Abstract

DNA mismatch repair (MMR) is an important postreplication process that eliminates mispaired or unpaired nucleotides to ensure genomic replication fidelity. In humans, Msh2-Msh6 and Msh2-Msh3 are the two mismatch repair initiation factors that recognize DNA lesions. While X-ray crystal structures exist for these proteins in complex with DNA lesions, little is known about their structures during the initial search along nonspecific double-stranded DNA, because they are short-lived and difficult to determine experimentally. In this study, various computational approaches were used to sidestep these difficulties. All-atom and coarse-grained simulations based on the crystal structures of Msh2-Msh3 and Msh2-Msh6 showed no translation along the DNA, suggesting that the initial search conformation differs from the lesion-bound crystal structure. We modeled probable search-mode structures of MSH proteins and showed, using coarse-grained molecular dynamics simulations, that they can perform rotation-coupled diffusion on DNA, which is a suitable and efficient search mechanism for their function and one predicted earlier by fluorescence resonance energy transfer and fluorescence microscopy studies. This search mechanism is implemented by electrostatic interactions among the mismatch-binding domain (MBD), the clamp domains, and the DNA backbone. During simulations, their diffusion rate did not change significantly with an increasing salt concentration, which is consistent with observations from experimental studies. When the gap between their DNA-binding clamps was increased, Msh2-Msh3 diffused mostly via the clamp domains while Msh2-Msh6 still diffused using the MBD, reproducing the experimentally measured lower diffusion coefficient of Msh2-Msh6. Interestingly, Msh2-Msh3 was capable of dissociating from the DNA, whereas Msh2-Msh6 always diffused on the DNA duplex. This is consistent with the experimental observation that Msh2-Msh3, unlike Msh2-Msh6, can overcome obstacles such as nucleosomes. Our models provide a molecular picture of the different mismatch search mechanisms undertaken by Msh2-Msh6 and Msh2-Msh3, despite the similarity of their structures.
In eukaryotes and prokaryotes, DNA mismatch repair (MMR) is an important postreplication process that eliminates mispaired or unpaired nucleotides to reduce the mutation rate and maintain genomic stability. The mammalian MMR pathway is also responsible for rectifying certain types of DNA damage. In humans, defects in the MMR system are the cause of hereditary nonpolyposis colorectal cancer, also known as Lynch syndrome.1,2 Defective MMR systems are also observed in 15–25% of sporadic tumors in a variety of tissues,3 myotonic dystrophy, fragile X syndrome, and Huntington’s disease.4−6
The MMR process has several stages, with the recognition, removal, and resynthesis steps conserved across all species with some differences. The search for mismatched bases is initiated by the mismatch recognition factor MutS or its homologue, which scans the DNA by forming an unstable ring around it that ultimately recognizes the mismatched bases.7 Upon addition of ATP, another MMR component, the MutL homologue, is then recruited to initiate the removal and resynthesis steps in the repair process.8−13
Binding of mispaired or unpaired bases by bacterial MutS and eukaryotic MutS homologues (MSH) is well-characterized.14−16 In humans, two MSH proteins are present, namely, Msh2-Msh6 and Msh2-Msh3 (also known as MutSα and MutSβ, respectively). Msh2-Msh6, a heterodimer of Msh2 and Msh6, recognizes small mismatches, such as a single-base mispair or one or two unpaired bases. The structure of Msh heterodimers shows that the mismatch-binding domain is responsible for recognizing DNA lesions. The MBD has a mixed α/β structure that contains 124, 157, and 137 residues in the Msh2, Msh6, and Msh3 units, respectively. The mismatch-binding domain of Msh2 (MBD2) is common to both structures, combined with either MBD6 or MBD3. Whereas in the Msh2-Msh3 dimer, both MBD regions are required for mismatch binding, in the Msh2-Msh6 dimer, only the MBD of Msh6 is required to bind lesions. MBD6 recognizes the mismatch size upon interacting with it. By contrast, Msh2-Msh3, a heterodimer of Msh2 and Msh3, recognizes relatively larger insertion–deletion loops (IDLs) of ≤15 nucleotides, although it can also recognize small mismatch sites of one or two unpaired bases.
Five crystal structures of human Msh2-Msh6 bound to a 15 bp oligomer containing a mismatch or a single-base insert reveal that the Msh2-Msh6 conformations were not altered appreciably when bound to each DNA substrate.15 A similar trend was observed in four crystal structures of human Msh2-Msh3 in complex with DNA containing insertion–deletion loops of two, three, four, or six unpaired nucleotides.16 Taken together, these structures indicate that the two proteins interact with DNA via a common mechanism.
Most structural and functional investigations of MMR have focused on the postrecognition stages. This includes ATP binding by MSH, its interaction with chromatin and chromatin-associated proteins, the interaction of Msh6 with proliferating cell nuclear antigen (PCNA), the role of Msh2-Msh3 in double-strand break repair and mutagenic expansion of trinucleotide repeats, and the regulation of MSH protein expression within the cell.17−20 In contrast to the post-recognition repair events, few studies have investigated the structural organization of the MMR initiation protein MSH in different states in its functional cycle, particularly the “search state” and the “sliding clamp state”, and how it is released from the DNA and recycles itself.
Real-time single-molecule fluorescence resonance energy transfer (smFRET) studies showed that, in its search mode, MutS forms a clamp on the DNA. Changes in FRET efficiency and distribution with length and time resolution indicated that MutS carries out mismatch search by translation-coupled rotation diffusion while in continuous contact with the duplex DNA.21,22 Using total internal reflection fluorescence microscopy (TIRFM), MutSα was also observed to perform translation-coupled rotation diffusion to maintain a constant register with the helical contour of the DNA.23−27
To date, the apo structures of human Msh2-Msh6 and Msh2-Msh3 have not been determined. The corresponding apo structure of MutS shows disorder of the DNA clamp domains.28 Such conformational flexibility suggests a way for the domains to “open up” to allow the loading and unloading of the MutS homologue dimer onto DNA. Real-time smFRET and fluorescence microscopy experiments confirmed that the complex formed by MutS binding to duplex DNA is short-lived and thus difficult to characterize experimentally.21−23 While providing indirect evidence for coupling between rotational diffusion and linear translocation along DNA, the current experimental strategies cannot directly reveal the translation-coupled rotation motion because of spatial resolution limitations.29
To understand the search-mode diffusion of human MMR initiation proteins Msh2-Msh6 and Msh2-Msh3 on DNA, we have modeled potential search-mode conformations that can diffuse on linear DNA. We show by coarse-grained molecular dynamics simulations that the modeled structures can perform rotation-coupled translation diffusion on DNA (i.e., the MMR proteins follow the major groove and thus perform a helical motion while diffusing along the linear DNA). This mechanism that is often called sliding dynamics and was found for various DNA-binding proteins30−35 is suitable for the effective search of mismatches present in the DNA. The rate of diffusion in one version of these models also remained unchanged with an increasing salt concentration, as observed in the case of bacterial MutS by real-time smFRET studies. We also show that a model with a large gap between the clamp regions of Msh2-Msh3 was capable of dissociating from the DNA during the search process. A similar model of Msh2-Msh6, however, did not dissociate from the DNA. This is consistent with the experimental observation that Msh2-Msh3, unlike Msh2-Msh6, can overcome obstacles such as nucleosomes.36 Our findings provide a molecular picture of the mismatch search mechanisms of both Msh2-Msh6 and Msh2-Msh3 and confer additional structural insights unavailable from the analysis of the static structures of the two proteins.
Materials and Methods
Crystal Structures of Msh2-Msh3 and Msh2-Msh6
The starting structures of this study were the X-ray crystal structures of DNA-bound heterodimers of human Msh2-Msh6 [Figure 1; Protein Data Bank (PDB) entry 2O8B] and Msh2-Msh3 (Figure 1S; PDB entry 3THX). Each structure is bound to a short DNA duplex containing unpaired nucleotides. We note that in both cases, the DNA structure was bent by ∼45° and adopted an arc shape with the unpaired nucleotides situated at the inflection point of the DNA. These unpaired nucleotides are recognized by MBD3 and MBD6, whereas the clamp domain of the monomers interacts with the inner side of the DNA arc. The C-terminal dimerization domains, however, do not interact with the DNA.
Figure 1.

Geometric parameters for characterizing the linear diffusion of Msh2-Msh6 along DNA. (A) Front view of Msh2-Msh6 bound to a DNA lesion with a G T mispair (PDB entry 2O8B). The lever, clamp, and mismatch-binding domains are indicated. Subunits Msh2 and Msh6 are colored light red and green, respectively; the DNA lesion is colored light gray. MBD2 and MBD6 are colored red and blue, respectively, and are also shown by surface representation. A nonspecific straight DNA placed at the DNA lesion position is colored dark gray. The distance, R, between the center of mass of MBD26 and the straight DNA molecule (straight dotted orange arrow) and rotation angle θ (curved dotted orange arrow) are shown. (B) Side view of Msh2-Msh6, indicating the translocation distance Z along the DNA.
Isolated MBD Dimers
On the basis of the assumption that the interaction of the MBD with canonical B-DNA differs from that with the lesion-bound complex seen in the crystal structures, we constructed dimers of the N-terminal regions of both Msh2-Msh3 and Msh2-Msh6. These substructures should bind to canonical DNA in an optimal way, without bias from the lesion-bound structures. From the crystal structure of Msh2-Msh3, only the MBD of each monomer (Msh2, residues 14–124; Msh3, residues 217–353) was retained to construct the MBD-only heterodimer MBD23. The MBD-only heterodimer MBD26 was constructed in a similar way (Msh2, residues 1–124; Msh6, residues 362–518). The N-terminal regions that precede the MBD regions, which are not seen in the crystal structures, were not modeled.
Structure of the Msh2-Msh6 Chimera
The structure of the chimera was constructed by replacing the MBD of Msh6 (Pro362–Asn534) with the MBD of Msh3 (Lys217–Asp353). Although the two domains have similar structures in regions contacting the DNA (comprising approximately 125 residues in both), MBD6 has a 45-residue random coil N-terminal tail, not seen in the Msh3 crystal structure. Even in the region close to the DNA, there are loops that do not correspond structurally. Thus, 81 Cα atoms were used by the “align” command in PyMOL to overlay the N-terminal domain of Msh3 on the N-terminal domain of Msh6 (root-mean-square deviation of 0.63 Å). The coordinates of the N-terminus of Msh6 were deleted up to, but not including, Tyr535. The coordinates of the aligned N-terminus of Msh3 were then pasted into the modified Msh26 coordinate file. Clashes involving seven side chains were fixed manually. The 7.9 Å gap between the C-terminus of the Msh3 domain (Asp353) and the new N-terminal residue (Tyr535) of Msh6 was joined by the subsequent 2000 steepest descent energy minimization step in GROMACS 5.1.137 with the AMBER99SB-ILDN force field.38 The energy-minimized structure was used as the basis for the coarse-grained simulations discussed in the results.
Identification of Possible Search-Mode Structures of Msh2-Msh6 and Msh2-Msh3 Using Normal-Mode Analysis
To date, the DNA-free structures of human Msh2-Msh6 and Msh2-Msh3 have not been determined. Biophysical studies confirm that MutS–DNA complexes are short-lived and their structures are challenging to trap experimentally, making direct observation of these structures difficult. In the DNA lesion-bound crystal structures of Msh2-Msh6 and Msh2-Msh3, both proteins interact tightly with the DNA using the MBDs and the lower clamp domains, leaving no gap between the protein and the DNA. These conformations mean that neither protein is likely to slide along DNA. Both atomistic and coarse-grained simulations of Msh2-Msh3/Msh6 dimers showed no translation along DNA when canonical B-DNA was docked with the crystal structures, in place of the bent mismatched DNA. We assumed, therefore, that the initial search structures of Msh2-Msh6 and Msh2-Msh3 are far more open to allow sliding along DNA. We used an elastic network model (ENM)-based algorithm (elNemo web server)39 to reveal the low-frequency collective modes (normal modes) of the individual monomers (Msh2, Msh3, and Msh6) to predict conformational changes that would create gaps in the dimer. Msh2-Msh3 and Msh2-Msh6, being heterodimers in which the amino acid sequences and conformations for Msh2, Msh3, and Msh6 monomers are not identical, are expected to have low-frequency collective modes. We predicted the conformational change of an individual monomer separately and then used them to build the probable “open” dimer conformation. Default values for the number of lowest-frequency normal modes to be computed (NMODES = 5) and the step size between minimum and maximum perturbation (DQSTEP = 20) were used to calculate the ENM models. Two additional variables, minimum and maximum perturbation, DQMIN and DQMAX, respectively, change the amplitude of displacement of the low-frequency motion in the models. To allow the required gap in the clamp region for model B and model C described below, we used (DQMIN = −200, DQMAX = 200) and (DQMIN = −400, DQMAX = 400) to build them. Conformations of the models built using these values allowed the docking of linear DNA and binding of the MBD domains without any steric clash with the clamp region. For all three monomers, the lowest-frequency mode predicted very little or no movement of the C-terminal dimerization domains, giving root-mean-square deviation (RMSD) values of <1 Å relative to the Msh2-Msh3 and Msh2-Msh6 crystal structures. The MBD regions also moved very little. The clamp regions, on the contrary, showed significant deviation from the structures observed in the crystal forms, with RMSD values of ∼13 Å (Figure 2). This is consistent with the observation that a wide range of conformations for the lever and clamp domains were seen in the five crystal structures for Msh2-Msh615 and the four crystal structures for Msh2-Msh3.16
Figure 2.
Structural comparison between the DNA-bound crystal conformation (model A) and the modeled conformations (model B and model C) of Msh2-Msh6 performing linear diffusion along DNA. The color scheme of the protein and DNA is the same as in Figure 1. The gap in the DNA-binding channel between MBD26 and the clamp domain increases in the following order: model A < model B < model C. This is shown by highlighting the distances between two residue pairs, one at the top and the other at the bottom of the clamp domain, and also the two angles at either side of the clamp domain.
Using the lowest-frequency mode obtained for each monomer, dimers consisting of Msh2 and Msh3 or Msh6 were constructed. The clamp regions of these structures exhibited a gap that could not accommodate straight DNA in a manner that enabled the MBD regions to bind DNA in a fashion similar to that seen in the simulations of the MBD dimers. To explore further conformations, the dimers were again submitted to the elNemo server. The lowest-frequency mode gave dimers for Msh2-Msh3 and Msh2-Msh6 with movement somewhat perpendicular to the long axis of the DNA, and a smaller movement parallel to this axis. This conformation, with a distance of 37 Å between the clamp domains (Figure 2, model B), allowed docking of linear DNA and binding of the MBD domains in a manner similar to that seen in the simulations of the MBD on DNA. The lowest-frequency modes for the monomers generated with a larger maximum displacement parameter were used to build Msh2-Msh3 and Msh2-Msh6 dimers with a distance of 51 Å between the clamp domains (Figure 2, model C). These dimers could dock to linear B-DNA with the MBD domains interacting with the DNA in a manner similar to the MBD simulations, without further modification.
Coarse-Grained Models for Protein and DNA
We used coarse-grained molecular dynamics (MD) simulations to study the dynamics of human Msh2-Msh6 and Msh2-Msh3 on DNA. Coarse-grained simulations access time scales that are long enough to achieve sampling of the diffusion process, which is extremely challenging in all-atom simulations. The proteins were modeled by two beads per residue located at the Cα and Cβ positions, except Gly, which has only Cα. Charged amino acids were modeled by placing a point charge of +1 (Lys and Arg) or −1 (Asp and Glu) on the Cβ bead. The DNA in the simulations was 100 bp double-stranded straight dsDNA in a canonical B form and was centered on and aligned with the Z-axis (the DNA geometry was adapted from DNA composed of poly-C and poly-G single-stranded chains). Each nucleotide was represented by three beads representing the phosphate (P), sugar (S), and nucleobase (B) moieties, which were positioned at the geometric center of each represented group. In the model, the phosphate bead bore a charge of −1.
The protein was modeled by a native topology-based model that used the Lennard-Jones (L-J) potential to represent native contact interactions, representing the inherent flexibility of the proteins, and a repulsive potential to prevent chain crossing.40−42 The interaction between the protein and the DNA was modeled using the Debye–Hückel approximation.35 Charged beads can participate in any nonspecific charge–charge interactions. Given that Msh proteins interact nonspecifically with DNA during the search process, electrostatic interactions are the main force driving their linear diffusion along DNA, whereas the contribution of hydrogen bonds, which may increase energetic ruggedness and thereby slow linear diffusion, is negligible. Excluded volume interactions were introduced between all of the protein and the DNA beads.
Coarse-Grained MD Simulations
The dynamics of Msh proteins moving along dsDNA was simulated with the Langevin equation. Each full-length protein was simulated by initially placing the DNA into the channel between the MBD and the clamp domain, as shown in Figure 2 and Figure S1. For the MBD dimer simulations, each dimer was initially placed close to the DNA. To retain the unimpaired native fold of the proteins during their diffusion along DNA, all simulations were run at low temperatures [maintained at 0.3 (reduced units)] at which the protein remains folded. We note that conformational fluctuations of the native state were found to have a minor effect on sliding.30 An implicit solvent model having a dielectric constant of 70 (water) was used. Salt concentrations were varied between 10 and 60 mM. We point out that the coarse-grained representation and the location of the charges on the Cα atom result in simulated salt concentrations that were effectively ∼2-fold higher than stated. The system was confined in a cubic box with periodic boundary conditions (PBC) and dimensions of 750 Å × 750 Å × 750 Å, where the DNA was aligned along the Z-axis. For all variants, we performed 20 simulations of 5 × 107 time steps to achieve equilibrium sampling. This coarse-grained model has been applied to address the diffusion and search mechanisms of various DNA-binding proteins on nonspecific DNA30,34,37,43,44 and was shown to capture some of their major experimental characteristics. The coarse-grained simulations for diffusion of proteins along DNA describe the molecular mechanism of diffusion of various DNA-binding proteins and their dependence on salt concentration.30,34,44 In particular, they successfully predicted the mechanism of linear diffusion and particularly the existence of coupling between translation and rotation.30,32,33,37 The coarse-grained simulations showed, in a manner consistent with experiments, how changing the asymmetry of the affinity of multidomain proteins to DNA affects the search kinetics.43,46−48 The consequence of mutating charged residues for the diffusion coefficients49 and for the rate of conversion from nonspecific to specific binding mode is also described by the coarse-grained model;50 nonetheless, a more elaborate model that includes conformational changes of both the protein and the DNA is required to address further aspects of this process.
Trajectory Analysis
In all of the simulations, the DNA was aligned along the Z-axis. The rotation angle of the clamps around the DNA, θ, was calculated as the rotation angle of the center of mass of the MBD in the X–Y plane (this definition is permitted because the protein did not undergo conformational changes during the simulations) (Figure 1). The position of the DNA with respect to the MBD was quantified by the distance R between the centers of mass of the MBD and the DNA (Figure 1). The D1 coefficient of one-dimensional (1D) diffusion was calculated from the mean square displacement (MSD) of the center of mass of the MBD moving along the DNA double helix:
where N is the trajectory length in time steps, n is the measurement window ranging from 1 to N, Δt is the time step interval, and Z is the location of the sliding clamp along the DNA. The linear diffusion coefficient, D1, was estimated from the slope of the MSD versus time, which was calculated between time frames 100 and 1000 for diffusion on DNA, because shorter time scales do not capture the slow diffusion process.
All-Atom Simulations
All-atom MD simulations were performed for the Msh2-Msh3 (PDB entry 3THX) and Msh2-Msh6 (PDB entry 2O8B) complexes using GROMACS 5.1.137 with the AMBER99SB-ILDN force field.38 Missing loops in all chains were built with Swiss-PDBViewer (version 4.00).51 The ADP molecules were not included in the simulations. The SPCE water model was used, with NaCl added to achieve a concentration of 125 mM. The solvated structures were minimized followed by equilibration runs (100 ps each) of the NVT and NPT ensembles. Simulations for the Msh2-Msh3 and Msh2-Msh6 crystal structure complexes were repeated three times, each run for 220 ns. In addition, we modeled the complexes of these proteins with nonspecific DNA by replacing the bent DNA in the two crystal structures with a 22-mer of ideal B-DNA with a sequence of GCATCGATCGGCTTCAGATGCG (3′ to 5′) and its complementary strand. To avoid clashes between the protein and the DNA, minor adjustments were made to side chains and one loop in each chain. Production MD simulations for each model were repeated four times, each lasting 320 ns.
Results
The DNA Lesion-Bound Conformation Shows No Sliding Movement along the DNA
To slide on DNA, the structures of proteins that embrace the DNA, such as Msh2-Msh6 or Msh2-Msh3, must include sufficient space to accommodate the DNA molecule (diameter of ∼20 Å). For example, the PCNA clamp has a ringlike structure with an inner diameter of ∼30 Å, which is sufficient to enable PCNA to linearly diffuse while encircling the DNA.37 However, the minimum distance between the DNA-binding domain [namely, the mismatch-binding domain (MBD)] and the clamp domain is ∼12 Å in the DNA-bound crystal structures of Msh2-Msh6 and Msh2-Msh3, which is insufficient for sliding or even diffusion.
To verify whether the crystal structure of the DNA lesion-bound conformation of Msh2-Msh3/Msh6 dimers can slide, a canonical 100 bp B-DNA was docked with the crystal structures to replace the bent mismatched DNA (model A, Figure 2) and coarse-grained simulations were performed. Multiple long coarse-grained simulations did not show any translation of the dimers along the DNA length (i.e., D1 = 0, independent of salt concentration), suggesting that the initial search conformation differs from the lesion-bound crystal structure (Figure 3). We note that various transcription factors and enzymes, either monomeric, dimeric, tetrameric, or multidomain, show diffusion along nonspecific DNA with a diffusion coefficient on the order of 0.01–1 μm/s2.30,32,34,37,43,44,52 Therefore, we hypothesize that the initial search structures of Msh2-Msh6 and Msh2-Msh3 are far more open than in model A to allow sliding along DNA. To build probable conformations of Msh2-Msh6 and Msh2-Msh3 capable of sliding on DNA and characterize their diffusion, we used an ENM-based algorithm to reveal the low-frequency collective modes (normal modes) of the individual monomers (Msh2, Msh3, and Msh6) to predict conformational changes that would create gaps in the dimer.
Figure 3.
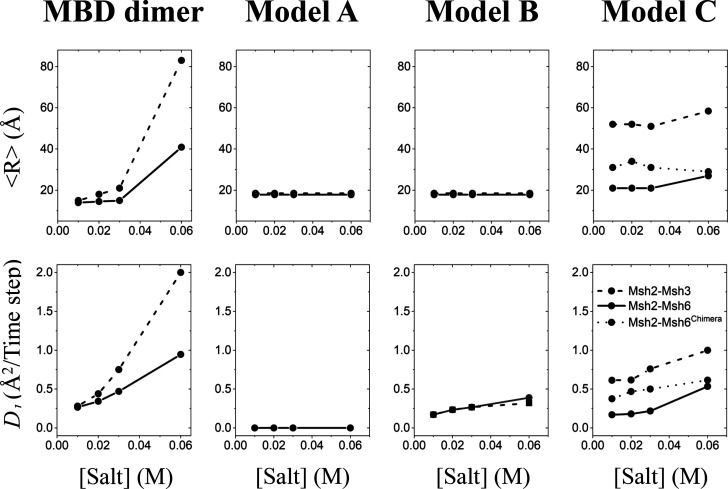
Effect of salt concentration on the interaction between DNA and (from left to right) an isolated MBD dimer, model A, model B, and model C as they diffuse along the DNA. The interaction between the MBD and nonspecific DNA is quantified by plotting the distance (R) between the center of mass of the MBD and that of the DNA. Average distances calculated from all trajectories are shown by black circles. Plots for Msh2-Msh6 and Msh2-Msh3 are compared in each model.
Characterization of MBD Diffusion along dsDNA: Comparing MBD26 and MBD23
We have shown, in the past, using coarse-grained simulations, that DNA-binding globular proteins slide along DNA using translation-coupled rotation, in which the protein follows the track defined by the DNA major groove.30,32,35 Here, we examined isolated MBD dimers (MBD23 and MBD26) to determine whether they can similarly diffuse along the DNA major groove.
Both dimers diffused bidirectionally while undergoing a translation-coupled rotation motion along the DNA (i.e., following the helical track of the DNA). The diffusion is found to be salt-dependent. At low salt concentrations, the dimers performed translation-coupled rotation diffusion to maintain a constant register with the helical contour of the DNA. Figure 3 (MBD dimer, top panel) shows that the average distance (⟨R⟩) between the center of mass of each MBD dimer and that of the DNA remained unchanged up to 0.03 M salt, but R increased at 0.06 M, indicating the occurrence of “hopping” and “three-dimensional (3D) diffusion” events. On the contrary, diffusion rate D1 increased with salt concentration (Figure 3, MBD dimer, bottom panel). Faster diffusion is expected at higher ionic strengths because of an increase in the frequency of microscopic DNA association–dissociation events that results from electrostatic screening between the phosphate backbone and the DNA-binding residues of the protein. MBD26 stayed closer to the DNA at higher salt concentrations compared with MBD23 (Figure 3, MBD dimer, top panel); MBD26 also showed much slower diffusion (lower D1 compared to that of MBD23) particularly at higher salt concentrations (Figure 3, MBD dimer, bottom panel). This difference between Msh2-Msh3 and Msh2-Msh6 can be rationalized by the stronger electrostatic interaction of MBD26-DNA compared with that of MBD23-DNA, as we will discuss below.
Rotational Motion of MBD Dimers around DNA Is Coupled with Their Translocation
If a sliding protein maintains continuous contact with the phosphate backbone, it will rotate 360° about the DNA approximately every 34 Å (10 bp), which is the helical pitch in a canonical B-DNA molecule. In this case, rotation along the helical path of the DNA enables the protein to continuously probe the base-pair content in the DNA major groove. Figure 4 shows coupling between rotation and translation as MBD26 and MBD23 diffuse on DNA. Panels A and D of Figure 4 show typical trajectories for MBD26 (top panel) and MBD23 (bottom panels) diffusing along the DNA. The trajectories were analyzed by following the location of the protein along the DNA axis (Z) and its rotation angle (θ) (Figure 1). The two simulations revealed ∼60 Å translocations of both dimers along the DNA (Figure 4A,D). Over that linear distance, the proteins also rotated around the DNA by an angle of ∼4π (Figure 4B,E).
Figure 4.
Observation of rotation-coupled diffusion in representative trajectories for the isolated MBD dimers MBD26 and MBD23 along nonspecific DNA simulated using coarse-grained molecular dynamics. (A and D) The trajectories show the translocation distance Z (red, left Y-axis) and the rotation angle θ (blue, right Y-axis) as a function of time at 0.01 M salt. The horizontal gray dashed lines indicate complete turns of the DNA (i.e., a distance of 34 Å along the DNA axis and a rotation of 2π). (B and E) At a low salt concentration of 0.01 M, both proteins are shown to rotate around the DNA while sliding and maintaining their interactions with the DNA (in the inset, the trace of the center of mass of the MBD dimer is colored green during sliding). The rotation (θ) vs translation (Z) linear scatter plots (average correlation between the rotation and translation motions of 0.98) indicate average slopes (⟨slope⟩ = 0.18) that are consistent with the geometrical properties of a B-DNA molecule (i.e., 2π/34 Å) indicating a strong coupling between protein translation and rotation and the helical pitch of dsDNA. (C and F) At a higher salt concentration of 0.03 M, linear diffusion events are shorter and more hopping events occur (trace of the MBD center of mass colored green). The rotation–translation scatter plots show weaker coupling of the rotation to the translation of the protein along DNA.
Panels B and E of Figure 4 show the rotation angle of the dimer against its initial position along the Z-axis at a low salt concentration of 0.01 M during a sliding event having a single trajectory. The plots show a clear linear relationship (correlation coefficient of 0.98) between rotation and translation with an average slope 2π/34 of 0.18 rad/Å. The inset figures show the paths taken by the center of mass of the dimer as it moves along the DNA surface during the sliding event, demonstrating that protein motion is coupled with the helical structure of the double-stranded DNA. When the salt concentration was increased to 0.03 M (Figure 4C,F), both the MBD26 and the MBD23 dimers showed translation along the DNA that was decoupled from the helical pitch of the major groove but still in the proximity of the surface of the DNA. The emergence of several parallel lines resulted from transient decoupling between the rotation angle and position, in which the protein may, for example, traverse between two neighboring grooves. Eventually, the translocation of both proteins along the DNA was also doubled at this salt concentration. However, at a high salt concentration of 0.06 M, both dimers performed mostly “hopping” and “3D diffusion” by moving away from the DNA (Figure 3, left panels).
In contrast to these observations, the search-mode diffusion coefficient of MutS is independent of salt concentration.21−23 This clearly indicates that, although the electrostatic interaction of MBD with DNA is crucial (illustrated in Figures 3 and 4), it is not the only factor influencing the search-mode diffusion of MSH proteins. Maintaining contact between the MBD and the DNA even at high salt concentrations is achieved with the help of the clamp domain that topologically traps the DNA duplex from the other side (Figure 1). Consequently, we modeled the full-length proteins by arranging the clamps in appropriate positions (Figure 2 and Figure S1; see Materials and Methods), which results in rotation-coupled diffusion.
Rotation-Coupled Diffusion of Full-Length Msh2-Msh6 and Msh2-Msh3 Heterodimers
The coarse-grained simulations based on the crystal structures of Msh2-Msh6 and Msh2-Msh3 bound to DNA did not show any rotation or lateral movement along nonspecific linear B-DNA (Figure 3). This observation confirmed our proposal that the initial search mode of the Msh proteins must be significantly more open than the conformations found in the crystal structures. The model structures (model B, Figure 2 and Figure S1) differ from the crystal conformations with respect to their DNA-binding domains, especially the clamp domain. Here, the rotation of the Msh2 and Msh6 clamp domains in the X–Y plane (where the elongated dimension of the DNA constitutes the Z-axis) from their crystal conformation to model B enhanced the space at the bottom of the DNA-binding channel of the dimer. This motion has also been predicted as a long-range allosteric communication mechanism.53,54 The displacement of the clamp domains also had a small component in the Z-direction. This allows the MBDs to align along the DNA groove during the simulation without creating a steric hindrance between the clamp domains and the DNA.
Msh2-Msh6 and Msh2-Msh3 dimers indeed display rotational motion about the DNA, while maintaining continuous contact with the DNA (Figure 3, top middle panel). Interestingly, diffusion rate D1 was independent of salt concentration (Figure 3, bottom middle panel), as observed experimentally for the MutS protein in search mode.21−23 In general, the ionic strength should not affect the diffusion of a protein during sliding that does not undergo “hopping” and “3D diffusion”, because microscopic ionic interactions between the protein and the DNA are continually preserved and shielded during movement.29,55 The function of Msh proteins suggests that they must act as a sliding protein during their mismatch search mode; the diffusion characteristics of the models showed that they serve this purpose well.
The coupling between rotation (θ) and translation along the Z-axis of full-length Msh2-Msh6 and Msh2-Msh3 at a low salt concentration (0.01 M) is shown in Figure 5 (top and bottom left panels). The two simulations show complete rotation of both proteins around DNA, during which they translocated ∼34 Å along the length of the DNA. There was excellent linear correlation (correlation coefficients of 0.98 and 0.97 for Msh2-Msh6 and Msh2-Msh3, respectively) between rotation and translation, with an average slope of 0.18 rad/Å. This value is very close to the value of 2π/34 Å for the helical pitch in canonical B-DNA (i.e., one turn per 10 bp rise). The position of the center of mass of the MBD6 and MBD3 (green dots in insets) shows the sliding of the protein along the DNA groove. With increasing salt concentrations, a subtle translocation of the protein on the DNA surface but not along the helical pitch was observed. Thus, at higher salt concentrations, the protein propagated along the Z-axis without rotating around the DNA. Sliding events at higher salt concentrations became more transient, and fast transitions of the MBD along the backbone toward an adjacent major groove became more frequent. The sliding events became significantly shorter and less committed to the DNA groove track, resulting in much weaker θ–Z correlations. Significantly, the diffusion rate did not increase considerably with an increasing salt concentration (Figure 3). All of these results may suggest a major difference in the mechanism of diffusion between MSH proteins and other globular DNA-binding proteins, such as various transcription factors and enzymes. In the case of a globular protein, they adopt a helical bound motion in the DNA major groove as they slide under low-salt conditions, while at higher salt concentrations, they sample major groove positions locally and rapidly translocate to other sites using the hopping mechanism.30,34,35,43 MSH proteins also translocate at higher salt concentrations, but to a much lesser extent. Perhaps their functional importance does not allow them to undergo rapid translocation via hopping at the expense of search fidelity that is achieved via sliding. Our models show that the topology of the protein plays a major role in achieving that goal.
Figure 5.

Comparison of rotation-coupled diffusion in representative trajectories for the Msh2-Msh6 (A and B) and Msh2-Msh3 (C and D) models along nonspecific DNA. (A and B) Msh2-Msh6 diffusion at a low salt concentration of 0.01 M. Both model B and model C are shown to rotate around the DNA while translocating and maintaining their interactions with the DNA (insets show a trace of the center of mass of the MBDs colored green during sliding). Rotation (θ) vs translation (Z) linear scatter plots (average correlation between the rotation and translation motions of 0.98) with average slopes of 0.18 and 0.17, respectively, indicate strong coupling between protein translation and rotation and the helical pitch of dsDNA. (C and D) Msh2-Msh3 diffusion at a low salt concentration of 0.01 M. Model B shows strong coupling between protein translation and rotation along the DNA (slope = 0.18; correlation = 0.97). For model C, the traces of the center of mass of the MBDs show that it dissociates from the DNA and that the protein interacts with the DNA using only the clamp domain (Figure 7). The θ vs Z scatter plot shows that rotation–translation coupling is weaker.
Role of the MBD and the Clamp Domain Electrostatics in MSH Diffusion on DNA
The observed differences in the affinity of MBD23 and MBD26 for DNA during simulations at different salt concentrations suggest that their binding affinities are largely electrostatic in nature. Furthermore, the sliding dynamics observed for MBD23 and MBD26 indicates that their electrostatic potential and geometry of the interface with DNA are similar to those of other DNA-binding proteins and support the rotation-coupled translation dynamics along DNA.32 While many DNA-binding proteins follow rotation-coupled translation diffusion along DNA, it is not a universal feature and some DNA-binding proteins33,37,56 (as well as other positively charged proteins32) follow diffusion where the translation along the DNA is uncoupled from rotation. Using the APBS plug-in to PyMOL, we see greater positive electrostatic potential associated with the DNA binding area of MBD6 relative to that of MBD3. This was further confirmed by calculating the energy of binding of MBD23 and MBD26 to DNA using APBS (Figure 6). The electrostatic binding affinities of MBD23 and MBD26 for DNA are approximately −6 and −47 kcal/mol, respectively, indicating much stronger electrostatic interactions between MBD26 and DNA compared to those of MBD23. This is consistent with the results of the coarse-grained simulations involving the MBD dimers that reflect a weaker dependence on salt concentration for MBD26 than for MBD23 (Figure 3). This is also consistent with the coarse-grained simulations of model C of Msh2-Msh3 and Msh2-Msh6 (Figures 5C and 7B), where the MBD23 region of Msh2-Msh3 dissociated from the DNA and remained associated with the DNA via the dimer’s clamp region. In contrast, the MBD26 region of Msh2-Msh6 did not dissociate from the DNA (Figures 5B and 7A).
Figure 6.

Differences in the electrostatic potential for the isolated MBD dimers of Msh2-Msh3 and Msh2-Msh6. The MBDs of Msh3 and Msh6 are shown with the electrostatic potential mapped on the solvent accessible surface of these domains, while the MBD of Msh2 is shown as a green cartoon. The DNA is also depicted as a cartoon. The range of the colors is from −3 kT/e (red) to 3 kT/e (blue). Electrostatic isosurfaces are computed using the Adaptive Poisson–Boltzmann Solver (APBS) plug-in in PyMOL with a probe radius of 1.4 Å. The calculated binding energies for the MBDs of Msh2-Msh3 and DNA were approximately −6 kcal/mol, compared to a value of approximately −47 kcal/mol for the Msh2-Msh6 dimer and DNA.
Figure 7.

Representative conformation of model C of Msh2-Msh6 and Msh2-Msh3 diffusing along the DNA in coarse-grained simulations. (A) The Msh2-Msh6 protein embraces the DNA using clamp domain of both Msh2 and Msh6. The MBD6 maintains constant contact with the DNA. (B) DNA dissociates from MBD3, and the Msh2-Msh3 protein interacts with the DNA using the clamp domains.
An Extended Gap in the Clamp Region of Msh2-Msh3, but Not Msh2-Msh6, Allows the Release of DNA
An interesting difference between the diffusion mechanisms of Msh2-Msh3 and Msh2-Msh6 is that only the former can bypass nucleosomes and other protein roadblocks on DNA, as revealed by single-molecule fluorescence microscopy.36 Avoidance of roadblocks implies dissociation from the DNA, which in turn suggests that the facilitated DNA scanning diffusion mechanism of Msh2-Msh3 includes both one-dimensional sliding and hopping. Our coarse-grained models showed that Msh2-Msh3 can indeed detach from DNA during simulation, whereas Msh2-Msh6 cannot.
We built more-extended conformations of Msh2-Msh6 and Msh2-Msh3 (model C), in which the gap between the clamp domains of the two monomers was longer than in model B (Figure 2 and Figure S1). During coarse-grained simulations, the Msh2-Msh3 version of model C showed quite different behavior relative to that of the Msh2-Msh6 version. The MBDs of Msh2-Msh3 separated completely from the DNA (Figures 3 and 5D); interaction with the DNA was instead maintained by the positively charged residues of the clamp region at low salt concentrations (Figure 7B). At high salt concentrations (0.06 M), Msh2-Msh3 did undergo dissociation–association events, or “hopping”. Although the isolated MBD23 dimer did remain associated with the DNA at low salt concentrations, it appears that the positively charged clamp regions in Msh2-Msh3 have greater affinity for the DNA than do the MBDs.
In contrast to the behavior of Msh2-Msh3, the MBDs of model C of Msh2-Msh6 did not dissociate from the DNA at lower salt concentrations, diffusing via the rotation-coupled translation mechanism [θ vs Z, slope of 0.17 rad/Å (Figures 3 and 5B)]. This behavior is consistent with the apparent greater electrostatic affinity of MBD26 for DNA, relative to MBD23, as discussed above. This difference in the affinity for DNA between the two molecules is consistent with the 3-fold larger diffusion rate obtained for Msh2-Msh3 versus that obtained for Msh2-Msh6 (Figure 3, bottom left panel). This is also consistent with experimental results that show a 4-fold greater diffusion coefficient for Msh2-Msh3 than for Msh2-Msh6.23,28
All-Atom Simulations of Msh2-Msh3 and Msh2-Msh6
As one might expect, full atomistic simulations of the Msh2-Msh3/Msh6 dimers in complex with their cognate DNA from crystal structures did not show any movement of the dimers relative to the DNA. Somewhat unexpected was the fact that neither Msh dimer showed any movement around or along nonspecific straight B-DNA. This lack of movement on straight DNA suggested that the initial search mode of these proteins was different from that observed in the crystal structures and prompted our attempts to model more open conformations using normal-mode analyses.
Results from these simulations were nevertheless informative. The energies of interaction between the clamp regions of Msh3 and Msh6 for both specific and nonspecific DNA were similar (Figure 8A), although the interaction of Msh3 clamp region with the DNA was somewhat stronger. In contrast, the energy of interaction between the clamp regions of Msh2-Msh3 differed noticeably from that of the Msh2-Msh6 dimer (Figure 8B). The clamp region of Msh2 interacted less favorably with the clamp region of Msh3 relative to Msh6 on the cognate mispaired DNA. This difference was even more pronounced on nonspecific DNA, where Msh2 hardly interacts with Msh3 in the clamp region. This would greatly facilitate opening the gap between Msh2 and Msh3 and contribute to the ability of the Msh2-Msh3 dimer to perform “hopping”, as seen in the coarse-grained simulations.
Figure 8.

Conformational stability of Msh2-Msh3 and Msh2-Msh6 from atomistic simulations. The energetics of interactions between some constituent domains is shown by the distributions of these energies of each dimer of either specific DNA (taken from the PDB of Msh2-Msh3 and Msh2-Msh6) or nonspecific DNA (B-DNA conformation) in four trajectories each of 320 ns. (A) Interaction between the clamp domains of Msh3 (red) or Msh6 (blue) and DNA. (B) Strength of interaction between the clamp domain of Msh3 or Msh6 and the clamp domain of Msh2 (blue or red, respectively).
Msh2-Msh6 Chimera
Brown et al.24 performed an experiment to elucidate the role of MBDs in roadblock bypassing. They prepared a chimeric version of Msh2-Msh6 in which MBD6 was replaced by MBD3, which demonstrated that MBD3 imparts roadblock bypass activity to chimeric Msh2-Msh6. They concluded that MBD3 is sufficient to allow MSH complex hopping. Similarly, we built a chimeric model C of Msh2-Msh6 (Msh2-Msh6Chimera) by replacing MBD6 with MBD3. Both mean distance R of the protein from the DNA and diffusion rate D1 of Msh2-Msh6Chimera lay between those of Msh2-Msh6 and Msh2-Msh3 (Figure 3, right panels), which is consistent with experiments.36 The properties of the chimera suggest that the difference between the electrostatics of MBD3 and MBD6 contributes to the different characteristics of Msh2-Msh3 compared to Msh2-Msh6.
Discussion and Conclusions
Various molecular features of the diffusion mechanism of many transcription factors or enzyme proteins when they diffuse along DNA while searching for their target site have been studied using coarse-grained molecular dynamics simulations. Here, we studied the diffusion of the two human mismatch repair proteins, aiming at understanding their diffusion along DNA and specifically under which conditions they diffuse while tracking the major groove (i.e., slide along DNA). Furthermore, our study was motivated by understanding the molecular origin of their different diffusion speed and different ability to bypass an obstacle while diffusing.36 Our initial coarse-grained simulations for both Msh2-Msh3 and Msh2-Msh6 heterodimers revealed that the crystal structure conformations for these complexes were not likely correct models for their initial search mode along DNA. Coarse-grained simulations using isolated MBD dimers from both systems showed the likely mode of interaction of the MBDs with canonical double-stranded DNA during this initial search. The MBD dimers followed the track of the major groove (i.e., moving spirally while diffusing along the DNA) that should be required for effective identification of mismatched base pairs. Normal-mode analysis of the crystal structures suggested structures in which the clamp regions were more open. This allowed us to model the dimers in conformations (model B and model C) that allowed the MBDs to interact with the DNA in the mode shown by the simulations of the MBDs but also prevented clashes between the protein clamp regions and the DNA. Coarse-grained simulations of model B demonstrated that it could move along DNA while the MBD regions tracked the major groove, in keeping with experimental observations. Model C showed that Msh2-Msh3 could dissociate from the DNA, while Msh2-Msh6 did not at lower salt concentrations. This is consistent with the ability of Msh2-Msh3 to bypass obstacles such as nucleosomes, while Msh2-Msh6 cannot. We propose, therefore, that the initial search mode is a conformation akin to model B, where transient dynamics allow Msh2-Msh3 to access a conformation akin to model C, enabling “hopping” and obstacle bypass. Msh2-Msh6, on the contrary, did not dissociate from the DNA even in the more open form, apparently due to the greater electrostatic affinity of MBD6 for DNA, based on our calculations of electrostatic potential. We note that model B and model C are possible conformations of the prerecognition state of MSH proteins when diffusing along nonspecific DNA. These models mostly serve to highlight the need of broadening the internal region for DNA binding in comparison to that found in the crystal structure with specific DNA, yet there is uncertainty regarding the molecular details of this conformational state.
The coarse-grained simulations suggest that Msh2-Msh6 performs a slower movement by keeping tighter contact with the DNA via the MBD of Msh6. Electrostatic interactions between the DNA-binding domains (MBD6, MBD3, and the lower clamp domain) and the DNA backbone play a major role in this facilitated diffusion. Because the main function of Msh2-Msh6 is to recognize a single mismatch, it must perform a more detailed search compared with Msh2-Msh3, as observed in our simulations. A 4-fold higher diffusion coefficient for Msh2-Msh3 compared with that of Msh2-Msh6 is comparable to that observed experimentally and can be explained by its weaker electrostatic affinity for DNA. The tighter electrostatic interactions of Msh2-Msh6 with DNA suggest that the coupling between rotation and translation during one-dimensional diffusion is stronger for Msh2-Msh6 than for Msh2-Msh3. Accordingly, the diffusion of Msh2-Msh3 is expected to be more sensitive to salt concertation, as was shown experimentally.36 Full-atom simulations suggest that the interactions between the clamp regions of Msh2-Msh3 are much weaker than those in Msh2-Msh6 when interacting with nonspecific DNA. This behavior may contribute to the ability of Msh2-Msh3 to open and dissociate from the DNA, although experimental results with the Msh2-Msh6 chimera suggest that the main ability to “hop” comes from only MBD3. This does not exclude, however, a contributing role to this ability from the clamp region of Msh3. Our study serves as another example for the linear diffusion of ring-shaped proteins along DNA33,37,57 and shows that the details of their structures govern the existence of coupled rotation–translation diffusion.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.0c00669.
Models A–C of Msh2-Msh3 (PDF)
This work was supported by the Kimmelman Center for Macromolecular Assemblies. Y.L. holds The Morton and Gladys Pickman professional chair in Structural Biology.
The authors declare no competing financial interest.
Supplementary Material
References
- Kolodner R. D. (1995) Mismatch repair: mechanisms and relationship to cancer susceptibility. Trends Biochem. Sci. 20, 397–401. 10.1016/S0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- Peltomäki P. (2005) Lynch syndrome genes. Fam. Cancer 4, 227–232. 10.1007/s10689-004-7993-0. [DOI] [PubMed] [Google Scholar]
- Peltomaki P. (2003) Role of DNA mismatch repair defects in the pathogenesis of human cancer. J. Clin. Oncol. 21, 1174–1179. 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- van den Broek W. J.; Nelen M. R.; Wansink D. G.; Coerwinkel M. M.; te Riele H.; Groenen P. J.; Wieringa B. (2002) Somatic expansion behaviour of the (CTG) n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch–repair proteins. Human molecular genetics 11, 191–198. 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- López Castel A.; Cleary J. D.; Pearson C. E. (2010) Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 11, 165. 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- McMurray C. T. (2010) Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 11, 786. 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R. D. (2016) A personal historical view of DNA mismatch repair with an emphasis on eukaryotic DNA mismatch repair. DNA Repair 38, 3. 10.1016/j.dnarep.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R. R.; Pluciennik A.; Burdett V.; Modrich P. L. (2006) DNA mismatch repair: functions and mechanisms. Chem. Rev. 106, 302–323. 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- Junop M. S.; Obmolova G.; Rausch K.; Hsieh P.; Yang W. (2001) Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell 7, 1–12. 10.1016/S1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- Habraken Y.; Sung P.; Prakash L.; Prakash S. (1998) ATP-dependent assembly of a ternary complex consisting of a DNA mismatch and the yeast MSH2-MSH6 and MLH1-PMS1 protein complexes. J. Biol. Chem. 273, 9837–9841. 10.1074/jbc.273.16.9837. [DOI] [PubMed] [Google Scholar]
- Mendillo M. L.; Hargreaves V. V.; Jamison J. W.; Mo A. O.; Li S.; Putnam C. D.; Woods V. L.; Kolodner R. D. (2009) A conserved MutS homolog connector domain interface interacts with MutL homologs. Proc. Natl. Acad. Sci. U. S. A. 106, 22223–22228. 10.1073/pnas.0912250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao P.; LeBlanc S. J.; Case B. C.; Elston T. C.; Hingorani M. M.; Erie D. A.; Weninger K. R. (2020) Recurrent mismatch binding by MutS mobile clamps on DNA localizes repair complexes nearby. Proc. Natl. Acad. Sci. U. S. A. 117, 17775–17784. 10.1073/pnas.1918517117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D. (2020) MutS sliding clamps on an uncertain track to DNA mismatch repair. Proc. Natl. Acad. Sci. U. S. A. 117, 20351–20353. 10.1073/pnas.2013560117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groothuizen F. S.; Winkler I.; Cristóvão M.; Fish A.; Winterwerp H. H.; Reumer A.; Marx A. D.; Hermans N.; Nicholls R. A.; Murshudov G. N.; et al. (2015) MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. eLife 4, e06744. 10.7554/eLife.06744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren J. J.; Pohlhaus T. J.; Changela A.; Iyer R. R.; Modrich P. L.; Beese L. S. (2007) Structure of the human MutSα DNA lesion recognition complex. Mol. Cell 26, 579–592. 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Gupta S.; Gellert M.; Yang W. (2012) Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 19, 72. 10.1038/nsmb.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groothuizen F. S.; Sixma T. K. (2016) The conserved molecular machinery in DNA mismatch repair enzyme structures. DNA Repair 38, 14–23. 10.1016/j.dnarep.2015.11.012. [DOI] [PubMed] [Google Scholar]
- Edelbrock M. A.; Kaliyaperumal S.; Williams K. J. (2013) Structural, molecular and cellular functions of MSH2 and MSH6 during DNA mismatch repair, damage signaling and other noncanonical activities. Mutat. Res., Fundam. Mol. Mech. Mutagen. 743, 53–66. 10.1016/j.mrfmmm.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T. T.; Hombauer H. (2016) Visualization of mismatch repair complexes using fluorescence microscopy. DNA Repair 38, 58–67. 10.1016/j.dnarep.2015.11.014. [DOI] [PubMed] [Google Scholar]
- Kim D.; Fishel R.; Lee J.-B. (2018) Coordinating multi-protein mismatch repair by managing diffusion mechanics on the DNA. J. Mol. Biol. 430, 4469–4480. 10.1016/j.jmb.2018.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C.; Cho W.-K.; Song K.-M.; Cook C.; Yoon T.-Y.; Ban C.; Fishel R.; Lee J.-B. (2011) MutS switches between two fundamentally distinct clamps during mismatch repair. Nat. Struct. Mol. Biol. 18, 379. 10.1038/nsmb.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.-K.; Jeong C.; Kim D.; Chang M.; Song K.-M.; Hanne J.; Ban C.; Fishel R.; Lee J.-B. (2012) ATP alters the diffusion mechanics of MutS on mismatched DNA. Structure 20, 1264–1274. 10.1016/j.str.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J.; Wang F.; Redding S.; Plys A. J.; Fazio T.; Wind S.; Alani E. E.; Greene E. C. (2012) Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc. Natl. Acad. Sci. U. S. A. 109, E3074–E3083. 10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quessada-Vial A.; van Oijen A. M. (2012) How DNA-repair proteins find their targets. Proc. Natl. Acad. Sci. U. S. A. 109, 18243–18244. 10.1073/pnas.1215845109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J.; Chowdhury A.; Surtees J. A.; Shimada J.; Reichman D. R.; Alani E.; Greene E. C. (2007) Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol. Cell 28, 359–370. 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradia S.; Acharya S.; Fishel R. (1997) The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell 91, 995–1005. 10.1016/S0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- Cho W. K.; Jeong C.; Kim D.; Chang M.; Song K. M.; Hanne J.; Ban C.; Fishel R.; Lee J. B. (2012) ATP Alters the Diffusion Mechanics of MutS on Mismatched DNA. Structure 20, 1264–1274. 10.1016/j.str.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obmolova G.; Ban C.; Hsieh P.; Yang W. (2000) Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407, 703. 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- Tafvizi A.; Huang F.; Fersht A. R.; Mirny L. A.; van Oijen A. M. (2011) A single-molecule characterization of p53 search on DNA. Proc. Natl. Acad. Sci. U. S. A. 108, 563–568. 10.1073/pnas.1016020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givaty O.; Levy Y. (2009) Protein sliding along DNA: dynamics and structural characterization. J. Mol. Biol. 385, 1087–1097. 10.1016/j.jmb.2008.11.016. [DOI] [PubMed] [Google Scholar]
- Bhattacherjee A.; Levy Y. (2014) Search by proteins for their DNA target site: 1. The effect of DNA conformation on protein sliding. Nucleic Acids Res. 42, 12404–12414. 10.1093/nar/gku932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigman L. S.; Levy Y. (2020) Protein Diffusion on Charged Biolymers: DNA versus Microtubule. Biophys. J. 118, 3008–3018. 10.1016/j.bpj.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt H. M.; Rozenberg H.; Daitchman D.; Levy Y. (2020) Does PCNA diffusion on DNA follow a rotation-coupled translation mechanism?. Nat. Commun. 11, 5000. 10.1038/s41467-020-18855-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazanov N.; Marcovitz A.; Levy Y. (2013) Asymmetric DNA-search dynamics by symmetric dimeric proteins. Biochemistry 52, 5335–5344. 10.1021/bi400357m. [DOI] [PubMed] [Google Scholar]
- Marcovitz A.; Levy Y. (2012) Sliding dynamics along DNA: a molecular perspective. Innovations in Biomolecular Modeling and Simulation 24, 236–262. 10.1039/9781849735056-00236. [DOI] [Google Scholar]
- Brown M. W.; Kim Y.; Williams G. M.; Huck J. D.; Surtees J. A.; Finkelstein I. J. (2016) Dynamic DNA binding licenses a repair factor to bypass roadblocks in search of DNA lesions. Nat. Commun. 7, 10607. 10.1038/ncomms10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitchman D.; Greenblatt H. M.; Levy Y. (2018) Diffusion of ring-shaped proteins along DNA: case study of sliding clamps. Nucleic Acids Res. 46, 5935–5949. 10.1093/nar/gky436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K.; Piana S.; Palmo K.; Maragakis P.; Klepeis J. L.; Dror R. O.; Shaw D. E. (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Struct., Funct., Genet. 78, 1950–1958. 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K.; Sanejouand Y.-H. (2004) ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 32, W610–W614. 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi C.; Nymeyer H.; Onuchic J. N. (2000) Topological and energetic factors: what determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J. Mol. Biol. 298, 937–953. 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- Levy Y.; Wolynes P. G.; Onuchic J. N. (2004) Protein topology determines binding mechanism. Proc. Natl. Acad. Sci. U. S. A. 101, 511–516. 10.1073/pnas.2534828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel J. K.; Levi M.; Raghunathan M.; Lammert H.; Hayes R. L.; Onuchic J. N.; Whitford P. C. (2016) SMOG 2: A Versatile Software Package for Generating Structure-Based Models. PLoS Comput. Biol. 12, e1004794. 10.1371/journal.pcbi.1004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuzman D.; Azia A.; Levy Y. (2010) Searching DNA via a ″Monkey Bar″ Mechanism: The Significance of Disordered Tails. J. Mol. Biol. 396, 674–684. 10.1016/j.jmb.2009.11.056. [DOI] [PubMed] [Google Scholar]
- Khazanov N.; Levy Y. (2011) Sliding of p53 along DNA can be modulated by its oligomeric state and by cross-talks between its constituent domains. J. Mol. Biol. 408, 335–355. 10.1016/j.jmb.2011.01.059. [DOI] [PubMed] [Google Scholar]
- Zandarashvili L.; Esadze A.; Vuzman D.; Kemme C. A.; Levy Y.; Iwahara J. (2015) Balancing between affinity and speed in target DNA search by zinc-finger proteins via modulation of dynamic conformational ensemble. Proc. Natl. Acad. Sci. U. S. A. 112, E5142–E5149. 10.1073/pnas.1507726112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandarashvili L.; Vuzman D.; Esadze A.; Takayama Y.; Sahu D.; Levy Y.; Iwahara J. (2012) Asymmetrical roles of zinc fingers in dynamic DNA-scanning process by the inducible transcription factor Egr-1. Proc. Natl. Acad. Sci. U. S. A. 109, E1724–1732. 10.1073/pnas.1121500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuzman D.; Levy Y. (2010) DNA search efficiency is modulated by charge composition and distribution in the intrinsically disordered tail. Proc. Natl. Acad. Sci. U. S. A. 107, 21004–21009. 10.1073/pnas.1011775107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leven I.; Levy Y. (2019) Quantifying the two-state facilitated diffusion model of protein-DNA interactions. Nucleic Acids Res. 47, 5530–5538. 10.1093/nar/gkz308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcovitz A.; Levy Y. (2013) Weak frustration regulates sliding and binding kinetics on rugged protein-DNA landscapes. J. Phys. Chem. B 117, 13005–13014. 10.1021/jp402296d. [DOI] [PubMed] [Google Scholar]
- Guex N.; Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723. 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Vuzman D.; Polonsky M.; Levy Y. (2010) Facilitated DNA search by multidomain transcription factors: cross talk via a flexible linker. Biophys. J. 99, 1202–1211. 10.1016/j.bpj.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.; Francis J.; Sharma M.; Law S. M.; Predeus A. V.; Feig M. (2016) Long-range signaling in MutS and MSH homologs via switching of dynamic communication pathways. PLoS Comput. Biol. 12, e1005159. 10.1371/journal.pcbi.1005159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S.; Law S. M.; Feig M. (2009) Deciphering the mismatch recognition cycle in MutS and MSH2-MSH6 using normal-mode analysis. Biophys. J. 96, 1707–1720. 10.1016/j.bpj.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J.; Plys A. J.; Visnapuu M. L.; Alani E.; Greene E. C. (2010) Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat. Struct. Mol. Biol. 17, 932–938. 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuculis L.; Abil Z.; Zhao H.; Schroeder C. M. (2016) TALE proteins search DNA using a rotationally decoupled mechanism. Nat. Chem. Biol. 12, 831. 10.1038/nchembio.2152. [DOI] [PubMed] [Google Scholar]
- Li H. C.; Doruker P.; Hu G.; Bahar I. (2020) Modulation of Toroidal Proteins Dynamics in Favor of Functional Mechanisms upon Ligand Binding. Biophys. J. 118, 1782–1794. 10.1016/j.bpj.2020.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.