Abstract
Objective:
Acute respiratory distress syndrome (ARDS) is frequently under recognized, and associated with increased mortality. Previously, we developed a model that utilized machine learning and natural language processing (NLP) of text from radiology reports to identify ARDS. The model showed improved performance in diagnosing ARDS when compared to a rule-based method. In this study, our objective was to externally validate the NLP model in patients from an independent hospital setting.
Design:
Secondary analysis of data across five prospective clinical studies.
Setting:
An urban, tertiary care, academic hospital
Patients:
Adult patients admitted to the medical intensive care unit and at-risk for ARDS
Interventions:
None
Measurements and Main Results:
The NLP model was previously derived and internally validated in burn, trauma, and medical patients at Loyola University Medical Center. Two machine learning models were examined with the following text features from qualifying radiology reports: (1) word representations (n-grams); (2) standardized clinical named entity mentions mapped from the National Library of Medicine Unified Medical Language System. The models were externally validated in a cohort of 235 patients at the University of Chicago Medicine, among which 110 (47%) were diagnosed with ARDS by expert annotation. During external validation, the n-gram model demonstrated good discrimination between ARDS and non-ARDS patients (c-statistic 0.78; 95% CI 0.72–0.84). The n-gram model had a higher discrimination for ARDS when compared to the standardized named entity model, although not statistically significant (c-statistic 0.78 vs 0.72, P=0.09). The most important features in the model had good face validity for ARDS characteristics but differences in frequencies did occur between hospital settings.
Conclusions:
Our computable phenotype for ARDS had good discrimination in external validation and may be used by other health systems for case-identification. Discrepancies in feature representation are likely due to differences in characteristics of the patient cohorts.
INTRODUCTION
Acute respiratory distress syndrome (ARDS) is a common condition that is associated with multiple organ failure, severe hypoxemia, and a high rate of mortality.1 ARDS is traditionally hard to recognize because of complex and heterogenous phenotypic representations.2 In addition, ARDS recognition requires extensive effort to integrate information from laboratory results, radiology reports, respiratory data, and disease characteristics in a timely manner.3–6 Methods for automated ARDS detection that utilize a keyword-based search on chest radiograph reports have previously been developed;7,8 however, they have not been updated to reflect current definitions of ARDS and do not include computed tomography (CT) reports. Additionally, these methods may not be applicable in other hospital settings due to variations in documentation, clinical practice, and patient case-mix. Prior work reported a high rate of false positives when these techniques were applied in an independent setting.9–11 The development of a generalizable model that can accurately identify ARDS onset irrespective of setting may facilitate research in the current digital era of electronic health record (EHR) data. Further, they may also guide clinical decision support for better and more timely recognition of ARDS at point of care.12,13
In a recent study, we developed a novel supervised machine learning model based on natural language processing (NLP) of text from radiology reports to identify ARDS patients.14 The model was developed in a cohort of 533 medical, trauma, and burn patients that were admitted to Loyola University Medical Center, among which 138 (26%) developed ARDS.5 The model demonstrated improved performance in ARDS identification compared to traditional methods (accuracy 77% vs. 67%, positive predictive value 55% vs. 42%) in a Loyola test cohort. Further, in a test dataset of 24-hour radiology reports within a qualifying PaO2/FiO2 ratio, the model had an area under the receiver operating characteristic curve (AUC) of 0.73 (95% CI: 0.61–0.85). However, the external applicability of the model was not measured because the model was derived and validated at a single center.
The aim of this study was to externally validate the Loyola NLP model for the prediction of ARDS in a separate, independent hospital setting. We hypothesized that the NLP model developed at Loyola will have good discrimination for ARDS in patients receiving mechanical ventilation at University of Chicago Medicine (UCM). We further hypothesized that utilizing standardized clinical terminology as input features would improve discrimination performance compared to using raw text-based features.
METHODS
Setting and study population
The analysis cohort at UCM comprised of mechanically ventilated intensive care unit (ICU) patients enrolled in three clinical trials (helmet ventilation ARDS,15 early mobilization,16 and Dexmedetomidine versus Propofol for ICU sedation [ClinicalTrials.gov Identifier: NCT01059929]) and two observational studies (delirium assessments during daily awakening of sedation17 and assessment of biomarkers in shock [IRB18–1163]). A total of 477 patient admissions were considered for the analytic cohort, out of which 338 had a qualifying PO2/FiO2 ratio of less than 300.5 Further inclusion criteria were: (1) patients received mechanical ventilation (n=316); (2) the qualifying PO2/FiO2 ratio occurred within 7 days of hospital admission (n=244); and (3) availability of chest radiology reports (radiograph and CTs) within 24 hours of qualifying PO2/FiO2 ratio (n=235) (Figure 1). The study was approved by the University of Chicago Institutional Review Board (IRB18–0119).
Figure 1:
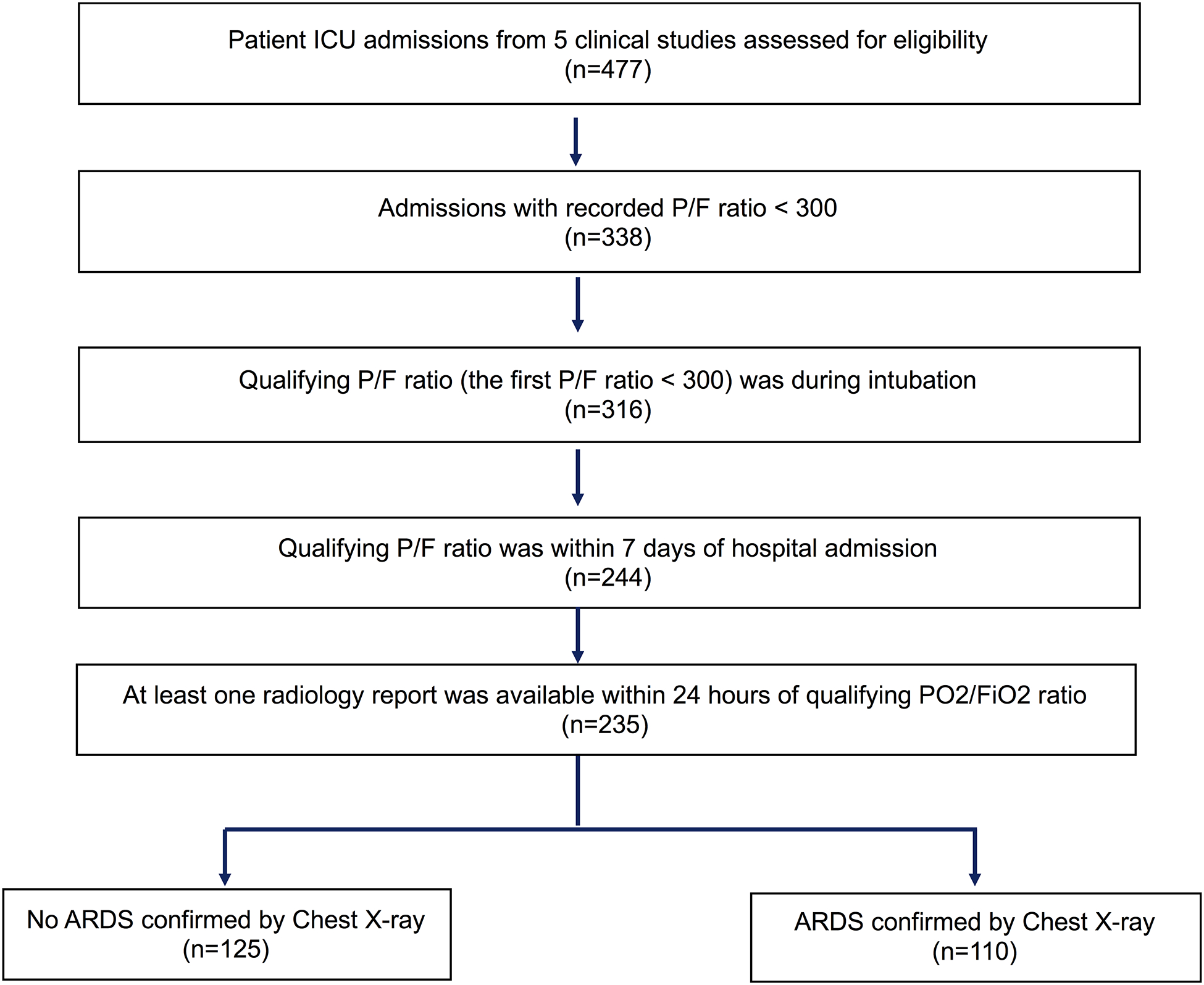
Workflow indicating patient selection at University of Chicago Medicine
Data sources
All radiology reports within a 24-hour period of qualifying oxygenation criteria were extracted from the electronic health record (EHR; Epic, Verona, WI). Other variables such as patient demographics and discharge disposition were collected from administrative data. All data were extracted from the Clinical Research Data Warehouse maintained by the Center for Research Informatics (CRI) at the University of Chicago.
Analysis Plan
The NLP model that was trained at Loyola was applied to a test dataset of chest radiology reports at UCM to predict ARDS. The primary outcome of ARDS at UCM was established by expert review of chest radiographs and chest CT images obtained during the entire hospital encounter for each patient. Expert review was performed by a board-certified pulmonary and critical care medicine physician and ARDS researcher (BP) and was in accordance with the Berlin definition of ARDS.5 Baseline characteristics were presented as means with standard deviations for continuous variables, medians with interquartile ranges for variables that are not likely to be normally distributed, and proportions for categorical variables. Descriptive statistics were used to assess differences at baseline between ARDS and non-ARDS patients, with t-test for comparing means, nonparametric Wilcoxon rank sum test for comparing medians, and chi-square tests for comparing of two or more proportions.
We considered validation of two distinct Loyola NLP model configurations based on the processing of text within radiology reports from UCM patients. In the first method, we constructed a feature matrix from text within radiology reports based on counts of word n-grams which are sequences of words of length n (e.g., unigram = “consolidation”; bigram = “bilateral consolidation”). We utilized unigram (n=1) representations and applied a term-frequency, inverse document-frequency normalization similar to prior work.14 Additional filtering included removing stop words and character normalization including removal of punctuation. The unigram features from the ARDS computable phenotype that was previously derived and internally validated at Loyola14 was applied to the UCM external validation cohort. As a comparison, we implemented a keyword-based search model that utilized joint occurrence of exact and synonym matches of ‘bilateral’ and ‘opacities’, as determined by Azzam et al., to predict ARDS. Explicit mentions of ARDS and pulmonary edema were also included in accordance with the methods in Azzam et al.7
In the second method, clinical radiology reports from UCM were processed using the clinical Text Analysis and Knowledge Extraction System (cTAKES, http://ctakes.apache.org) to map text to named entity mentions related to diseases/disorders, signs/symptoms, findings, procedures, anatomical sites etc., that were derived from the Unified Medical Language System (UMLS).18 Entity mentions were mapped to a concept unique identifier (CUI) followed by negation analyzation using the cTAKES negation module as illustrated in prior work.14 For instance, the term “consolidation” is a finding and mapped to the CUI “C0239027”, while the term “bilateral” is a spatial concept mapped to CUI “C0238767”. Other examples of CUI-based mappings for radiology notes are illustrated in a recent review.19 The CUI features from the Loyola model were also applied to the UCM external validation cohort. We created separate support vector machine classifiers for both the text-based and CUI-based features. The following parameters were applied from the trained model: (1) linear kernel; (2) regularization parameter with degree of tolerance (C)=1; (3) balanced class weight. Platt scaling was applied to convert the binary classifications into predicted probabilities for ARDS.20 Further details on derivation of both models are available in Afshar et al,14 and the model is available at https://github.com/AfsharJoyceInfoLab/ARDS_Classifier.
Model performance
We utilized AUC as our main metric to assess the discrimination of both the CUI-based and the text-based models for identifying ARDS. AUC comparisons between the models were performed using the DeLong’s method.21 To further evaluate performance in a clinical context, we compared area under the precision-recall curve as well as sensitivity, specificity, positive predictive value, and negative predictive value measures of both models at various thresholds of predicted probabilities.
Best practices for reporting the validation of prediction models were followed using the Transparent Reporting of multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) (checklist in Supplementary Table 1). All analyses were performed using Python Version 3.6.2 and R version 3.6.2 (R Project for Statistical Computing) with P < 0.05 indicating statistical significance.
RESULTS
Patient Characteristics
Among 235 patients in the UCM validation cohort, 110 (47%) had a confirmed diagnosis of ARDS. In comparison to patients without ARDS, patients with ARDS had a lower mean age (55 years vs. 60 years, P = 0.02), had a lesser proportion of black race (57% vs 72%, P = 0.04), had a lower mean P/F ratio (138 vs. 190, P < 0.001), had a longer median hospital length of stay (14 days vs 9 days, P < 0.001), had a longer median ICU length of stay (8 days vs. 4 days, P < 0.001), and had a greater proportion who died in-hospital (34% vs 18%, P = 0.007). There was no difference between the median number of qualifying chest radiograph reports (4 vs 4, P = 0.446), and the median number of qualifying CT reports (1 vs. 0, P = 0.823) between patients with ARDS and those without ARDS. The most frequent diagnoses in the ARDS group were sepsis/infection (48%) and hepatitis/liver-failure (12%).
Model Performance
Figure 2 depicts the AUC for both text-based and CUI-based models. As observed, the text-based model demonstrated a higher AUC in comparison to the CUI-based model, although this difference was not statistically significant (0.78 [95% CI: 0.72–0.84] vs. 0.72 [95% CI: 0.66–0.78], P=0.09). The text-based model also had a higher area under the precision-recall curve than the CUI-based model (0.73 vs 0.68), as illustrated in Supplementary Figure 1. The text-based model performance dropped when utilizing only text from the first radiology report (AUC: 0.65 [95% CI: 0.58–0.72]) as well as using only using CT reports (AUC 0.60 [95% CI: 0.49–0.70]). Performance for the text-based model was equivalent between using both CT and chest radiograph reports and only using chest radiograph reports (AUC for both 0.78 [95% CI 0.72–0.84]) vs. AUC using only chest radiograph 0.79 [95% CI, 0.73–0.85]).
Figure 2:
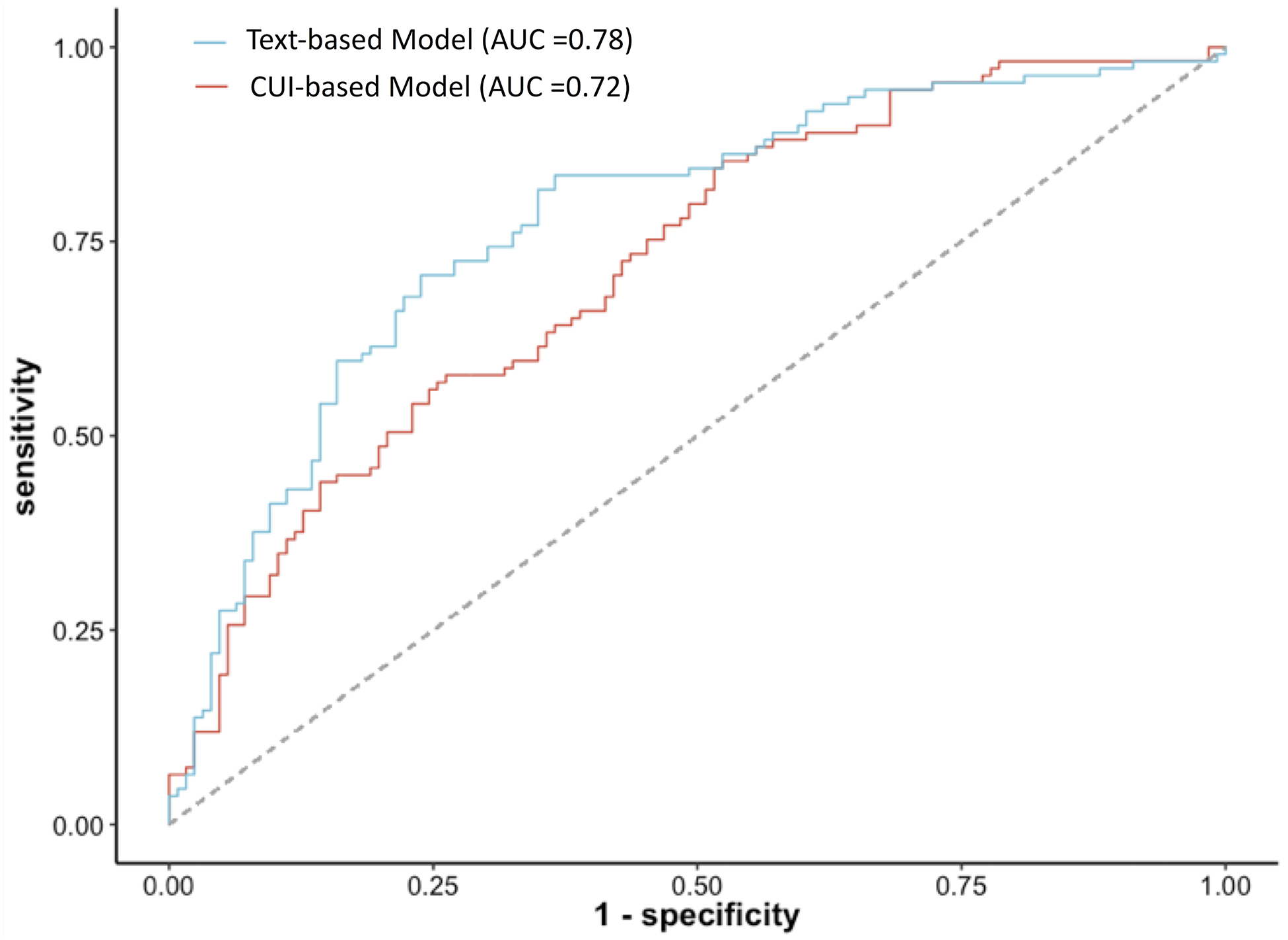
ROC curves for both CUI- and Text-based models along with corresponding AUCs
Table 2 compares different prediction metrics (sensitivity, specificity, positive and negative predictive values) between the text-based and CUI-based models. At a specificity of approximately 88%, the text-based model showed higher sensitivity (42% vs 37%), higher positive predictive value (77% vs. 73%), and a higher negative predictive value (64% vs. 62%) compared to the CUI-based model. Conversely, at a sensitivity of around 72%, the text-based model reported higher specificity (72% vs 57%), positive predictive value (69% vs 59%), and a higher negative predictive value (75% vs. 69%).
Table 2:
Sensitivity, specificity, positive, and negative predictive values for Text and CUI-based models. Bolded lines are referenced in text.
| Model | Threshold | Sensitivity | Specificity | Positive Predictive Value | Negative Predictive Value |
|---|---|---|---|---|---|
| CUI-based model | ≥ 0.1 | 98% | 16% | 50% | 91% |
| ≥ 0.2 | 88% | 41% | 57% | 80% | |
| ≥ 0.3 | 71% | 57% | 59% | 69% | |
| ≥ 0.4 | 57% | 75% | 66% | 67% | |
| ≥ 0.5 | 46% | 80% | 67% | 63% | |
| ≥ 0.6 | 37% | 88% | 73% | 62% | |
| ≥ 0.7 | 24% | 94% | 79% | 59% | |
| ≥ 0.8 | 8% | 97% | 75% | 55% | |
| ≥ 0.9 | 1% | 100% | 100% | 54% | |
| Text-based model | ≥ 0.1 | 95% | 32% | 55% | 87% |
| ≥ 0.2 | 72% | 72% | 69% | 75% | |
| ≥ 0.3 | 42% | 89% | 77% | 64% | |
| ≥ 0.4 | 19% | 96% | 81% | 58% | |
| ≥ 0.5 | 8% | 98% | 75% | 55% | |
| ≥ 0.6 | 3% | 100% | 100% | 54% |
We further compared prediction metrics between the text-based model and the keyword rule-based model. At a similar threshold for specificity (32% vs. 31%), the text-based model had a higher sensitivity (95% vs. 93%), similar positive predictive values (55 % vs 54%) and a higher negative (predictive value (87% vs 83%) than the keyword rule-based search model.
Figure 3 depicts the calibration plots across deciles of predicted probabilities corresponding to both the text-based and CUI-based models in the UCM cohort. The text-based model calibration (Figure 3a) had an intercept of 1.24 [95% CI: 0.97–1.52] and a slope of 1.31 [95% CI: 0.90–1.72] (P <0.001 for H0: slope=1, intercept=0). The CUI-based model calibration (Figure 3b) had an intercept of 0.43 [95% CI: 0.14–0.73] and a slope of 0.72 [95% CI: 0.65–0.78] (P=0.002 for H0: slope=1, intercept =0). The distribution of predicted probabilities for both models are illustrated in Supplementary Figure 2.
Figure 3:
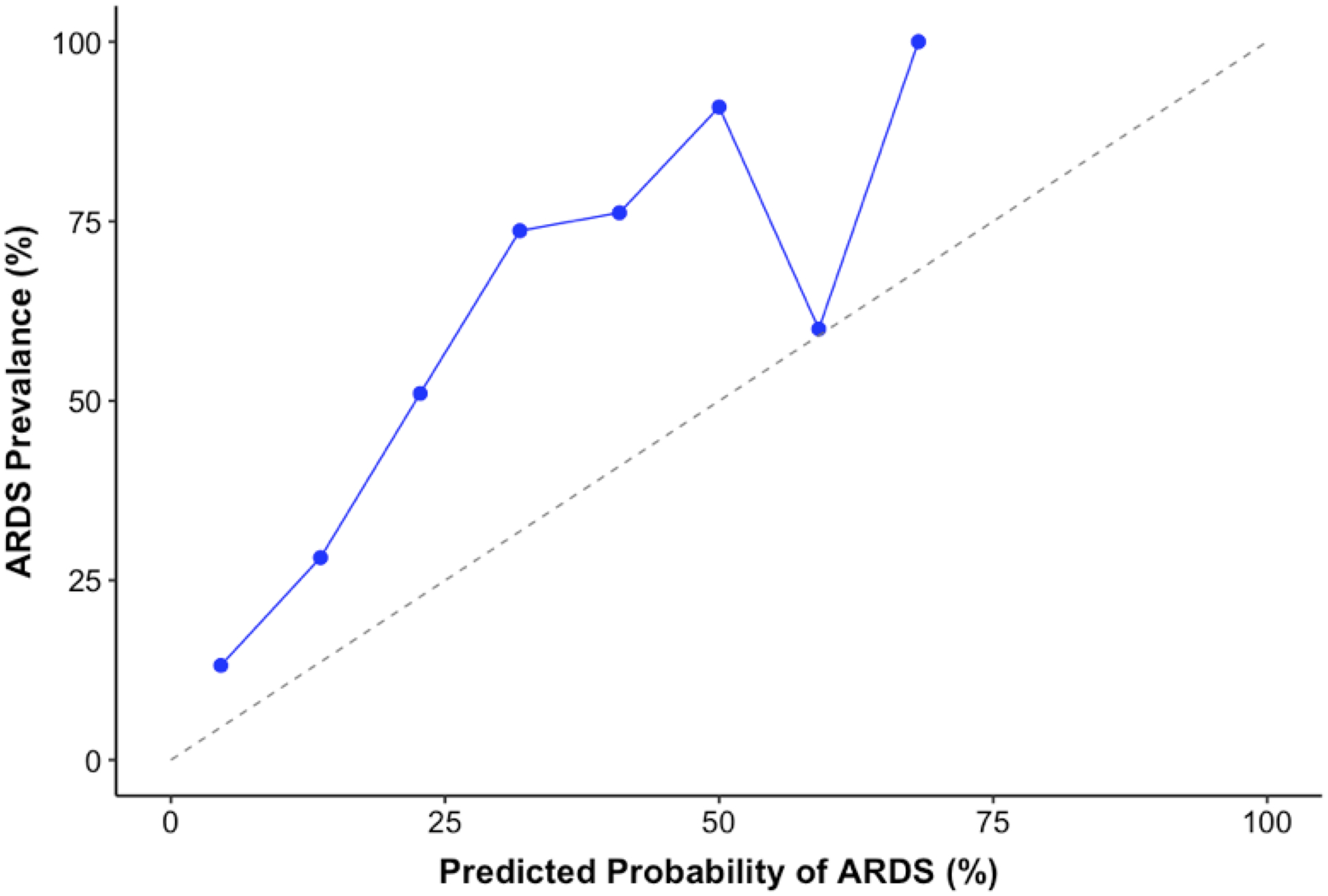
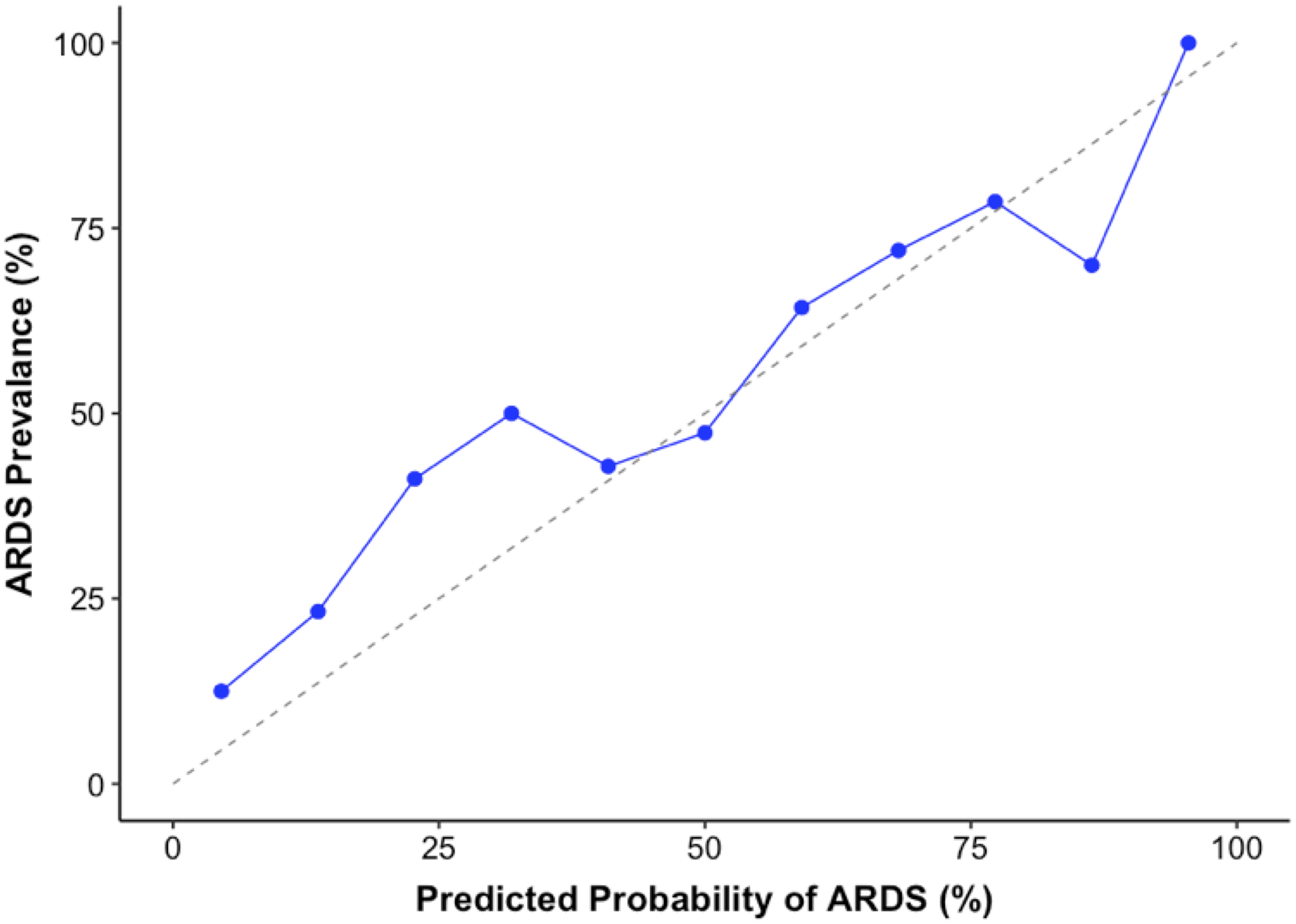
Calibration plots demonstrating alignment between predicted probability of ARDS from (a) the text-based model and (b) the CUI-based model against true rate of outcome in the validation cohort at UCM.
Supplementary Figure 4 highlights the frequencies of the top text (unigram) features, based on weights returned by SVM in the original machine learning model, for both the derivation (Loyola) and external validation (UCM) cohort. Comparisons are shown for the top 25 positive features, i.e., features predictive of ARDS (Supplementary Figure 4a), and top 25 negative features, i.e., features not predictive of ARDS (Supplementary Figure 4b). Most of the important unigram features are represented well in both UCM and Loyola, underlining the power of the model in utilizing features consistent across medical practice. Some important disparities in frequencies between sites for the positive features include “subclavian”, “diffuse”, and “lung”, which are over-represented in Loyola. Examples of differences in frequencies between sites for the negative features include “suspicious” that is over-represented in UCM and “consolidations” and “infectious” that are over-represented in Loyola.
DISCUSSION
We externally validated two models that incorporate the information in radiology reports to identify patients with ARDS. The models developed at Loyola were: (a) text-based model that used textual features to identify ARDS, and (b) CUI-based model to account for semantic ambiguities and lexical variation by using UMLS-based named entity terms for identifying ARDS. In the validation cohort of 235 patients from UCM, the text-based model showed improved performance over the CUI-based model in discriminating ARDS from non-ARDS patients (although the improvement was not statistically significant), while the CUI-model demonstrated better calibration in an external setting.
Several studies have highlighted the challenges in reliable recognition of ARDS in patients.11 For example, Sjoding et al. recently sought to measure agreement between physicians in ARDS diagnosis.22 In a cohort of 205 patients with hypoxic respiratory failure, the study reported moderate inter-clinician reliability (a kappa of 0.50), in diagnosing ARDS.22 Most of the disagreement was explained by different interpretations of chest imaging studies. A follow-up study suggested that clinician disagreement over-diagnosis could be further attributed to the wide spectrum of risk factors and measures of lung injury observed in ARDS patients.23 It is also well known that ARDS manifests with heterogenous phenotypes and etiologies that vary with cohort differences such as trauma versus medical patients.2,24 These imperfect clinical criteria have likely contributed to the poor recognition of ARDS and a need for automated systems to augment recognition for providers.
While machine learning algorithms have been shown to improve detection of findings in radiology images and reports, the application for identification or resolution of ARDS is in its infancy.19,25 Rule-based models that focus on searching keywords within chest radiograph reports have been proposed for ARDS recognition.7 However, such models have shown high false positive rates in external validation.11 A recent study demonstrated that keyword-based sniffer algorithms are limited in utility because of poor specificity.10 This is presumably a result of variation in terminology within hospital systems.6 Models that incorporate NLP methods have been developed to identify patients with ARDS.9 However, similar to keyword-based models, these NLP models utilized older American European Consensus Conference definitions, did not incorporate CT reports, and only considered text and not standardized medical terminology.9,10 In recent work, we developed a unigram-based linear SVM model using a mixed cohort of medical and surgical patients that was referenced against the Berlin definition and incorporated CT reports.14 The model achieved an AUC of 0.73 in internal validation using 24-hour radiology reports.
At UCM, we applied the same model using the same inclusion criteria. In our cohort of 235 medical and surgical patients at UCM, the Loyola text-based model, consisting of both unigram and bigram features, identified ARDS patients with an AUC of 0.78. Similar model performance was achieved at Loyola thereby demonstrating generalizability to identify ARDS across multiple settings. The text-based model also demonstrated better sensitivity and specificity than the CUI-based model at various thresholds. The choice of an operational threshold is dependent on whether the model is to be used as an initial screening tool (with minimal false negative rates), or as a diagnostic tool to enable intervention (with minimal false positive rates to avoid alarm fatigue).
Analysis of the unigram features that were important predictors for ARDS revealed similarities between sites. Features with high frequency for predicting ARDS at both Loyola and UCM included “bilateral”, “opacities”, and “edema”. These are also hallmark morphologic features of ARDS and therefore offer good face validity for our model. Features such as “nodular”, “administered”, and “mildly” were negative predictors for ARDS and frequently encountered at both Loyola and UCM. The negative features are also indicative of non-pathologic chest imaging. Our results also show medical centers frequently use the same words to express radiographic characteristics of ARDS and non-ARDS patients.
Several reasons may exist for the text-based model outperforming the CUI-based model in detecting ARDS in the UCM cohort. For example, UMLS-derived CUI features are limited in accounting for synonyms for words such as ‘bilateral’, which are characteristic of ARDS and certain text words like ‘bibasilar’ do not have a CUI. This loss in granularity may have occurred when mapping the raw text to CUIs because the CUI model was derived on 1504 unique CUI features compared to the raw text model with over 6000 unique features. The differences in case mix could be another factor. Approximately 60% of the Loyola cohort consisted of burn and trauma patients with inhalation injuries and venous access away from facial features,14 whereas the UCM cohort did not have any burn or trauma patients. Accordingly, terms such as “subclavian” and “bibasilar” have relatively lower representation in the UCM cohort. These results indicate that standardizing to medical vocabularies such as CUIs for detecting ARDS may not be necessary and variability in language is likely not contributing to the machine learning model’s ability to identify ARDS. It is possible that text-to-CUI mapping within cTAKES for ARDS-related terms may be suboptimal. The text-based model also requires less pre-processing in comparison to the CUI-based model, thereby underlining a more pragmatic application.
Even though the text model performed well in detecting ARDS in an external setting, the calibration was not optimal, with the model under-predicting the case-rate of ARDS at all predicted probability thresholds. From the calibration plot, we can deduce that the calibration-at-large correspondence was poor and likely due to differences in case mix between Loyola and UCM. For example, the Loyola cohort is more likely to observe conditions that mimic ARDS symptoms such as lung contusions due to their trauma/burn patient population, as opposed to the medical ICU patients in the UCM cohort. Further, the prevalence rate at Loyola was about half of UCM’s prevalence rate. The calibration for the text model had a linear slope indicating that moderate shrinkage would be required for the model to be effective at UCM. This highlights the difficulty in operationalizing the text-based model without further re-calibration to account for variations in patient population. Notably, the CUI model demonstrated better calibration than the text-based model. This suggests that the CUI terms provide better fit when there is heterogeneity in case-mix and ARDS prevalence rates across settings, but at the expense of lower discrimination for ARDS possibly due to loss in granularity of medical terminology. The very close match between predicted probabilities and actual prevalence of ARDS indicate that the CUI-based model may be implemented in a hospital without need for re-calibration.
The choice between the two models is dependent on the goals within a hospital setting. If the objective is to utilize the model as a screening tool without implementing cTAKES, then the text-based model is proficient at ARDS discrimination. However, if the motivation is to acquire highly accurate estimates of ARDS risk without need for further calibration, then the CUI-based model is the better choice.
Our model provides a pragmatic approach at detecting ARDS that is feasible at point-of-care. Once deployed, all radiology reports collected within 24 hours of a patient meeting the oxygenation criteria can be utilized by our model to determine risk of ARDS. Early recognition of patients at-risk for ARDS using our model can facilitate timely intervention and rescue. Some factors that need to be considered before deploying our model are as follows: (1) hospital infrastructure must facilitate feeding radiology reports to our machine learning model, (2) hospital infrastructure must facilitate interfacing with the model using Python to determine predicted probabilities for ARDS, and (3) a preliminary study must be conducted to account for possible calibration of the model to account for cohort differences, including prevalence.
Our study has several limitations. First, due to the retrospective study design and use of a single reviewer, there may be misclassification bias in our annotation of ARDS and non-ARDS cases. In particular, cases with cardiogenic edema or opacities that are not typical of ARDS are important patient populations where ARDS determination is more difficult. Second, we utilized the Berlin definition of ARDS which has shown limited reproducibility among clinicians. However, we utilized both chest radiographs and CT scans as confirmation; therefore, providing a high degree of confidence in ARDS diagnosis. Additionally, our study is based on data from randomized controlled trials that represent a high prevalence of ARDS and excludes patients not meeting a time frame for oxygenation criteria and not having available radiology notes. Determining the impact of the model in a more general population across all intensive care unit patients is a priority for future studies. Finally, we only validated a linear-SVM model utilizing Platt scaling for approximating predicted probabilities.20,26 Other models, such as neural networks or models that incorporate structured data (images, vitals, laboratory results, etc.), information from all clinical notes and reports, or patient characteristics (underlying conditions, risk factors, etc.) may potentially lead to better identification of ARDS patients but also raises model complexity and hampers pragmatic application.
CONCLUSION
We externally validated a computable phenotype for ARDS using NLP in a separate health system with a different prevalence of ARDS as well as different types of patients. To our knowledge, this is the first study that externally validates an NLP-based model for ARDS identification in accordance with Berlin criteria. Our model may help providers and health systems to more comprehensively and expeditiously identify cases of ARDS, and may serve as a tool for recruitment into clinical trials for patients with ARDS.
Supplementary Material
Table 1:
Patient Characteristics of ARDS Validation Cohort
| Patient Characteristics | Patients with ARDS (n=110) | Patients without ARDS (n=125) | P-value |
|---|---|---|---|
| Age, years, mean (sd) | 55 (16) | 60 (16) | 0.020 |
| Female, n (%) | 56 (51) | 67 (54) | 0.779 |
| Race, n (%) | |||
| Black | 63 (57) | 90 (72) | 0.040 |
| White | 40 (36) | 27 (22) | |
| Other | 7 (6) | 8 (6) | |
| Primary Diagnosis, n (%) | |||
| Acute Respiratory Failure | 15 (14) | 32 (26) | 0.001 |
| Cardiovascular Disease/ CHF | 2 (2) | 6 (5) | |
| Chronic Lung Disease | 4 (4) | 13 (11) | |
| Hepatitis/Liver Failure | 13 (12) | 6 (5) | |
| Kidney Failure | 2 (2) | 4 (3) | |
| Malignancy | 10 (9) | 8 (7) | |
| Sepsis/Infection | 53 (48) | 33 (27) | |
| Other | 11 (10) | 22 (18) | |
| P/F ratio, mean (sd) | 138 (68) | 190 (71) | <0.001 |
| Number of qualifying X-ray reports, median (IQR) | 4 (3–5) | 4 (3–5) | 0.446 |
| Number of qualifying CT reports, median (IQR) | 1 (0–1) | 0 (0–1) | 0.823 |
| Length of hospital stay, days, median (IQR) | 14 (8–19) | 9 (5–15) | <0.001 |
| Length of ICU stay, days, median (IQR) | 8 (5–11) | 4 (3–8) | <0.001 |
| In-hospital deaths, n (%) | 37 (34) | 22 (18) | 0.007 |
Conflicts & Disclosures:
Dr. Mayampurath is supported by a career development award from the National Heart, Lung, and Blood Institute (K01HL148390). Dr. Churpek is supported by an R01 from NIGMS (R01 GM123193). Dr. Afshar is supported by a career development award from the National Institute on Alcohol Abuse and Alcoholism (K23 AA024503). Dr. Mayampurath has performed consulting services for Litmus Health (Austin, TX). Dr. Churpek has a patent pending (ARCD. P0535US.P2) for risk stratification algorithms for hospitalized patients and has received research support from EarlySense (Tel Aviv, Israel).
REFERENCES
- 1.Bellani G, Laffey JG, Pham T, et al. Epidemiology, Patterns of Care, and Mortality for Patients With Acute Respiratory Distress Syndrome in Intensive Care Units in 50 Countries. JAMA. 2016;315(8):788–800. doi: 10.1001/jama.2016.0291 [DOI] [PubMed] [Google Scholar]
- 2.Calfee CS, Janz DR, Bernard GR, et al. Distinct Molecular Phenotypes of Direct vs Indirect ARDS in Single-Center and Multicenter Studies. Chest. 2015;147(6):1539–1548. doi: 10.1378/chest.14-2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernard GR, Artigas A, Brigham KL, et al. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med. 1994;149(3):818–824. doi: 10.1164/ajrccm.149.3.7509706 [DOI] [PubMed] [Google Scholar]
- 4.Howard AE, Courtney-Shapiro C, Kelso LA, Goltz M, Morris PE. Comparison of 3 Methods of Detecting Acute Respiratory Distress Syndrome: Clinical Screening, Chart Review, and Diagnostic Coding. Am J Crit Care. 2004;13(1):59–64. [PubMed] [Google Scholar]
- 5.Acute Respiratory Distress Syndrome: The Berlin Definition. JAMA. 2012;307(23):2526–2533. doi: 10.1001/jama.2012.5669 [DOI] [PubMed] [Google Scholar]
- 6.Matthay MA, Zemans RL, Zimmerman GA, et al. Acute Respiratory Distress Syndrome. Nat Rev Dis Primers. 2019;5(1):18. doi: 10.1038/s41572-019-0069-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azzam HC, Khalsa SS, Urbani R, et al. Validation Study of an Automated Electronic Acute Lung Injury Screening Tool. Journal of the American Medical Informatics Association. 2009;16(4):503–508. doi: 10.1197/jamia.M3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herasevich V, Yilmaz M, Khan H, Hubmayr RD, Gajic O. Validation of an electronic surveillance system for acute lung injury. Intensive Care Med. 2009;35(6):1018–1023. doi: 10.1007/s00134-009-1460-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yetisgen-Yildiz M, Bejan C, Wurfel M. Identification of Patients with Acute Lung Injury from Free-Text Chest X-Ray Reports In: Proceedings of the 2013 Workshop on Biomedical Natural Language Processing. Sofia, Bulgaria: Association for Computational Linguistics; 2013:10–17. [Google Scholar]
- 10.McKown AC, Brown RM, Ware LB, Wanderer JP. External Validity of Electronic Sniffers for Automated Recognition of Acute Respiratory Distress Syndrome. J Intensive Care Med. January 2017:885066617720159. doi: 10.1177/0885066617720159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solti I, Cooke CR, Xia F, Wurfel MM. Automated Classification of Radiology Reports for Acute Lung Injury: Comparison of Keyword and Machine Learning Based Natural Language Processing Approaches. Proceedings (IEEE Int Conf Bioinformatics Biomed). 2009;2009:314–319. doi: 10.1109/BIBMW.2009.5332081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrickson CM, Calfee CS. A new frontier in ARDS trials: phenotyping before randomisation. The Lancet Respiratory Medicine. 2019;7(10):830–831. doi: 10.1016/S2213-2600(19)30175-4 [DOI] [PubMed] [Google Scholar]
- 13.Constantin J-M, Jabaudon M, Lefrant J-Y, et al. Personalised mechanical ventilation tailored to lung morphology versus low positive end-expiratory pressure for patients with acute respiratory distress syndrome in France (the LIVE study): a multicentre, single-blind, randomised controlled trial. The Lancet Respiratory Medicine. 2019;7(10):870–880. doi: 10.1016/S2213-2600(19)30138-9 [DOI] [PubMed] [Google Scholar]
- 14.Afshar M, Joyce C, Oakey A, et al. A Computable Phenotype for Acute Respiratory Distress Syndrome Using Natural Language Processing and Machine Learning. AMIA Annu Symp Proc. 2018;2018:157–165. [PMC free article] [PubMed] [Google Scholar]
- 15.Patel BK, Wolfe KS, Pohlman AS, Hall JB, Kress JP. Effect of Noninvasive Ventilation Delivered by Helmet vs Face Mask on the Rate of Endotracheal Intubation in Patients With Acute Respiratory Distress Syndrome: A Randomized Clinical Trial. JAMA. 2016;315(22):2435–2441. doi: 10.1001/jama.2016.6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schweickert WD, Pohlman MC, Pohlman AS, et al. Early physical and occupational therapy in mechanically ventilated, critically ill patients: a randomised controlled trial. The Lancet. 2009;373(9678):1874–1882. doi: 10.1016/S0140-6736(09)60658-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel SB, Poston JT, Pohlman A, Hall JB, Kress JP. Rapidly Reversible, Sedation-related Delirium versus Persistent Delirium in the Intensive Care Unit. Am J Respir Crit Care Med. 2014;189(6):658–665. doi: 10.1164/rccm.201310-1815OC [DOI] [PubMed] [Google Scholar]
- 18.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association : JAMIA. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai T, Giannopoulos AA, Yu S, et al. Natural Language Processing Technologies in Radiology Research and Clinical Applications. Radiographics. 2016;36(1):176–191. doi: 10.1148/rg.2016150080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Platt JC. Probabilistic Outputs for Support Vector Machines and Comparisons to Regularized Likelihood Methods In: Advances in Large Margin Classifiers. MIT Press; 1999:61–74. [Google Scholar]
- 21.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 22.Sjoding MW, Hofer TP, Co I, Courey A, Cooke CR, Iwashyna TJ. Interobserver Reliability of the Berlin ARDS Definition and Strategies to Improve the Reliability of ARDS Diagnosis. Chest. 2018;153(2):361–367. doi: 10.1016/j.chest.2017.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sjoding MW, Hofer TP, Co I, McSparron JI, Iwashyna TJ. Differences between Patients in Whom Physicians Agree and Disagree about the Diagnosis of Acute Respiratory Distress Syndrome. Annals ATS. 2018;16(2):258–264. doi: 10.1513/AnnalsATS.201806-434OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinha P, Calfee CS. Phenotypes in acute respiratory distress syndrome: moving towards precision medicine. Current Opinion in Critical Care. 2019;25(1):12. doi: 10.1097/MCC.0000000000000571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choy G, Khalilzadeh O, Michalski M, et al. Current Applications and Future Impact of Machine Learning in Radiology. Radiology. 2018;288(2):318–328. doi: 10.1148/radiol.2018171820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niculescu-Mizil A, Caruana R. Predicting Good Probabilities With Supervised Learning. In: Proceedings of the 22nd International Conference on Machine Learning.; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


