Abstract
Early and correct diagnosis of inflammatory rheumatic diseases (IRD) poses a clinical challenge due to the multifaceted nature of symptoms, which also may change over time. The aim of this study was to perform protein expression profiling of four systemic IRDs, systemic lupus erythematosus (SLE), ANCA-associated systemic vasculitis (SV), rheumatoid arthritis (RA), and Sjögren’s syndrome (SS), and healthy controls to identify candidate biomarker signatures for differential classification. A total of 316 serum samples collected from patients with SLE, RA, SS, or SV and from healthy controls were analyzed using 394-plex recombinant antibody microarrays. Differential protein expression profiling was examined using Wilcoxon signed rank test, and condensed biomarker panels were identified using advanced bioinformatics and state-of-the art classification algorithms to pinpoint signatures reflecting each disease (raw data set available at https://figshare.com/s/3bd3848a28ef6e7ae9a9.). In this study, we were able to classify the included individual IRDs with high accuracy, as demonstrated by the ROC area under the curve (ROC AUC) values ranging between 0.96 and 0.80. In addition, the groups of IRDs could be separated from healthy controls at an ROC AUC value of 0.94. Disease-specific candidate biomarker signatures and general autoimmune signature were identified, including several deregulated analytes. This study supports the rationale of using multiplexed affinity-based technologies to reflect the biological complexity of autoimmune diseases. A multiplexed approach for decoding multifactorial complex diseases, such as autoimmune diseases, will play a significant role for future diagnostic purposes, essential to prevent severe organ- and tissue-related damage.
Keywords: proteomics, antibody microarray, autoimmune diseases, whole blood
Introduction
Inflammatory rheumatic diseases (IRD) are heterogeneous syndromes that are classified based on clinical phenotypes and key disease markers. Systemic erythematosus lupus (SLE), rheumatoid arthritis (RA), Sjögren’s syndrome (SS), and antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis (SV) represent four IRDs, which, if left untreated, can lead to severe and sometimes permanent disability, increased morbidity, and premature mortality.1,2 Diagnosis at an early stage plays a crucial role in establishing proper disease monitoring and enabling therapeutic interventions to prevent or minimize organ- and tissue-related damage. However, clinical diagnosis remains a challenge due to fluctuating symptoms over time, including a wide repertoire of manifestations such as fatigue, joint and muscle pain, and inflammation symptoms, which are shared among several IRDs, and also with other conditions mimicking IRDs, e.g., infections, malignancies, etc. In addition, a patient can be affected by more than one autoimmune disease at the same time (such as concurrent Sjögren’s syndrome in SLE and RA patients), which confers an increased risk of misdiagnosis and/or underdiagnosis.3−5 Current tools for clinical diagnosis include the combined information generated from clinical, laboratory, and imaging findings, where the presence of various autoantibodies, such as antinuclear antibodies (ANA), anticyclic citrullinated peptides (aCCP), rheumatoid factor (RF), ANCA (including antiproteinase 3 (anti-PR3) and antimyeloperoxidase (anti-MPO)) antibodies, anti-double-stranded antibodies (anti-dsDNA), anti-Ro/SSA, and anti-LA/SSB, constitutes important biomarkers in the diagnostic routine of SLE, RA, SS, and SV.6−9 However, a positive result for an autoantibody may not be exclusive for one disease, and the use of single markers has not reached the high levels of specificity as required.5,10−12 Identification of new blood-based biomarkers for correct and early diagnosis is of high clinical relevance to enable early therapeutic interventions, thereby saving both lives and cost for society. Considering that underlying disease biology is still unclear, panels of disease-specific markers may provide important insights on key disease-specific molecular alterations. Previous studies have shown that high-performance proteomic technologies, such as recombinant antibody microarrays, which offer a multiplexed approach, reflect the complexity of multifactorial diseases better.13−17 Using this approach, candidate biomarker panels indicative for SLE, systemic sclerosis, and SLE disease activity have been identified.15,17,18 The aim of this study was to perform protein expression profiling of the IRDs SLE, RA, SS, and SV and of healthy controls (H) and to identify candidate biomarker signatures for classification. To this end, a total of 316 serum samples collected from patients with autoimmune disease and from healthy controls were analyzed on 394-plex antibody microarrays. Using this methodology, we showed for the first time that classification of IRD could be achieved at high accuracy. These results highlight the power of using a multiplexed approach for decoding multifactorial complex diseases, such as IRDs, which may play a significant role for future diagnostic purposes.
Experimental Procedures
Clinical Samples
This retrospective study included a total of 316 serum samples collected from healthy controls (n = 77) and patients diagnosed with an IRD (n = 239). All samples were collected from Departments of Rheumatology and Nephrology at Skåne University Hospital (Malmö or Lund). Patients were diagnosed with either SLE (n = 39), RA (n = 45), SS (n = 73), or SV (n = 82) and were considered, according to their specific clinical criteria, to be in an active (n = 198) or inactive (n = 41) disease when samples were collected. For SLE patients, disease activity was defined using the SLEDAI-2000-score19 (mean score 7, range 1–19). All patients with RA had fulfilled the 1987 American College of Rheumatology criteria for RA,20 and had active disease, with a median CRP of 31 mg/L (interquartile range 13–55). The majority of RA patients were positive for anti-CCP (86%) and RF (84%). ANCA specificity in SV patients was defined according to anti-MPO (n = 40) or anti-PR3 status (n = 42) and clinical activity according to the BVAS score.21 All Sjögren samples were collected from patients that fulfilled the 2002 American-European Consensus Group criteria22 for primary SS. As controls, serum samples from healthy individuals with no previous history of autoimmune disease were used. Within the IRD cohort, the mean age was 59 years and the female to male ratio was 168:82, whereas the mean age in healthy controls was 60 years and the female to male ratio was 66:11 (Table 1). Ethical approval for the study was granted by the Regional Ethics Review Board in Lund, Sweden.
Table 1. Demographic Data of the Patients Diagnosed with an Inflammatory Rheumatic Disease SLE, RA, SS, or SV and Healthy Controls (H).
| inflammatory
rheumatic diseases |
healthy controls | |||||
|---|---|---|---|---|---|---|
| parameter | SLE | RA | SS | SV | H | total |
| no. of samples | n = 39 | n = 45 | n = 73 | n = 82 | n = 77 | n = 316 |
| female:male ratio | 33:6 | 32:13 | 71:2 | 32:50 | 66:11 | 234:83 |
| mean age years (range) | 51 (29–77) | 65 (38–85) | 61 (24–85) | 60 (11–83) | 50 (18–81) | 60 (11–85) |
Antibody Microarray Production and Analysis
A total of 394 recombinant scFv antibodies were selected from in-house designed phage display libraries23,24 (Supporting Information Table S1). Of these, 379 of the scFv antibodies were directed against 161 (mainly immunoregulatory) antigens. The remaining 15 scFv antibodies were directed against 15 short amino acid motifs (4–6 amino acids long), denoted CIMS antibodies25 (Supporting Information Table S2). For some analytes, more than one scFv antibody clone (n = 2–9) targeting different epitopes was chosen to minimize the risk of impaired antibody activity followed by epitope masking during the sample labeling process. All scFv antibodies were produced according to standardized protocols in 15 mL of E. coli cultures and purified from the cell periplasmic space using the MagneHis Protein Purification System (Promega, Madison, WI) and a KingFisher96 Robot (Thermo Fisher Scientific, Waltham, MA). Buffer exchange to PBS was performed using a Zeba 96-well desalt spin plate (Pierce). Concentration and purity of the scFvs were determined using a Nanodrop at 280 nm (NanoDrop Technologies, Wilmington) and SDS-PAGE analysis (InVitrogen, Carlsbad, CA). Production of 26 × 28 spot subarrays was generated by a noncontact printer (SciFlexarrayer S11, Scenion, Berlin, Germany). Briefly described, single droplets (300 pL) of scFv antibody solutions, PBS (blank) or biotinylated BSA (position marker), were printed on black polymer Maxisorp slides (NUNC A/S, Roskilde, Denmark) and allowed to absorb to the surface. Antibody microarrays were analyzed, as previously described.26 In brief, biotinylated samples were added to individual subarrays, and bound proteins were detected using Alexa-647-labeled streptavidin. Slides were scanned at 635 nm using the LS Reloaded laser scanner (Tecan) at a fixed laser scanning setting of 150 PMT gain.
Data Preprocessing
Data preprocessing was performed as follows. In brief, spot signal intensities were quantified using Immunovia Quant software, v1.0 (Immunovia AB, Lund, Sweden). Signal intensities with local background subtraction were used for data analysis. Each data point represented the mean value of three technical replicate spots, unless any replicate cross-validation (CV) exceeded 15%, in which case the worst-performing replicate was eliminated and the average value of the two remaining replicates was used. The data were normalized using a two-step strategy. First, the data were normalized according to the day-to-day variation using the “subtract by group mean” approach, as previously described,27 and secondly, a modified semiglobal normalization was used to minimize array-to-array variations. In this approach, 15% of the antibodies displaying the lowest CV values over all samples were identified and used to calculate a scaling factor, as previously described.28,29 Quality control and visualization of potential outliers was performed using Qlucore Omice Explorer 3.1 software (Qlucore AB, Lund, Sweden). The raw array data set is available at https://figshare.com/s/3bd3848a28ef6e7ae9a9.
Data Analysis
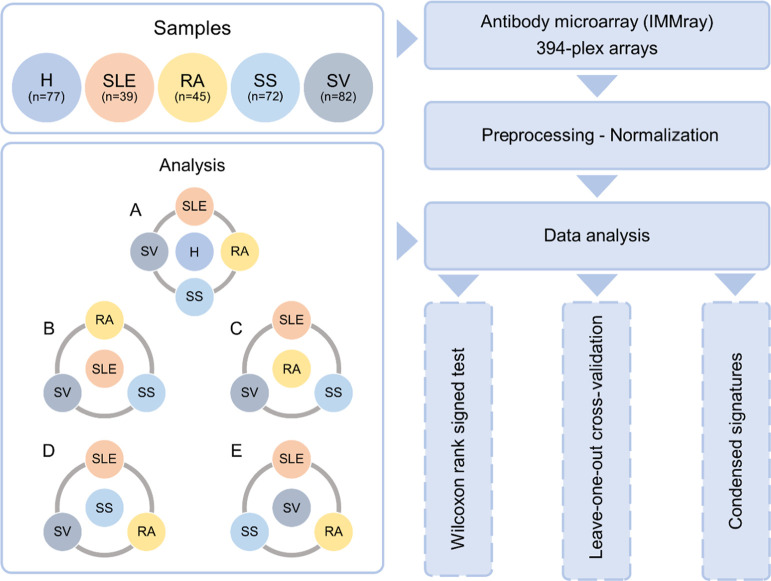
A schematic outline of the experimental analysis process is demonstrated in Figure 1. For differential analysis, leave-one-out cross-validation (LOO CV), and signature development, one group (H, SLE, RA, SS, or SV) was set against the remaining groups. Analysis 1A in Figure 1 refers to the identification of a general IRD signature, where healthy controls (H) were set against the IRDs, meaning H versus SLE+RA+SS+SV. When performing analysis within the IRD group (Figure 1B–E), each individual disease was set against the group of the remaining three diseases, as follows: (B) SLE versus RA+SS+SV, (C) RA versus SLE+SS+SV, (D) SS versus SLE+RA+SV, and (E) SV versus SLE+RA+SS.
Figure 1.
Schematic outline of the antibody microarray process applied on serum samples from systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), Sjögren syndrome (SS), ANCA-associated vasculitis (SV), and healthy controls (H). For each analysis (Wilcoxon, leave-one-out cross-validation, and signature development), each group was set against the remaining samples, i.e., (A) H versus SLE+RA+SS+SV, (B) SLE versus RA+SS+SV, (C) RA versus SLE+SS+SV, (D) SS versus SLE+RA+SV, and (E) SV versus SLE+RA+SS.
Up or downregulated proteins were identified using Wilcoxon signed rank test (q < 0.05) and p-values were adjusted with the Benjamini and Hochberg method.30 Venn diagrams were created at http://bioinformatics.psb.ugent.be/webtools/Venn/. For supervised classification analysis, a linear support vector machine (SVM) (cost parameter = 1) combined with a LOO CV algorithm was used to evaluate the predictive performance of a model. In the LOO CV procedure, one sample was removed, and the remaining samples were used to train the model. The left-out sample was then used to test the model, and the process was repeated until every sample had been used as a test sample. A decision value for each excluded sample was generated, corresponding to the distance to the hyper plane and a receiver operating characteristic (ROC) curve was constructed. The area under the curve (AUC) was then calculated and used as a measure of the prediction performance of the classifier.
Identification of Disease-Specific Signatures
To define a condensed biomarker signature for the differential profiling analysis, a ranking procedure combined with two levels of K-fold cross-validation loops was used (Supporting Information Figure S1). For each individual analysis (H versus SLE+RA+SS+SV etc.), the output was a list of proteins ranked according to how important they were in classification analysis. In short, in the first level of K-fold cross-validation, the ranking of the proteins was defined using an inner loop. Here, the data set was randomly divided into training and validation set 15 times and then repeated 5 times (5-fold cross-validation strategy). In the end, proteins were ranked according to their average importance, resulting in a ranking list. In the next level of K-fold cross-validation, an outer loop was used to test the biomarker signatures of a given length. The final condensed biomarker signature, of a given size, was then assembled using all ranking lists analyzed in the outer loop. For details, see the Supporting Information Materials and Methods section and Figure S1.
Results
The aim of this study was to perform differential protein expression profiling of IRDs and healthy controls and to identify condensed biomarker signatures for disease classification. To this end, a total of 316 serum samples, collected from healthy controls (n = 77) and patients diagnosed with SLE (n = 39), RA (n = 45), SV (n = 82), or SS (n = 73), were analyzed on 394-plex antibody microarrays. One sample collected from a patient with Sjögren’s syndrome was removed from analysis due to technical reasons. One antibody, targeting Keratin-19, failed during the printing process and was removed from further analysis, though two clones targeting the same antigen remained. Altogether, a total of 315 samples and 393 antibodies were used for final data analysis, differential profiling, and signature development. Visualization of the data set in Qlucore revealed no differences in relation to array block, sample labeling day, assay day, or scanning positions, suggesting that any technical variations had successfully been removed during normalization.
Differential Protein Expression Profiling of Healthy and Autoimmune Serum Samples
In the first step of analysis, we wanted to investigate if a signature reflecting IRD (including SLE, RA, SS, and SV) could be identified. Altogether, we were able to demonstrate that the IRD samples could be distinguished from healthy controls and that a biomarker signature, indicative for IRD indeed could be identified. Using an SVM analysis combined with LOO CV, including all antibodies (n = 393), the IRDs could be separated from healthy controls with an ROC AUC value of 0.94 (Figure 2A). Since LOO CV analysis utilizes all antibodies for classification, further analysis was performed to investigate whether healthy and autoimmune samples could be classified using a smaller set of antibodies. Using a ranking procedure (see Methods section), the 40 best-performing antibodies were selected (Supporting Information Table S3), which were able to classify IRD and healthy controls by a predictive AUC value of 0.93. These results clearly show that these IRDs can be differentiated from healthy controls using a protein signature, which could potentially pave the way for a diagnostic test of IRDs.
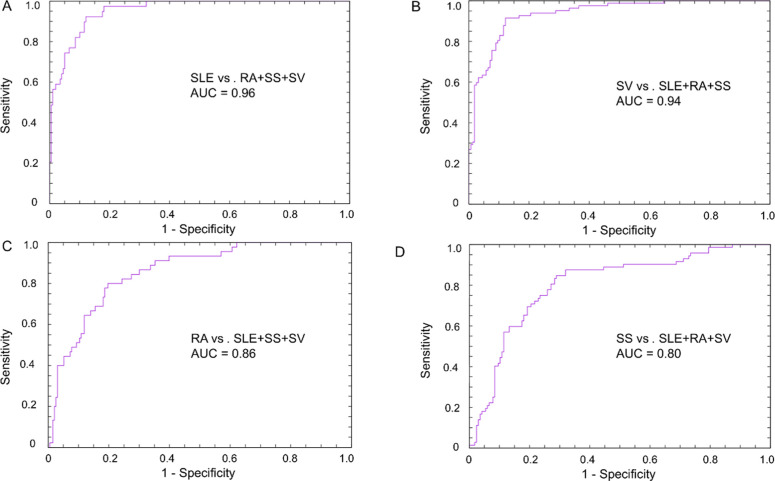
Figure 2.
(A) ROC curve including AUC value generated from leave-one-out cross-validation analysis on healthy versus autoimmune diseases (SLE, RA, SS, and SV). (B) Heatmap from supervised analysis including the top 18 differentially expressed analytes, represented by 25 scFv clones (Wilcoxon analysis q < 0.05) between healthy (yellow bars) and inflammatory rheumatic diseases (blue bars), which include SLE, RA, SS, and SV. Individual clone suffixes are shown in brackets.
Next, we were interested in which analytes were downregulated among the IRDs. Using Wilcoxon, a total of 77 analytes, targeted by 114 antibodies, were found to be differentially expressed (q < 0.05) between IRDs and healthy controls. Among the upregulated analytes, some of the most interesting immunoregulatory analytes included apolipoprotein A1, IL-6, IL-12, TNF-α, IL-16, osteopontin, PRKCZ, and DLG4, whereas antibodies targeting C3, IL-4, VEGF, KKCC1-1, and SPDLY-1 were found among the downregulated analytes. A heatmap including the top 25 antibodies and their corresponding analytes revealed some separation of the two groups (Figure 2B, Supporting Information Table S4). Supported by the fact that separation of IRD from healthy controls could be achieved using two different approaches, though Wilcoxon signed rank test is a nonparametric test and relies on multiple testing, whereas the K-fold cross-validation is an algorithm within machine learning to estimate the prediction error, we compared the lists including the top 25 antibodies with the 40-plex signature panel. Some overlapping could be observed including antibodies targeting the analytes C3, C4, RPS6KA2, KCC2B-3C5, and UBC9. Altogether, these results indicated that a general IRD signature may indeed be present, involving upregulation of several analytes with immunoregulatory functions.
Differential Protein Expression Profiling of SLE, RA, SS, and SV
Considering that many autoimmune diseases display similar symptoms, making clinical diagnosis challenging, we turned our focus toward the IRDs (Figure 1). Herein, a total of four groups were formed as follows: (B) SLE versus RA+SS+SV, (C) RA versus SLE+SS+SV, (D) SS versus SLE+RA+SV, and (E) SV versus SLE+RA+SS. Leave-one-out cross-validation analysis, including all antibodies, showed that the classification of, respectively, IRD-type could be achieved at high accuracies, as presented by ROC AUC values ranging from 0.96 to 0.80 (Figure 3). The best separation was achieved for SLE with an ROC AUC value of 0.96 (Figure 3A) followed by SV and RA, which were classified at ROC AUC values of 0.94 (Figure 3B) and 0.86 (Figure 3C), respectively, whereas SS demonstrated an ROC AUC value of 0.80 (Figure 3D).
Figure 3.
ROC curves with AUC values generated from LOO CV analysis, representing (A) SLE versus RA+SS+SV, (B) SV versus SLE+RA+SS, (C) RA versus SLE+SS+SV, and (D) SS versus SLE+RA+SV.
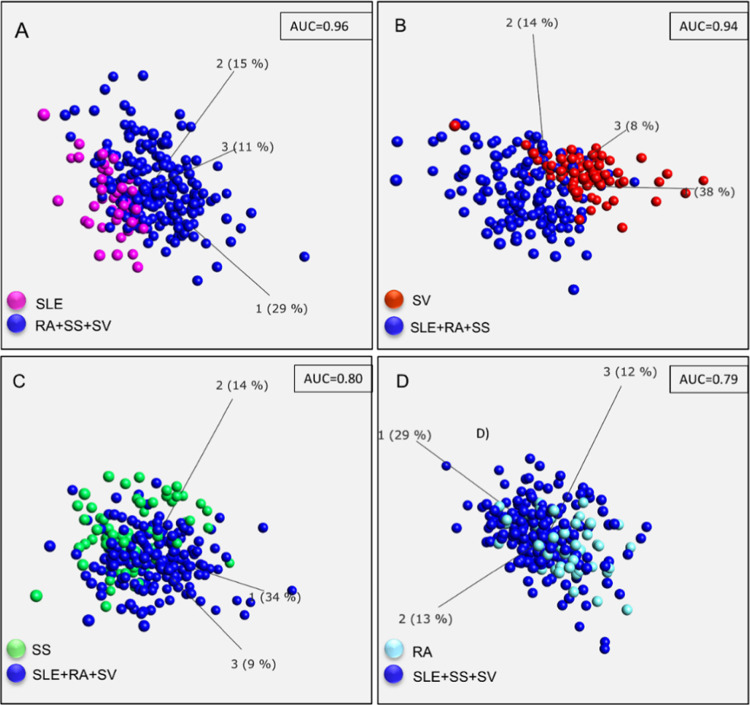
Again, we were interested in if the different groups could be separated using shorter biomarker signatures. Condensed biomarker signatures for SLE, RA, SS, and SV, respectively, were identified using the same procedure, as described previously (Supporting Information Table S3). Herein, using the disease-specific signatures, SLE was again found to be classified with highest accuracy (AUC = 0.96) followed by SV (AUC = 0.94), SS (AUC = 0.80), and RA (AUC = 0.79). Principal component analysis (PCA) plots of the obtained condensed biomarker signatures are presented in Figure 4.
Figure 4.
PCA plots of supervised analysis based on 40-plex biomarker panels representing SLE (A), SV (B), SS (C), and RA (D).
A closer look at these four disease-specific signatures revealed that antibodies targeting analytes such as C3, C4, apolipoprotein A1, and factor B were present on more than one list (Supporting Information Table S3). However, analytes unique for each signature were also identified, such as Lewis x and TNF-a in SLE, PRKCZ and PTK6 in RA, IL-8 and RANTES in SS, and C1q and IL-18 in SV, which could indicate the presence of disease-specific markers. Altogether, by applying 394-plex antibody microarrays interfaced with stringent data analysis, 40-plex antibody signatures capable of classifying the autoimmune diseases SLE, RA, SS, and SV at high predictive powers were pinpointed.
To further explore the serum proteomes of SLE, RA, SS, and SV, differentially expressed analytes for respective disease type were identified (Wilcoxon. q < 0.05) (Supporting Information Table S4). In total, the highest number of differentially expressed analytes was found for SV (n = 326 antibodies targeting 160 analytes) followed in a decreasing order by SS (n = 207 antibodies targeting 127 analytes), SLE (n = 127 antibodies targeting 85 analytes), and RA (n = 114 antibodies targeting 81 analytes).
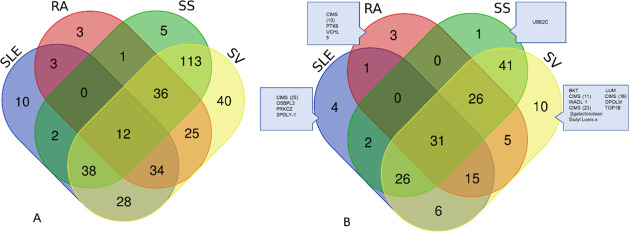
Considering the complexity of underlying molecular alterations in IRD and that both common and disease-specific alterations would be of interest, we investigated the amount of overlap. First, we investigated the overlap based on an antibody level, i.e., relating to the specific clones, irrespective of which analytes they targeted. This revealed a major overlap (Figure 5A), which was not surprising, considering the high number of antibodies generated from the differential analysis.
Figure 5.
Venn diagrams representing the overlap of variables generated from differential analysis (Wilcoxon signed rank test, q < 0.05) for SLE, RA, SS, and SV. Since an analyte may be targeted by more than one antibody diagram, (A) represents the overlap of antibodies, whereas (B) represents the overlap on an analyte level. Disease-specific analytes are outlined in (B). Diagram was created at http://bioinformatics.psb.ugent.be/webtools/Venn/.
A summary including the top 25 antibodies and their specific targets for each disease is presented in Supporting Information Table S4. Out of those top 25 lists, most analytes within SLE, RA, and SS were found to be upregulated (15, 21, and 25 respectively), whereas the opposite, e.g., downregulation of most analytes (n = 23) was observed in SV. Accordingly, the overlap with the condensed biomarker signatures for respective diseases was also investigated, which revealed some overlap. Altogether, these results indicated that biologic events, including deregulation of specific analytes for each disease type, could be identified, which may indicate different pathogenetic routes and which could potentially be used to further understand the complexity behind disease progression and for further diagnostic tools.
Discussion
Autoimmune diseases today pose a global health issue, affecting millions of people around the globe, and there is an urgent need for refined clinical tools for early and differential diagnosis.31 Diffuse, general symptoms, such as fatigue, inflammation, and joint pain, which also change in severity over time, shared among several diseases, make clinical diagnosis challenging. In this study, candidate biomarker signatures for the inflammatory rheumatic diseases RA, SLE, SS, and SV were identified. Altogether, the results showed that LOO CV analysis including all antibodies (n = 393) could accurately classify individual IRDs at AUC values ranging between 0.96 and 0.80 (Figure 3). In addition, panels including 40 antibodies could still classify the autoimmune diseases at high accuracy, with AUC values ranging between 0.96 and 0.79 (Figure 4). These results show that using a multiplexed approach to reflect the pathogenetic complexity in rheumatic disorders looks very promising and is a venue to continue and explore to identify new targets for early and differential diagnosis of autoimmune diseases. There is no doubt that there is a large call for better biomarkers in autoimmune diseases. Blood-based biomarkers constitute a simple noninvasive approach, suitable for both discovery biomarker analysis as for the clinical setting and constitute a major ground within the autoimmune community research. Although a few biomarkers have been found as early manifestations of the disease, such as the presence of antinuclear antibodies (ANAs) in SLE,32,33 aCCP in RA,34,35 and anti-SSA/B in SS,36 many biomarkers display too low specificity and/or sensitivity and are used one-by-one or too few in concert to reflect the complexity of the disease.16,37 Biomarkers for differential diagnosis are difficult to identify and refined tools for correct and early diagnosis are of urgent need to prevent severe organ- and tissue-related damage. This study utilized an antibody microarray platform targeting mainly immunoregulatory proteins, which seems to have an advantage when it comes to identifying levels of proteomic changes within systemic autoimmune disorders, as previously demonstrated by the delivery of candidate biomarker signatures for classification of SLE, systemic sclerosis, and SLE disease activity.13,17,18,29,38,39
Based on classification analysis, SLE and SV were found to be the ones most readily separated from the others (AUC = 0.96 and AUC = 0.94 respectively), while RA and SS were a bit more difficult to separate (AUC = 0.86 and AUC = 0.80, respectively) (Figures 3 and 4). This may partly be explained by the fact that Sjögren’s syndrome may overlap in patients with SLE and RA, and similar pathogenic mechanisms have been suggested.3,4 To our knowledge, samples in this study were collected from patients diagnosed with primary SS. RA, which has the highest prevalence of the IRDs investigated in this study, is a heterogeneous condition with complex pathogenesis. This may be reflected by the overlap of the biomarker signature with that of other disorders. Analyzing the serum proteome in patients with primary but also secondary SS, RA, and SLE would indeed be of great value for decoding underlying molecular pathways and of importance from a diagnostic and therapeutic perspective.
The low number of samples used in this study confers a limiting factor since an independent data set for validation was not included. The use of supervised learning algorithms may pose a problem when they are applied in small data sets due to the risk of overfitting, which may lead to poor performance in new sample sets.40,41 Considering this, the approach used for feature extraction and subsequent generation of condensed signatures in this study was carefully selected to avoid the risk of overtraining. Ultimately, a short signature with high predictive power may always be preferred from a logistical and cost-effective view. However, there is always a trade-off between the length of the signature and performance, which is why in this first study, we compromised to include 40 antibodies in the final consensus list. Also, the high number of antibodies most likely reflects that pinpointed diseases do share similar pathogenic pathways, and thus a higher number of antibodies for differential diagnosis may be necessary from this perspective. This assumption may also be supported by the major overlap of analytes observed from the differential analysis (Wilcoxon) (Figure 5 and Supporting Information Table S4), which further stresses the significance of larger data sets to achieve even more stringent analysis.
Based on the differential protein expression analysis, only a small number of disease-specific analytes were found (Supporting Information Table S4, Figure 5B). The complement system is highly involved in the pathogenesis of autoimmune diseases,42 and the major overlap of analytes may suggest similar molecular mechanisms underlying disease progression in autoimmunity. Only one analyte, UBEC2, was found uniquely in SS. UBEC2, is a member of the ubiquitin-conjugating enzyme family, which is involved in the process of destruction of mitotic cyclins and for cell cycle progression.43,44 Interestingly, Ro52 has previously been identified as an E3 ubiquitin ligase, whose increased expression may lead to increased apoptosis and promote autoreactivity as in the generation of Ro52 autoantibodies.45 Compared to the other IRDs, most analytes were found to be downregulated among SV samples, which could explain the high number of differentially expressed analytes within this group. The reason for this difference, however, can only be speculated on, but may indicate that the underlying molecular events taking place in systemic vasculitis are different from the other three diseases. Further studies with bigger sample sets stratified by disease phenotype may help to clarify the underlying role of disease-specific analytes and to aid in the search for novel candidate biomarkers for therapeutic strategies.
In this study, several analytes involved in immunoregulatory response were found to be deregulated among the IRDs compared to the healthy controls (Supporting Information Table S4). One of the upregulated analytes was TNF-α, which has already been shown to be a useful therapeutic target for treatment with biological TNF inhibitors, especially in RA.46,47 Other analytes included the proinflammatory cytokine IL-6, which is also highly interesting from a therapeutic perspective. Monoclonal antibodies that block the IL-6 receptor have been shown to be effective in the treatment of RA48 and large vessel vasculitis.49 The level of osteopontin has previously been demonstrated to be elevated in SLE patients, which we could confirm in this study. Osteopontin has been suggested to be associated with SLE development and a potential marker for SLE activity and organ damage.50 Altogether, these data suggest that a more general autoimmune signature may be present, including several already known and novel markers that may play significant roles in autoimmunity. In addition, the finding of a candidate biomarker signature for classification of IRDs from healthy controls, which is also supported from other studies,15 further strengthens the potential of using our antibody microarray platform for biomarker discovery in autoimmune diseases. A future tool, capable of functioning as a sensor for autoimmune diseases, resulting in the transferral of patients to the right instance, would be of high significance for early and correct diagnosis.
The four systemic IRDs analyzed in this study were chosen based on that although they share some clinical symptoms and autoimmune features, the phenotype and, in particular, the long-term disease course differ substantially. In addition, three of them, e.g., SLE, RA, and SS, are among the most common autoimmune diseases. SV is not that common, though associated with a poor prognosis if untreated. In future studies, it would however be interesting to include other relevant types of immunological diseases and/or nonautoimmune inflammatory conditions such as septic arthritis, scleroderma, multiple sclerosis, and spondyloarthritis. Furthermore, samples from patients with early, clinically undifferentiated disease should be investigated. This would give an opportunity to identify more relevant markers for differential diagnosis and would, even more, reflect the everyday challenge faced at the diagnostic routine at the clinic. A special focus is needed on the clinical challenge of how to differentiate severe autoimmune diseases from nonautoimmune inflammatory conditions, which could be pivotal for early therapeutic interventions. Of note, today, effective treatments are missing for some IRDs, i.e., Sjögren’s syndrome. Better diagnostics would open new and better combinations of therapy, which would decrease the risk for severe organ- and tissue-related damage and increase the quality of life for the patients.
In this study, we conclude that a general IRD biomarker signature could be delineated and that individual IRDs could be classified at high accuracies using a multiplexed microarray. These results together with previous studies15,17,18 suggest that the use of a multiplexed approach is highly suitable for decoding multifactorial diseases such as autoimmune diseases and will play a significant role for future purposes of early diagnosis, essential to prevent severe organ- and tissue-related damage.
Acknowledgments
The authors thank Emelie Noltorp for laboratorial assistance. This study was funded by the Swedish Research Council, the Swedish Rheumatism Association, Alfred Österlund′s Foundation, the Anna-Greta Crafoord Foundation, the Greta and Johan Kock′s Foundation, the King Gustav V′s 80th Birthday Foundation, Lund University Hospital, and the Medical Faculty of Lund University.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00657.
Description of the method for defining condensed biomarker panels; schematic view on data processing for condensed biomarker signatures (Figure S1); list of scFV antibodies included in the study (Table S1); list of CIMS antibodies (Table S2); condensed scFv antibody panels (Table S3); top differentially expressed analytes (Table S4) (PDF)
Author Contributions
◆ These two authors contributed equally to this work.
M.O, CW, and AIE are coinventors of pending patent application(s) submitted by Immunovia AB. C.K. is an employee at Immunovia AB. T.H., A.A.B., E.T., and C.T. have no potential conflict of interests to disclose.
The authors declare the following competing financial interest(s): M.O, CW and AIE are co-inventors of pending patent application(s) submitted by Immunovia AB. CW has also held a part time employment as CTO at Immunovia concering technology development, i.e. work separate from this project. CK is an employee at Immunovia AB. TH, AAB, ET and CT have no potential conflict of interests to disclose.
Supplementary Material
References
- Thomas S. L.; Griffiths C.; Smeeth L.; Rooney C.; Hall A. J. Burden of mortality associated with autoimmune diseases among females in the United Kingdom. Am. J. Public Health 2010, 100, 2279–2287. 10.2105/AJPH.2009.180273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh S. J.; Rau L. M. Autoimmune diseases: a leading cause of death among young and middle-aged women in the United States. Am. J. Public Health 2000, 90, 1463–1466. 10.2105/AJPH.90.9.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoussakis M. N.; Georgopoulou C.; Zintzaras E.; Spyropoulou M.; Stavropoulou A.; Skopouli F. N.; Moutsopoulos H. M. Sjogren’s syndrome associated with systemic lupus erythematosus: clinical and laboratory profiles and comparison with primary Sjogren’s syndrome. Arthritis Rheum. 2004, 50, 882–891. 10.1002/art.20093. [DOI] [PubMed] [Google Scholar]
- Toro-Domínguez D.; Carmona-Saez P.; Alarcon-Riquelme M. E. Shared signatures between rheumatoid arthritis, systemic lupus erythematosus and Sjogren’s syndrome uncovered through gene expression meta-analysis. Arthritis Res. Ther. 2014, 16, 489. 10.1186/s13075-014-0489-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A.; Radfar L.; Lewis D.; Grundahl K.; Stone D. U.; Kaufman C. E.; Rhodus N. L.; Segal B.; Wallace D. J.; Weisman M. H.; Venuturupalli S.; Kurien B. T.; Lessard C. J.; Sivils K. L.; Scofield R. H. Previous diagnosis of Sjogren’s Syndrome as rheumatoid arthritis or systemic lupus erythematosus. Rheumatology 2016, 55, 1195–1201. 10.1093/rheumatology/kew023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander E. L.; Hirsch T. J.; Arnett F. C.; Provost T. T.; Stevens M. B. Ro(SSA) and La(SSB) antibodies in the clinical spectrum of Sjogren’s syndrome. J. Rheumatol. 1982, 9, 239–246. [PubMed] [Google Scholar]
- Falk R. J.; Jennette J. C. Anti-neutrophil cytoplasmic autoantibodies with specificity for myeloperoxidase in patients with systemic vasculitis and idiopathic necrotizing and crescentic glomerulonephritis. N. Engl. J. Med. 1988, 318, 1651–1657. 10.1056/NEJM198806233182504. [DOI] [PubMed] [Google Scholar]
- Rekvig O. P. Anti-dsDNA antibodies as a classification criterion and a diagnostic marker for systemic lupus erythematosus: critical remarks. Clin. Exp. Immunol. 2015, 179, 5–10. 10.1111/cei.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervaert J. W.; Goldschmeding R.; Elema J. D.; van der Giessen M.; Huitema M. G.; van der Hem G. K.; The T. H.; von dem Borne A. E.; Kallenberg C. G. Autoantibodies against myeloid lysosomal enzymes in crescentic glomerulonephritis. Kidney Int. 1990, 37, 799–806. 10.1038/ki.1990.48. [DOI] [PubMed] [Google Scholar]
- Haller-Kikkatalo K.; Alnek K.; Metspalu A.; Mihailov E.; Metskula K.; Kisand K.; Pisarev H.; Salumets A.; Uibo R. Demographic associations for autoantibodies in disease-free individuals of a European population. Sci. Rep. 2017, 7, 44846 10.1038/srep44846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E. M.; Feltkamp T. E.; Smolen J. S.; Butcher B.; Dawkins R.; Fritzler M. J.; Gordon T.; Hardin J. A.; Kalden J. R.; Lahita R. G.; Maini R. N.; McDougal J. S.; Rothfield N. F.; Smeenk R. J.; Takasaki Y.; Wiik A.; Wilson M. R.; Koziol J. A. Range of antinuclear antibodies in “healthy” individuals. Arthritis Rheum. 1997, 40, 1601–1611. 10.1002/art.1780400909. [DOI] [PubMed] [Google Scholar]
- Wandstrat A. E.; Carr-Johnson F.; Branch V.; Gray H.; Fairhurst A. M.; Reimold A.; Karp D.; Wakeland E. K.; Olsen N. J. Autoantibody profiling to identify individuals at risk for systemic lupus erythematosus. J. Autoimmun. 2006, 27, 153–160. 10.1016/j.jaut.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Borrebaeck C. A.; Sturfelt G.; Wingren C. Recombinant antibody microarray for profiling the serum proteome of SLE. Methods Mol. Biol. 2014, 1134, 67–78. [DOI] [PubMed] [Google Scholar]
- Borrebaeck C. A.; Wingren C. Transferring proteomic discoveries into clinical practice. Expert Rev. Proteomics 2009, 6, 11–13. 10.1586/14789450.6.1.11. [DOI] [PubMed] [Google Scholar]
- Carlsson A.; Wuttge D. M.; Ingvarsson J.; Bengtsson A. A.; Sturfelt G.; Borrebaeck C. A. K.; Wingren C. Serum Protein Profiling of Systemic Lupus Erythematosus and Systemic Sclerosis Using Recombinant Antibody Microarrays. Mol. Cell. Proteomics 2011, 10, M110.005033 10.1074/mcp.M110.005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. S.; Banha J.; Penque D.; Costa L.; Conrads T. P.; Cahill D. J.; O’Brien J. K.; Rooney M. E. Diagnostic and prognostic biomarker discovery strategies for autoimmune disorders. J. Proteomics 2010, 73, 1045–1060. 10.1016/j.jprot.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Petersson L.; Dexlin-Mellby L.; Bengtsson A. A.; Sturfelt G.; Borrebaeck C. A.; Wingren C. Multiplexing of miniaturized planar antibody arrays for serum protein profiling--a biomarker discovery in SLE nephritis. Lab Chip 2014, 14, 1931–1942. 10.1039/C3LC51420J. [DOI] [PubMed] [Google Scholar]
- Delfani P.; Sturfelt G.; Gullstrand B.; Carlsson A.; Kassandra M.; Borrebaeck C. A.; Bengtsson A. A.; Wingren C. Deciphering systemic lupus erythematosus-associated serum biomarkers reflecting apoptosis and disease activity. Lupus 2017, 26, 373–387. 10.1177/0961203316669240. [DOI] [PubMed] [Google Scholar]
- Gladman D. D.; Ibanez D.; Urowitz M. B. Systemic lupus erythematosus disease activity index 2000. J Rheumatol 2002, 29, 288–291. 10.3899/jrheum.080776. [DOI] [PubMed] [Google Scholar]
- Arnett F. C.; Edworthy S. M.; Bloch D. A.; McShane D. J.; Fries J. F.; Cooper N. S.; Healey L. A.; Kaplan S. R.; Liang M. H.; Luthra H. S.; et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988, 31, 315–324. 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- Mukhtyar C.; Lee R.; Brown D.; Carruthers D.; Dasgupta B.; Dubey S.; Flossmann O.; Hall C.; Hollywood J.; Jayne D.; Jones R.; Lanyon P.; Muir A.; Scott D.; Young L.; Luqmani R. A. Modification and validation of the Birmingham Vasculitis Activity Score (version 3). Ann. Rheum. Dis. 2009, 68, 1827–1832. 10.1136/ard.2008.101279. [DOI] [PubMed] [Google Scholar]
- Vitali C.; Bombardieri S.; Jonsson R.; Moutsopoulos H. M.; Alexander E. L.; Carsons S. E.; Daniels T. E.; Fox P. C.; Fox R. I.; Kassan S. S.; Pillemer S. R.; Talal N.; Weisman M. H. European Study Group on Classification Criteria for Sjogren’s, S., Classification criteria for Sjogren’s syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann. Rheum. Dis. 2002, 61, 554–558. 10.1136/ard.61.6.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderlind E.; Strandberg L.; Jirholt P.; Kobayashi N.; Alexeiva V.; Aberg A. M.; Nilsson A.; Jansson B.; Ohlin M.; Wingren C.; Danielsson L.; Carlsson R.; Borrebaeck C. A. Recombining germline-derived CDR sequences for creating diverse single-framework antibody libraries. Nat. Biotechnol. 2000, 18, 852–856. 10.1038/78458. [DOI] [PubMed] [Google Scholar]
- Säll A.; Walle M.; Wingren C.; Muller S.; Nyman T.; Vala A.; Ohlin M.; Borrebaeck C. A. K.; Persson H. Generation and analyses of human synthetic antibody libraries and their application for protein microarrays. Protein Eng., Des. Sel. 2016, 29, 427–437. 10.1093/protein/gzw042. [DOI] [PubMed] [Google Scholar]
- Olsson N.; Wingren C.; Mattsson M.; James P.; O’Connell D.; Nilsson F.; Cahill D. J.; Borrebaeck C. A. Proteomic analysis and discovery using affinity proteomics and mass spectrometry. Mol. Cell. Proteomics 2011, 10, M110.003962 10.1074/mcp.M110.003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog P.; Ohlsson M.; Ferno M.; Ryden L.; Borrebaeck C. A. K.; Wingren C. Tumor tissue protein signatures reflect histological grade of breast cancer. PLoS One 2017, 12, e0179775 10.1371/journal.pone.0179775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfani P.; Dexlin Mellby L.; Nordstrom M.; Holmer A.; Ohlsson M.; Borrebaeck C. A.; Wingren C. Technical Advances of the Recombinant Antibody Microarray Technology Platform for Clinical Immunoproteomics. PLoS One 2016, 11, e0159138 10.1371/journal.pone.0159138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson A.; Wingren C.; Kristensson M.; Rose C.; Ferno M.; Olsson H.; Jernstrom H.; Ek S.; Gustavsson E.; Ingvar C.; Ohlsson M.; Peterson C.; Borrebaeck C. A. Molecular serum portraits in patients with primary breast cancer predict the development of distant metastases. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 14252–14257. 10.1073/pnas.1103125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson A.; Wuttge D. M.; Ingvarsson J.; Bengtsson A. A.; Sturfelt G.; Borrebaeck C. A.; Wingren C. Serum protein profiling of systemic lupus erythematosus and systemic sclerosis using recombinant antibody microarrays. Mol. Cell. Proteomics 2011, 10, M110.005033 10.1074/mcp.M110.005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y.; Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., Ser. B 1995, 57, 289–300. [Google Scholar]
- Cooper G. S.; Bynum M. L.; Somers E. C. Recent insights in the epidemiology of autoimmune diseases: improved prevalence estimates and understanding of clustering of diseases. J. Autoimmun. 2009, 33, 197–207. 10.1016/j.jaut.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuckle M. R.; McClain M. T.; Rubertone M. V.; Scofield R. H.; Dennis G. J.; James J. A.; Harley J. B. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med 2003, 349, 1526–1533. 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- Eriksson C.; Kokkonen H.; Johansson M.; Hallmans G.; Wadell G.; Rantapaa-Dahlqvist S. Autoantibodies predate the onset of systemic lupus erythematosus in northern Sweden. Arthritis Res. Ther. 2011, 13, R30. 10.1186/ar3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gaalen F. A.; Linn-Rasker S. P.; van Venrooij W. J.; de Jong B. A.; Breedveld F. C.; Verweij C. L.; Toes R. E.; Huizinga T. W. Autoantibodies to cyclic citrullinated peptides predict progression to rheumatoid arthritis in patients with undifferentiated arthritis: a prospective cohort study. Arthritis Rheum. 2004, 50, 709–715. 10.1002/art.20044. [DOI] [PubMed] [Google Scholar]
- Visser H.; le Cessie S.; Vos K.; Breedveld F. C.; Hazes J. M. How to diagnose rheumatoid arthritis early: a prediction model for persistent (erosive) arthritis. Arthritis Rheum. 2002, 46, 357–365. 10.1002/art.10117. [DOI] [PubMed] [Google Scholar]
- Theander E.; Jonsson R.; Sjostrom B.; Brokstad K.; Olsson P.; Henriksson G. Prediction of Sjogren’s Syndrome Years Before Diagnosis and Identification of Patients With Early Onset and Severe Disease Course by Autoantibody Profiling. Arthritis Rheumatol. 2015, 67, 2427–2436. 10.1002/art.39214. [DOI] [PubMed] [Google Scholar]
- Mohan C.; Assassi S. Biomarkers in rheumatic diseases: how can they facilitate diagnosis and assessment of disease activity?. Br. Med. J. 2015, 351, h5079 10.1136/bmj.h5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson J.; Larsson A.; Sjoholm A. G.; Truedsson L.; Jansson B.; Borrebaeck C. A.; Wingren C. Design of recombinant antibody microarrays for serum protein profiling: targeting of complement proteins. J. Proteome Res. 2007, 6, 3527–3536. 10.1021/pr070204f. [DOI] [PubMed] [Google Scholar]
- Petersson L.; Coen M.; Amro N. A.; Truedsson L.; Borrebaeck C. A.; Wingren C. Miniaturization of multiplexed planar recombinant antibody arrays for serum protein profiling. Bioanalysis 2014, 6, 1175–1185. 10.4155/bio.13.342. [DOI] [PubMed] [Google Scholar]
- Aliferis C. F.; Statnikov A.; Tsamardinos I. Challenges in the analysis of mass-throughput data: a technical commentary from the statistical machine learning perspective. Cancer Inf. 2007, 2, 133–162. [PMC free article] [PubMed] [Google Scholar]
- Han H.; Li X. L. Multi-resolution independent component analysis for high-performance tumor classification and biomarker discovery. BMC Bioinformatics 2011, 12, S7. 10.1186/1471-2105-12-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.; Daha M. R.; Kallenberg C. G. The complement system in systemic autoimmune disease. J. Autoimmun. 2010, 34, J276–286. 10.1016/j.jaut.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Pickart C. M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Weissman A. M. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2001, 2, 169–178. 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Espinosa A.; Zhou W.; Ek M.; Hedlund M.; Brauner S.; Popovic K.; Horvath L.; Wallerskog T.; Oukka M.; Nyberg F.; Kuchroo V. K.; Wahren-Herlenius M. The Sjogren’s syndrome-associated autoantigen Ro52 is an E3 ligase that regulates proliferation and cell death. J. Immunol. 2006, 176, 6277–6285. 10.4049/jimmunol.176.10.6277. [DOI] [PubMed] [Google Scholar]
- Feldmann M.; Maini R. N. Anti-TNF alpha therapy of rheumatoid arthritis: what have we learned?. Annu. Rev. Immunol. 2001, 19, 163–196. 10.1146/annurev.immunol.19.1.163. [DOI] [PubMed] [Google Scholar]
- Lipsky P. E.; van der Heijde D. M.; St. Clair E. W.; Furst D. E.; Breedveld F. C.; Kalden J. R.; Smolen J. S.; Weisman M.; Emery P.; Feldmann M.; Harriman G. R.; Maini R. N. Anti-Tumor Necrosis Factor Trial in Rheumatoid Arthritis with Concomitant Therapy Study, G., Infliximab and methotrexate in the treatment of rheumatoid arthritis. Anti-Tumor Necrosis Factor Trial in Rheumatoid Arthritis with Concomitant Therapy Study Group. N. Engl. J. Med. 2000, 343, 1594–1602. 10.1056/NEJM200011303432202. [DOI] [PubMed] [Google Scholar]
- Smolen J. S.; Beaulieu A.; Rubbert-Roth A.; Ramos-Remus C.; Rovensky J.; Alecock E.; Woodworth T.; Alten R. Investigators, O., Effect of interleukin-6 receptor inhibition with tocilizumab in patients with rheumatoid arthritis (OPTION study): a double-blind, placebo-controlled, randomised trial. Lancet 2008, 371, 987–997. 10.1016/S0140-6736(08)60453-5. [DOI] [PubMed] [Google Scholar]
- Stone J. H.; Tuckwell K.; Dimonaco S.; Klearman M.; Aringer M.; Blockmans D.; Brouwer E.; Cid M. C.; Dasgupta B.; Rech J.; Salvarani C.; Schett G.; Schulze-Koops H.; Spiera R.; Unizony S. H.; Collinson N. Trial of Tocilizumab in Giant-Cell Arteritis. N. Engl. J. Med. 2017, 377, 317–328. 10.1056/NEJMoa1613849. [DOI] [PubMed] [Google Scholar]
- Kaleta B. Role of osteopontin in systemic lupus erythematosus. Arch. Immunol. Ther. Exp. 2014, 62, 475–482. 10.1007/s00005-014-0294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.