cellobiose | + | - | + | + | + | + | D | + | - | - | + | ND | + | + | + | + | - |
| Rhamnose | + | - | - | - | + | ND | D | ND | - | - | + | + | + | + | - | + | - |
| Raffinose | + | + | ND | + | + | + | D | + | - | - | + | + | + | + | - | ND | w |
| Melezitose | ND | - | ND | - | + | ND | D | ND | - | - | ND | ND | + | + | - | + | w |
| Mannitol | + | - | - | - | + | ND | D | ND | - | - | - | ND | + | - | - | + | - |
| Melibiose | + | w | - | + | + | + | D | ND | - | - | + | ND | ND | + | + | ND | - |
+, Positive; –, negative; W, weakly positive; D,different among strains; ND, no data.
A, acetic acid; S, succinic acid; L,lactate.
Data are from Kaneuchi et al.(1976), Holdeman et al.(1974), Bernalier et al.(1996), Simmering et al.(2002), Lorowitz et al.(1984), Rieu-Lesme et al.(1996), Liu et al.(2008), Furuya et al. (2010), Park et al. (2012), Park et al. (2013), Moore et al. (1976), Lagkouvardos et al. (2016), Durand et al. (2017), Shin et al. (2018), Paek et al. (2019), Ghimire et al. (2020), Kim et al. (2020).
Genomic and comparative genomic analysis of the members of Blautia
The high performance and efficiency of next-generation sequencing technology have allowed novel insights into the whole genome of several bacteria.35 Bioinformatics is widely used to analyze bacterial genome information, enabling the research direction shift from phenotypic assessments to genomic evaluations and even prediction of potential probiotic functions.36 Compared with the abundance of well-known probiotics such as Bifidobacterium and Lactobacillus species and of their genomic data, fewer Blautia species have been isolated, so information on their genome is limited. Currently, there are 12 isolated Blautia species with a total of 195 genome assemblies according to the NCBI database. The genome size varies greatly and ranges from 3.17 to 6.07 Mb with a median of 3.49 Mb. The median GC content is 44.25%, and the median protein count is 3205 (These data were obtained from the NCBI database on September 1, 2020, and detailed information is presented in Table 3).
Table 3.
The genomic characteristics of Blautia.
| Species | Number of genome assembly | Median total length (Mb) | Median GC (%) | Median protein count |
|---|---|---|---|---|
| Blautia wexlerae | 81 | 4.07 | 41.20 | 3572 |
| Blautia obeum | 50 | 3.66 | 41.60 | 3302 |
| Blautia luti | 5 | 3.96 | 43.80 | 3469 |
| Blautia hansenii | 4 | 3.17 | 41.10 | 2916 |
| Blautia schinkii | 8 | 3.67 | 43.20 | 3165 |
| Blautia hydrogenotrophica | 3 | 3.53 | 44.90 | 3225 |
| Blautia producta | 12 | 6.07 | 45.70 | 5300 |
| Blautia marasmi | 2 | 6.02 | 46.30 | 5055 |
| Blautia coccoides | 3 | 5.97 | 45.60 | 5156 |
| Blautia faecis | 6 | 4.53 | 42.80 | 3929 |
| Blautia glucerasea | 3 | 4.31 | 43.00 | 3738 |
| Blautia massiliensis | 18 | 3.49 | 44.10 | 3072 |
Data were obtained from NCBI database on September 1, 2020.
For a better understanding of the differences among different species of Blautia, comparative genomic analysis was performed on the 74 strains of Blautia, the assembly level of genome sequences of which was more than half (Table S1). Based on the orthologous genes, a phylogenetic tree was constructed to evaluate the evolution of different species (Figure 2). The phylogenetic tree showed that Blautia was a paraphyletic group, and all strains were derived from the same ancestor. Different strains within the same species were clustered together except Blautia hansenii and Blautia obeum.
Figure 2.
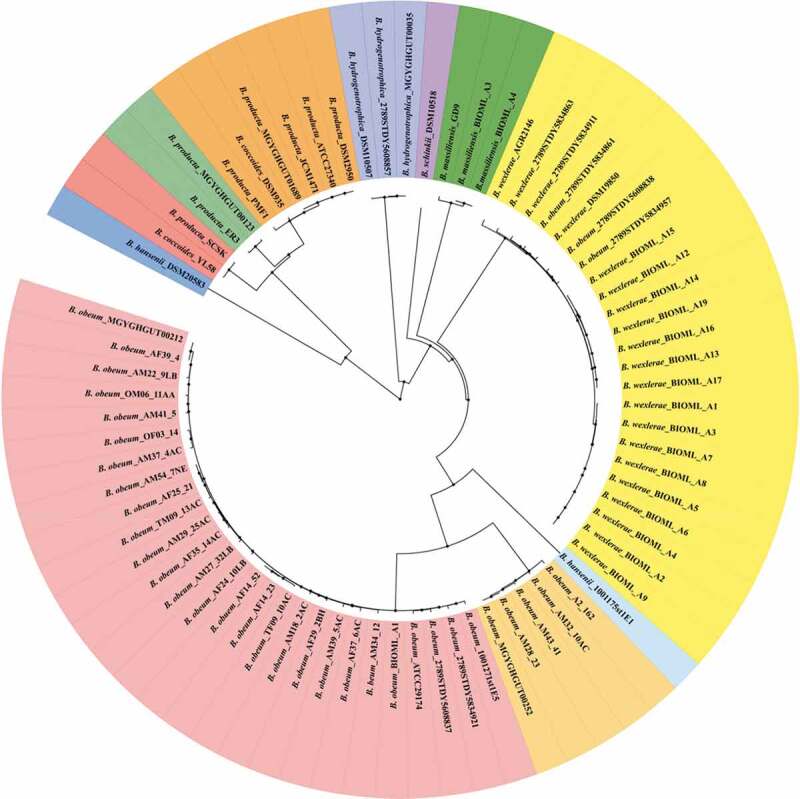
Phylogenetic relationships of different species of Blautia based on the orthologous genes. The phylogenetic tree was constructed using the python script3 and visualized using iTOL. Tree nodes are depicted by filled circles. The genomes of 74 Blautia strains were obtained from NCBI database
The Pan-genome is defined as the collection of all genes in a bacterial species, including core genome in all strains and dispensable genome in some strains. The open or closed characteristics of pan-genome could reflect the species diversity in genetic composition.37 As shown in Figure 3a, the number of pan-genome increased with the addition of Blautia genome, and the total number of pan-genome reached 26,728 when the 74th genome was added to the calculations. The pan-genome curve showed an upward trend, indicating that Blautia has an open pan-genome.38 In contrast, the core genes curve showed a downward trend, and gradually stabilized at 488 when the 74th genome was added.39 The core genes and unique genes of Blautia were displayed with a Venn diagram (Figure 3b). The number of core genes was 606, while the number of unique genes ranged from 3 to 995. Notedly, Blautia schinkii DSM10518 has the largest number of unique genes, which may be related to the isolation source. Because B. schinkii DSM10518 was the only strain isolated from rumen, and the other strains were isolated from human or mouse feces.
Figure 3A.
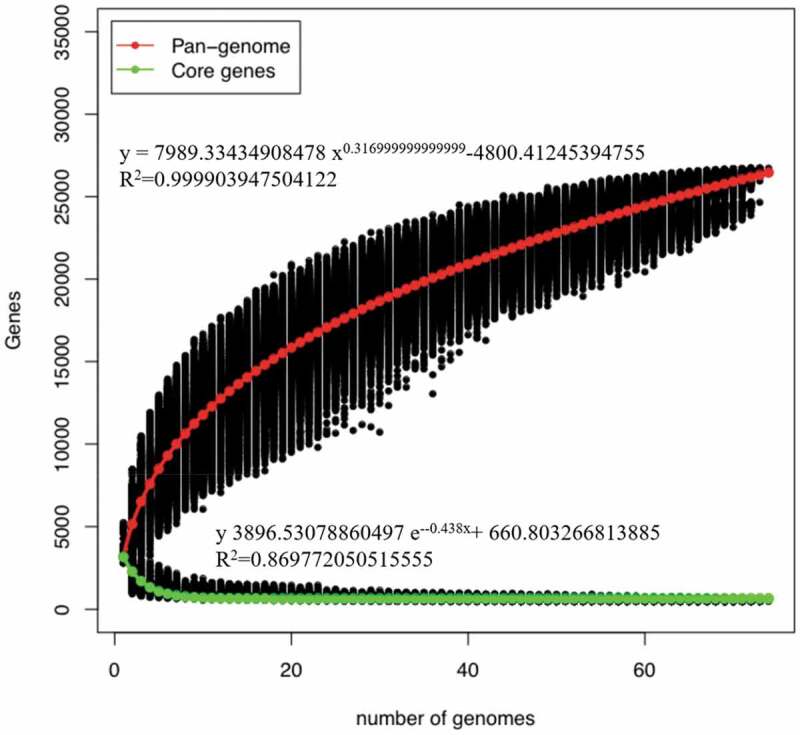
Pan-genome and core genes of Blautia.
Figure 3B.
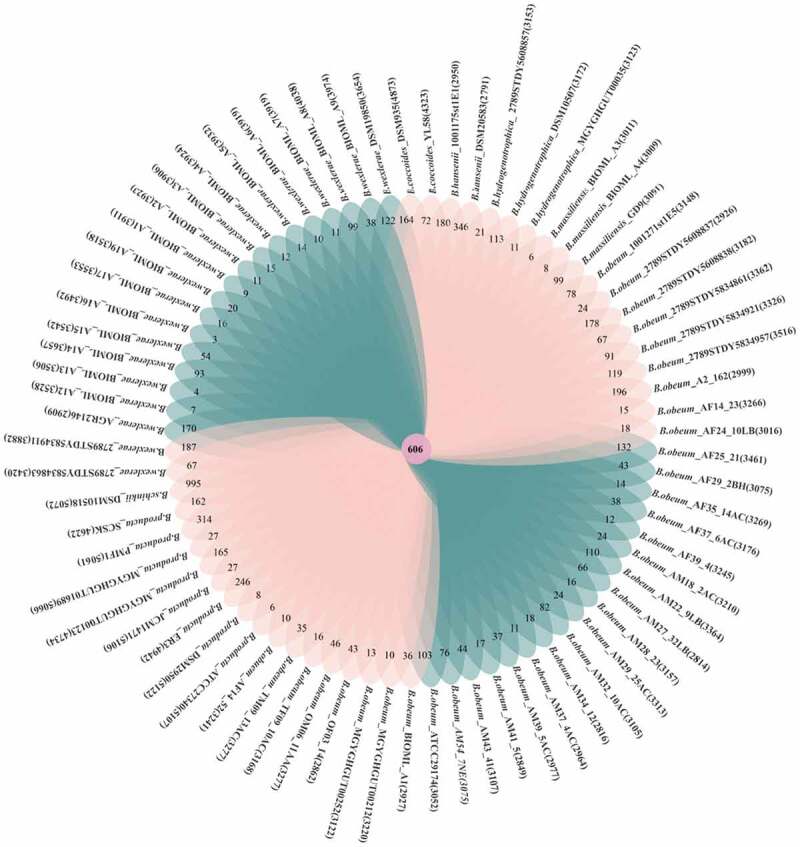
Venn diagram displaying core genes and unique genes of Blautia strains
Figure 3C.
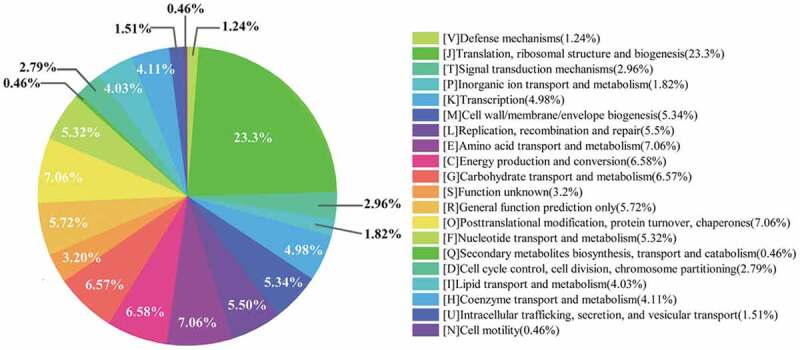
Functional annotations of the core genes using the COG database
Figure 3D.
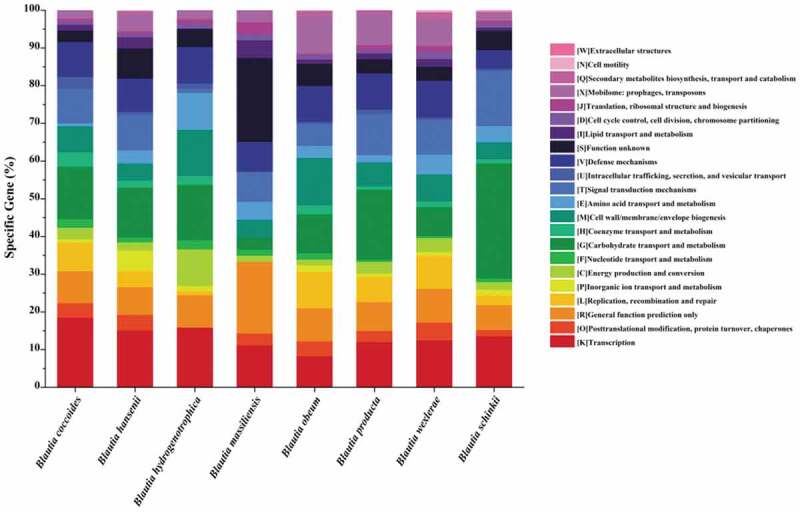
Functional assignment of the unique genes based using the COG database
Based on the clusters of orthologous genes (COG), the core genes of Blautia were annotated and evaluated (Figure 3c). The functional genes were roughly divided into 19 categories and were most enriched in [J] Translation, ribosomal structure, and biogenesis (23.30%). The genes for ribosome structure and biogenesis were usually associated with environmental stresses, such as acid, oxidative, heat, and salt stresses.40–43 Thus, it may be beneficial for Blautia to survive in the harsh conditions in the host gastrointestinal tract due to the large number of stress-related genes.44 Compared with most lactic acid bacteria, the proportion of genes related to [G] Carbohydrate transport and metabolism was relatively small in Blautia (6.57%), indicating that Blautia may have weak abilities to metabolize carbohydrates.
Compared with other species, Blautia obeum, Blautia producta, and Blautia wexlerae possess more unique genes encoding prophages and transposons (Figure 3d). Prophages allowed bacteria to acquire the antibiotic resistance and enhance the adaption to environment and adhesion ability, which could help the bacteria to cope with multiple adverse environments and colonize in mammals.45,46 Remarkably, Blautia schinkii isolated from the rumen showed prominent carbohydrate metabolism ability. It is well known that rumen microbiota present in ruminants showed effective utilization of carbohydrates such as fiber and starch in feed, and the special ecological niche would endow strains with unique functions.47
Effects of diet, age, and geography on the abundance of Blautia
Diet is the main factor that drives the composition and metabolic activities of intestinal microbiota, as different types and quantities of diet and the balance between major nutrients have a significant effect on the intestinal microbes.48 The traditional Japanese cooking method Washoku, which contains fermented food prepared with the nonpathogenic fungus koji, is thought to be closely related to Japanese longevity. One study reported that koji contains a large number of glycosylceramides and showed that the addition of 1% purified glycosylceramides as a prebiotic in the diet of mice for 1 week could improve the abundance of B. coccoides in the intestinal tract of mice, reduce their blood sugar level, and upregulate their renal glandular hormone level. B. coccoides was found to degrade glycosylceramides to ceramides and then metabolize the ceramides to fatty acids and sphingoid bases, which were absorbed by the intestine and produced a beneficial effect.49 The addition of dietary fiber extracted from corn (F-FOPs) to the diet of mice fed a high-fat (HF) diet significantly increased the abundance of Blautia in the mouse feces. Compared with mice on the HF diet, those on the F-FOPs + HF diet showed losses of body and tissue weight, and the results revealed a negative correlation between the abundance of Blautia and the markers of obesity-related metabolic disorders.50 The enrichment of Blautia has also been observed in the feces of healthy adult dogs whose diet was supplemented with potato fiber.51 Similarly, addition of 20% freeze-dried soy milk to rats fed an HF diet led to an increase in the abundance of Blautia in the rat feces. The soy milk was found to regulate the serum high-density lipoprotein-cholesterol level in the rats and enhance the gene expression of tight junction proteins (ZO-1 and occlusion protein) and inflammation-related proteins (IL-1β, IL-10, and Foxp3) in their colon.52 Dietary protein and prebiotics from various sources also affect the intestinal microbiota. In one study, 30 female rats were divided into six groups and fed casein or soy protein isolate each with cellulose, raffinose, or fructooligosaccharide (FOS). The results showed that dietary protein from both sources could change the acetic acid concentration and the abundance of Lactobacillus in the rat feces, but FOS increased the abundance of Blautia regardless of the dietary protein source.53 Consistently, another study reported a significant increase in the abundance of Blautia in the feces of mice fed FOS.7 An increase in the cecal abundance of B. productus (previously as Ruminococcus productus) was observed in rats fed 3% difructose anhydride III for 2 weeks, accompanied by a decrease in the cecal pH and an increase in the content of short-chain fatty acids (SCFAs). The study also showed that intestinal acidification in rats may inhibit the formation of secondary bile acids.54 Furthermore, another study in which a 45-year-old male volunteer consumed 600 mg of omega-3 daily for 14 days, the volunteer’s overall intestinal microbial diversity decreased, accompanied particularly with reduced Faecalibacterium abundance and significantly increased Blautia abundance.55 In addition to diet, the way people eat also shapes the gut microbiota. Compared with the ordinary diet, the alternating diet and the self-service diet can enhance the abundances of Blautia and Ruminococcus in the gut microbiota, in addition to inducing changes in some host metabolism-related parameters.56 With the development of whole-genome sequencing, future studies can examine how various diets regulate the metabolic activities of Blautia and improve host health.
Significant changes occur in the gut microbiota during transitions between various stages of life (i.e., from childhood to adulthood to old age). A study that monitored the intestinal microbes of children from 2 weeks to 13 years of age found that B. coccoides was rarely present in the intestinal microbiota of children younger 6 months, but was more common in those older than 12 months.57 A similar phenomenon was also observed in commercial diet-fed pigs and rats. The researchers found that the abundance of fecal microorganisms continued to change with the age of pigs, accompanied by an increase in the proportion of Blautia.58 In one study, middle-aged rats and young rats fed the same egg protein diet for 14 days showed significantly different intestinal microbiome at the end of 14 days. Particularly, the middle-aged rats’ relative abundance of Blautia was increased.59 Another study that analyzed the cecal microbiome of specific pathogen-free chicks on days 14, 28, and 42 by sequencing of the V3-V4 region of 16S rRNA gene on the Illumina MiSeq platform revealed that the abundance of Blautia increased with the age of chicks.60 A cross-sectional study of the stool samples of 367 healthy Japanese subjects aged 0–104 years using high-throughput sequencing of V3-V4 region of 16S rRNA gene reported that the intestinal microbiota of Japanese adults (21–69 years old) contained high abundance of Blautia and Bifidobacterium and low abundance of Bacteroides.5 In addition, compared with the adults, the elderly showed reduced microbiome diversity and abundance of individual microbes, including a lower Blautia abundance. This phenomenon may be related to an age-related decline in immune function known as immunosenescence, accompanied by many age-related conditions that involve chronic low-level inflammation.61
A recent study analyzed the microbial community characteristics in the stool samples of 303 school-age children from urban or rural areas of five countries in temperate and tropical regions of Asia. The intestinal microbiota of the children was divided into two groups, Prevotella (P type) and Bifidobacterium/Bacteroides (BB type). The gut microbiota of children in China, Japan, Taiwan, and other temperate regions was mostly BB type, whereas that of children in Thailand, Indonesia, and other tropical places was mostly P type. Notably, Blautia was significantly enriched in the BB-type intestinal microbiota, accounting for 10% of the total BB-type bacterial composition but only 5% of the total P-type bacterial composition.6 Schnorr et al.62 found a difference in the intestinal microbial composition between Hadza and Italians, characterized by a lower Blautia abundance in Hadza. Differences in the human gut microbiota were also noted between different altitudes and geographies. Sequencing of the fecal microbiota of 208 Tibetans from six regions based on the analysis of the operational taxonomic units revealed Blautia to be the dominant genus in the human gut microbiota across all six regions. Further principal component analysis showed that the intestinal microbiota of Tibetans changed significantly with the increase in altitude, body mass index, and age; specifically, the abundance of facultative anaerobes increased. These findings suggest that the intestinal microbiota play an important role in regulating altitude and geographical adaptability.63 One study pointed out that the predominant intestinal genera in Japanese people were Bifidobacterium and Clostridium; that in the American, Chinese, French, and Spanish people was Bacteroides; and that in Australians was Blautia.64 Reportedly, differences in human gut microbial diversity between geographical locations are largely related to heredity, lifestyle, and diet.65 Interestingly, Blautia was reported of having strong taxonomic association in twin inheritance.66 To identify the differences in intestinal microbial communities between human and animal hosts, a study collected fecal samples from seven hosts, including human, pig, cattle, deer, dog, cat, and chicken, and sequenced the V6 region of the 16S rRNA genes. Two hundred high-resolution taxonomic units in Blautia were identified using oligotyping, and the Blautia oligotypes could accurately identify different host sources, suggesting that the genus has host specificity and host preference.67
Physiological functions of Blautia
Biotransformation of bioactive substances by Blautia
Human intestinal bacteria belonging to the genera Prevotella and Xylanibacter can degrade dietary components such as cellulose and xylan that are not digested by the host, increase the content of SCFAs in feces, promote food digestion, and maximize energy intake.68 In recent years, research on the biotransformation and metabolism of herbal plants and functional foods by Blautia has attracted research attention.
Polymethoxyflavones (PMFs) are flavonoids isolated from Kaempferia and citrus fruits and have anticancer, anti-inflammatory, antiviral, and anticoagulant properties, and other biological functions.69–72 Studies have shown that the strain Blautia sp. MRG-PMF1 has a hydrolytic effect on aryl methyl ether functional groups by converting 5,7-dimethoxyflavone (5,7-DMF) and 5,7,4-trimethoxyflavone (5,7,4-TMF) into bioactive chrysin and apigenin, respectively. This strain also possessed deglycosylation ability, whereby it can metabolize isoflavones, flavones, and flavonoids into corresponding aglycones.8 Wu et al.73 found that Blautia sp. MRG-PMF1 can also biotransform icariin under anaerobic conditions and metabolize it to its hydrolyzates icariin and desmethylicaritin, which were reported to exhibit estrogenic effects by acting on estrogen receptors, in addition to significant antilipogenic activity. Another study revealed that this strain can also metabolize curcumin to demethylcurcumin and bisdescurcumin.74 Compared with curcumin, demethylcurcumin has been found to be highly toxic to human HCT116 colon cancer cells, whereas synthetic desmethylcurcumin has been found to exert better neuroprotective and anti-inflammatory effects.75–77 Furthermore, the strain Blautia sp. AUH-JLD56 has been shown to specifically and efficiently biotransform arctiin or arctigenin to (−)-3′-desmethyl arctigenin, which possesses good free radical-scavenging activity.78 B. glucerasei sp. nov. HFTH-1 T produces a specific extracellular glucosylceramide enzyme that hydrolyzes glucosylceramide into functional substances with specific preventive effect against colon cancer (Table 4).18 It should be pointed out that some biotransformation by Blautia may not be beneficial and could potentially even be harmful. Certain Blautia species could perform 7-α-dehydroxylation of primary bile acids and convert them to secondary bile acids such as lithocholic acid and deoxycholic acid, which have been reported as colon cancer-inducing carcinogens and are found in higher concentrations in the feces of patients with ulcerative colitis and dysplasia or cancer.79
Table 4.
Biotransformation of bioactive substances by Blautia.
 |
Blautia is also involved in the process of polyphenol deglycosylation and lignan catabolism. In general, bacterial metabolism in the intestine does not involve oxygen but rather reductions and hydrolysis, resulting in the formation of nonpolar low molecular weight products. In the course of flavonoid conversion, the reactions catalyzed by Blautia include demethylation, dehydroxylation, O-and C- deglycosylation and c-ring cleavage,80 which may be due to its corresponding enzymes, such as β-glucosidases and O-glycosidase.81,82 By analyzing the key metabolic pathways and enzymes related to biotransformation, it is possible to predict whether the bacteria can undergo the biotransformation of specific bioactive substances. Thus, the exploration of biotransformation by Blautia is essential for the development of new enzymes and bioactive metabolites for supplementation in food and provide valuable perspective for the metabonomics research of human intestinal microbiome.83,84
Relationships between Blautia and host health
Intestinal microbiota is a complex ecosystem that is linked to the development of host disease, drug metabolism, immune system regulation, and other processes.85 Blautia, as a dominant genus in the intestinal microbiota, has a significant correlation with host physiological dysfunctions, such as obesity, diabetes, cancer, and various inflammatory diseases.
Blautia and the secondary metabolites
Secondary metabolites are biologically active compounds produced by microorganisms during growth and metabolism and widely used in antibacterial and anticancer drugs, herbicides, and insecticides, which were also an important source of microbial drug development.86,87 According to the categories, there were more than 20 kinds of secondary metabolites, such as polyketides (PKS), non-ribosomal peptides (NRPS), lantipeptides/lantibiotics, bacteriocins, and terpenes.88 As early as 1980, bacteriocins produced by bifidobacteria were reported to possess antibacterial activity against pathogenic microorganisms such as Listeria monocytogenes, Clostridium perfringens, and Escherichia coli.89 Nisin, which is produced by Lactococcus lactis, is used as a natural food preservative.90 According to the chemical structures and mechanisms of action, bacteriocins are divided into four classes, and sactipeptides and lanthipeptides are post-translationally modified antibacterial peptides belonging to class I bacteriocins.91
Blautia usually has the ability to produce bactericins. Through the annotation of secondary metabolites using the antismash database, 74 strains of Blautia were annotated into 7 categories and a total of 261 secondary metabolic biosynthesis gene clusters (BGCs) including NRPS, sactipeptide, lanthipeptide, bacteriocin, lassopeptide, betalactone and transat-pks (Figure 4). NRPs, sactipeptide, lanthipeptide were generally distributed in all the strains (Table S2). NRPs and PKs were among the most profuse families of secondary metabolites with diverse functions, including siderophores involved in iron scavenging, pigments that provide protection against an array of stress factors, as well as nutrient acquisition, chemical communication, and defense responses.92,93 Azevedo et al. found that both Blautia schinkii DSM 10518 and Blautia sp. SF-50 possess a gene cluster that encodes sactipeptide, the structural peptide sequence of which belongs to the protein family TIGR04065. Blautia sp. SF-50 also possesses a gene cluster that encodes lanthipeptide.94 Hatziioanou et al. found that B. obeum A2-162 possess a new antibiotic – nisin O, which contains four structural peptides and an unusual leader peptide sequence and showed antibacterial activity against C. perfringens in the presence of trypsin.95 In one study, commensal bacteria CBBPSCSK comprising C. bolteae, B. producta, Bacteroides sartorii, and Parabacteroides distasonis were isolated from mouse feces and were shown to demonstrate colonization resistance to vancomycin-resistant enterococci (VRE).96 When the four strains were co-cultured with VRE, only B. producta was found to inhibit the growth of VRE by secreting a lantibiotic to exert a bacteriostatic effect. Notably, lanthipeptide has a narrow antibacterial spectrum and specifically targeted VRE without affecting other commensal bacteria. Thus, B. producta may serve as a potential probiotic to prevent the infection and transmission of antibiotic-resistant conditional pathogens.97
Figure 4.
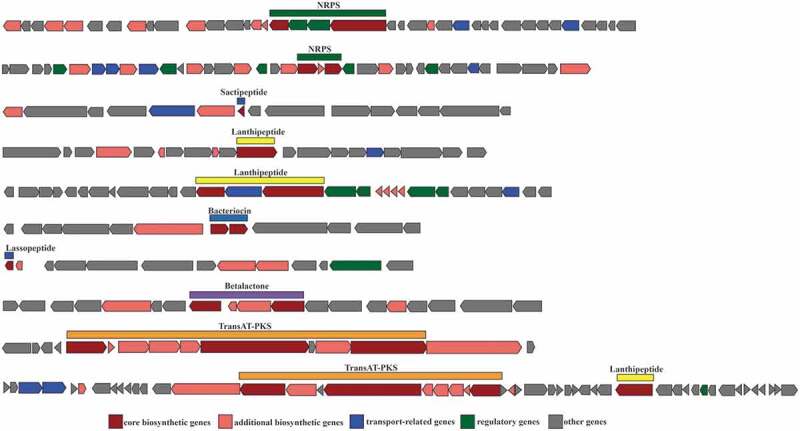
Schematic representation of 10 secondary metabolic biosynthetic gene clusters (BGCs) extracted from 74 Blautia genomes
Based on the above reports and analysis of secondary metabolites, we believe that the ability to produce bacteriocins gives Blautia the potential to inhibit the colonization of pathogenic bacteria in the intestine, and it can also affect the composition of intestinal microbiota. In particular, B. obeum and B. producta can inhibit the proliferation of C. perfringens and vancomycin-resistant enterococci, which makes it possible to become potential probiotics and exert probiotic functions.
Blautia and the obesity-related diseases
In recent decades, the prevalence of obesity-related metabolic syndromes such as type 2 diabetes has dramatically increased worldwide, which has, in turn, increased the risk of atherosclerosis, nonalcoholic fatty liver disease, certain cancers, and other diseases. Recent studies have suggested that the intestinal microbiota play an important role in obesity and related diseases.98 In a population-based cross-sectional study, the researchers investigated the relationship of visceral fat accumulation and body mass index in Japanese men and women aged 20–76 years with the intestinal microbiota stratified by sex. Blautia was found to be the only genus whose abundance showed a significant negative relationship with visceral fat accumulation in Japanese people, regardless of sex.99 Consistently, one study observed a change in the composition of intestinal microbiota and an increase in the abundance of Blautia in overweight/obese patients with nonalcoholic fatty liver who consumed a low-calorie and high-protein diet for 3 weeks.100 In another study, obese children, regardless of the presence of nonalcoholic steatohepatitis, showed a higher intestinal abundance of Bacteroides and a lower abundance of Firmicutes, accompanied by reduced abundances of Blautia and Faecalibacterium.101 A study evaluating the effect of Panax ginseng on the weight loss of middle-aged obese women in Korea showed that Blautia was a dominant genus in the intestinal microbiota of women in the effective weight loss group but not in those in the ineffective weight loss group.102 A previous study evaluating the effects of berberine and metformin on the intestinal microbiota of rats with HF diet-induced obesity demonstrated that berberine and metformin altered the overall structure of the intestinal microbiota; more specifically, a reduction in the diversity of intestinal microbiota but a significant increase in the abundance of SCFAs-producing Blautia were observed.103 In another study, the abundance of Blautia was decreased significantly in children with diabetes compared with healthy children.104 Inoue et al.105 revealed that Blautia abundance was significantly reduced in Japanese patients with type 2 diabetes compared with the control group, indicating a negative correlation between Blautia abundance and the levels of fasting plasma glucose and hemoglobin A1C (HbA1c). A previous study showed that the insulin signaling pathway and glycosylation/glycogen metagenesis were upregulated in diabetic patients, and this upregulation was positively correlated with the HbA1c level. Metformin and a specially designed herbal formula (AMC) were used to treat patients with type 2 diabetes for 12 weeks and significant decreases in hyperglycemia and hyperlipidemia were observed, in addition to changes in the structure of intestinal microbiota; specifically, the abundance of Blautia was increased, which was associated with the improvements in glucose and lipid homeostasis.106 Blautia is a common acetic acid producer in the intestine, which may inhibit insulin signaling and fat accumulation in adipocytes by activating the G protein-coupled receptors GPR41 and GPR43, in turn promoting the metabolism of unbound lipids and glucose in other tissues and thus alleviating obesity-related diseases.107 A cross-sectional study showed that Blautia, especially B. luti and B. wexlerae, probably help to reduce the inflammation associated with obesity-related complications. Therefore, these new findings will provide valuable information for the microbiota-based strategies for early prevention of obesity-related complications in the future.108
Blautia and inflammatory diseases
Vibrio cholerae usually causes human diarrhea, but people have different susceptibility to the pathogen, which may be driven by the interpersonal microbiome variation. Alavi et al. found that the intestinal microbiota of cholera patients were significantly different from that of healthy individuals, in which Blautia obeum showed a significant negative correlation with the colonization of Vibrio cholerae. Further studies showed that the genes encoding bile salt hydrolases (BSH) in B. obeum genome could reduce the expression of tcpA gene in V. cholera, inhibit its colonization and alleviate diarrhea.109 Allogeneic blood/bone marrow transplantation (allo-BMT) is a vital therapy for patients with leukemia, lymphoma, and other cancer-related hematological malignancies, but its biggest drawback is that the donor’s immune system recognizes the recipient’s organs as foreign, leading to graft-versus-host disease (GVHD).110 A study with biomarker analysis revealed that increased diversity of intestinal microbiota in patients undergoing allo-BMT, particularly increased abundance of symbiotic bacteria of the Blautia genus, is associated with a reduction in lethal GVHD and an increase in overall survival.111 A few studies have also reported the relationship of decreased abundance of Blautia with ileal pouch–anal anastomosis and liver cirrhosis.112,113 Blautia, as a genus of commensal obligate anaerobic bacteria, plays an important role in maintaining environmental balance in the intestine and preventing inflammation by upregulating intestinal regulatory T cells and producing SCFAs.114 Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis, is a nonspecific chronic intestinal inflammatory disease of unknown etiology, whose incidence has been increasing in many developing countries in recent decades.115 Analysis of fecal and mucosal microbial communities in IBD patients and healthy people has revealed that Blautia abundance is significantly reduced in the cecal mucosal microbiota of patients with CD.116 A similar decrease in Blautia abundance has also been reported in the mucosal adherent microbiota of colorectal cancer patients.117 In addition, the abundances of Faecalibacterium prausnitzii and Blautia have been reported to be reduced in the gut microbiota of patients with colitis-related cancer compared with that in healthy individuals. The study showed that Blautia and F. prausnitzii accounted for a lower proportion in the intestinal mucosa-associated microbiota of sporadic cancer patients than in that of healthy individuals, but were abundant in the extratumor microenvironment.118 F. prausnitzii has been shown to exert anti-inflammatory effects on colitis by blocking NF-kB expression and IL-8 secretion, as well as inducing colonization resistance against pathogens; these findings suggest that F. prausnitzii and Blautia exert protective effects against carcinogenesis.119,120
The above research shows that the abundance of Blautia is negatively correlated with some diseases. However, higher abundance of Blautia was found in the fecal microbiota of irritable bowel syndrome and ulcerative colitis patients compared with healthy individuals.121,122 A greater abundance of Blautia has also been reported in early breast cancer patients with increasing clinical stage and tissue pathological classification (Table 5).123 These conflicting conclusions may raise the question that whether Blautia is good for human health. In fact, most of these reports focused on the genus level, and did not conduct in-depth studies at the species or even strain levels. We must avoid drawing general conclusions at the genus level. There may be differences in the composition of Blautia at the species level, and different species of Blautia may exert beneficial or adverse effects on human health.
Table 5.
Reported roles of Blautia in host health
| Species/strain | Functions | Significant findings | Reference (author,year) |
|---|---|---|---|
| B. schinkii DSM 10518 | Antibacterial (bacteriocins) |
B. schinkii DSM 10518 had a gene cluster encoding Sactipeptide | Azevedo et al., 201594 |
| Blautia sp. Strain SF-50 | Blautia sp. Strain SF-50 had the genes cluster encoding both Sactipeptide and lanthipeptide | ||
| B. obeum A2-162 | Antibacterial (bacteriocins) |
A new lantibiotic nisin O isolated from B. obeum A2-162 showed antibacterial activity against Clostridium perfringens | Hatziioanou et al., 201795 |
| B. producta | Antibacterial (bacteriocins) |
Commensal bacterias CBBPSCSK containing B. producta demonstrated colonization resistance to vancomycin-resistant enterococci (VRE) | Caballero et al., 201796 |
| B. producta | Antibacterial (bacteriocins) |
B. producta inhibited the growth of VRE by secreting a lantibiotic similar to nisin-A to exerts bacteriostatic effect | Kim et al., 201997 |
| Blautia | Visceral fat accumulation (VFA) | Regardless of gender, Blautia was the only genus which significantly negatively related to VFA | Ozato et al., 201999 |
| Blautia | obese | For overweight/obese patients with nonalcoholic fatty liver, the intestinal microbiota of the patients changed and the abundance of Blautia increased | Pataky et al., 2016100 |
| Blautia | obese | Obese children showed higher levels of Bacteroides and lower levels of Firmicutes, accompanied by reduced the number of Blautia and Faecalibacterium | Zhu et al., 2013101 |
| Blautia | obese | A study evaluating the effect of Panax ginseng showed that Blautia was dominant in the intestinal microbiota of the effective weight loss group than the ineffective weight loss group | Song et al., 2014102 |
| Blautia | obese | Berberine and metformin alter the overall structure of the intestinal microbiota of obese rats, increasing the abundance of SCFAs – producing bacteria Blautia | Zhang et al., 2015103 |
| B. luti and B. wexlerae | obese | In obesity, the decrease of B.luti and B.wexlerae species in intestinal ecosystem may lead to metabolic inflammation and insulin resistance | Benitez-Paez et al, 2020108 |
| Blautia | diabetes | Compard to the healthy children, the abundance of Blautia significantly decreased in children with diabetes | Murri et al., 2013104 |
| Blautia | diabetes | Compard to the control group, Blautia was significantly reduced in patients with type 2 diabetes | Inoue et al., 2017105 |
| Blautia spp | diabetes | Metformin and specially designed herbal formula (AMC) could change the structure of intestinal microbiota and increase the abundance of Blautia | Tong et al., 2018106 |
| Blautia | graft-versus-host disease (GVHD) | In allogeneic blood/bone marrow transplantation (allo BMT) patients, the abundance of Blautia was associated with a reduction in lethal GVHD and an increase in overall surviva | Jenq et al., 2015111 |
| Blautia | ileal pouch-anal anastomosis | The abundance of Blautia decreased in ileal pouch anal anastomosi | Tyler et al., 2013112 |
| Blautia | liver cirrhosis | The abundance of Blautia decreased in liver cirrhosis | Kakiyama et al., 2013113 |
| Blautia | Inflammatory Bowel Disease(IBD) | Compared to the healthy people, the cecal mucosal microbiota Blautia was significantly reduced in patients with Crohn’s disease (CD) | Chen et al., 2014116 |
| Blautia | colorectal cancer (CRC) | There was a decrease in Blautia in the mucosal adherent microbiota of colorectal cancer patients | Chen et al., 2012117 |
| Blautia obeum | Cholera Infection | Blautia Obeum could disable the pathogenic mechanism of Vibrio cholerae, preventing it from colonizing the gut | Alavi etal, 2020109 |
In summary, Blautia abundance has a close relationship with various diseases, but literature on the accurate evidence of this relationship is currently limited. Despite some conflicting findings and lack of clarity on the potential mechanism of Blautia in various diseases, Blautia abundance can still be used as a potential tool for the early diagnosis or treatment of related diseases.
Cross-feeding of Blautia with other microorganisms
When the metabolites produced by bacteria from dietary components act as substrates to support the growth of other species, it is called cross-feeding.124 Cross-feeding is an important interaction between anaerobic bacteria in the intestinal microbiota that can affect their metabolic pathways and contribute to their stability and productivity.125
As a genus of anaerobic bacteria, the cross-feeding of Blautia with other bacteria also contributes to metabolic regulation to some extent. A study found that by using 0.2% resistant starch as an energy source, R. bromii produces formic acid, ethanol, and acetic acid in approximately equal molar ratios on RUM-RS medium.9 However, batch co-culture with acetogenic bacteria B. hydrogenotrophica on starch resulted in the disappearance of formic acid with an increase in acetic acid levels. RNA sequencing was used to further study the interspecific interactions to detect gene expression in continuous co-cultures of R. bromii and B. hydrogenotrophica. Transcriptome analysis revealed the upregulation of B. hydrogenotrophica genes involved in the Wood–Ljungdahl pathway in addition to a 10-gene cluster responsible for increased branched-chain amino acid fermentation in the co-culture. Cross-feeding between formic acid-producing species and acetic acid-producing species may play an important role in the formation of SCFAs in the colon and contribute to the massive production of acetic acid.9 As a key hydrogen-consuming anaerobic microorganism, B. hydrogenotrophica has been reported to mediate regulation related to the coexistence of anaerobic respiratory pathways. B. hydrogenotrophica consumes H2 and CO2 via the Wood–Ljungdahl pathway to produce acetic acid – a pathway significantly activated when coexisting with Bifidobacterium bifidum. Bi. bifidum serves as a special carbohydrate-fermenting species and produces CO2, which is a fixed substrate in the Wood–Ljungdahl pathway. Thus, the changes observed in the Wood–Ljungdahl pathway in B. hydrogenotrophica may be the result of cross-feeding by Bi. bifidum.126
Conclusions
As a dominant genus of intestinal microbiota, Blautia plays certain roles in metabolic diseases, inflammatory diseases, and biotransformation. However, most of the properties of this genus are linked with its potential probiotic functions, and the causal relationship between Blautia abundance and diseases is not yet clear. In addition, there is a disparity in association of the genus with human diseases (less Blautia in sufferers of diabetes/obesity, less in CRC and CD but more in IBD). We could not overgeneralize the effects of Blautia at a genus level and should consider the importance of species – and perhaps strain differences. Whether Blautia plays a direct regulatory role in diseases requires further intervention studies and more detailed evidence. Meanwhile, there is no clear evidence to support its application in the clinical setting and food industries, and its safety in humans requires further verification. As strictly anaerobic bacteria, Blautia requires harsh cultivation conditions with rigorous operation methods. To date, only a few strains of Blautia have been isolated, and no description of the characteristics of this genus appears in the Bergey’s manual. Moreover, it still has not attracted adequate attention from the researchers, and its genomic information remains limited. Moreover, Blautia has host specificity and the oligotypes could accurately identify different host sources. Thus, more Blautia strains need to be isolated from different hosts to characterize the physiological properties of this genus and sequence their genomes to clarify the connection between Blautia and host health for a comprehensive understanding. Given the role of Blautia in the metabolic regulation in host, the use of prebiotics, such as some oligosaccharides, as substrates to promote the proliferation of Blautia also remains to be explored to elucidate its probiotic functions.
Supplementary Material
Funding Statement
This work was supported by National Natural Science Foundation of China (Grant No. 31972086, 31801530), National First-Class Discipline Program of Food Science and Technology (JUFSTR20180102) and Collaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Stanley D, MS G, SE D, VR H, TM C, RJ H, RJ M.. Identification of chicken intestinal microbiota correlated with the efficiency of energy extraction from feed. Vet Microbiol. 2013;164(1–2):85–21. doi: 10.1016/j.vetmic.2013.01.030. [DOI] [PubMed] [Google Scholar]
- 2.Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the genetic basis of fibrolytic specialization by lachnospiraceae and ruminococcaceae in diverse gut communities. Diversity. 2013;5(3):627–640. doi: 10.3390/d5030627. [DOI] [Google Scholar]
- 3.Chakravarthy SK, Jayasudha R, Prashanthi GS, Ali MH, Sharma S, Tyagi M, Shivaji S. Dysbiosis in the gut bacterial microbiome of patients with uveitis, an inflammatory disease of the eye. Indian J Microbiol. 2018;58(4):457–469. doi: 10.1007/s12088-018-0746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khattab MSA, Abd El Tawab AM, MT F. Isolation and characterization of anaerobic bacteria from frozen rumen liquid and its potential characterizations. Int J Dairy Sci. 2017;12(1):47–51. doi: 10.3923/ijds.2017.47.51. [DOI] [Google Scholar]
- 5.Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, Abe F, Osawa R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016;16(1):1–12. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakayama J, Watanabe K, Jiang J, Matsuda K, Chao SH, Haryono P, La-ongkham O, Sarwoko MA, Sujaya IN, Zhao L, et al. 2015. Diversity in gut bacterial community of school-age children in Asia. Sci Rep-Uk. 5(1):8397. doi: 10.1038/srep08397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao B, Gu J, Li D, Cui S, Zhao J, Zhang H, Chen W. Effects of different doses of fructooligosaccharides (fos) on the composition of mice fecal microbiota, especially the bifidobacterium composition. Nutrients. 2018;10(8):1105. doi: 10.3390/nu10081105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim M, Kim N, Han J. Metabolism of kaempferia parviflora polymethoxyflavones by human intestinal bacterium bautia sp. MRG-PMF1. J Agric Food Chem. 2014;62(51):12377–12383. doi: 10.1021/jf504074n. [DOI] [PubMed] [Google Scholar]
- 9.Laverde Gomez JA, Mukhopadhya I, Duncan SH, Louis P, Shaw S, Collie-Duguid E, Crost E, Juge N, Flint HJ. Formate cross-feeding and cooperative metabolic interactions revealed by transcriptomics in co-cultures of acetogenic and amylolytic human colonic bacteria. Environ Microbiol. 2019;21(1):259–271. doi: 10.1111/1462-2920.14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C, Finegold SM, Song Y, Lawson PA. Reclassification of clostridium coccoides, ruminococcus hansenii, ruminococcus hydrogenotrophicus, ruminococcus luti, ruminococcus productus and ruminococcus schinkii as blautia coccoides gen. nov., comb. nov., blautia hansenii comb. nov., blautia hydrogenotrophica comb. nov., blautia luti comb. nov., blautia producta comb. nov., blautia schinkii comb. nov. and description of blautia wexlerae sp. nov., isolated from human faeces. Int J Syst Evol Micr. 2008;58(8):1896–1902. doi: 10.1099/ijs.0.65208-0. [DOI] [PubMed] [Google Scholar]
- 12.Kaneuchi CJ, Benno Y, Mitsuoka T. Clostridium coccoides, a new species from the feces of mice. Int J Syst Bacteriol. 1976;26(4):482–486. doi: 10.1099/00207713-26-4-482. [DOI] [Google Scholar]
- 13.Lillian V H, Moore WEC. New genus, coprococcus, twelve new species, and emended descriptions of four previously described species of bacteria from human feces. Int J Syst Bacteriol. 1974;24(2):260–277. doi: 10.1099/00207713-24-2-260. [DOI] [Google Scholar]
- 14.Bernalier A, Willems A, Leclerc M, Rochet V, Collins MD. Ruminococcus hydrogenotrophicus sp. nov., a new H2/CO2-utilizing acetogenic bacterium isolated from human feces. Arch Microbiol. 1996;166(3):176–183. doi: 10.1007/s002030050373. [DOI] [PubMed] [Google Scholar]
- 15.Simmering R, Taras D, Schwiertz A, Le Bla G, Gruhl B, Lawson PA, Collins MD, Blaut M. Ruminococcus luti sp. nov., isolated from a human faecal sample. Syst Appl Microbiol. 2002;25(2):189–193. doi: 10.1078/0723-2020-00112. [DOI] [PubMed] [Google Scholar]
- 16.Ezaki T, Li N, Hashimoto Y, Miura H, Yamamoto H. 16S ribosomal DNA sequences of anaerobic cocci and proposal of ruminococcus hansenii comb. Nov. And ruminococcus productus comb. nov. Int J Syst Bacteriol. 1994;44(1):130–136. doi: 10.1099/00207713-44-1-130. [DOI] [PubMed] [Google Scholar]
- 17.Lesme FR, Morvan B, Collins MD, Fonty G, Willems A. A New H2/CO2-using acetogenic bacterium from the rumen: description of Ruminococcus schinkii sp. nov. FEMS Microbiol Lett. 1996;140(2):281–286. doi: 10.1016/0378-1097(96)00195-4. [DOI] [PubMed] [Google Scholar]
- 18.Furuya H, Ide Y, Hamamoto M, Asanuma N, Hino T. Isolation of a novel bacterium, Blautia glucerasei sp. nov., hydrolyzing plant glucosylceramide to Ceramide. Arch Microbiol. 2010;192(5):365–372. doi: 10.1007/s00203-010-0566-8. [DOI] [PubMed] [Google Scholar]
- 19.Park SK, Kim MS, Roh SW, Bae JW. Blautia stercoris sp. nov., isolated from human faeces. Int J Syst Evol Micr. 2012;62(4):776–779. doi: 10.1099/ijs.0.031625-0. [DOI] [PubMed] [Google Scholar]
- 20.Park SK, Kim MS, Bae JW. Blautia faecis sp. nov., isolated from human faeces. Int J Syst Evol Micr. 2013;63(Pt_2):599–603. doi: 10.1099/ijs.0.036541-0. [DOI] [PubMed] [Google Scholar]
- 21.Moore WEC, Johnson JL, Holdeman LV. Emendation of bacteroidaceae and butyrivibrio and descriptions of desulfornonas gen.nov. and ten new species in the genera desulfomonas, butyrivibrio, eubacterium, czostridium, and ruminococcus. Int J Syst Bacteriol. 1976;26(2):238–252. doi: 10.1099/00207713-26-2-238. [DOI] [Google Scholar]
- 22.Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier-Kolthoff JP, Kumar N, Bresciani A, Martínez I, Just S, Ziegler C, et al. 2016. The mouse intestinal bacterial collection (mibc) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat Microbiol. 1(10). doi: 10.1038/nmicrobiol.2016.131. [DOI] [PubMed] [Google Scholar]
- 23.Durand GA, Pham T, Ndongo S, Traore SI, Dubourg G, Lagier JC, Michelle C, Armstrong N, Fournier P, Raoult D, et al. 2017. Blautia massiliensis sp. nov., isolated from a fresh human fecal sample and emended description of the genus Blautia. Anaerobe. 43:47–55. doi: 10.1016/j.anaerobe.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Traore SI, Azhar EI, Yasir M, Bibi F, Fournier PE, Jiman-Fatani AA, Delerce J, Cadoret F, Lagier JC, Raoult D. 2017. Description of ‘Blautia Phocaeensis’ sp. nov. and ‘lachnoclostridium edouardi’ sp. nov., isolated from healthy fresh stools of Saudi Arabia bedouins by culturomics. New Microbes and New Infections. 19:129–131. doi: 10.1016/j.nmni.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pham TPT, Cadoret F, Alou MT, Brah S, Diallo BA, Diallo A, Sokhna C, Delerce J, Fournier PE, Million M, et al. 2017. Urmitella Timonensis’ gen. nov., sp. nov., ‘Blautia marasmi’ sp. nov., ‘Lachnoclostridium pacaense’ sp. nov., ‘Bacillus marasmi’ sp. nov. and ‘Anaerotruncus rubiinfantis’ sp. nov., isolated from stool samples of undernourished African children. New Microbes and New Infections. 17:84–88. doi: 10.1016/j.nmni.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pham TPT, Cadoret F, Alou MT, Brah S, Diallo BA, Diallo A, Sokhna C, Delerce J, Fournier PE, Million M, et al. 2017. ‘Marasmitruncus massiliensis’ gen. nov., sp. nov., ‘Clostridium culturomicum’ sp. nov., ‘Blautia provencensis’ sp. nov., ‘Bacillus caccae’ sp. nov. and ‘Ornithinibacillus massiliensis’ sp. nov., isolated from stool samples of undernourished African Children. New Microbes and New Infections. 19:38–42. doi: 10.1016/j.nmni.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin NR, Kang W, Tak EJ, Hyun DW, Kim PS, Kim HS, Lee JY, Sung H, Whon TW, Bae JW. Blautia hominis sp. nov., isolated from human faeces. Int J Syst Evol Micr. 2018;68(4):1059–1064. doi: 10.1099/ijsem.0.002623. [DOI] [PubMed] [Google Scholar]
- 28.Paek J, Shin Y, Kook JK, Chang YH. Blautia argi sp. nov., a new anaerobic bacterium isolated from dog faeces. Int J Syst Evol Micr. 2019;69(1):33–38. doi: 10.1099/ijsem.0.002981. [DOI] [PubMed] [Google Scholar]
- 29.Ghimire S, Wongkuna S, Kumar R, Nelson E, Christopher-Hennings J, Scaria J. 2020. Genome sequence and description of Blautia brookingsii SG772 sp. nov., a novel bacterial species isolated from human faeces. New Microbes and New Infections. 34:100648. doi: 10.1016/j.nmni.2019.100648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JS, Park J-E, Lee KC, Choi S-H, Oh BS, Yu SY, Eom MK, Kang SW, Han K-I, Suh MK, et al. 2020. Blautia faecicola sp. nov., isolated from faeces from a healthy human. Int J Syst Evol Micr. 70(3):2059–2065. doi: 10.1099/ijsem.0.004015. [DOI] [PubMed] [Google Scholar]
- 31.Arumugam M, Raes J, Pelletier E, Paslier DL, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. 2011. Enterotypes of the human gut microbiome. Nature. 473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Touyama M, Jin JS, Kibe R, Hayashi H, Benno Y. Quantification of Blautia wexlerae and Blautia luti in human faeces by real-time PCR using specific primers. Benef Microbes. 2015;6(4):583–590. doi: 10.3920/BM2014.0133. [DOI] [PubMed] [Google Scholar]
- 33.Geerligs G, Aldrich HC, Harder W, Diekert G. Isolation and characterization of a carbon monoxide utilizing strain of the acetogen Peptostreptococcus productus. Arch Microbiol. 1987;148(4):305–313. doi: 10.1007/BF00456709. [DOI] [Google Scholar]
- 34.Lorowitz WH, Bryant MP. Peptostreptococcus productus Strain that grows rapidly with co as the energy source. Appl Environ Microb. 1984;47(5):961–964. doi: 10.1128/AEM.47.5.961-964.1984. PMID: 6430231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamada M, Hase S, Sato K, Toyoda A, Fujiyama A, Sakakibara Y. Whole genome complete resequencing of bacillus subtilis natto by combining long reads with high-quality short reads. PLoS One. 2014;9(10):e109999. doi: 10.1371/journal.pone.0109999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang J, Yang B, Ross RP, Stanton C, Zhao J, Zhang H, Chen W. 2020. Comparative genomics of pediococcus pentosaceus isolated from different niches reveals genetic diversity in carbohydrate metabolism and immune system. Front Microbiol. 11:253. doi: 10.3389/fmicb.2020.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15(6):589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 38.Bosi E, Monk JM, Aziz RK, Fondi M, Nizet V, Palsson BØ. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. P Natl Acad Sci USA. 2016;113(26):E3801–E3809. doi: 10.1073/pnas.1523199113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhardwaj T, Somvanshi P. 2017. Pan-genome analysis of Clostridium botulinum reveals unique targets for drug development. Gene. 623:48–62. doi: 10.1016/j.gene.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W, Wang Y, Li K, Kwok L, Liu W, Zhang H. Short communication: modulation of fatty acid metabolism improves oxygen tolerance of Bifidobacterium Animalis ssp. lactis Probio-M8. J Dairy Sci. 2020;103(10):8791–8795. doi: 10.3168/jds.2019-18049. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Xiao Z, Zou Q, Fang J, Wang Q, Yang X, Gao N. Ribosome profiling reveals genome- wide cellular translational regulation upon heat stress in escherichia coli. Genom Proteom Bioinf. 2017;15(5):324–330. doi: 10.1016/j.gpb.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Len ACL, Harty DWS, Jacques NA. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology. 2004;150(5):1339–1351. doi: 10.1099/mic.0.27008-0. [DOI] [PubMed] [Google Scholar]
- 43.Wang D, Hao Z, Zhao J, Jin Y, Huang J, Zhou R, Wu C. 2019. Comparative physiological and transcriptomic analyses reveal salt tolerance mechanisms of Zygosaccharomyces rouxii. Process Biochem. 82:59–67. doi: 10.1016/j.procbio.2019.04.009. [DOI] [Google Scholar]
- 44.Li Q, Chen Q, Ruan H, Zhu D, He G. Isolation and characterisation of an oxygen, acid and bile resistant Bifidobacterium Animalis subsp. lactis Qq08. J Sci Food Agr. 2010;90(8):1340–1346. doi: 10.1002/jsfa.3942. [DOI] [PubMed] [Google Scholar]
- 45.Shen J, Zhou J, Xu Y, Xiu Z. Prophages contribute to genome plasticity of Klebsiella pneumoniae and may involve the chromosomal integration of ARGs in CG258. Genomics. 2020;112(1):998–1010. doi: 10.1016/j.ygeno.2019.06.016. [DOI] [PubMed] [Google Scholar]
- 46.Chen D, Huang T, Wang Q, Bai C, Yang L. 2020. Analysis on the virulomes and resistomes of multi-drug resistance clinical Escherichia coli isolates, as well as the interactome with gut microbiome. Microb Pathogenesis. 148:104423. doi: 10.1016/j.micpath.2020.104423. [DOI] [PubMed] [Google Scholar]
- 47.Parmar NR, Nirmal Kumar JI, Joshi CG. 2015. Deep insights into carbohydrate metabolism in the rumen of Mehsani buffalo at different diet treatments. Genomics Data. 6:59–62. doi: 10.1016/j.gdata.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol Res. 2012;69(1):52–60. doi: 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 49.Hamajima H, Matsunaga H, Fujikawa A, Sato T, Mitsutake S, Yanagita T, Nagao K, Nakayama J, Kitagaki H. Japanese traditional dietary fungus koji Aspergillus oryzae functions as a prebiotic for Blautia coccoides through glycosylceramide: japanese dietary fungus koji is a new prebiotic. SpringerPlus. 2016;5(1):1321. doi: 10.1186/s40064-016-2950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, Bindels LB, Segura Munoz RR, Martínez I, Walter J, Ramer-Tait AE, Rose DJ. Disparate metabolic responses in mice fed a high-fat diet supplemented with maize-derived non-digestible feruloylated oligo- and polysaccharides are linked to changes in the gut microbiota. PLoS One. 2016;11(1):e146144. doi: 10.1371/journal.pone.0146144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panasevich MR, Kerr KR, Dilger RN, Fahey GC, Guérin-Deremaux L, Lynch GL, Wils D, Suchodolski JS, Steiner JM, Dowd SE, et al. 2015. Modulation of the faecal microbiome of healthy adult dogs by inclusion of potato fibre in the diet. Brit J Nutr. 113(1):125–133. doi: 10.1017/S0007114514003274. [DOI] [PubMed] [Google Scholar]
- 52.Lee SM, Han HW, Yim SY. Beneficial effects of soy milk and fiber on high cholesterol diet-induced alteration of gut microbiota and inflammatory gene expression in rats. Food Funct. 2015;6(2):492–500. doi: 10.1039/C4FO00731J. [DOI] [PubMed] [Google Scholar]
- 53.Bai G, Ni K, Tsuruta T, Dietary Casein NN. Soy protein isolate modulate the effects of raffinose and fructooligosaccharides on the composition and fermentation of gut microbiota in rats. J Food Sci. 2016;81(8):H2093–H2098. doi: 10.1111/1750-3841.13391. [DOI] [PubMed] [Google Scholar]
- 54.Minamida K, Kaneko M, Ohashi M, Sujaya IN, Sone T, Wada M, Yokota A, Hara H, Asano K, Tomita F. Effects of difructose anhydride (DFA III) administration on bile acids and growth of DFA III-assimilating bacterium ruminococcus productus on rat intestine. J Biosci Bioeng. 2005;99(6):548–554. doi: 10.1263/jbb.99.548. [DOI] [PubMed] [Google Scholar]
- 55.Noriega BS, Sanchez-Gonzalez MA, Salyakina D, Coffman J. 2016. Understanding the impact of omega-3 rich diet on the gut microbiota. Case Rep Med. 2016:3089303. doi: 10.1155/2016/3089303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaakoush NO, Martire SI, Raipuria M, Mitchell HM, Nielsen S, Westbrook RF, Morris MJ. Alternating or continuous exposure to cafeteria diet leads to similar shifts in gut microbiota compared to chow diet. Mol Nutr Food Res. 2017;61(1):1–9. doi: 10.1002/mnfr.201500815. [DOI] [PubMed] [Google Scholar]
- 57.Endo A, Pӓrtty A, Kalliomӓki M, Isolauri E, Salminen S. 2014. Long-term monitoring of the human intestinal microbiota from the 2nd week to 13 years of age. Anaerobe. 28:149–156. doi: 10.1016/j.anaerobe.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 58.Kima HB, Borewicza K, Whiteb BA, Singera RS, Sreevatsana S, Tuc ZJ, Isaacson RE. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet Microbiol. 2011;153(1–2):124–133. doi: 10.1016/j.vetmic.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 59.Zhu Y, Li H, Xu X, Li C, Zhou G. The gut microbiota in young and middle-aged rats showed different responses to chicken protein in their diet. BMC Microbiol. 2016;16(1):281. doi: 10.1186/s12866-016-0895-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xi Y, Shuling N, Kunyuan T, Qiuyang Z, Hewen D, ChenCheng G, Tianhe Y, Liancheng L, Xin F. 2019. Characteristics of the intestinal flora of specific pathogen free chickens with age. Microb Pathogenesis. 132:325–334. doi: 10.1016/j.micpath.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 61.Vaiserman AM, Koliada AK, Marotta F. 2017. Gut microbiota: a player in aging and a target for anti-aging intervention. Ageing Res Rev. 35:36–45. doi: 10.1016/j.arr.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 62.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, et al. 2014. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 5(1):3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lan D, Ji W, Lin B, Chen Y, Huang C, Xiong X, Fu M, Mipam TD, Ai Y, Zeng B, et al. 2017. Correlations between gut microbiota community structures of Tibetans and geography. Sci Rep-Uk. 7(1):16982. doi: 10.1038/s41598-017-17194-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta VK, Paul S, Geography DC. 2017. Ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 8:1162. doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang J, Guo Z, Xue Z, Sun Z, Zhang M, Wang L, Wang G, Wang F, Xu J, Cao H, et al. 2015. A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. Isme J. 9(9):1979–1990. doi: 10.1038/ismej.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, Deelen P, Vatanen T, Schirmer M, Smeekens SP, et al. 2016. The effect of host genetics on the gut microbiome. Nat Genet. 48(11):1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- 67.Eren AM, Sogin ML, Morrison HG, Vineis JH, Fisher JC, Newton RJ, McLellan SL. A single genus in the gut microbiome reflects host preference and specificity. Isme J. 2015;9(1):90–100. doi: 10.1038/ismej.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 69.Horikawa T, Shimada T, Okabe Y, Kinoshita K, Koyama K, Miyamoto K, Ichinose K, Takahashi K, Aburada M. Polymethoxyflavonoids from kaempferia parviflora induce adipogenesis on 3T3-L1 preadipocytes by regulating transcription factors at an early stage of differentiation. Biol Pharm Bull. 2012;35(5):686–692. doi: 10.1248/bpb.35.686. [DOI] [PubMed] [Google Scholar]
- 70.Wongsrikaew N, Kim H, Vichitphan K, Cho SK, Han J. Antiproliferative activity and polymethoxyflavone composition analysis of kaempferia parviflora extracts. J Korean Soc Appl Biol Chem. 2012;55(6):813–817. doi: 10.1007/s13765-012-2175-5. [DOI] [Google Scholar]
- 71.Sae-wong C, Tansakul P, Tewtrakul S. Anti-inflammatory mechanism of Kaempferia parviflora in murine macrophage cells (RAW 264.7) and in experimental animals. J Ethnopharmacol. 2009;124(3):576–580. doi: 10.1016/j.jep.2009.04.059. [DOI] [PubMed] [Google Scholar]
- 72.Li S, Lo C, Ho C. Hydroxylated polymethoxyflavones and methylated flavonoids in sweet orange (citrus sinensis) peel. J Agr Food Chem. 2006;54(12):4176–4185. doi: 10.1021/jf060234n. [DOI] [PubMed] [Google Scholar]
- 73.Wu H, Kim M, Han J. Icariin metabolism by human intestinal microflora. Molecules. 2016;21(9):1158. doi: 10.3390/molecules21091158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burapan S, Kim M, Han J. Curcuminoid demethylation as an alternative metabolism by human intestinal microbiota. J Agr Food Chem. 2017;65(16):3306–3311. doi: 10.1021/acs.jafc.7b00943. [DOI] [PubMed] [Google Scholar]
- 75.Tamvakopoulos C, Dimas K, Sofianos ZD, Hatziantoniou S, Han Z, Liu Z, Wyche JH, Metabolism PP. Anticancer activity of the curcumin analogue, dimethoxycurcumin. Clin Cancer Res. 2007;13(4):1269–1277. doi: 10.1158/1078-0432.CCR-06-1839. [DOI] [PubMed] [Google Scholar]
- 76.Pinkaew D, Changtam C, Tocharus C, Govitrapong P, Jumnongprakhon P, Suksamrarn A, Tocharus J. Association of neuroprotective effect of Di-O-Demethylcurcumin on Aβ25-35-induced neurotoxicity with suppression of NF-κB and activation of Nrf2. Neurotox Res. 2016;29(1):80–91. doi: 10.1007/s12640-015-9558-4. [DOI] [PubMed] [Google Scholar]
- 77.Pinkaew D, Changtam C, Tocharus C, Thummayot S, Suksamrarn A, Tocharus J. 2015. Di-O-demethylcurcumin protects SK-N-SH cells against mitochondrial and endoplasmic reticulum-mediated apoptotic cell death induced by Aβ25-35. Neurochem Int. 80:110–119. doi: 10.1016/j.neuint.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 78.Liu MY, Li M, Wang XL, Liu P, Hao QH, Yu XM. Study on human intestinal bacterium Blautia sp. AUH-JLD56 for the conversion of arctigenin to (–)-3ʹ-desmethylarctigenin. J Agr Food Chem. 2013;61(49):12060–12065. doi: 10.1021/jf403924c. [DOI] [PubMed] [Google Scholar]
- 79.Vaughn BP, Kaiser T, Staley C, Hamilton MJ, Reich J, Graiziger C, Singroy S, Kabage A, Sadowsky MJ, Khoruts A. 2019. A pilot study of fecal bile acid and microbiota profiles in inflammatory bowel disease and primary sclerosing cholangitis. Clin Exp Gastroenterol. 12:9–19. doi: 10.2147/ceg.S186097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Braune A, Blaut M. Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes. 2016;7(3):216–234. doi: 10.1080/19490976.2016.1158395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Michlmayr H, Kneifel W. β-Glucosidase activities of lactic acid bacteria: mechanisms, impact on fermented food and human health. FEMS Microbiol Lett. 2014;352(1):1–10. doi: 10.1111/1574-6968.12348. [DOI] [PubMed] [Google Scholar]
- 82.Braune A, Engst W, Blaut M. Identification and functional expression of genes encoding flavonoid O- and C-glycosidases in intestinal bacteria. Environ Microbiol. 2016;18(7):2117–2129. doi: 10.1111/1462-2920.12864. [DOI] [PubMed] [Google Scholar]
- 83.González-Sarrías A, Espín JC, Tomás-Barberán FA. 2017. Non-extractable polyphenols produce gut microbiota metabolites that persist in circulation and show anti-inflammatory and free radical-scavenging effects. Trends Food Sci Tech. 69:281–288. doi: 10.1016/j.tifs.2017.07.010. [DOI] [Google Scholar]
- 84.Woting A, Clavel T, Loh G, Blaut M. Bacterial transformation of dietary lignans in gnotobiotic rats. FEMS Microbiol Ecology. 2010;72(3):507–514. doi: 10.1111/j.1574-6941.2010.00863.x. [DOI] [PubMed] [Google Scholar]
- 85.Xu X, Xu P, Ma C, Tang J, Gut Microbiota ZX. host health, and polysaccharides. Biotechnol Adv. 2013;31(2):318–337. doi: 10.1016/j.biotechadv.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 86.Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3(10):777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 87.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41(W1):W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Adamek M, Alanjary M, Sales-Ortells H, Goodfellow M, Bull AT, Winkler A, Wibberg D, Kalinowski J, Ziemert N. Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis species. BMC Genomics. 2018;19(1):1–15. doi: 10.1186/s12864-018-4809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martinez FAC, Balciunas EM, Converti A, Cotter PD, Oliveira RPD. Bacteriocin production by Bifidobacterium spp. A review. Biotechnol Adv. 2013;31(4):482–488. doi: 10.1016/j.biotechadv.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 90.Zacharof MP, Lovitt RW. 2012. Bacteriocins produced by lactic acid bacteria a review article. APCBEE Procedia. 2:50–56. doi: 10.1016/j.apcbee.2012.06.010. [DOI] [Google Scholar]
- 91.Zhong Z, He B, Li J, Li Y. Challenges and advances in genome mining of ribosomally synthesized and post-translationally modified peptides (RiPPs). Synthetic and Systems Biotechnology. 2020;5(3):155–172. doi: 10.1016/j.synbio.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao M, Yuan J, Zhang R, Dong M, Deng X, Zhu C, Li R, Shen Q. 2018. Microflora that harbor the NRPS gene are responsible for Fusarium wilt disease-suppressive soil. Appl Soil Ecol. 132:83–90. doi: 10.1016/j.apsoil.2018.08.022. [DOI] [Google Scholar]
- 93.Dror B, Jurkevitch E, Cytryn E. 2020. State-of-the-art methodologies to identify antimicrobial secondary metabolites in soil bacterial communities-A review. Soil Biol Biochem. 147:107838. doi: 10.1016/j.soilbio.2020.107838. [DOI] [Google Scholar]
- 94.Azevedo AC, Bento CBP, Ruiz JC, Queiroz MV, Mantovani HC. Distribution and Genetic Diversity of Bacteriocin Gene Clusters in Rumen Microbial Genomes. Appl Environ Microb. 2015;81(20):7290–7304. doi: 10.1128/aem.01223-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hatziioanou D, Gherghisan-Filip C, Saalbach G, Horn N, Wegmann U, Duncan SH, Flint HJ, Mayer MJ, Narbad A. Discovery of a novel lantibiotic nisin O from Blautia obeum A2-162, isolated from the human gastrointestinal tract. Microbiology-Sgm. 2017;163(9):1292–1305. doi: 10.1099/mic.0.000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Caballero S, Kim S, Carter RA, Leiner IM, Sušac B, Miller L, Kim GJ, Ling L, Pamer EG . 2017. Cooperating commensals restore colonization resistance to vancomycin-resistant enterococcus faecium. Cell Host Microbe. 21(5):592–602. doi: 10.1016/j.chom.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim SG, Becattini S, Moody TU, Shliaha PV, Littmann ER, Seok R, Gjonbalaj M, Eaton V, Fontana E, Amoretti L, et al. 2019. Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature. 572(7771):665–669. doi: 10.1038/s41586-019-1501-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tilg H, Gut Microbiome KA. Obesity, and metabolic dysfunction. J Clin Invest. 2011;121(6):2126–2132. doi: 10.1172/jci58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ozato N, Saito S, Yamaguchi T, Katashima M, Tokuda I, Sawada K, Katsuragi Y, Kakuta M, Imoto S, Ihara K, et al. 2019. Blautia Genus associated with visceral fat accumulation in adults 20-76 years of age. Npj Biofilms Microbi. 5(1):1–9. doi: 10.1038/s41522-019-0101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pataky Z, Genton L, Spahr L, Lazarevic V, Terraz S, Gaïa N, Rubbia-Brandt L, Golay A, Schrenzel J, Pichard C. Impact of hypocaloric hyperproteic diet on gut microbiota in overweight or obese patients with nonalcoholic fatty liver disease: a pilot study. Digest Dis Sci. 2016;61(9):2721–2731. doi: 10.1007/s10620-016-4179-1. [DOI] [PubMed] [Google Scholar]
- 101.Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. Characterization of gut microbiomes in nonalcoholic steatohepatitis (nash) patients: a connection between endogenous alcohol and nash. Hepatology. 2013;57(2):601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 102.Song MY, Kim BS, Kim H. Influence of Panax ginseng on obesity and gut microbiota in obese middle-aged Korean women. J Ginseng Res. 2014;38(2):106–115. doi: 10.1016/j.jgr.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang X, Zhao Y, Xu J, Xue Z, Zhang M, Pang X, Zhang X, Zhao L. 2015. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci Rep. 5(1). doi: 10.1038/srep14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, Queipo-Ortuño MI. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11(1):46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Inoue R, Ohue-Kitano R, Tsukahara T, Tanaka M, Masuda S, Inoue T, Yamakage H, Kusakabe T, Hasegawa K, Shimatsu A, et al. 2017. Prediction of functional profiles of gut microbiota from 16s rrna metagenomic data provides a more robust evaluation of gut dysbiosis occurring in japanese type 2 diabetic patients. J Clin Biochem Nutr. 61(3):217–221. doi: 10.3164/jcbn.17-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tong XL, Xu J, Lian FM, Yu XT, Zhao YF, Xu LP, Zhang MH, Zhao XY, Shen J, Wu SP, et al. 2018. Structural alteration of gut microbiota during the amelioration of human type 2 diabetes with hyperlipidemia by metformin and a traditional chinese herbal formula: a multicenter, randomized, open label clinical trial. Mbio. 9(3):e2317–e2392. doi: 10.1128/mBio.02392-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, Terasawa K, Kashihara D, Hirano K, Tani T, et al. 2013. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Benitez-Paez A, Gomez DPE, Lopez-Almela I, Moya-Perez A, Codoner-Franch P, Sanz Y. Depletion of Blautia species in the microbiota of obese children relates to intestinal inflammation and metabolic phenotype worsening. mSystems. 2020;5(2):2. doi: 10.1128/mSystems.00857-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alavi S, Mitchell JD, JY C, Liu R, Macbeth JC, Hsiao A. Interpersonal gut microbiome variation drives susceptibility and resistance to cholera infection. Cell. 2020;181(7):1533–1546. doi: 10.1016/j.cell.2020.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Paul S, Tsai HL, Lowery P, Fuchs EJ, Luznik L, Bolanos-Meade J, Swinnen LJ, Shanbhag S, Wagner-Johnston N, Varadhan R, et al. 2020. Allogeneic haploidentical blood or marrow transplantation with post-transplantation cyclophosphamide in chronic lymphocytic leukemia. Biol Blood Marrow Tr. 26(3):502–508. doi: 10.1016/j.bbmt.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, Littmann ER, Ling L, Gobourne AC, Miller LC, et al. 2015. intestinal blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Tr. 21(8):1373–1383. doi: 10.1016/j.bbmt.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tyler AD, Knox N, Kabakchiev B, Milgrom R, Kirsch R, Cohen Z, McLeod RS, Guttman DS, Krause DO, Silverberg MS. Characterization of the gut-associated microbiome in inflammatory pouch complications following ileal pouch-anal anastomosis. PLoS One. 2013;8(9):e66934. doi: 10.1371/journal.pone.0066934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, et al. 2013. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 58(5):949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim CH, Park J, Gut Microbiota-Derived KM. Short-chain fatty acids, t cells, and inflammation. Immune Netw. 2014;14(6):277–288. doi: 10.4110/in.2014.14.6.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, et al. 2012. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 142(1):46–54. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 116.Chen L, Wang W, Zhou R, Ng SC, Li J, Huang M, Zhou F, Wang X, Shen B, Kamm MA, et al. 2014. Characteristics of fecal and mucosa-associated microbiota in chinese patients with inflammatory bowel disease. Medicine. 93(8):e51. doi: 10.1097/MD.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human Intestinal Lumen and Mucosa-Associated Microbiota in Patients with Colorectal Cancer. PLoS One. 2012;7(6):e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Richard ML, Liguori G, Lamas B, Brandi G, da Costa G, Hoffmann TW, Di Simone MP, Calabrese C, Poggioli G, Langella P, et al. 2018. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes. 9(2):131–142. doi: 10.1080/19490976.2017.1379637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. 2008. Faecalibacterium Prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of crohn disease patients. Proc Natl Acad Sci U S A. 105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Benus RF, Harmsen HJ, Welling GW, Spanjersberg R, Zijlstra JG, Degener JE . 2010. van der Werf TS. Impact of Digestive and Oropharyngeal Decontamination On the Intestinal Microbiota in Icu Patients. Intens Care Med. 36(8):1394–1402. doi: 10.1007/s00134-010-1826-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rajilić Stojanović M, Biagi E, Heilig HGHJ, Kajander K, Kekkonen RA, Tims S, de Vos WM. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141(5):1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 122.Nishino K, Nishida A, Inoue R, Kawada Y, Ohno M, Sakai S, Inatomi O, Bamba S, Sugimoto M, Kawahara M, et al. 2018. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J Gastroenterol. 53(1):95–106. doi: 10.1007/s00535-017-1384-4. [DOI] [PubMed] [Google Scholar]
- 123.Luu TH, Michel C, Bard JM, Dravet F, Nazih H, Bobin-Dubigeon C. intestinal proportion of blautia sp. is associated with clinical stage and histoprognostic grade in patients with early-stage breast cancer. Nutr Cancer. 2017;69(2):267–275. doi: 10.1080/01635581.2017.1263750. [DOI] [PubMed] [Google Scholar]
- 124.Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microb. 2006;72(5):3593–3599. doi: 10.1128/aem.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sieber JR, McInerney MJ, Gunsalus RP. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol. 2012;66(1):429–452. doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- 126.Plichta DR, Juncker AS, Bertalan M, Rettedal E, Gautier L, Varela E, Manichanh C, Fouqueray C, Levenez F, Nielsen T, et al. 2016. Transcriptional interactions suggest niche segregation among microorganisms in the human gut. Nat Microbiol. 1(11). doi: 10.1038/nmicrobiol.2016.152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


