Introduction
Liquid biopsy in cancer is a revolutionary diagnostic concept defined by the analysis of biological material of tumor origin that extravasate to body fluids. Most common liquid biopsy studies use circulating tumor cells (CTCs) or circulating tumor-derived factors, in particular, circulating tumor DNA (ctDNA) (Alix-Panabières and Pantel, 2016; Perakis and Speicher, 2017). In the last 5 years, cutting-edge technologies such as next-generation sequencing (NGS) or digital PCR (dPCR) have been applied to detect blood-based, tumor-specific biomarkers such as CTCs and ctDNA (Husain and Velculescu, 2017; Pantel and Alix-Panabières, 2019). The quantification of circulating DNA molecules or CTCs showed per se prognostic value in many cancers (Haber and Velculescu, 2014). Besides this immediate analysis, another advantage of these biomarkers resides in the possibility of testing specific mutations, methylation profiles, and other DNA patterns (cfDNA and CTCs) and alternatively, proteins and the possibility of generating patient-derived xenografts (PDX) from the most aggressive cells in the tumor that putatively could initiate metastatic outgrowth (CTCs). Furthermore, the development of high sensitivity and specificity techniques enabled the identification of minimal residual disease (MRD) in cancer patient's follow-up blood samples (Pantel and Alix-Panabières, 2019). Complementary to these biomarkers, extracellular vesicles (EVs) are emerging as powerful biomarkers to provide information about the tumor and the systemic changes occurring during the disease. EVs are a broad and heterogenous group of vesicles secreted by almost any kind of cell that display a wide range of sizes (30 nm−5 μm in diameter Witwer and Théry, 2019) and are composed of a lipid bilayer enclosing nucleic acids, proteins, lipids, metabolites (Colombo et al., 2014). EVs are considered as a mechanism of cell-cell communication regulating paracrine and distal cell communication (Tkach and Théry, 2016). According to this, EVs have been detected in most biological fluids (Wiklander et al., 2019). The isolation of EVs allows for the subsequent analysis of their content that is defined by the cell of origin of the vesicle. Due to their heterogeneous content (protein, nucleic acids, lipids, metabolites, etc.), their ubiquitous production by body cells and detection in most biological fluids, circulating EVs could be useful for specific or multiplatform analyses to provide an accurate evaluation of cancer disease at early time points, during progression, therapy and post-treatment facilitating the detection of minimal residual disease and relapse anticipation (LeBleu and Kalluri, 2020).
EV-Shed DNA, Characteristics and Considerations
Since 2010, several studies have shown the presence of different DNA species in EVs such as ssDNA (Balaj et al., 2011), dsDNA (Kahlert et al., 2014; Thakur et al., 2014) and mtDNA (Cai et al., 2016; Lötvall et al., 2016). Although it is widely accepted that EVs carry DNA, the origins, localization and biological properties of this DNA are not entirely understood (Malkin and Bratman, 2020). Some works support that DNA secretion in EVs is an integral mechanism of clearing cytosolic DNA to maintain cellular homeostasis and avoid senescence and apoptosis (Takahashi et al., 2017). Another report suggests that DNA is a danger-associated signal secreted by dying cells promoting inflammation and anti-tumor immune responses (Kitai et al., 2017). A role for EV-associated DNA in priming and conferring protection to dendritic cells against pathogen infection has also been described (Torralba et al., 2018). Regarding the mechanism involved in DNA secretion on EVs, although microvesicles and exosomes have been proposed to carry DNA, a detailed analysis of the involved mechanism is missing. In a landmark study, Coffey and colleagues found that small EVs did not contain detectable amounts of dsDNA (Jeppesen et al., 2019). Surprisingly, they found that dsDNA was associated with non-vesicular particles that can be co-isolated in the same fraction than small EVs.
Regardless of their origin and mechanism, it appears clear that most of the EV-shed DNA is located on the outside surface of the EVs (Lazaro-Ibanez et al., 2019). However, after DNAse treatment, still a small proportion of DNA remains inside the vesicles (Thakur et al., 2014; Lazaro-Ibanez et al., 2019). Surface-associated DNA confers increased zeta-potential to vesicles, favors exosome-cell adhesion and contributes to the vesicle internalization in the target cell (Fischer et al., 2016; Lötvall et al., 2016; Németh et al., 2017).
Interestingly, all chromosomes are represented in EV-shed DNA with no specific regions overrepresented (Lazaro-Ibanez et al., 2019) suggesting the absence of a sequence-specific loading of DNA species in vesicles. Despite the lower amount of EV-enclosed DNA compared to EV surface-associated DNA, inner DNA displays a better quality and performance in NGS (Lazaro-Ibanez et al., 2019). Remarkably, compared to cfDNA, the reported average fragment length is longer in EVs. Thus, together with a nucleosome-associated pattern, fragments up to 4 kb are observed in exosome-derived DNA (Vagner et al., 2018; Lazaro-Ibanez et al., 2019; Malkin and Bratman, 2020) and chromosomal DNA has been found in large EVs such as oncosomes in prostate cells and in plasma of prostate cancer patients (Vagner et al., 2018).
Several works have reported that EV-shed DNA allows the detection of mutations that reliably reflects the mutational state in the tumor of origin (Kahlert et al., 2014; Thakur et al., 2014; Vagner et al., 2018; García-Romero et al., 2019; Garcia-Silva et al., 2019; Kunz et al., 2019). As a consequence of this, the use of EV-derived DNA for liquid biopsy purposes has begun to be exploited (Table 1). The first studies with plasma-derived EVs focused on the analysis of EGFR in lung cancer and glioblastoma (Figueroa et al., 2017; Krug et al., 2017; Castellanos-Rizaldos et al., 2018) and KRAS in pancreatic cancer (Allenson et al., 2017; Yang et al., 2017). More recently, predictive detection of BRAF mutation in circulating EV-DNA from pediatric central nervous system tumors has been described (García-Romero et al., 2019) together with the use of this mutation in seroma obtained post-lymphadenectomy in melanoma patients as a measure of minimal residual disease (Garcia-Silva et al., 2019). These and other studies demonstrate that the analysis of circulating EV-DNA can provide a prognostic value in diverse cancer types.
Table 1.
Summary of the main reports performing DNA-EV analyses for liquid biopsy tests.
| References | Body fluid | Tumor type | Method | EV type | DNAase | Yield | DNA size | Biomarkers/platform |
|---|---|---|---|---|---|---|---|---|
| Krug et al. (2017) | Plasma | NSCLC | Exo-Elution™ Plus extraction kit | Small and large EVs | No | ND | ND | EGFR T790M by NGS |
| Castellanos-Rizaldos et al. (2018) | Plasma | NSCLC | Exo-Elution™ Plus extraction kit | Small and large EVs | No | ND | ND | EGFR mutations by NGS |
| Allenson et al. (2017) | Plasma | Pancreatic cancer | Ultracentrifugation | Small EVs | No | ND | ND | KRAS mutations by ddPCR |
| Yang et al. (2017) | Serum | Pancreatic cancer | Ultracentrifugation | Small EVs | No | ND | ND | KRAS and TP53 mutations by ddPCR |
| San Lucas et al. (2016) | Pleural fluid | Pancreatic cancer | Ultracentrifugation | Small EVs | No | ND | >10kb | Tumor profiling by Illumina Hiseq2500 |
| Figueroa et al. (2017) | CSF | Glioblastoma | Ultracentrifugation | Small EVs | Yes | ND | ND | EGFRvIII mutation by QPCR |
| Garcia-Silva et al. (2019) | Lymphatic exudate | Melanoma | Exo-Elution™ Plus extraction kit | Small and large EVs | No | ND | ND | BRAF V600E by QPCR |
| Hur et al. (2019) | BALF | NSCLC | Ultracentrifugation | Small EVs | No | ND | ND | EGFR mutations by QPCR |
| Song et al. (2019) | Pleural effusion fluid | NSCLC | Exo-Elution™ Plus extraction kit | Small and large EVs | No | ND | ND | Targeted-NGS |
| Lee et al. (2018) | Urine | Bladder cancer | ExoQuick-TC | Small EVs | No | ND | ND | Mutation analysis by target capture sequencing /CNV by whole genome sequencing |
CFS, cerebrospinal fluid; BALF, bronchoalveolar lavage fluid; NSCLC, non-small cell lung cancer; PDAC, pancreatic ductal adenocarcinoma; ND, not determined.
An Overview of cfDNA-Based Liquid Biopsies
Compared to EV-DNA, cfDNA has been much more extensively used in liquid biopsies. cfDNA is DNA freely circulating in the bloodstream. It is estimated that circulating tumor DNA might only be <0.01% of the total cfDNA (Pantel and Alix-Panabières, 2019). It is mainly originated during the apoptotic process, although senescence and necrosis also influence cfDNA release kinetics (Rostami et al., 2020). As a consequence of its origin, it is a highly fragmented nucleic acid.
cfDNA-based liquid biopsy can be used to detect specific tumor aberrations such as point mutations, loss of heterozygosity (LOH) or amplifications (Kerachian et al., 2019). These specific tumor genetic alterations (or a panel of them) are analyzed by targeting DNA sequencing techniques such as digital PCR (dPCR), beads–emulsion–amplification–magnetics (BEAMing), safe-sequencing system (Safe-SeqS), cancer personalized profiling by deep sequencing (CAPP-Seq), and tagged-amplicon deep sequencing (TAmSeq) (Heitzer et al., 2019). Alternatively, a non-targeted next-generation sequencing (NGS), commonly a whole-genome/exome sequencing can also be applied to cfDNA in order to extract information about copy number alterations (CNAs), DNA rearrangements or detection of subclonal mutations. The biggest drawback of this approach is the lower overall sensitivity (above 1–5%) and the need for higher concentrations of ctDNA whereas targeted DNA sequencing techniques can reach a limit of detection (LOD) minor than 0.01% (Pantel and Alix-Panabières, 2019). However, ultra-sensitive detection in plasma cfDNA samples can be achieved by analyzing hundreds to thousands of mutations previously identified by tumor genotyping (Wan et al., 2020) or by using genome-wide mutational integration (Zviran et al., 2020).
The most frequent application of cfDNA-based liquid biopsy is the monitoring of therapy response and the analysis of minimal residual disease (MRD) in clinical samples in order to anticipate the relapse of the disease before its detection with current clinical techniques (e.g., CT or PET/CT, multiparametric flow cytometry, immunohistochemistry) (Cheng et al., 2019; Pantel and Alix-Panabières, 2019). In colon cancer, the positive detection of mutations in cfDNA efficiently demonstrated prognostic value in a cohort of 230 colorectal cancer patients in which cfDNA positive status was associated to inferior recurrence-free survival (Tie et al., 2016). In breast cancer, the use of ddPCR on cfDNA samples to validate previously identified chromosome rearrangements reached a sensitivity of 93% and a specificity of 100% (Olsson et al., 2015). Targeted capture sequencing analysis of ctDNA defined MRD-associated mutations and even predict the genetic events of subsequent metastatic relapse more accurately than sequencing of the primary breast cancer (Garcia-Murillas et al., 2019). In a landmark study in lung cancer, Swanton and colleagues developed a NGS-based mutational panel of 12–30 SNVs, with addition of clinical and radiological evaluations. The detection of SNVs in ctDNA seemed to correlate with clinical evidence of NSCLC relapse with a ctDNA detection threshold <0.1% (Abbosh et al., 2017). These and other studies (Reinert et al., 2016; Chaudhuri et al., 2017; Schøler et al., 2017) demonstrate how targeted and genome-wide DNA sequencing techniques provide an accurate information during disease monitoring.
cfDNA VS. EV-NA, Who is the Winner?
It should be first noticed that when defining blood-derived cfDNA it is unclear the physical state/source of it. It can be effectively circulating DNA, or circulating in association with protein complexes or even associated to EVs (Jahr et al., 2001; Thierry et al., 2016). Importantly, standard protocols for cfDNA isolation do not exclude EV-shed DNA although neither concentrate nor select for it as other approaches do.
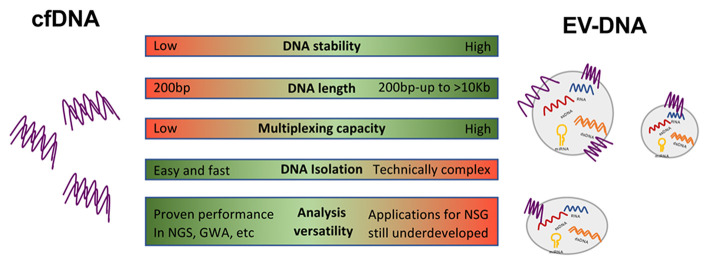
An overview of the differences between EV-DNA and cfDNA are summarized in Figure 1. EV-DNA is more stable than cfDNA whose half-life is estimated in <2 h (Cheng et al., 2016) forcing fast protocols of sample collection. On the contrary, EVs and their cargo show a considerable long-term stability in body fluids that facilitate their analysis in biobanked samples (Kalra et al., 2013). In addition, the fragmented nature of cfDNAs makes it difficult to generate a reliable genomic characterization for NGS, which in turn requires barcode and deep sequencing (Mouliere et al., 2011). On the other side, the presence of long EV-DNA fragments, that in cases display a chromatin structure favors amplification and better performance in NGS (Lazaro-Ibanez et al., 2019). Nevertheless, to date, most liquid biopsy studies focused on GWA and non-targeted sequencing have been performed in cfDNA such as methylation analyses (Sprang et al., 2020), DNA fragmentation patterns (Cristiano et al., 2019), or microbiome-DNA signatures (Poore et al., 2020) probably due to the delay in the application of those techniques to EV-based liquid biopsy.
Figure 1.
Advantages and disadvantages of EV-associated DNA and cfDNA. Circulating cell-free (cfDNA) shows a half-life of 2 h. However, extracellular vesicles (EVs) and associated content are stable for longer times. EVs display a wide range of DNA fragment sizes compared to highly fragmented cfDNA. Multiple EV cargo allows for polyvalent multiplexed analysis. DNA isolation is easy and fast for cfDNA but is technically more complex and time-consuming in the case of EVs. Non-targeted sequence-based analyses in EV-DNA are still underdeveloped while cfDNA has proven remarkable performance in next-generation sequencing techniques (NSG) and studies based in gene-wide analysis (GWA).
Another advantage of exosomes and other EVs-based liquid biopsy is the possibility of multiplexing analyses of DNA with other EV cargo such as miRNA, long-non coding RNA, proteins, etc., that can provide a highly accurate information about the disease and will facilitate personalized and time-shaped medicine (LeBleu and Kalluri, 2020).
Few studies provide insights about the comparison of both cfDNA and EV-DNA in plasma. In a cohort of 84 non-small cell lung cancer patients, the combined use of exosomal-RNA and ctDNA sequencing improved the detection of EGFR (epidermal growth factor receptor)-activating mutations up to 98 vs. 84% for ctDNA alone (Krug et al., 2017). Remarkably, the best improvement was observed in the group of patients with intrathoracic metastatic disease known to have low levels of cfDNA. Moreover, the combined use of EV-RNA/DNA (EV-NA) together with cfDNA overcame the limited abundance of the EGFR T790M mutation and other EGFR mutations and obtained improved sensitivity and specificity than cfDNA alone (Castellanos-Rizaldos et al., 2018, 2019). These studies highlight the enhanced performance of combining EV-associated nucleic acids together with cfDNA vs. cfDNA analysis alone.
Following with the improved performance of EV-DNA tests, in a recent study, the assessment of Rhesus D genotype and gender by coupling quantitative PCR to EV-DNA and cfDNA isolation from maternal blood gave successful results with both approaches although only EV-DNA achieved a 100% of sensitivity and specificity for both assays (Yaşa et al., 2020).
An important drawback in EV-DNA tests is the lack of standardized isolation methods compared to cfDNA that implies additional steps of optimization, that are technically more complex and required clinical validation (Théry et al., 2006; Royo et al., 2020).
There is Future Beyond Plasma! Use of Circulating EV in Other Biofluids
The secretion of EVs is a universal process and consequently EVs have been detected, isolated and analyzed in many biological fluids such as plasma, urine, cerebrospinal fluid (CSF) or bronchoalveolar lavage fluid (BALF) suggesting that these fluids can be explored for EV-based liquid biopsy tests (González and Falcón-Pérez, 2015; Gui et al., 2015; Broggi et al., 2019; Carnino et al., 2019; Kim et al., 2020). Until now, most of the studies based on other biofluids than blood have been focused on the quantification of exosomes or specific protein biomarkers. However, the analysis of mutations suggests that other fluids could be enriched in EV-associated DNA compared to plasma providing increase sensitivity. This is the case for lymphatic exudate also called exudative seroma in melanoma (Garcia-Silva et al., 2019). A combined approach isolating EV-associated nucleic acids together with cfDNA yield 600 times more copies of BRAFwt in this biofluid than in plasma. Thus, the limit of detection and sensitivity of the technique can be improved by choosing a fluid anatomically close to the tumor that would be enriched in tumor-derived EVs and consequently in EV-NA cargo. Another example of such an improvement is the analysis of CSF as an alternative to the scarce detection of biomarkers in blood for brain neoplasms. Thus, in glioblastoma patients, the use of CSF-derived EVs yield high specificity for the detection of tumor-associated EGFR amplifications (Figueroa et al., 2017). Additionally, profiling of BRAFV600E mutation in exudative seroma-derived EV-NA obtained after lymphadenectomy efficiently detected minimal residual disease and showed a strong prognostic value in a small cohort of stage III melanoma patients (Garcia-Silva et al., 2019). This study implies that malignancies such as breast cancer, melanoma, or others in which lymphadenectomy is performed could benefit from testing seroma-derived EV-NA for detection of minimal residual disease. It has been also proposed that NGS using EVs isolated from pleural fusions and urine could effectively replace tissue-based NGS in cases where there is a shortage of tissue (Lee et al., 2018; Song et al., 2019). Recent studies have demonstrated that EVs successfully isolated from BALF of lung cancer patients contain an abundant amount of dsDNA, and that liquid biopsy for EGFR genotyping using BALF is tissue-specific and extremely sensitive compared to cfDNA analysis (Hur et al., 2019; Kim et al., 2020). In summary, these works drive attention to the relevance of the selection of the ideal fluid as a source of EVs. The anatomical proximity to the tumor or disease and the intrinsic EV concentration certainly impact in the yield of the isolation process and consequently in the sensibility of the tests.
Concluding Remarks
Compared to other types of liquid biopsy biomarkers, EV-DNA tests are in their early steps. However, they could provide additional analytical power to more established diagnostic methods due to the relatively abundant cargo of nucleic acids displaying increased stability compared to cfDNA. Although a few studies suggest that EV-NA-based tests could perform slightly better than cfDNA tests, more comparative analyses will be required to evaluate EV-NA capacity to overcome or complement cfDNA-based liquid biopsy. Combined EV-DNA/RNA with cfDNA approaches could amplify disease information and improved the specificity and the limit of detection in the tests, for example in the detection of some tumor mutations known to be underrepresented in cfDNA. On the other hand, there is a need for increased sensitivity that could boost non-invasive early detection and also enhance the analysis of minimal residual disease. Due to the heterogeneity of EV cargo, multiplexed assays combining DNA analysis with additional information on other biomolecules could also serve for a more personalized medicine. In addition, alternative biofluids are coming on the scene demonstrating improved performance than blood-based tests due to their enrichment in tumor-specific EVs. Finally, together with targeted-sequencing techniques, the development of diverse types of GWA and the capacities of machine learning to combine multiple layers of information, will definitively expand the applicability of EV-DNA-based liquid biopsy.
Author Contributions
HP and SG-S conceived and designed the manuscript. SG-S, MG, and HP wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the reviewer and editor for their helpful comments.
Footnotes
Funding. The authors gratefully acknowledged the support of the following sources of funding: H.P. RETOS SAF2017-82924-R (AEI/10.13039/501100011033/FEDER-UE), Fundación AECC: LABAE19027PEIN, Fundación Ramón Areces, and Translational NeTwork for the CLinical application of Extracellular VesicleS, TeNTaCLES, RED2018-102411-T (AEI/10.13039/501100011033). MC contra el Cancer foundation, ISCIII (CP19/00140, PI18/00295).
References
- Abbosh C., Birkbak N. J., Wilson G. A., Jamal-Hanjani M., Constantin T., Salari R., et al. (2017). Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451. 10.1038/nature22364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix-Panabières C., Pantel K. (2016). Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 6, 479–491. 10.1158/2159-8290.cd-15-1483 [DOI] [PubMed] [Google Scholar]
- Allenson K., Castillo J., San Lucas F. A., Scelo G., Kim D. U., Bernard V., et al. (2017). High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 28, 741–747. 10.1093/annonc/mdx004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaj L., Lessard R., Dai L., Cho Y. J., Pomeroy S. L., Breakefield X. O., et al. (2011). Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2:180. 10.1038/ncomms1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broggi M. A. S., Maillat L., Clement C. C., Bordry N., Corthesy P., Auger A., et al. (2019). Tumor-associated factors are enriched in lymphatic exudate compared to plasma in metastatic melanoma patients. J. Exp. Med. 216, 1091–1107. 10.1084/jem.20181618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Wu G., Jose P. A., Zeng C. (2016). Functional transferred DNA within extracellular vesicles. Exp. Cell Res. 349, 179–183. 10.1016/j.yexcr.2016.10.012 [DOI] [PubMed] [Google Scholar]
- Carnino J. M., Lee H., Jin Y. (2019). Isolation and characterization of extracellular vesicles from Broncho-alveolar lavage fluid: a review and comparison of different methods. Respir. Res. 20:240. 10.1186/s12931-019-1210-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos-Rizaldos E., Grimm D. G., Tadigotla V., Hurley J., Healy J., Neal P. L., et al. (2018). Exosome-based detection of EGFR T790M in plasma from non-small cell lung cancer patients. Clin. Cancer Res. 24, 2944–2950. 10.1158/1078-0432.CCR-17-3369 [DOI] [PubMed] [Google Scholar]
- Castellanos-Rizaldos E., Zhang X., Tadigotla V. R., Grimm D. G., Karlovich C., Raez L. E., et al. (2019). Exosome-based detection of activating and resistance EGFR mutations from plasma of non-small cell lung cancer patients. Oncotarget 10, 2911–2920. 10.18632/oncotarget.26885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri A. A., Chabon J. J., Lovejoy A. F., Newman A. M., Stehr H., Azad T. D., et al. (2017). Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 7, 1394–1403. 10.1158/2159-8290.cd-17-0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Su L., Qian C. (2016). Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget 7, 48832–48841. 10.18632/oncotarget.9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F. T., Lapke N., Wu C. C., Lu Y. J., Chen S. J., Yu P. N., et al. (2019). Liquid biopsy detects relapse five months earlier than regular clinical follow-up and guides targeted treatment in breast cancer. Case Rep. Oncol. Med. 2019:6545298. 10.1155/2019/6545298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Raposo G., Thery C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289. 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- Cristiano S., Leal A., Phallen J., Fiksel J., Adleff V., Bruhm D. C., et al. (2019). Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389. 10.1038/s41586-019-1272-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa J. M., Skog J., Akers J., Li H., Komotar R., Jensen R., et al. (2017). Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro Oncol. 19, 1494–1502. 10.1093/neuonc/nox085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S., Cornils K., Speiseder T., Badbaran A., Reimer R., Indenbirken D., et al. (2016). Indication of horizontal DNA gene transfer by extracellular vesicles. PLoS ONE 11:e0163665. 10.1371/journal.pone.0163665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Murillas I., Chopra N., Comino-Méndez I., Beaney M., Tovey H., Cutts R. J., et al. (2019). Assessment of molecular relapse detection in early-stage breast cancer. JAMA Oncol. 5, 1473–1478. 10.1001/jamaoncol.2019.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Romero N., Carrión-Navarro J., Areal-Hidalgo P., Ortiz de Mendivil A., Asensi-Puig A., Madurga R., et al. (2019). BRAF V600E detection in liquid biopsies from pediatric central nervous system tumors. Cancers 12:66. 10.3390/cancers12010066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Silva S., Benito-Martin A., Sanchez-Redondo S., Hernandez-Barranco A., Ximenez-Embun P., Nogues L., et al. (2019). Use of extracellular vesicles from lymphatic drainage as surrogate markers of melanoma progression and BRAF (V600E) mutation. J. Exp. Med. 216, 1061–1070. 10.1084/jem.20181522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E., Falcón-Pérez J. M. (2015). Cell-derived extracellular vesicles as a platform to identify low-invasive disease biomarkers. Expert Rev. Mol. Diagn. 15, 907–923. 10.1586/14737159.2015.1043272 [DOI] [PubMed] [Google Scholar]
- Gui Y., Liu H., Zhang L., Lv W., Hu X. (2015). Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 6, 37043–37053. 10.18632/oncotarget.6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber D. A., Velculescu V. E. (2014). Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 4, 650–661. 10.1158/2159-8290.cd-13-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer E., Haque I. S., Roberts C. E. S., Speicher M. R. (2019). Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 71–88. 10.1038/s41576-018-0071-5 [DOI] [PubMed] [Google Scholar]
- Hur J. Y., Lee J. S., Kim I. A., Kim H. J., Kim W. S., Lee K. Y. (2019). Extracellular vesicle-based EGFR genotyping in bronchoalveolar lavage fluid from treatment-naive non-small cell lung cancer patients. Transl. Lung Cancer Res. 8, 1051–1060. 10.21037/tlcr.2019.12.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain H., Velculescu V. E. (2017). Cancer DNA in the circulation: the liquid biopsy. JAMA 318, 1272–1274. 10.1001/jama.2017.12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahr S., Hentze H., Englisch S., Hardt D., Fackelmayer F. O., Hesch R. D., et al. (2001). DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61, 1659–1665. [PubMed] [Google Scholar]
- Jeppesen D. K., Fenix A. M., Franklin J. L., Higginbotham J. N., Zhang Q., Zimmerman L. J., et al. (2019). Reassessment of exosome composition. Cell 177, 428–445 e418. 10.1016/j.cell.2019.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert C., Melo S. A., Protopopov A., Tang J., Seth S., Koch M., et al. (2014). Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 289, 3869–3875. 10.1074/jbc.C113.532267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra H., Adda C. G., Liem M., Ang C. S., Mechler A., Simpson R. J., et al. (2013). Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 13, 3354–3364. 10.1002/pmic.201300282 [DOI] [PubMed] [Google Scholar]
- Kerachian M. A., Poudineh A., Thiery J. P. (2019). Cell free circulating tumor nucleic acids, a revolution in personalized cancer medicine. Crit. Rev. Oncol. Hematol. 144:102827. 10.1016/j.critrevonc.2019.102827 [DOI] [PubMed] [Google Scholar]
- Kim I. A., Hur J. Y., Kim H. J., Lee S. E., Kim W. S., Lee K. Y. (2020). Liquid biopsy using extracellular vesicle-derived DNA in lung adenocarcinoma. J. Pathol. Transl. Med. 54, 453–461. 10.4132/jptm.2020.08.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitai Y., Kawasaki T., Sueyoshi T., Kobiyama K., Ishii K. J., Zou J., et al. (2017). DNA-containing exosomes derived from cancer cells treated with topotecan activate a STING-dependent pathway and reinforce antitumor immunity. J. Immunol. 198, 1649–1659. 10.4049/jimmunol.1601694 [DOI] [PubMed] [Google Scholar]
- Krug A. K., Enderle D., Karlovich C., Priewasser T., Bentink S., Spiel A., et al. (2017). Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann. Oncol. 29:2143 10.1093/annonc/mdx765 [DOI] [PubMed] [Google Scholar]
- Kunz F., Kontopoulou E., Reinhardt K., Soldierer M., Strachan S., Reinhardt D., et al. (2019). Detection of AML-specific mutations in pediatric patient plasma using extracellular vesicle-derived RNA. Ann. Hematol. 98, 595–603. 10.1007/s00277-019-03608-y [DOI] [PubMed] [Google Scholar]
- Lazaro-Ibanez E., Lasser C., Shelke G. V., Crescitelli R., Jang S. C., Cvjetkovic A., et al. (2019). DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J. Extracell Vesicles 8:1656993. 10.1080/20013078.2019.1656993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu V. S., Kalluri R. (2020). Exosomes as a multicomponent biomarker platform in cancer. Trends Cancer 6, 767–774. 10.1016/j.trecan.2020.03.007 [DOI] [PubMed] [Google Scholar]
- Lee D. H., Yoon H., Park S., Kim J. S., Ahn Y. H., Kwon K., et al. (2018). Urinary exosomal and cell-free DNA detects somatic mutation and copy number alteration in urothelial carcinoma of bladder. Sci. Rep. 8:14707. 10.1038/s41598-018-32900-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötvall G. S., Jang S. C., Yin Y., Lässer C., Lötvall J. (2016). Human mast cells release extracellular vesicle-associated-DNA. Matters. 10.19185/matters.201602000034 [DOI] [Google Scholar]
- Malkin E. Z., Bratman S. V. (2020). Bioactive DNA from extracellular vesicles and particles. Cell Death Dis. 11:584. 10.1038/s41419-020-02803-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouliere F., Robert B., Arnau Peyrotte E., Del Rio M., Ychou M., Molina F., et al. (2011). High fragmentation characterizes tumour-derived circulating DNA. PLoS ONE 6:e23418. 10.1371/journal.pone.0023418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh A., Orgovan N., Sódar B. W., Osteikoetxea X., Pálóczi K., Szabó-Taylor K., et al. (2017). Antibiotic-induced release of small extracellular vesicles (exosomes) with surface-associated DNA. Sci. Rep. 7:8202. 10.1038/s41598-017-08392-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson E., Winter C., George A., Chen Y., Howlin J., Tang M. H., et al. (2015). Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol. Med. 7, 1034–1047. 10.15252/emmm.201404913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel K., Alix-Panabières C. (2019). Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat. Rev. Clin. Oncol. 16, 409–424. 10.1038/s41571-019-0187-3 [DOI] [PubMed] [Google Scholar]
- Perakis S., Speicher M. R. (2017). Emerging concepts in liquid biopsies. BMC Med. 15:75. 10.1186/s12916-017-0840-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poore G. D., Kopylova E., Zhu Q., Carpenter C., Fraraccio S., Wandro S., et al. (2020). Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 579, 567–574. 10.1038/s41586-020-2095-1 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Reinert T., Schøler L. V., Thomsen R., Tobiasen H., Vang S., Nordentoft I., et al. (2016). Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 65, 625–634. 10.1136/gutjnl-2014-308859 [DOI] [PubMed] [Google Scholar]
- Rostami A., Lambie M., Yu C. W., Stambolic V., Waldron J. N., Bratman S. V. (2020). Senescence, necrosis, and apoptosis govern circulating cell-free DNA release kinetics. Cell Rep. 31:107830. 10.1016/j.celrep.2020.107830 [DOI] [PubMed] [Google Scholar]
- Royo F., Théry C., Falcón-Pérez J. M., Nieuwland R., Witwer K. W. (2020). Methods for separation and characterization of extracellular vesicles: results of a worldwide survey performed by the ISEV rigor and standardization subcommittee. Cells 9:1955. 10.3390/cells9091955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Lucas F. A., Allenson K., Bernard V., Castillo J., Kim D. U., Ellis K., et al. (2016). Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann. Oncol. 27, 635–641. 10.1093/annonc/mdv604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schøler L. V., Reinert T., Ørntoft M. W., Kassentoft C.G., Árnadóttir S. S., Vang S., et al. (2017). Clinical implications of monitoring circulating tumor dna in patients with colorectal cancer. Clin. Cancer Res. 23, 5437–5445. 10.1158/1078-0432.ccr-17-0510 [DOI] [PubMed] [Google Scholar]
- Song Z., Cai Z., Yan J., Shao Y. W., Zhang Y. (2019). Liquid biopsies using pleural effusion-derived exosomal DNA in advanced lung adenocarcinoma. Transl. Lung Cancer Res. 8, 392–400. 10.21037/tlcr.2019.08.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang M., Paret C., Faber J. (2020). CpG-islands as markers for liquid biopsies of cancer patients. Cells 9:1820. 10.3390/cells9081820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A., Okada R., Nagao K., Kawamata Y., Hanyu A., Yoshimoto S., et al. (2017). Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 8:15287. 10.1038/ncomms15287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur B. K., Zhang H., Becker A., Matei I., Huang Y., Costa-Silva B., et al. (2014). Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 24, 766–769. 10.1038/cr.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Amigorena S., Raposo G., Clayton A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 3:Unit 3.22. 10.1002/0471143030.cb0322s30 [DOI] [PubMed] [Google Scholar]
- Thierry A. R., El Messaoudi S., Gahan P. B., Anker P., Stroun M. (2016). Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 35, 347–376. 10.1007/s10555-016-9629-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie J., Wang Y., Tomasetti C., Li L., Springer S., Kinde I., et al. (2016). Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 8:346–392. 10.1126/scitranslmed.aaf6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach M., Théry C. (2016). Communication by extracellular vesicles: where we are and where we need to go. Cell 164, 1226–1232. 10.1016/j.cell.2016.01.043 [DOI] [PubMed] [Google Scholar]
- Torralba D., Baixauli F., Villarroya-Beltri C., Fernández-Delgado I., Latorre-Pellicer A., Acín-Pérez R., et al. (2018). Priming of dendritic cells by DNA-containing extracellular vesicles from activated T cells through antigen-driven contacts. Nat. Commun. 9:2658. 10.1038/s41467-018-05077-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner T., Spinelli C., Minciacchi V. R., Balaj L., Zandian M., Conley A., et al. (2018). Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell Vesicles 7:1505403. 10.1080/20013078.2018.1505403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J. C. M., Heider K., Gale D., Murphy S., Fisher E., Mouliere F., et al. (2020). ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci. Transl. Med. 12:aaz8084. 10.1126/scitranslmed.aaz8084 [DOI] [PubMed] [Google Scholar]
- Wiklander O. P. B., Brennan M., Lötvall J., Breakefield X. O., El Andaloussi S. (2019). Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 11:eaav8521. 10.1126/scitranslmed.aav8521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer K. W. Théry C. Extracellular vesicles or exosomes? On primacy, precision, popularity influencing a choice of nomenclature. J. Extracell. Vesic. (2019) 8:1648167 10.1080/20013078.2019.1648167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Che S. P., Kurywchak P., Tavormina J. L., Gansmo L. B., Correa de Sampaio P., et al. (2017). Detection of mutant KRAS and TP53 DNA in circulating exosomes from healthy individuals and patients with pancreatic cancer. Cancer Biol. Ther. 18, 158–165. 10.1080/15384047.2017.1281499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaşa B., Sahin O., Öcüt E., Seven M., Sözer S. (2020). Assessment of fetal rhesus D and gender with cell-free DNA and exosomes from maternal blood. Reprod. Sci. 10.1007/s43032-020-00321-4 [DOI] [PubMed] [Google Scholar]
- Zviran A., Schulman R. C., Shah M., Hill S. T. K., Deochand S., Khamnei C. C., et al. (2020). Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 26, 1114–1124. 10.1038/s41591-020-0915-3 [DOI] [PMC free article] [PubMed] [Google Scholar]