Abstract
Abstract
Corona Virus Disease (COVID-19) has spread globally quickly, and has resulted in a large number of causalities and medical resources insufficiency in many countries. Reverse-transcriptase polymerase chain reaction (RT-PCR) testing is adopted as biopsy tool for confirmation of virus infection. However, its accuracy is as low as 60-70%, which is inefficient to uncover the infected. In comparison, the chest CT has been considered as the prior choice in diagnosis and monitoring progress of COVID-19 infection. Although the COVID-19 diagnostic systems based on artificial intelligence have been developed for assisting doctors in diagnosis, the small sample size and the excessive time consumption limit their applications. To this end, this paper proposed a diagnosis prototype system for COVID-19 infection testing. The proposed deep learning model is trained and is tested on 2267 CT sequences from 1357 patients clinically confirmed with COVID-19 and 1235 CT sequences from non-infected people. The main highlights of the prototype system are: (1) no data augmentation is needed to accurately discriminate the COVID-19 from normal controls with the specificity of 0.92 and sensitivity of 0.93; (2) the raw DICOM image is not necessary in testing. Highly compressed image like Jpeg can be used to allow a quick diagnosis; and (3) it discriminates the virus infection within 6 seconds and thus allows an online test with light cost. We also applied our model on 48 asymptomatic patients diagnosed with COVID-19. We found that: (1) the positive rate of RT-PCR assay is 63.5% (687/1082). (2) 45.8% (22/48) of the RT-PCR assay is negative for asymptomatic patients, yet the accuracy of CT scans is 95.8%. The online detection system is available: http://212.64.70.65/covid.
Graphic Abstract
Keywords: Coronavirus disease (COVID-19), Computed tomography, Deep learning, Computer-aided efficient detection, Diagnosis model, Asymptomatic patients
Introduction
The outbreak of novel coronavirus 2019 (COVID-19) has rapidly spread to almost every country in the world [9]. To date (SEP 9th 2020), there have been 27, 775, 319 confirmed cases and 902, 665 death cases all around the world. This epidemic has caused the global health organizations to raise concern for public health due to coronavirus outbreak and to seek viable methods for detecting COVID-19 as soon as possible.
Currently, the most common diagnosis of COVID-19 is still based on real-time reverse-transcriptase polymerase chain reaction (RT-PCR), and is once adopted as golden standard for confirmation of infection. However, the sensitivity if RT-PCR is unsatisfying with the lowest accuracy of only 60% [2, 10, 17]. Therefore, the current solution is to conduct multiple tests in consecutive time to increase the true positive ratio of the test. On the contrary, medical-imaging-based methods such as chest computed tomography (CT) and Chest X-ray (CXR), can be a more reliable way of confirming positive COVID-19 patients.
On the one hand, CXR imaging is inexpensive, swift and universal but unable to detect acute pneumonia in previous studies [4, 6, 28]. On the other hand, CT imaging is now clinically adoptable as the principal way to confirm positive or suspected-positive COVID-19 cases [11]. Early studies [12] have analyzed and compared both the effectiveness and limitation of both methods in the diagnosis and management of COVID-19 patients. [13] has analyzed 1099 confirmed COVID-19 inpatients and outpatients in the hospital. During the CT scan when the patients are admitted to the hospital, 86.2% showed abnormal symptoms. The most common symptoms of COVID-19 patients from CT image are ground glass opacity (56.4%) and bilateral patchy shadows (51.8%). The study manifested that a patient with pneumonia due to COVID-19 may show lung abnormalities on chest CT, yet the initial RT-PCR test is not necessary positive. Chest CT achieves higher sensitivity for diagnosis of COVID-19 as compared with initial RT-PCR from pharyngeal swab samples. It has been reported that the sensitivity of chest CT in suggesting COVID-19 is 97% [2].
Nevertheless, non-uniformly results by CT and RT-PCR results are commonly observed, especially on patients recovered from the virus inflection, who are shown to have negative RT-PCR results. To alleviate the problem, CT examination is shown to be necessary for diagnosis. According to the latest guideline of Diagnosis and Treatment of Pneumonitis Caused by 2019-nCoV (trial 7th version) published by the Chinese government [25], if the imaging features are discovered by CT during an early stage, it can be confirmed as a clinically diagnosed case. Besides, recent studies [3, 7, 26] also rely on the chest CT to develop accurate and efficient method for diagnosing COVID-19.
Most of the recent works have employed deep learning models identify and localize the areas infected by COVID-19 [1, 5, 8, 15, 20, 22–24, 30]. Among them, Wang et al. [29] proposed a method based on the inception transfer learning to detect the patients with COVID-19, which extracts COVID-19’s graphical features and the external testing data accuracy is . The team collected 1065 CT images, including 325 images with COVID-19 and 740 images with the other typical viral pneumonia. Fu et al. [12] have trained the model on ResNet network, which obtains the accuracy of . The study collected five types of images, including COVID-19 pneumonia (50 cases), non-COVID-19 viral pneumonia (52 cases), bacterial pneumonia (53 cases), pulmonary tuberculosis (54 cases), and normal lung (100 cases). Zheng et al. [34] developed a detection for COVID-19 deep learning approach with weak label, which utilizes 3D CT volumes to predict the probability of COVID-19 infectious and the commendable performance benefits from the 3D deep neural network. The authors have trained 499 CT volumes and the accuracy is . Ying et al. [33] first extracted the main regions of lungs and then designed a Details Relation Extraction neural network (DER-Net) to train the model. Finally, the team developed a deep learning-based CT diagnosis system with the detection time of 30s. 88 chest CT scans patients with COVID-19, 101 patients infected with bacteria pneumonia and 86 healthy persons are collected to finish the classification task. Jin et al. [16] presented an artificial intelligence (AI) method via CT imaging for assisting the screening of COVID-19. This research reduced the burden of radiologists and implemented a model for segmentation and classification simultaneously. Li et al. [21] developed an framework to detect patients with COVID-19. Their works have employed the RT-PCR as the ground truth even if the rule is unreasonable with lower accuracy. More recently, Shi et al. [27] reviewed the artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. The study enrolled patients with COVID-19 and patients, suffering from other diseases, including bacterial pneumonia, viral pneumonia, Influenza-A, community-acquired pneumonia, non-pneumonia and Normal. Finally, their study reported the classification accuracies by several popular deep learning models.
On the whole, the aforementioned methods suffer from the following deficiencies: 1) The training dataset consists of a small sample size. For example, there are only 275 scanned positive with COVID-19 used in [32]. Although data augmentation techniques are adopted, the training model is inclined to be over-fitting, which in term will influence the capability of generalization; 2) Unbalanced training samples will bias the classifier to the category with significantly more samples; 3) The time required for diagnosis is often lengthy.
To facilitate online testing of COVID-19 symptoms through CT images, this paper proposes a deep learning method to classify and localize areas infected by coronavirus in chest CT images. The main contributions of the proposed method are as follows: 1) No data augmentation is required to accurately discriminate the COVID-19 from normal controls with the specificity of 0.92 and sensitivity of 0.93; 2) The raw DICOM image is not necessary for testing. A highly compressed image like Jpeg can be used to allow a quick diagnosis; 3) It discriminates against the virus infection within 6s and thus allows an online test with the lightest cost. Furthermore, we applied our model on 48 asymptomatic patients diagnosed with COVID-19; 4) On a large patient cohort, the proposed method achieves substantial performance.
The rest of the paper is organized as follows: in Sect. 2, the material and method of this study is introduced in detail; in Sect. 3, the classification performances of our model on CT images is evaluated; Conclusion is drawn in Sect. 4.
Materials and Methods
Training Data Acquisition on COVID-19 Patients and Non-COVID-19
The COVID-19 CT dataset utilized in this study are from Huangpi Hospital of Traditional Chinese Medicine, Wuhan, China. The dataset contains both medical imaging data and clinical data. Medical imaging data are presented as lung window CT of inhospitable patients. This retrospective study has been approved by the ethics committee of First Affiliated Hospital of Guangxi Medical University and the requirement for written informed consent was waived (Approval No. 2020 (KY-E-082)).
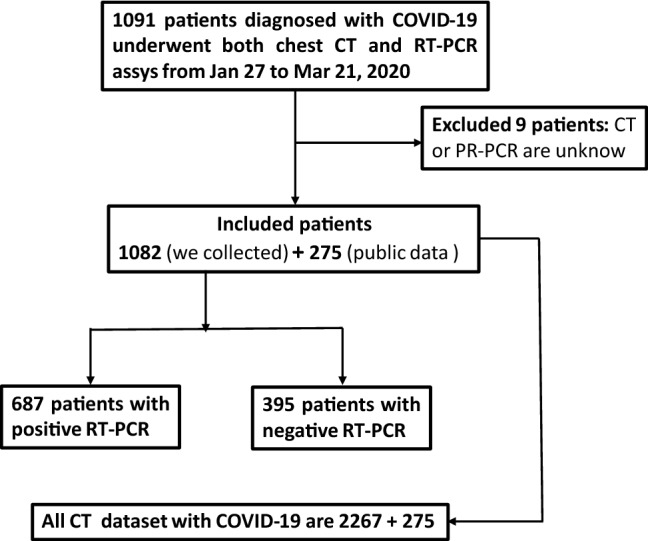
The dataset contains chest CT exams and clinical laboratory information from 1091 patients with COVID-19 between Jan 27, 2020 to Mar 21, 2020. The clinical symptoms involved temperature and blood pressure, fever, dry cough, fatigue, headache, chest distress, pharyngeal pain, etc. All laboratory results (including routine blood tests, high-sensitivity C-reactive protein measurement, antibody, etc.) were also recorded. All the patients were confirmed by RT-PCR. Characteristics of the included 1082 patients (exclude 9 patients of unknown age and gender) are indicated in Table 1.
Table 1.
Statistics on the collected dataset on 1082 COVID-19 patients
| Ages (years) | Number of Patients/Percentage |
|---|---|
| Mean age | 50.4 |
| 0-9 | 8 / 0.7% |
| 10-19 | 15/ 1.4% |
| 20-29 | 83/ 7.7% |
| 30-39 | 177/ 16.4% |
| 40-49 | 231/ 21.4% |
| 50-59 | 243/ 22.5% |
| 60-69 | 194/ 17.9% |
| 70-79 | 98/ 9.1% |
| 80-89 | 29/ 2.7% |
| 90-99 | 4/ 0.4% |
| Gender | – |
| Male | 584/ 54.0% |
| Female | 498/ 46.0% |
| RT-PCR assay | – |
| Positive | 687/ 63.5% |
| Negative | 395/ 36.5% |
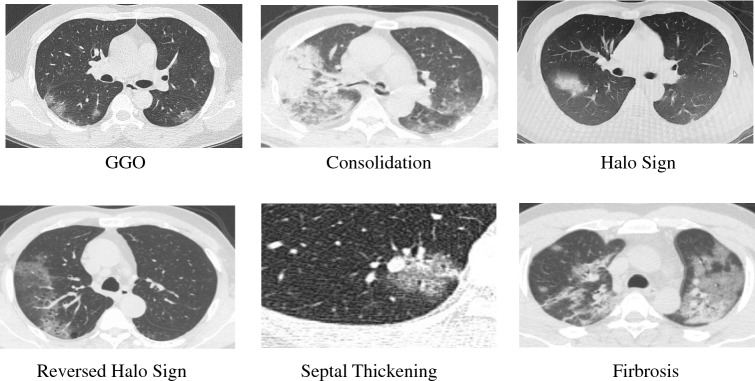
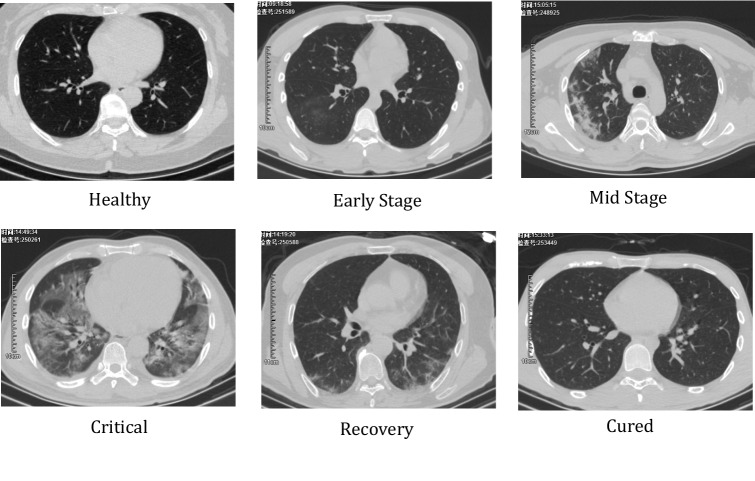
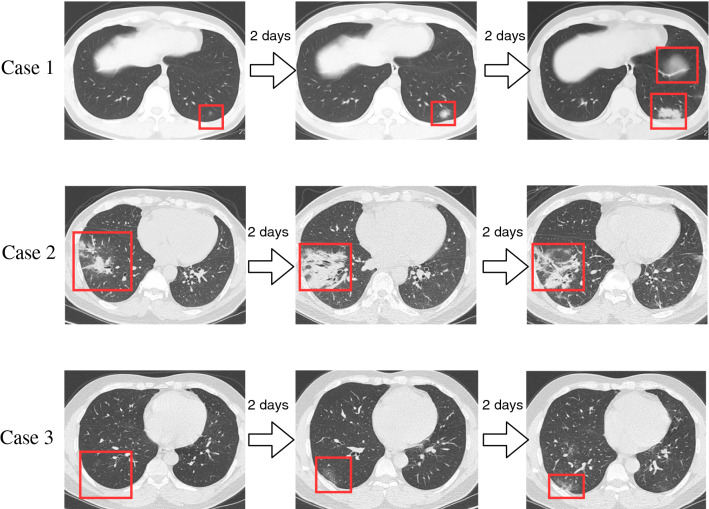
All chest CT examinations were conducted on GE BRIVO CT 325 or Siemens SOMATOM Definition Flash CT Scanners. The statistics on the collected CT images on different types are summarized in Fig. 1. Each volumetric exam contains about CT slices with a varying slice-thickness from 1.3mm to 3mm. The reconstruction matrix of CT image is pixels with in-plane pixel spatial resolution of . Every patient who is infected or suspected of being infected with covid-19 will undergo one or more lung CT scans during the course of the hospitalization. The average number of lung CT scans for each patient is 2.1, with a range from 1 to 5. In our experiment, we considered the CT images for each patient, taken in different time, as independent samples, because they independently represented the image characteristics a affected by COVID-19, as shown in Fig. 2. The training and testing dataset are divided disjointedly. Therefore, the CT image with COVID-19 in force is 2267. Figure 3 demonstrates the examples of three cases with COVID-19 scanning CT in three different periods (two days interval).
Fig. 1.
Illustrative examples on different image types, manifestoed in CT
Fig. 2.
Different stages of COVID-19
Fig. 3.
Three cases diagnosed with COVID-19. The figure intuitively indicates the change of the lesions of patients over time
Testing Data Collection on COVID-19 Patients and Non-COVID-19
To verify the robustness of the model, two independent datasets are collected and tested. The first dataset is reported and analyzed in [32], which contains 275 positive cases and 195 normal cases. The second dataset is collected from the radiology department in the First Affiliated Hospital of Guangxi Medical University hospitals, Guangxi, China. The chest CT examinations were conducted on the GE LightSpeed VCT Scanner, GE’s Revolution CT 256-Slice Scanner, Siemens SOMATOM Definition Flash CT Scanner or Siemens SOMATOM Perspective CT Scanners. The scanning parameters are as follows: the reconstruction matrix of lung window, ; slice-thickness, 1.3mm-2mm. It consists of 40 normal samples in the CT examination physical examination for COVID-19.
The final dataset consists of 1357 () exams for positive samples and 1235 () negative samples (i.e. Non-COVID-19).
Classification COVID-19 Patients via Deep Learning Model
Image Preprocessing
Preprocessing is important to lower both the intensive computational cost of deep classification model during training and testing phase, as well as to facilitate realtime online diagnosis process. First, the raw DICOM images are compressed from 300 to 60 M, which drastically reduce the input size of the model. Second, we sorted the slices of each patient from top to bottom according to the serial number, and randomly selected 6 images from COVID-19 patients while 12 images from Non-COVID patient. Those slice covering the lung area are selected. Finally, all data is normalized to remove the variations caused by the multi-sourced collection. Fourthly, while ensuring the integrity of the lungs, we cropped each image up and down by 50 pixels to reduce the impact of the background. The reason why we do not perform lung segmentation on the selected images is to avoid removing the lesion area at the lung boundaries. Finally, we resized the image to as input for the base-model which is ResNet50.
Deep Learning Model
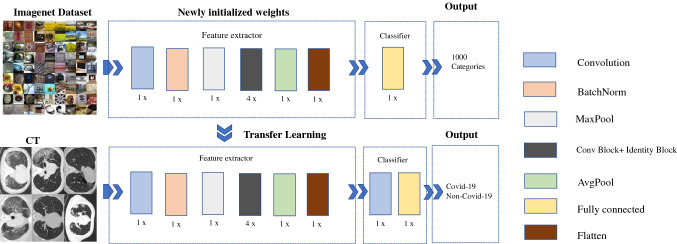
Our model is developed on the Keras platform based on ResNet50 architecture. To alleviate the tendency of over-fitting, to speed up the convergence and to improve the robustness of the model, the model is pre-trained on the ImageNet dataset and then finetuned in our dataset. The proposed model contains a feature extractor and a classifier. In particular, the structure of the feature extractor remains unchanged. The classifier is supplemented by an additional convolutional layer to reducing the output category to match our classes (COVID-19 and non-COVID-19). The network is retrained on imagenet for assuring a promising performance. Stochastic gradient descent (SGD) optimizer is employed for the optimizer and its learning rate is set at 0.001. To avoid over-fitting, we employed a dropout after the fully-connected layer. Moreover, the training is early stopped after 50 epochs. The architecture of the proposed model is exhibited in Fig. 4.
Fig. 4.
Architecture of the proposed model
Experiments
Once the deep learning model is trained, new test samples can be processed quickly. The average processing time for each CT slice is 9.857 milliseconds on a workstation (GPU NVIDIA GeForce GTX 1070 8 GB, RAM 48 GB, and Intel Core (TM) i7-7700 CPU 3.6GHz).
Experimental Setting and Performance Evaluation
The performance of the model is evaluated via training set, validation set and testing set. Table 2 indicates the distribution of the dataset. The distribution of the study population is demonstrated in Fig. 5.
Table 2.
Summary of training, validation and testing datasets
| Non-COVID-19 | COVID-19 | Total | |
|---|---|---|---|
| Training set | 600 | 1267 | 1867 |
| Validation set | 400 | 1000 | 1400 |
| Testing set | 235 | 275 | 510 |
Fig. 5.
Schematic diagram of the distribution of patients
Five widely used metrics, including the accuracy (ACC), sensitivity (SENS), specificity (SPE), intersection over union (IoU), the area under the receiver operating characteristic curve (AUC) and F1-score (F1), were employed to measure the performance of the method. These qualitative metrics were defined as follows:
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
| 6 |
where TP, FP, TN, FN, P, R M and N represented true positive, false positive, true negative and false negative, precision, recall, the number of positive and negative samples, respectively. We also plotted the receiver operating characteristic curve (ROC) for each method. Then estimate the area under the ROC curve (AUC).
Classification of COVID-19 from Normal Controls
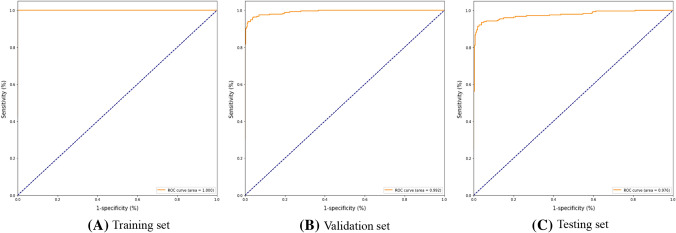
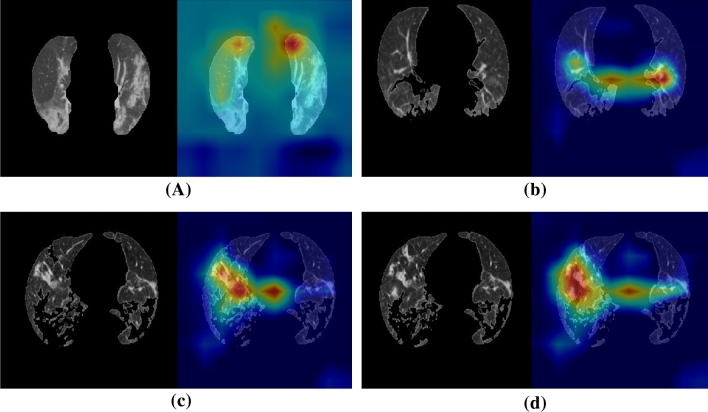
The experimental numerical results are illustrated in Table 3. The receiver operating characteristic (ROC) curve is plotted in Fig. 6. Four examples of the localization of the lesions are illustrated in Fig. 7.
Table 3.
The quantitative results in classification of COVID-19 and Non-COVID-19
| Metrics | VGG Tr./Va. | VGG Te. | GoogLeNet Tr./Va. | GoogLeNet Te. | Ours Tr./Va. | Ours Te. |
|---|---|---|---|---|---|---|
| ACC | 0.99/0.98 | 0.92 | 0.99/0.98 | 0.92 | 1.00/0.96 | 0.93 |
| SENS | 1.00/0.98 | 0.92 | 0.99/0.99 | 0.92 | 0.99/0.95 | 0.93 |
| SPE | 0.99/0.98 | 0.92 | 0.99/0.98 | 0.91 | 0.99/0.95 | 0.92 |
| IoU | 0.99/0.97 | 0.85 | 0.98/0.97 | 0.85 | 0.99/0.91 | 0.85 |
| F1 | 0.99/0.98 | 0.92 | 0.99/0.98 | 0.92 | 0.99/0.95 | 0.92 |
| AUC | 0.99/0.98 | 0.92 | 0.99/0.98 | 0.92 | 1.00/0.99 | 0.93 |
Comparison of Vgg16 with GoogLeNet and ResNet50.
*Tr. denotes the result on the training set.
*Va. denotes the result on the validation set.
*Te. denotes the result on the testing set
Fig. 6.
COVID-19 and Non-COVID-19 detection results evaluated via the receiver operating characteristic (ROC) curve
Fig. 7.
Four examples of the localization of the lesions for the COVID-19 patients
Recognition of the Asymptomatic Patients Confirmed as COVID-19
It has been reported that there are a rising number of asymptomatic patients confirmed with COVID-19 [18, 31]. According to the latest guideline, asymptomatic infection refers to those who have no relevant clinical symptoms (such as fever, cough, sore throat, etc.), yet are positive for RT-PCR or antibody tests in specimens such as the respiratory tract. Asymptomatic infections are not diagnosed patients, because they are not new infections. Asymptomatic people can also become a source of infection, with a certain risk of transmission.
The presence of unilateral ground-glass opacities in asymptomatic patients with COVID-19 pneumonia, in other words, asymptomatic patients probably exit CT changes before symptom onset [19]. This finding suggests that CT is a sensitive modality with which to detect COVID-19 pneumonia, even in asymptomatic. Therefore, CT images of COVID-19 asymptomatic patients have certain imaging characteristics, and chest CT scans have important diagnostic value in the close contact with infected people.
In some patients, it may be negative for the RT-PCR, whereas early on the CT tomography typical lung consolidation [14]. Consequently, a comparative experiment about the detection accuracy between RT-PCR and CT imaging is performed. Experimental summary of asymptomatic patients with COVID-19 as shown in Table. 4.
Table 4.
Summary of asymptomatic patients with COVID-19
| Detection method | of Patients (number) | Total | Accuracy | ||
|---|---|---|---|---|---|
| Positive | Negative | ||||
| CT image | Positive | 46 | 0 | 46 | 0.96 |
| Negative | 2 | 0 | 2 | ||
| RT-PCR | Positive | 26 | 0 | 26 | 0.54 |
| Negative | 22 | 0 | 22 | ||
Conclusion and Discussion
This paper proposes a classification and localization method for CoronaVirus infected areas on a large patient cohort. The proposed method has the following vital advantages compared to the previous related works. (1) No data augmentation is needed to accurately discriminate the COVID-19 from normal controls with the specificity of 0.92 and sensitivity of 0.93; (2) The raw DICOM image is compressed to allow a quicker diagnosis procedure and online testing; (3) On 48 asymptomatic patients diagnosed with COVID-19, our method has shown substantial performance.
Extensive experiments indicate that: (1) The positive rates of RT-PCR assay in our collected patients are 63.5% (687/1082); (2) With the diagnosis results of clinical indicators, The joint detection of RT-PCR and CT as ground truth, the classification results for COVID-19 and Non-COVID-19 that the accuracy, sensitivity, specificity, IoU, F1-score and AUC are 93%, 93%, 92%, 85%, 92% and 93% respectively; (3) For 48 asymptomatic patients, 45.8% (22/48) of the RT-PCR assay is negative in early diagnosis, and the accuracy of CT scans is 95.8% (46/48).
In our further work, we aim to detect asymptomatic patients. Statistical analysis of clinical indicators of asymptomatic patients, combined with imaging characteristics to find the differences between such patients and symptomatic patients.
Acknowledgements
This work was partially supported by the Key-Area Research and Development of Guangdong Province under Grant (2020B010166002, 2020B1111190001), the National Natural Science Foundation of China (61472145, 61771007), Guangdong Natural Science Foundation (2017A030312008), and the Health & Medical Collaborative Innovation Project of Guangzhou City (201803010021, 202002020049).
Compliance with ethical standards
Conflict of interest
Each author have participated in (a) concept and design, or analysis and mining of the data; (b) drafting the paper or revising it critically for important intellectual content; and (c) approval of the final version. The manuscript has not been submitted to another journal or other publication, nor has it been reviewed here. The author has no relationship with any organization that has direct or indirect economic interest in the subject discussed in this article. All authors are associated with organizations that have direct or indirect economic interest in the topics discussed in this article.
Footnotes
Ziwei Zhu, Guihua Tao and Tingting Dan have contributed equally to this study and are considered as joint first authors.
Contributor Information
Zhang Xingming, Email: cszxm@scut.edu.cn.
Hongmin Cai, Email: hmcai@scut.edu.cn.
References
- 1.A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering, (2020). 10.1016/j.eng.2020.04.010 [DOI] [PMC free article] [PubMed]
- 2.Ai Tao, Yang Zhenlu, Hou Hongyan, Zhan Chenao, Chen Chong, Lv Wenzhi, Tao Qian, Sun Ziyong, Xia Liming. Correlation of chest ct and rt-pcr testing for coronavirus disease 2019 (covid-19) in china: A report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arabi Sina, Vaseghi Golnaz, Heidari Zahra, Shariati Laleh, Amin Bahareh, Rashid Harunor, Haghjooy Javanmard haghayegh (2020) Clinical characteristics of covid-19 infection in pregnant women: a systematic review and meta-analysis. medRxiv. 10.1101/2020.04.05.20053983
- 4.Cai H, Yang Z, Cao X, Xia W, Xu X. A new iterative triclass thresholding technique in image segmentation. IEEE Transactions on Image Processing. 2014;23(3):1038–1046. doi: 10.1109/TIP.2014.2298981. [DOI] [PubMed] [Google Scholar]
- 5.Chen Jun, Wu Lianlian, Zhang Jun, Zhang Liang, Gong Dexin, Zhao Yilin, Hu Shan, Wang Yonggui, Hu Xiao, Zheng Biqing, Zhang Kuo, Wu Huiling, Dong Zehua, Xu Youming, Zhu Yijie, Chen Xi, Yu Lilei, Yu Honggang (2020) Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. medRxiv. 10.1101/2020.02.25.20021568 [DOI] [PMC free article] [PubMed]
- 6.Cheng X, Cai Hongmin, Zhang Yue, Xu Bo, Su Weifeng (2015) Optimal combination of feature selection and classification via local hyperplane based learning strategy. BMC Bioinformatics, 16. 10.1186/s12859-015-0629-6 [DOI] [PMC free article] [PubMed]
- 7.Chung Michael, Bernheim Adam, Mei Xueyan, Zhang Ning, Huang Mingqian, Zeng Xianjun, Cui Jiufa, Wenjian Xu, Yang Yang, Fayad Zahi A, Jacobi Adam, Li Kunwei, Li Shaolin, Shan Hong. Ct imaging features of 2019 novel coronavirus (2019-ncov) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Souza Tiago H, Nadal José A, Nogueira Roberto J N, Pereira Ricardo M, Brandão Marcelo B. Clinical manifestations of children with covid-19: A systematic review. Pediatric Pulmonology. 2020;55(8):1892–1899. doi: 10.1002/ppul.24885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Director WHO. General’s opening remarks at the media briefing on covid-19-11 March 2020. Geneva: World Health Organization; 2020. [Google Scholar]
- 10.Fang Yicheng, Zhang Huangqi, Xie Jicheng, Lin Minjie, Ying Lingjun, Pang Peipei, Ji Wenbin. Sensitivity of chest ct for covid-19: Comparison to rt-pcr. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leiwen Fu, Wang Bingyi, Yuan Tanwei, Chen Xiaoting, Ao Yunlong, Fitzpatrick Thomas, Li Peiyang, Zhou Yiguo, Zou Huachun. Clinical characteristics of coronavirus disease 2019 (covid-19) in china: A systematic review and meta-analysis. Journal of Infection. 2020;80(6):656–665. doi: 10.1016/j.jinf.2020.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Min, Yi Shuang-Lian, Zeng Yuanfeng, Ye Feng, Li Yuxuan, Dong Xuan, Ren Yan-Dan, Luo Linkai, Pan Jin-Shui, Zhang Qi (2020) Deep learning-based recognizing covid-19 and other common infectious diseases of the lung by chest ct scan images. medRxiv. 10.1101/2020.03.28.20046045
- 13.Guan Wei-jie, Zheng-yi Ni YuHu, Liang Wen-hua, Chun-quan Ou, He Jian-xing, Liu Lei, Shan Hong, Lei Chun-liang, Hui David SC, Bin Du, Li Lan-juan, Zeng Guang, Yuen Kwok-Yung, Chen Ru-chong, Tang Chun-li, Wang Tao, Chen Ping-yan, Xiang Jie, Li Shi-yue, Wang Jin-lin, Liang Zi-jing, Peng Yi-xiang, Wei Li, Liu Yong, Ya-hua Hu, Peng Peng, Wang Jian-ming, Liu Ji-yang, Chen Zhong, Li Gang, Zheng Zhi-jian, Qiu Shao-qin, Luo Jie, Ye Chang-jiang, Zhu Shao-yong, Zhong Nan-shan. Clinical characteristics of coronavirus disease 2019 in china. New England Journal of Medicine. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Peikai, Liu Tianzhu, Huang Lesheng, Liu Hailong, Lei Ming, Wangdong Xu, Xiaolu Hu, Chen Jun, Liu Bo. Use of chest ct in combination with negative rt-pcr assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 2020;295(1):22–23. doi: 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin Cheng, Chen Weixiang, Cao Yukun, Xu Zhanwei, Tan Zimeng, Zhang Xin, Deng Lei, Zheng Chuansheng, Zhou Jie, Shi Heshui, Feng Jianjiang (2020) Development and evaluation of an ai system for covid-19 diagnosis. medRxiv. 10.1101/2020.03.20.20039834 [DOI] [PMC free article] [PubMed]
- 16.Jin Shuo, Wang Bo, Xu Haibo, Luo Chuan, Wei Lai, Zhao Wei, Hou Xuexue, Ma Wenshuo, Xu Zhengqing, Zheng Zhuozhao, Sun Wenbo, Lan Lan, Zhang Wei, Mu Xiangdong, Shi Chenxi, Wang Zhongxiao, Lee Jihae, Jin Zijian, Lin Minggui, Jin Hongbo, Zhang Liang, Guo Jun, Zhao Benqi, Ren Zhizhong, Wang Shuhao, You Zheng, Dong Jiahong, Wang Xinghuan, Wang Jianming, Xu Wei (2020) Ai-assisted ct imaging analysis for covid-19 screening: Building and deploying a medical ai system in four weeks. medRxiv. 10.1101/2020.03.19.20039354
- 17.Kanne Jeffrey P, Little Brent P, Chung Jonathan H, Elicker Brett M, Ketai Loren H. Essentials for radiologists on covid-19: An update–radiology scientific expert panel. Radiology. 2020;296(2):E113–E114. doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Gwang-un, Kim Min-Jae, Ra Sang Hyun, Lee Jeongsoo, Bae Seongman, Jung Jiwon, Kim Sung-Han. Clinical characteristics of asymptomatic and symptomatic patients with mild covid-19. Clinical Microbiology and Infection. 2020;26(7):948.e1–948.e3. doi: 10.1016/j.cmi.2020.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Elaine Y P, Ng Ming-Yen, Khong Pek-Lan. Covid-19 pneumonia: what has ct taught us? The Lancet. Infectious diseases. 2020;20(4):384–385. doi: 10.1016/s1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Kaiyan, Chen Dian, Chen Shengchong, Feng Yuchen, Chang Chenli, Wang Zi, Wang Nan, Zhen Guohua (2020) Radiographic findings and other predictors in adults with covid-19. medRxiv. 10.1101/2020.03.23.20041673 [DOI] [PMC free article] [PubMed]
- 21.Li Lin, Qin Lixin, Zeguo Xu, Yin Youbing, Wang Xin, Kong Bin, Bai Junjie, Yi Lu, Fang Zhenghan, Song Qi, Cao Kunlin, Liu Daliang, Wang Guisheng, Qizhong Xu, Fang Xisheng, Zhang Shiqin, Xia Juan, Xia Jun. Using artificial intelligence to detect covid-19 and community-acquired pneumonia based on pulmonary ct: Evaluation of the diagnostic accuracy. Radiology. 2020;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Yang, Liu Jingyu, Tang Zhenyu, Lei Baiying. Deep spatial-temporal feature fusion from adaptive dynamic functional connectivity for mci identification. IEEE Transactions on Medical Imaging. 2020;39(9):2818–2830. doi: 10.1109/TMI.2020.2976825. [DOI] [PubMed] [Google Scholar]
- 23.Yang Li Yu, Liu Wei-Gang Cui, Guo Yu-Zhu, Huang Hui, Zhong-Yi Hu. Epileptic seizure detection in eeg signals using a unified temporal-spectral squeeze-and-excitation network. IEEE Transactions on Neural Systems and Rehabilitation Engineering. 2020;28(4):782–794. doi: 10.1109/TNSRE.2020.2973434. [DOI] [PubMed] [Google Scholar]
- 24.Li Yang, Zhang Xian-Rui, Zhang Bin, Lei Meng-Ying, Cui Wei-Gang, Guo Yu-Zhu. A channel-projection mixed-scale convolutional neural network for motor imagery eeg decoding. IEEE Transactions on Neural Systems and Rehabilitation Engineering. 2019;27(6):1170–1180. doi: 10.1109/TNSRE.2019.2915621. [DOI] [PubMed] [Google Scholar]
- 25.General Office of National Health Committee et al. Office of state administration of traditional chinese medicine. Notice on the issuance of a program for the diagnosis and treatment of novel coronavirus (2019-nCoV) infected pneumonia (trial version 6)[text in Chinese, (2020)
- 26.Pan Feng, Ye Tianhe, Sun Peng, Gui Shan, Liang Bo, Li Lingli, Zheng Dandan, Wang Jiazheng, Hesketh Richard L, Yang Lian (2020) Time course of lung changes on chest ct during recovery from, et al (2019) novel coronavirus (covid-19) pneumonia. Radiology 10.1148/radiol.2020200370 [DOI] [PMC free article] [PubMed]
- 27.Shi Feng, Wang Jun, Shi Jun, Wu Ziyan, Wang Qian, Tang Zhenyu, He Kelei, Shi Yinghuan, Shen Dinggang (2020) Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19. IEEE Reviews in Biomedical Engineering, pages 1–1. 10.1109/RBME.2020.2987975 [DOI] [PubMed]
- 28.Wang Jinhua, Yang X, Cai Hongmin, Tan W, Jin Cangzheng, Li L (2016) Discrimination of breast cancer with microcalcifications on mammography by deep learning. Scientific Reports, 6. 10.1038/srep27327 [DOI] [PMC free article] [PubMed]
- 29.Wang Shuai, Kang Bo, Ma Jinlu, Zeng Xianjun, Xiao Mingming, Guo Jia, Cai Mengjiao, Yang Jingyi, Li Yaodong, Meng Xiangfei, Xu Bo (2020) A deep learning algorithm using ct images to screen for corona virus disease (covid-19). medRxiv. 10.1101/2020.02.14.20023028 [DOI] [PMC free article] [PubMed]
- 30.Wang Shuo, Zha Yunfei, Li Weimin, Qingxia Wu, Li Xiaohu, Niu Meng, Wang Meiyun, Qiu Xiaoming, Li Hongjun, He Yu, Gong Wei, Bai Yan, Li Li, Zhu Yongbei, Wang Liusu, Tian Jie (2020) A fully automatic deep learning system for covid-19 diagnostic and prognostic analysis. European Respiratory Journal. 10.1183/13993003.00775-2020 [DOI] [PMC free article] [PubMed]
- 31.Yang Rongrong, Gui Xien, Xiong Yong. Comparison of Clinical Characteristics of Patients with Asymptomatic vs Symptomatic Coronavirus Disease 2019 in Wuhan. China. JAMA Network Open. 2020;3(5):e2010182–e2010182. doi: 10.1001/jamanetworkopen.2020.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Xingyi, He Xuehai, Zhao Jinyu, Zhang Yichen, Zhang Shanghang, Xie Pengtao March (2020) Covid-ct-dataset: A ct scan dataset about covid-19. arXiv e-prints, page arXiv:2003.13865
- 33.Ying Song, Zheng Shuangjia, Li Liang, Zhang Xiang, Zhang Xiaodong, Huang Ziwang, Chen Jianwen, Zhao Huiying, Wang Ruixuan, Chong Yutian, Shen Jun, Zha Yunfei, Yang Yuedong (2020) Deep learning enables accurate diagnosis of novel coronavirus (covid-19) with ct images. medRxiv. 10.1101/2020.02.23.20026930 [DOI] [PMC free article] [PubMed]
- 34.Zheng Chuansheng, Deng Xianbo, Fu Qiang, Zhou Qiang, Feng Jiapei, Ma Hui, Liu Wenyu, Wang Xinggang (2020) Deep learning-based detection for covid-19 from chest ct using weak label. 10.1101/2020.03.12.20027185