Abstract
Phenotypic analysis of Caenorhabditis elegans has greatly advanced our understanding of the molecular mechanisms implicated in the aging process as well as in age-related pathologies. However, conventional high-resolution imaging methods and survival assays are labor-intensive and subject to operator-based variations and decreased reproducibility. Recent advances in microfluidics and automated flatbed scanner technologies have significantly improved experimentation by eliminating handling errors and increasing the sensitivity in measurements. Here, we introduce a medium-throughput microfluidic platform, which efficiently positions and immobilizes single worms through pressurization for high resolution imaging. Worms are sorted based on select imaging criteria, and subsequently transferred into multi-well plates for automated lifespan assessment. To illustrate the applicability of this method, we imaged α-synuclein deposits in a C. elegans model of Parkinson’s Disease (PD). We found that age synchronized individuals expressing human α-synuclein vary greatly in the quantity and size of intracellular α-synuclein foci at early stages in life. Subsequent lifespan analysis of the individuals, however, did not reveal any correlation between the number or extent of α-synuclein deposits and subsequent lifespan. These studies suggest that the observed natural variations in α-synuclein deposits found in C. elegans models of PD do not originate from inherent differences in the fitness of the organism or contribute to alterations in lifespan.
Keywords: C. elegans, Microfluidics, Lifespan, α-synuclein
1. Introduction
C. elegans is a powerful system for elucidating phenotypes associated with aging and modeling the pathophysiology of age-related neurodegenerative diseases [1,2]. C. elegans strains expressing human disease-causing proteins display phenotypes relevant to the human pathology including protein aggregation, cell toxicity and movement dysfunction [3]. These transgenic worms have been instrumental in the identification and functional evaluation of genetic modifiers and neuroprotective compounds. C. elegans models of PD overexpress human α-synuclein in either body wall muscle cells [4,5] or dopaminergic neurons [6,7] and develop motor deficits or neuronal loss, respectively. These phenotypes are tightly correlated to the age-dependent intracellular accumulation of α-synuclein into inclusions, similar to human pathological inclusions [8]. However, it is not known if or how accumulation of the disease-causing protein impacts the aging process, and whether the quantity and/or timing of aggregate deposition correlates with life expectancy in humans or C. elegans.
Microfluidic systems have been pivotal in increasing the throughput, reproducibility and sensitivity in C. elegans microscopic observations. Such systems have enabled non-invasive, reversible, on-chip immobilization for high resolution imaging at the subcellular level [9–14] and are often complemented with sorting capabilities [15]. Immobilization is achieved by various techniques including confinement in tapered channels or by deformable membrane, CO2 exposure, cooling or entrapment in a thermo-sensitive gel matrix [9,10,12]. Other advanced methods have also been developed to enable automated monitoring of C. elegans lifespan in large groups of individuals [16,17]. These methods employ flatbed scanners and are able to capture even subtle differences in survival curves [18].
In this study, we combined and simplified features of existing platforms to develop a dual system capable of testing whether early-life individual variability in patterns of α-synuclein inclusions can serve as a predictor for the remaining lifespan of the organism. The platform incorporates a microfluidic chip which accomplishes the tasks of worm positioning and brief confinement for high resolution imaging followed by worm recovery or disposal, in a medium-throughput manner. The biochip allows single worm loading in an imaging chamber where pressure-driven immobilization is achieved. This method is neither limiting to a specific worm age or size, nor compresses the worm, as in the case of devices where tapered channels or deformable membrane, respectively were used. Moreover, this device does not require local temperature or air composition control. Recovered worms are placed in multi well plates and transferred in the automated flatbed scanner for lifespan monitoring as individuals.
Previous studies aimed to investigate the potential connections between α-synuclein deposits in dopaminergic neurons, neuronal cell death and lifespan of worms failed to reveal any correlation [19]. This result might be explained by the fact that no neuronal α-synuclein inclusions were detected, potentially due to the extremely small size of C. elegans neurons. Deposits of human α-synuclein fused to the yellow fluorescent protein (YFP) under the control of the unc-54 promoter in body wall muscles of C. elegans, on the other hand, can be detected as early as the second day of hatching [5,20]. We exploited this second C. elegans PD model and imaged individuals from an aged-synchronized population. We quantified their α-synuclein accumulation in early adult stages and subsequently monitored their lifespan using an automated lifespan platform. We observed large inter-individual variations in the number of α-synuclein inclusions and found this number to steadily increase with age. Notably, neither the number nor the relative intensity of α-synuclein deposits in the body wall muscle cells of young individuals appeared to correlate with their lifespan. These results suggest that the α-synuclein aggregation pattern is not linked to the aging process as manifested by the time of death. Moreover, we show that our combined system can serve as a reliable tool for deep phenotyping of cellular and subcellular features subject to interindividual variability and for studying their involvement in lifespan determination.
2. Methods
2.1. C. elegans maintenance
Wild type (N2) and NL5901 (pkIs2386 [unc-54:α-synuclein:YFP + unc-119(+)]) strains were provided by the Caenorhabditis Genetics Center (University of Minnesota). Worms were synchronized by hypochlorite bleaching and eggs allowed to hatch overnight in M9 buffer. The larvae were subsequently cultured at 20 °C on 90 mm Petri dishes with nematode growth media (NGM) supplemented with streptomycin (0.1 mg/ml) and nystatin (0.01 mg/ml) and seeded with E. coli strain OP50–1. Once the L4 stage was reached, worms were placed on NGM plates supplemented with 50 μМ fluoro-2′-deoxyuridine (FUdR) to prevent growth of progeny.
2.2. Microfluidic chip fabrication
We designed the flow and control layers of the microfluidic chip in AutoCAD (Autodesk, Inc). Chrome masks (Soda Lime Glass, 90 mm thick, low reflect) for both layers were fabricated at the Lurie Nanofabrication Facility of University of Michigan using a mask maker (Heidelberg μPG 501 Mask Maker, Heidelberg Instruments). We created the master molds for the worm flow and control layers by patterning SU-8–2025 photoresist (MicroChem) according to manufacturer’s recommendations to achieve a 30 μm and 40 μm thick layers respectively, on bare 100 mm silicon wafers. Both molds were coated with trichlorosilane (Sigma-Aldrich) in a vacuum desiccator overnight. We degassed and fully cured a 60 μm thick PDMS worm flow layer (spin-coated of 25 g of PDMS prepolymer mixture Sylgard 184, Dow Corning; 20:1; 900 rpm, 100 rpm/s for 5 s). We degassed, partially cured (25 min at 80 °C) and peeled off a 5 mm PDMS control layer (45 g of PDMS mixture 5:1) and then bonded it to a 20 μm thick PDMS membrane [spin-coated 25 g of PDMS mixture 20:1 at 900 rpm (100 rpm/s) for 10 s]. Using a dissecting microscope, we aligned the control and worm flow layers and bonded them using oxygen plasma treatment. Inlet and outlet holes were made by a reusable biopsy punch (World Precision Instruments; inner diameter 0.58 mm, outer diameter 0.96 mm). The combined PDMS layers were finally bonded onto a glass coverslip (24 × 60 mm #1.5, Fisherbrand).
2.3. System operation
Age-synchronized worms were washed off the plates and suspended in M9 solution, loaded in a 25 ml syringe barrel and injected in the microfluidic chip under air-pressurized flow (~30 psi). We interfaced all off- and on-chip hardware components to a desktop computer via a MATLAB-supported data acquisition board (BNC-2110, National Instruments). MATLAB (MathWorks, Natick, MA, U.S.A.) routines were custom-designed (available upon request) and contained five basic elements: waiting for worm’s entrance to the detection zone, worm confinement, confirmation of head-to-tail orientation, image acquisition and permission for the worm to exit before returning to the initial state. Image analysis was performed after the experiment was finalized.
2.4. Automated image acquisition and analysis
Fluorescence images were acquired with an upright microscope (BX61, Olympus) equipped with a Photometrics Coolsnap HQ2 cooled CCD camera, a 10 × objective lens (0.3 NA, UPLFLN, Olympus), two optical shutters (one for fluorescence and one for white light), an excitation filter (500/20 nm) and emission filter (535/30 nm), a z-stage system (NanoScanZ NZ400CE Nano-positioning Piezo Z Stage, Prior Scientific) and a X-CITE® Exacte light source equipped with a closed feedback-loop. Based on DIC micrographs of the first worm positioned in the imaging chamber, we defined the initial focal plane which remained the same throughout the experiment as no big variations in posture were observed among serially confined worms. For image acquisition, the exposure time was tuned to 160 ms to avoid extensive animal movement and the z-resolution was set to 2.5 μm, totaling 30 slices for each z-stack. For post-experimental image analysis, out-of-focus frames were discarded. A threshold (70% of maximum intensity) was applied to in-focus z-slices to determine the number and fluorescence intensity of foci. Fluorescence was calculated as the average intensity in the inclusion region, subtracted by the average intensity of the background (a larger region of the animal with no YFP fluorescence).
2.5. Statistical analysis
Data from raw images were extracted with MATLAB and analyzed for statistical significance using one-way ANOVA followed by Bonferroni’s multiple comparisons test. Relationships between time of death and imaging data were analyzed for statistical significance using linear regression. Lifespan data were displayed as Kaplan-Meier curves and were analyzed for statistical significance with log-rank (Mantel-Cox). These statistical analyses were done with GraphPad Prism (GraphPad Software, San Diego, CA, U.S.A.).
2.6. Lifespan analysis
For automated lifespan assays, worms were recovered from the microfluidic chip and singled out into 24-well plates containing 5 ml of solid NGM media supplemented with streptomycin (0.1 mg/ml), nystatin (0.01 mg/ml) and FUdR 50 μМ, and seeded with 50 μl E. coli OP50–1. The plates were arranged on a modified Epson v700 scanner in a 20 °C incubator and held in place by a rubber mat [17]. Plates were imaged every 20 min for thirty days.
Images were analyzed using the Worm Browser software developed for the automated lifespan system [17]. Analysis includes specifying the location of individual plates on the scanner, detecting individual worms, and tracking worm movement. The resulting data are time of death calls for each individual worm, based on the cessation of movement. All plates were annotated by hand to ensure that non-worm objects were excluded. Additionally, the time of death calls for the first and last 10% of worms on each plate were manually checked as these time points are more error prone. The final lifespans were calculated using the egg lay as day zero.
3. Results
3.1. System design and automated operation
To track intracellular α-synuclein inclusions, we used a previously established C. elegans transgenic line, which expresses human α-synuclein-YFP in the body wall muscles [5]. To live-image individual worms, quantify their α-synuclein deposition and foci formation, and individually dispense the worms for subsequent lifespan measurements, we modified a previously developed microfluidic-based platform [10] (Fig. 1a). In the new design, the main features include a newly added worm pushing channel to accelerate worm loading and unloading and an imaging chamber where worms are serially immobilized followed by a 3-way outlet, in the place of the previous 2-way outlet architecture (Supplementary Fig. 1a). Two of the three channels serve for worm unloading; the flushing channel, which is also a new feature, is used to manually unclog the imaging chamber. It serves, for instance, to reposition worms that enter the chamber in a head-to-tail orientation or separate worms that enter the imaging chamber at the same time. The waste channel is used to discard unwanted worms while the collection channel is used to recover imaged worms and dispense them individually into multi-well plates for lifespan assessment.
Fig. 1.
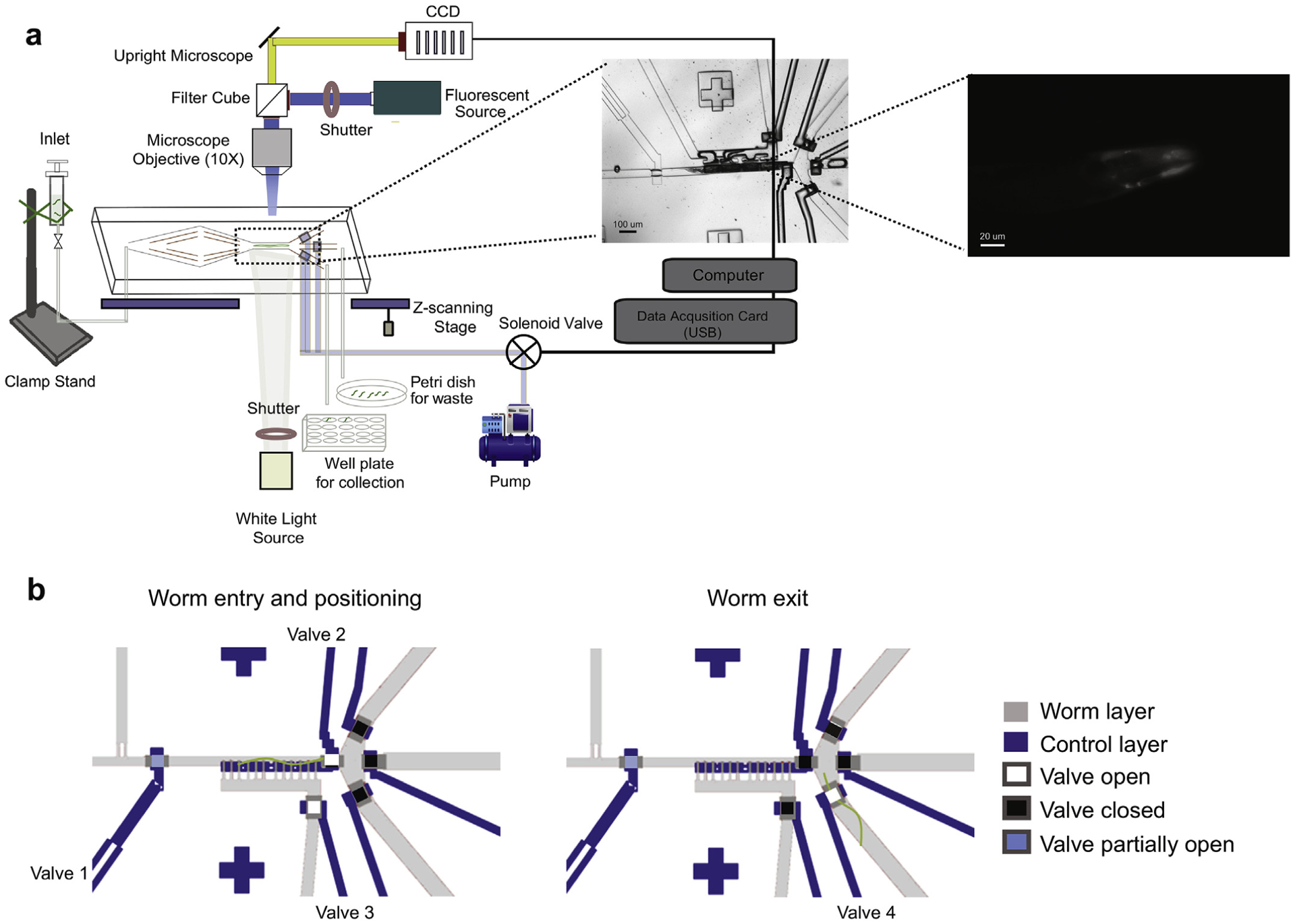
Operation of the automated microfluidic-based platform for C. elegans imaging and dispensing. (a) Age-synchronized worms were loaded from a liquid culture into the biochip (enhanced view, biochip) and serially immobilized in the imaging chamber by a valve-controlled, air pressurized flow. Worms that were positive for YFP signal (enhanced view, head region of worm) were z-stack imaged and then directed in the collection outlet, followed by recovery in individual wells of 24-well plates. The entire process including on-chip worm handling, head region recognition, focusing and image capturing, is software-controlled. (b) Schematic of the valve control sequence during on-chip worm manipulation. Valve profile for a single worm entering and positioned in the imaging chamber (left). Valve 1 (flow regulator valve) is always partially closed to prevent multiple worms from entering. Valve 2 (image chamber control valve) is open to generate a pressure gradient for positioning the worm into the imaging chamber. Valve 3 (flow balancing valve) creates pressure adjustments for avoiding damage to the animals during confinement. Once the worm is immobilized in the imaging chamber, all the outlet control valves are closed to eliminate flow fluctuation. Valve profile for an animal exiting the imaging chamber and guided in the collection channel (right). Outlet control valve 4 is open to allow the worm to exit the imaging chamber and enter the collection channel. Once the worm leaves the imaging chamber the valves return to the worm entry and positioning state.
To conduct the experiments, we loaded synchronized worms from a suspension into the biochip by a constant pressure-driven flow, applied synergistically by the worm inlet and the worm pushing channel (Supplementary Fig. 1a). To drive single worms into the imaging chamber, we closed the valve-controlled outlet channels while keeping the side-positioning channels open (Fig. 1b). We then pushed the worm against the side wall of the imaging chamber by applying a pressure gradient through the side-positioning channels. This caused the valves to close via the air pressure-driven deflection of a membrane, which is located between the worm and flow control layers (Supplementary Fig. 1b). At the same time, the pressure gradient prevented a second worm from entering the chamber. Since the extent of immobilization proved to be adequate for the short term (up to 30 s) imaging of worms, we did not implement the motion-stopping cooling feature of the initial system [10]. The modest mechanical stress that worms experience during the brief confinement in the biochip does not affect their lifespan (Supplementary Fig. 2a). After imaging, we released the respective worm from the imaging chamber by opening one of the exit valves (Fig. 1b). Worms that were too small or had no detectable fluorescence were discarded. However, these losses were minimal (~5%). Air pressurization, valve opening/closing, worm recognition upon entering the imaging chamber and image acquisition were controlled by custom-designed MATLAB routines.
As a proof-of-concept, we first tested the capability of our automated system in identifying discrete α-synuclein-YFP foci. We wrote an image recognition algorithm, which enabled us to identify the worm’s orientation and discard the worms entering the trap with a tail-to-head orientation by activating the flush channel. Properly oriented immobilized worms were imaged and a dense z-stack of images was acquired from the head region. Our software then processed these images, and distinguished the bright foci, which correspond to α-synuclein inclusions, from both background fluorescence and diffuse YFP signals present in the body wall muscles. This is performed through an evaluation procedure that we termed minimal counting method (MCM), which identifies all inclusion objects bigger than 2 μm2 and with a fluorescent intensity greater than 70% of the highest intensity value detected in the worm head region (Fig. 2). By using this system, we were able to process ~70 worms/h. Most of the experimental time was consumed by scanning the entire worm and saving images to disk. All worms received the same handling in the automated platform and images were analyzed uniformly using the same criteria, thus reducing noise and biases introduced by manual operation.
Fig. 2.

Image analysis flow. Worms expressing α-synuclein-YFP in the head region. Z-projection of maximum signal from 26 image slices of a representative worm shown in left panel. Foci of bright (threshold set at 70% of maximum intensity) expression were identified (middle panel). These foci were then filtered by size (>2 μm2) and only foci present in the body wall muscle cells were analyzed further. Right panel shows in red outlines the overlay of the foci identified for analysis, on original image.
To verify the inclusion detection accuracy, we manually analyzed the stacked images with a commercially available imaging software (Metamorph, Molecular Devices). The detection accuracy (total worms with correctly identified inclusions over total worms imaged) was 90% (data not shown).
3.2. Characterization of α-synuclein inclusion patterns in young adults
Previous studies using permanent agarose pad worm mounting, manual image acquisition and analysis revealed that the number of α-synuclein-YFP inclusions in the body wall muscles of worms increased with age [5]. Our platform enabled us to timely process ~200 animals at Day 1, Day 2 and Day 4 of adulthood in a less stressful and more consistent microenvironment without limiting their utility for downstream applications such as survival analysis. Moreover, photobleaching of YFP reporters or other artifacts owing to an operator’s handling were minimized by avoiding ‘feature-seeking’ steps during manual focusing.
Our analysis confirmed the previous results and showed a steady increase in the number of α-synuclein-YFP foci as the worms aged (Fig. 3a). Specifically, the number of α-synuclein inclusions increased from a mean of 2.6 ± 1.8 foci at Day 1, to 4.8 ± 2.5 at Day 2, to 7 ± 3.9 at Day 4. This increase was largely due to the increase in the maximal number of inclusions found in individual animals (Fig. 3b–d). Specifically, the number of YFP-labeled foci within a population ranged from 2 to 8 per individual for Day 1 adults, from 2 to 14 foci per individual for Day 2 adults and from 2 to 18 foci per individual for Day 4 adults. It is possible that these age-dependent, inter-individual differences in α-synuclein accumulation within a cohort have stochastic sources. More importantly though, these differences might reflect variabilities in the onset and penetrance of late onset pathological phenotypes and potentially modulate the lifespan of individuals.
Fig. 3.
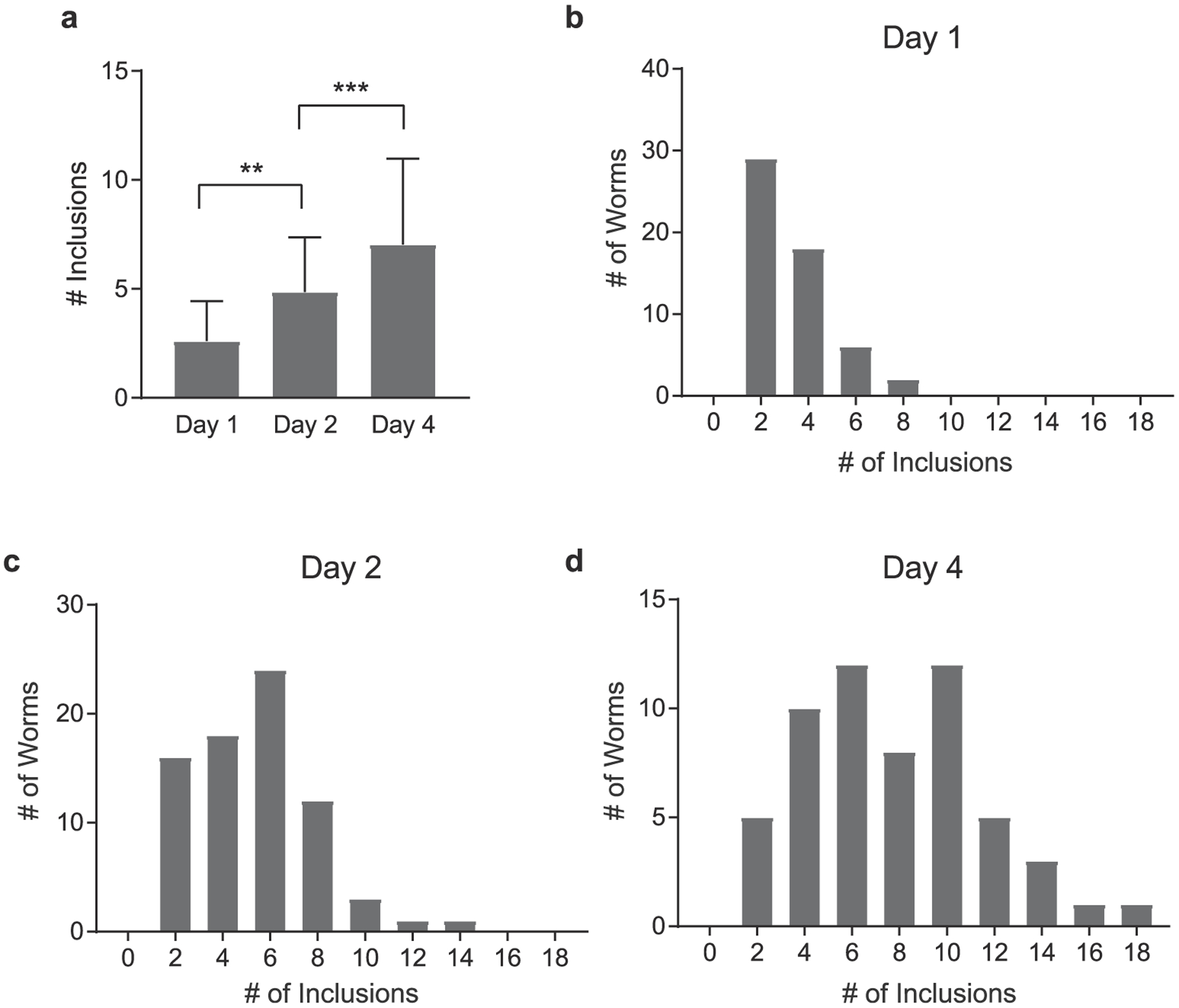
Profile of α-synuclein inclusions. (a) Quantification of the number of inclusions of a-synuclein-YFP expressing worms at Day 1, Day and Day 2 of adulthood. **, p < 0.01; ***, p < 0.001 (one-way ANOVA with Bonferroni correction). Data represent mean ± SD. (b)–(d) Distribution of the number of identified α-synuclein inclusions per animal, in age-synchronized populations imaged on Day 1, Day 2 and Day 4 of adulthood.
3.3. Relationship between α-synuclein inclusions and lifespan
Increased expression of α-synuclein has been linked to the rapid age-associated progression of the disease in both humans and C. elegans [6,21,22] To test whether a correlation exists between the number of α-synuclein inclusions observed in early adulthood and the lifespan of C. elegans, we recovered individuals from the biochip after imaging, transferred them into individual wells of 24-well plates, and placed them in an automated imaging and analysis platform - the C. elegans “lifespan machine” [17] - for the remainder of their lifetime (Supplementary Fig. 3). The “lifespan machine” has repeatedly been shown to serve as a reliable tool for C. elegans lifespan assessment by generating information on the worm position through successive time frames and identifying the time of death by the cessation of postural changes. We confirmed that groups of age-synchronized α-synuclein-YFP worms that were monitored at different time points (Day 1, Day 2 or Day 4 of adulthood) had indistinguishable mean lifespans (Supplementary Fig. 2b). This result indicated that their brief passage through the fluidic environment of the biochip at different stages during early adulthood did not generate variations in their time of death.
We then focused our downstream analysis on worms imaged at Day 2 and Day 4 of adulthood, when we recorded the highest variability in the number of fluorescent foci per individual (Fig. 3c–d). We first asked whether individuals with increased numbers of YFP-labeled foci were at a greater risk of early death compared to individuals with fewer foci. As shown in Fig. 4a and b, we were unable to find any distinct association between these two variables. We next examined the fluorescence intensity of the foci by employing the 70% of maximum intensity threshold (Fig. 4c and d). In one experiment, we found a significant (r2 = 0.7) negative correlation between the averaged intensity per inclusion and time of death. However, when we repeated the experiment, this result was not reproduced (r2 = 0.01). No association between these two variables (intensity versus time of death) in either experimental replicate was observed, even when we removed the 70% intensity correction (Fig. 4e and f). From these results, we concluded that variabilities in inclusion patterns among young, age-synchronized individuals do not serve as reliable markers of longevity.
Fig. 4.
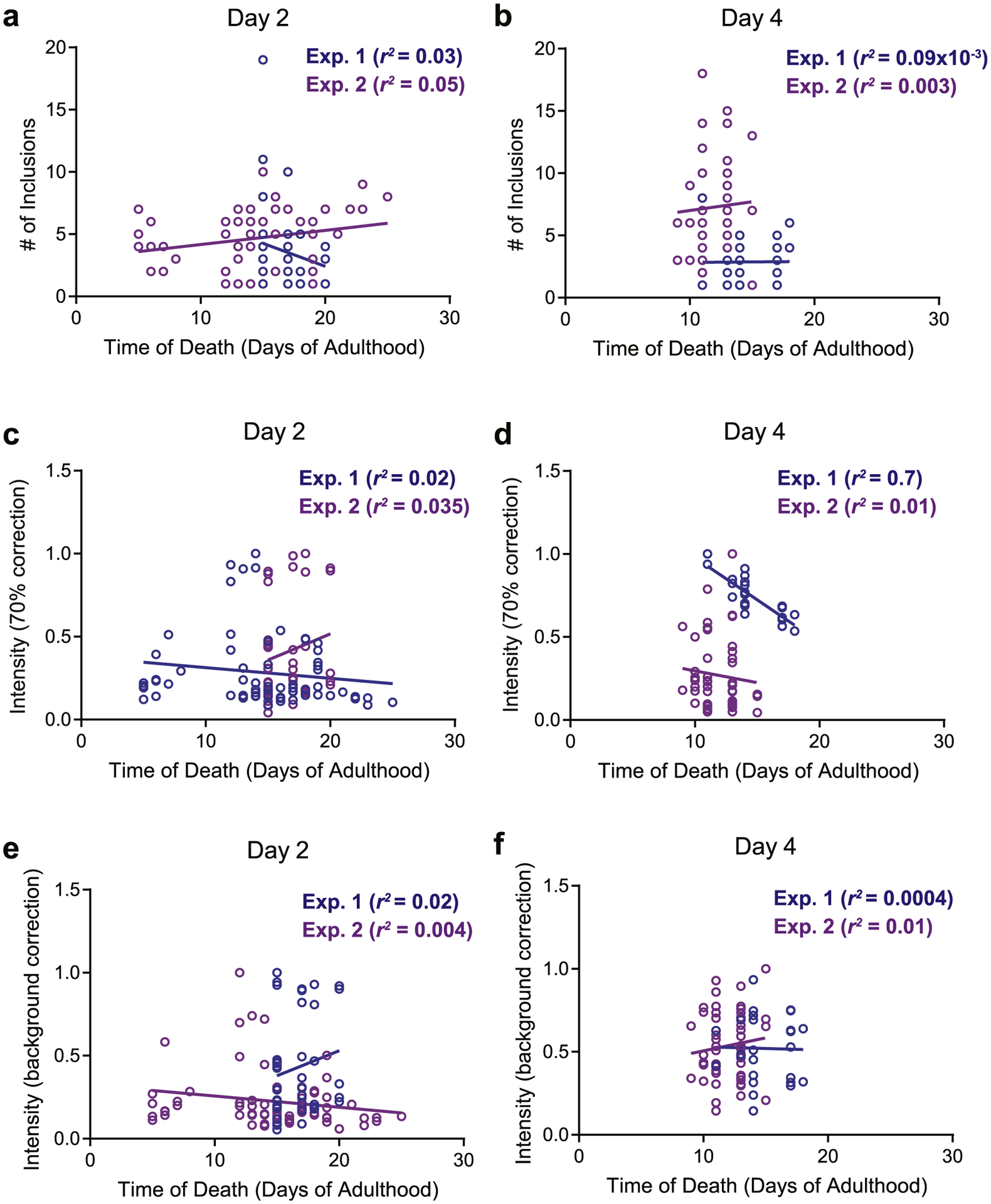
α-synuclein inclusions and lifespan. Relationships between the time of death of individuals (symbols) imaged at Day 2 or Day 4 of adulthood and (a), (b) number of inclusions; (c), (d) average intensity per inclusion after a 70% correction; (e), (f) average intensity per inclusion after background correction. Two biological replicates were tested for each association.
4. Discussion
In this study, we developed a microfluidic system capable of acquiring high-resolution images of subcellular features in individual worms whose lifespan was subsequently determined in the automated “lifespan machine”. Our microfluidic-based chip has several advantages over previously established microfluidic platforms: i) a simplified architecture, which enables short-term, pressure-based confinement of single worms of various ages; ii) a high worm-trapping and recovery efficiency (95%); and iii) a compatibility with all typical microscopy setups without the need for temperature or gas control features. Moreover, its use in combination with subsequent lifespan assessment in a fully automated C. elegans “lifespan machine” significantly diminishes operator-based variations and generates reliable physiological data. This technological synergy is thus suitable for detection of subtle phenotypic differences potentially linked to the aging process.
To test this new setup, we investigated whether early-stage α-synuclein accumulation correlates with organismal death. This correlation would be expected if α-synuclein deposits affected essential processes, for instance by sequestering proteins critical for cell survival, or were reflective of a defect in the organism’s proteostasis network early in life. To address this question, we used transgenic C. elegans, which accumulate α-synuclein in inclusions at very early stages in life [5]. Our analysis confirmed that individuals of an age-synchronized population of worms display large variations in the number of deposits as young adults. Although numbers of deposits increased with age, we failed to detect any clear correlation with the lifespan of the organism. These results suggest that differences in the formation of α-synuclein inclusions in worms do not trigger processes that impact longevity. It is unclear whether this result is of general nature or more specific to C. elegans, given that α-synuclein inclusions in this transgenic system only form in the body wall muscle cells. However, previous studies using α-synuclein overexpression in dopaminergic neurons only, also failed to show any correlation, suggesting that this effect might not be cell type specific [19]. Moreover, the molecular nature of α-synuclein inclusions that accumulate in C. elegans models [23] or, for that matter, in most other organisms, including PD patients, have not been characterized in detail, making it challenging to draw general conclusions. In either case, we can conclude that C. elegans PD models remain useful for identifying effectors and interventions that affect the deposition of α-synuclein aggregates but not for studying the consequences of α-synuclein inclusions in lifespan. The approach, however that we established here is widely applicable to answer a range of different research questions as it provides a medium-throughput methodology for individualized lifespan determination after detailed morphological and/or fluorescence-based microscopic analysis.
Supplementary Material
Acknowledgements
We would like to thank Robert Heller and Johnathon Gallias for IT support with establishing the “lifespan machine”, Gregg Sobo-cinski for assistance with the microscope set up and members of the Jakob lab for helpful discussion. Worm and bacterial strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Funding
This work was supported by the National Institute of Health (grant numbers GM122506 and AG046799) and the Priority Program SPP 1710 of the Deutsche Forschungsgemeinschaft (Schw823/ 3-2) awarded to U.J. and NIH T32 Career Training in the Biology of Aging grants (AG000114) awarded to D.B and E.Q.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tma.2020.04.001.
References
- [1].Apfeld J, Alper S, What can we learn about human disease from the nematode C. elegans? Methods Mol. Biol 1706 (2018) 53–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dimitriadi M, Hart AC, Neurodegenerative disorders: insights from the nematode Caenorhabditis elegans, Neurobiol. Dis 40 (1) (2010) 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Alexander AG, Marfil V, Li C, Use of Caenorhabditis elegans as a model to study Alzheimer’s disease and other neurodegenerative diseases, Front. Genet 5 (2014) 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hamamichi S, Rivas RN, Knight AL, et al. , Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson’s disease model, Proc. Natl. Acad. Sci. U. S. A 105 (2) (2008) 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].van Ham TJ, Thijssen KL, Breitling R, et al. , C. elegans model identifies genetic modifiers of alpha-synuclein inclusion formation during aging, PLoS Genet. 4 (3) (2008), e1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cao S, Gelwix CC, Caldwell KA, et al. , Torsin-mediated protection from cellular stress in the dopaminergic neurons of Caenorhabditis elegans, J. Neurosci 25 (15) (2005) 3801–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lakso M, Vartiainen S, Moilanen AM, et al. , Dopaminergic neuronal loss and motor deficits in Caenorhabditis elegans overexpressing human alpha-synuclein, J. Neurochem 86 (1) (2003) 165–172. [DOI] [PubMed] [Google Scholar]
- [8].Lewy FH, Paralysis agitans. I. Pathologische Anatomie, Lewandowsky’s Handbuch der Neurologie, 3. Band: Spez Neurologie II, Springer, Berlin, 1912, pp. 920–933. [Google Scholar]
- [9].Chokshi TV, Ben-Yakar A, Chronis N, CO2 and compressive immobilization of C. elegans on-chip, Lab Chip 9 (1) (2009) 151–157. [DOI] [PubMed] [Google Scholar]
- [10].Chung K, Crane MM, Lu H, Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans, Nat. Methods 5 (7) (2008) 637–643. [DOI] [PubMed] [Google Scholar]
- [11].Cornaglia M, Krishnamani G, Mouchiroud L, et al. , Automated longitudinal monitoring of in vivo protein aggregation in neurodegenerative disease C. elegans models, Mol. Neurodegener 11 (2016) 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Krajniak J, Lu H, Long-term high-resolution imaging and culture of C. elegans in chip-gel hybrid microfluidic device for developmental studies, Lab Chip 10 (14) (2010) 1862–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lee H, Kim SA, Coakley S, et al. , A multi-channel device for high-density target-selective stimulation and long-term monitoring of cells and subcellular features in C. elegans, Lab Chip 14 (23) (2014) 4513–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mondal S, Hegarty E, Martin C, et al. , Large-scale microfluidics providing high-resolution and high-throughput screening of Caenorhabditis elegans poly-glutamine aggregation model, Nat. Commun 7 (2016) 13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Midkiff D, San-Miguel A, Microfluidic technologies for high throughput screening through sorting and on-chip culture of C. elegans, Molecules 24 (23) (2019) 4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mathew MD, Mathew ND, Ebert PR, WormScan: a technique for high-throughput phenotypic analysis of Caenorhabditis elegans, PloS One 7 (3) (2012), e33483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Stroustrup N, Ulmschneider BE, Nash ZM, et al. , The Caenorhabditis elegans lifespan machine, Nat. Methods 10 (7) (2013) 665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stroustrup N, Anthony WE, Nash ZM, et al. , The temporal scaling of Caenorhabditis elegans ageing, Nature 530 (7588) (2016) 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Apfeld J, Fontana W, Age-dependence and aging-dependence: neuronal loss and lifespan in a C. elegans model of Parkinson’s disease, Biology 7 (1) (2017).). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bodhicharla R, Nagarajan A, Winter J, et al. , Effects of alpha-synuclein overexpression in transgenic Caenorhabditis elegans strains, CNS Neurol. Dis-ord. - Drug Targets 11 (8) (2012) 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Farrer M, Kachergus J, Forno L, et al. , Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications, Ann. Neurol 55 (2) (2004) 174–179. [DOI] [PubMed] [Google Scholar]
- [22].Singleton AB, Farrer M, Johnson J, et al. , alpha-Synuclein locus triplication causes Parkinson’s disease, Science 302 (5646) (2003) 841. [DOI] [PubMed] [Google Scholar]
- [23].Laine RF, Sinnige T, Ma KY, et al. , Fast fluorescence lifetime imaging reveals the aggregation processes of alpha-synuclein and polyglutamine in aging Caenorhabditis elegans, ACS Chem. Biol 14 (7) (2019) 1628–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


