Abstract
Objective:
Suicide death is a highly preventable, yet growing, worldwide health crisis. To date, there has been a lack of adequately powered genomic studies of suicide, with no sizable suicide death cohorts available for study. To address this limitation, we conducted the first comprehensive genomic analysis of suicide death using previously unpublished genotype data from a large, population-ascertained cohort.
Methods:
The analysis sample consisted of 3,413 population-ascertained cases of European ancestry and 14,810 ancestrally matched controls. Analytical methods included principal components analysis for ancestral matching and adjusting for population stratification, linear mixed model genome-wide association testing (conditional on genetic relatedness matrix), gene and gene set enrichment testing, polygenic score analyses, as well as SNP heritability and genetic correlation estimation using LD score regression.
Results:
GWAS identified two genome-wide significant loci (6 SNPs, p<5×10–8, including rs34399104, rs35518298, rs34053895, rs66828456, rs35502061, and rs35256367). Gene-based analyses implicated 22 genes on chromosomes 13, 15, 16, 17, and 19 (q<0.05). Suicide death heritability was estimated at h2SNP = .25 (SE = .04), and .16 (.02) when converted to a liability scale. Notably, suicide polygenic scores were significantly predictive. Polygenic scores for several other psychiatric disorders and psychological traits were also predictive, particularly behavioral disinhibition and major depressive disorder.
Conclusions:
In this report, we identify multiple genome-wide significant loci/genes and demonstrate polygenic score prediction of suicide death case-control status, adjusting for ancestry, in independent training and test sets. Additionally, we report that suicide death cases have increased genetic risk for behavioral disinhibition, major depression, depressive symptoms, autism spectrum disorder, psychosis, and alcohol use disorder relative to controls.
INTRODUCTION
Suicide death is a behavioral event which reflects a complex, heritable phenotype with diverse clinical antecedents and environmental risk factors.1 The estimated annual age-adjusted suicide death prevalence rate is approximately 0.014% in the United States,2 and this rate has been steadily increasing since 2000.3 Suicide is now ranked the second leading cause of death for all persons 15–24 years old in the United States.4 As such, suicide is currently considered a major public health challenge,5 which has spurred research into its etiology and clinical prediction.6,7
While it is established that suicide is significantly heritable,8 well-powered genomic research on the topic has been largely limited to the study of suicide-related behaviors rather than the ultimate phenotype of suicide death. And though results from well-powered GWAS of suicidal behavior have advanced our understanding of the process,9 the vast majority of suicidal behavior does not result in suicide death.1,10 Thus, suicidal behavior represents a less severe and likely more heterogeneous phenotype than suicide death, characteristics which may adversely affect statistical power to detect genetic associations. Conversely, the unambiguous phenotype of suicide death avoids several confounds inherent in the study of suicidal behavior or ideation, and also focuses study on one of the single most critical contemporary public health outcomes. Previous genomic studies of suicidal behavior have also been limited by narrow ascertainment, studying only individuals with specific diagnoses (e.g., mood disorders, psychotic disorders) in order to maximize severity and accommodate post hoc study designs. We avoid this limitation here, instead using population-based sampling with ascertainment wholly independent of any co-occurring diagnoses; thus improving the representativeness of cases to the corresponding population and, consequently, the generalizability of results.
Suicide death, like its associated clinical antecedents including schizophrenia and depression, has a highly multifactorial, and likely highly polygenic, etiology.9,11 Currently, the scientific literature lacks well-powered studies of suicide death in relation to molecular genetic risk for any medical or psychiatric diagnoses, and no robust polygenic scores have yet been developed for the critical outcome of suicide death. There have been, however, a few modestly powered GWAS of suicide death published,12,13 the largest of which combined two independent Japanese cohorts totaling 746 completed suicide cases.13 Further, a recent UK Biobank GWAS of suicidal behavior has conducted preliminary polygenic analyses of suicide death using a very modest number completed suicide cases (n = 127).14 Of note, these smaller studies reported evidence of polygenicity in both suicidal behavior and suicide death, but with widely varying SNP-based heritability estimates, from h2SNP = 4.615 and 7.6%14 for suicidal behavior not including death, to h2SNP = 35–48% for suicide death.13 The current study marks a notable advance from earlier research, as it uses the world’s largest DNA databank of suicide death, merged with a massive bank of electronic medical record and sociodemographic data,16,17 to comprehensively model common variant genetic, and clinical phenotypic, precursors of suicide death.
This study adds to the growing body of genomic research on suicide,13,14 providing the first adequately powered genome-wide association study of suicide death. Analyses integrated data on modes of suicide death, medical and psychiatric diagnostic (ICD-10) codes,18 and medical and psychiatric polygenic scores to comprehensively model common variant genetic risk for suicide death. Secondary analyses of sex differences were conducted due to the substantial sex differences in suicide death rates and modes of suicide death.19,20 Reliable and significant prediction of case-control status was achieved, adjusting for ancestry, using both (1) a novel polygenic score for suicide death and (2) polygenic scores for a range of comorbid psychiatric and medical risk factors, particularly behavioral disinhibition, major depressive disorder, and depressive symptoms. Additionally, gene set enrichment, SNP heritability, and genetic correlations were examined in this unique, unpublished data resource. We conclude with a brief discussion of the relevance of our findings to the broader field and promising future research directions.
METHODS
Sample Ascertainment
Cases
In collaboration with the centralized, statewide Utah Office of Medical Examiner (OME), the authors obtained DNA samples from ~6000 persons who died by suicide. The centralized OME and conservative determination helped to maximize the accuracy of suicide case status.21 Suicide cause-of-death determination results from a detailed investigation of the scene of the death and circumstances of death, determination of medical conditions by full autopsy, review of medical and other public records concerning the case, interviews with survivors, and standard toxicology workups. Suicide determination is traditionally made quite conservatively due to its impact on surviving relatives.
DNA from suicide deaths were extracted from whole blood using the Qiagen Autopure LS automated DNA extractor (www.qiagen.com). Genotyping is described below. Data collection and genotyping for the Utah Suicide Study is ongoing. To date, 4,381 samples have been genotyped in two waves as determined by the date of sample receipt and the availability of genotyping funds (for detailed information on sample collection see Supplementary Methods S1. After quality control procedures and ancestry analysis, data are comprised of 3,413 Utah suicide deaths. The Utah population is primarily Northwestern European in ancestry, a relatively genetically homogeneous group with low inbreeding across generations, comparable to the rest of the United States.22
Controls
Generation Scotland.
Controls closely matching the Northern European ancestry of the cases were obtained from previously curated datasets in the UK. Wave 1 analysis included 3,623 founder controls from the population-based Generation Scotland Scottish Family Health Study.23 The Generation Scotland Scottish Family Health Study (N > 24,000) constitutes an ancestrally comparable population-based cohort for comparison with the suicide decedents in Utah. To eliminate confounding arising from intra-dataset relatedness, only the 3,623 founders from the Generation Scotland dataset were used in analyses.
UK10K.
A total of 11,049 UK10K controls24 were analyzed in wave 2 and GWAS analyses of both waves. This second control cohort is comprised of approximately 4000 genomes from the UK along with 6000 exomes from UK individuals with selected health phenotypes. We chose these data due to the extensive phenotyping and characterization of any medical conditions present, and to avoid choosing a cohort of entirely psychiatrically and medically healthy individuals. 4,000 highly phenotyped “super control” samples were supplied from the TwinsUK study from King’s College London and the Avon Longitudinal Study of Parents and Children. UK10K was a collaborative project to examine obesity, autism, schizophrenia, familial hypercholesterolemia, thyroid disorders, learning disabilities, ciliopathies, congenital heart disease, coloboma, neuromuscular disorders, and rare disorders including severe insulin resistance. Analyses of this control cohort included all diagnostic groups. While this inclusion may have been conservative due to possible associations of chronic disease with suicide risk, determination of effects of specific disease states was beyond the scope of the current study. Genotyping and sequencing procedures for UK10K have been previously described24 (http://www.uk10k.org) and all molecular genetic data from UK10K were filtered to the hard call variants present in our suicide death cohort prior to imputation of all cohorts simultaneously.
1000 Genomes Reference Panel.
The CEU population from the 1000 Genomes Project25 (1KG), which includes only Utah residents carefully screened for Northwestern European ancestry, was utilized as a model for excluding ancestrally discordant suicide and control samples. These CEU data were downloaded from the 1000 Genomes Project public repository. Unrelated individuals in the CEU provide a compelling, albeit small, ancestrally matched control resource (n = 99). A variety of candidate control samples were assessed via PCA for ancestral comparability to CEU and decedent data, with UK10K and Generation Scotland founder data representing the closest match.
Utah controls would be an ideal match for the suicide cases, but as with many GWAS, local controls were not readily available at the sample size needed for GWAS. CEPH ancestry 1KG were a useful comparison group to assess the likelihood that UK controls were an appropriate match for the cases. In addition (and described in more detail below), we performed a Generation Scotland controlUK10K control GWAS and subsequently eliminated any SNPs from the case-control analysis that evidenced signal between the control cohorts. This was performed to minimize the possibility of false positives in the case control GWAS due to population and geographic stratification across cohorts.
Genotyping and Quality Control
Suicide cases were genotyped using Illumina Infinium PsychArray platform measuring 593,260 single nucleotide polymorphisms (SNPs). Generation Scotland samples were genotyped using Illumina OmniExpress SNP GWAS and exome chip, measuring 700,000 and 250,000 SNPs, respectively.23 UK10K samples were whole genome sequenced24 and variants were extracted to match the available QC’d hard-called variants in the suicide cases. Genotypes were subsequently imputed in all cases and controls (details of imputation are presented in Analytics, below). Both case and control datasets resulted from population-based ascertainment. Cryptic relatedness was modeled via the derivation of genomic relatedness matrices. Genotyping quality control was performed using SNP clustering in Illumina Genome Studio https://www.illumina.com/techniques/microarrays/array-data-analysis-experimentaldesign/genomestudio.html). SNPs were retained if the GenTrain score was > 0.5 and the Cluster separation score was > 0.4. SNPs were converted to HG19 plus strand. SNPs with >5% missing genotypes were removed. Samples with a call rate < 95% were removed.
Prior to case-control GWAS, a control-control GWAS was conducted (using the same methods described in Data Analysis: Genome-Wide Association Testing, below) in order to detect differences between control groups (all variants both pre- and post-imputation with GWAS control-control q values <.10). For example, chromosome 4 TLR variants are often filtered from analyses involving Scottish controls due to population stratification. 167 variants in the control-control comparison were then filtered from subsequent case-control analyses. For the purposes of future meta-GWAS analyses and because the MHC is relevant to psychiatric risk, we included MHC-associated filtered variants in a second version of summary statistics.
Data Analysis
Principal Component Analysis (PCA)
Supplementary Figure S1 shows 1000 Genomes superpopulations and suicide case/control samples, both included and excluded, plotted by the top two principal components (PCs). Approximately 20% of the population-based suicide cases had a significant degree of non-Northwestern European ancestry (chiefly of admixed ancestry) and were excluded from analyses. The variation explained by top four PCs was reduced 7.2-fold. The top four PCs explain 0.89% of variation before sample filtering and 0.12% of variation after filtering if calculated on pruned genotypes. For adequate statistical power, we examined only cases of Northern European ancestry. However, it is clear from Figure S1 (a) and (c) that the cohort was comprised of multiple ancestries and that research on suicide death in non-European ancestries will reflect an important step beyond this first study.
PCA was performed on control, suicide, and 1000 Genomes cohorts after linkage disequilibrium (LD) pruning at a 0.2 threshold. To exclude ancestrally heterogeneous samples, the top four principal components (defined as those components which accounted for > 0.1% of the genotype variance) were used to establish PC centroid limits centered around 1000 Genomes CEU data such that 99% of the CEU data fell within the limits. Only suicide and control samples also falling within these limits were considered ancestrally homogenous and were included in the association study. The ancestry PCA was performed using RaMWAS, a Bioconductor package written by our analytical team, which comprises a complete toolset for high dimensional genomic analyses.26,27
Imputation
European ancestry cases and controls were well-matched to 1000 Genomes CEPH. The Haplotype Reference Consortium is comprised in part by UK controls used here so we elected to impute genotypes based on the 1000 Genomes reference panel using minimac328 and Eagle.29 SNPs were imputed jointly from a common SNP list based on variant overlap across case and control datasets. SNPs with ambiguous strand orientation, >5% missing calls, or Hardy-Weinberg equilibrium p < 0.001 were excluded. SNPs with minor allele frequency below 0.01 or imputation R2 < 0.5 or average imputation call rate < 0.9 were excluded after imputation. Genomic data were handled using PLINK.30,31 Final GWAS analysis was performed on 7,519,308 variants passing quality control.
Genome-wide Association Testing
A Linear Mixed Model (LMM) algorithm tested variant association with suicide death and provided follow-up examination of significant hits for linkage disequilibrium and gene set enrichment. GWAS were performed using GEMMA,32 a computationally efficient and open-source LMM algorithm for GWAS that models population stratification remaining after PCA by use of genomic relatedness matrices. Sex was not included as a covariate in GWAS analyses due to the association of suicide with sex status at a ratio of approximately 4:1 males to females. GWAS with hard call-only and then with imputed data were examined separately to assess potential population stratification unique to our imputed GWAS. Prior to case-control GWAS, control-control GWAS was implemented to filter signal likely due to population stratification in the controls. To assess the likelihood that observed results may be due to chance, we assessed FDR at 5%.
Gene and Gene Set Enrichment and Functional Mapping
SNP results from the GWAS were then mapped to genes within 1kb of the SNP. These genes were examined for gene set enrichment and LD using FUMA33 and GREAT.34 FUMA annotates SNPs, uses MAGMA to identify associated genes (of approximately 18,612), and provides gene and gene pathway enrichment analysis (of approximately 10,649 pathways). GREAT analyzes the functional significance of sets of cis-regulatory regions by modeling the genome regulatory landscape using multiple information sources and can determine the functional domain of intergenic variants. GREAT improves upon the identification of genes associated with non-coding genomic regions through statistically rigorous incorporation of more distal binding sites from 20 ontologies. The GWAS catalog (https://www.ebi.ac.uk/gwas/) includes studies and associations if they: include a primary GWAS analysis from >100,000 SNPs with SNP-trait p-value <1.0×10–5 in the overall (initial GWAS + replication) population. The most significant SNP from each independent locus is extracted.
Polygenic Risk Scores, SNP Heritability (h2), and Genetic Correlations (rG)
Discovery GWAS summary statistics for phenotypes were compiled to score each cohort for polygenic risk. A polygenic score (PS) for suicide death was derived using PRSice 2.035 and summary statistics from a 10-fold cross validation procedure to avoid overfitting. To elaborate, k-folds cross-validation is a well-established method36 allowing a single dataset to serve as both training and testing data for the purpose of suicide death polygenic score development and validation. We conservatively set the p-value threshold for predicting case status based on the data to 1.0. This eliminated overfitting arising from choosing the threshold based on the phenotype.
Using related methods, we calculated polygenic scores for several psychiatric and psychological traits in the current dataset. Of several thousand medical and psychological GWAS now available as training weights for calculating polygenic scores, only GWAS with N>10,000 individuals and >1,000 cases (or for population-based studies, adequate base rates) were selected for these analyses. These generally included the largest medical and psychiatric GWAS. When multiple versions of GWAS were available for the same phenotype (for example, neuroticism or depression), we selected the most comprehensive (see the online GWAS Atlas at http://atlas.ctglab.nl/). PRSice 2.0 was used to calculate individual PS for 59 phenotypes with estimated risk allele effect sizes for each discovery sample trait. A PS is traditionally calculated as a weighted sum score, where a score for an individual in the target sample is calculated by the summation of each SNP multiplied by the effect size of that SNP in the discovery GWAS. Based on the cross-disorder psychiatric genomics findings to date, we hypothesized significant positive prediction of suicide with PS for depressive symptoms, depressive disorders, behavioral disinhibition, schizophrenia, autism, loneliness, child IQ, alcohol use, and neuroticism.
Linkage Disequilibrium SCore regression (LDSC)37,38 was used to calculate the observed scale common variant h2 using summary statistics from a logistic regression model with five ancestry covariates and pruning related samples at 0.05 p̂ from IBD. LDSC was also used to calculate common variant molecular genetic correlations (rG) with psychiatric and medical phenotypes. Finally, we performed secondary analyses to characterize genetic predictors and clinical antecedents of suicide (Supplementary Methods S2) and mode of suicide death (Supplementary Methods S3). Specifically, in these secondary analyses, we conducted (1) epidemiological association tests between four sufficiently prevalent/powered modes of death (i.e., gun, overdose, asphyxiation, and violent trauma) and 30 ICD-10 derived clinical antecedents, and (2) association tests the suicide polygenic score with modes of death, adjusting for five ancestry covariates in multivariate regressions.
Sex Differences
As suicide rates and modes of suicide death are characterized by substantial sex differences,19,20 we performed secondary epidemiological and genomic analyses to characterize sex differences in mode of death and clinical antecedents. These sex stratified analyses mirrored the full sample analyses described above, including (1) sex-stratified epidemiological association tests between four sufficiently prevalent/powered modes of death (i.e., gun, overdose, asphyxiation, and violent trauma) and 30 ICD-10 derived clinical antecedents, and (2) sex stratified association tests the suicide polygenic score with modes of death, adjusting for five ancestry covariates in multivariate regressions. We constrained these exploratory analyses to only those medical diagnoses with frequencies high enough in either females or males to provide decent power for testing and report false discovery rate (FDR) corrected p-values; nonetheless, it is worth noting that power for these secondary analyses of sex differences was limited by N, which was restricted to cases-only and stratified by sex and mode of death.
RESULTS
Genome-wide Association: Positional Evidence
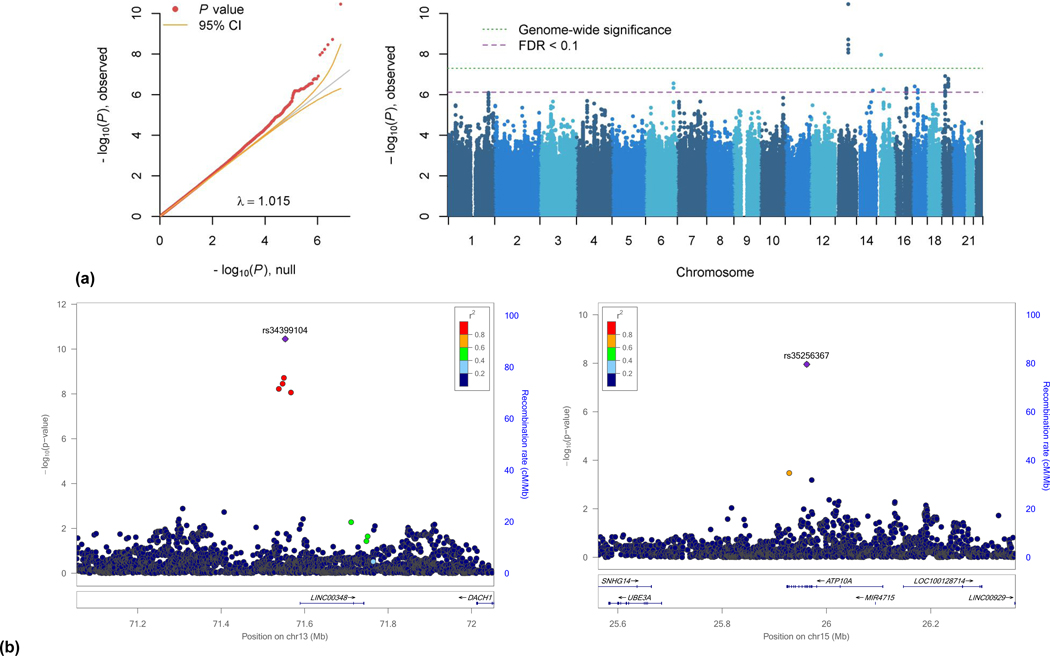
A total of six variants from two loci met genome-wide criteria for statistical association with suicide death (p < 5×10−8). An additional 52 variants were nominally significant at q < 0.1 and mapped to 22 genes (λ = 1.015, Figure 1 and Table 1).39,40 All results on the full cohort are derived from analyses adjusting for effects of ancestry. Genes associated with top genomic regions are presented in Supplementary Table S1. The significant chromosome 13 and 15 regions were supported by additional positive results that were suggestive but below threshold. The large number of signals in the SNP-based tests prompted quality control analyses varying the degree of LD pruning prior to PCA for the purposes of sensitivity analysis. Results and respective λ’s were consistent across these analyses. Supplementary Figures S2-S8 present additional plots of the top signals in each of eight regions.
Figure 1. GWAS Results.
(a) QQ and Manhattan plots from GWAS of suicide death. Y-axes for both plots reflect observed p-values. The x-axis on the qq-plot is the number of significant p-values expected under H0, and the x-axis on the Manhattan plot maps each chromosome. The purple dashed line indicates FDR corrected nominal statistical significance, the green dotted line representing the threshold for genome-wide significance with Bonferroni correction. 57 SNPs met threshold for nominal significance and 6 met genome-wide significance. (b) Regional plots of genome-wide significant loci on chromosomes 13 and 15.
Table 1.
Genome-wide significant loci from GWAS of death by suicide.
| Chr | SNP | Imputation R2 | A1 | A2 | AF Cases | AF Controls | OR | 95% CI | p value | FDR | Nearest Gene | Function | CADD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13 | rs34399104 | 0.75 | T | C | 0.027 | 0.015 | 1.769 | 1.48–2.11 | 3.54×10−11 | 2.7×10−4 | LINC00348 | Intergenic | 19.86 |
| 13 | rs35518298 | 0.81 | T | C | 0.024 | 0.014 | 1.726 | 1.43–2.07 | 1.92×10−9 | 7.2×10−3 | LINC00348 | Intergenic | 0.33 |
| 13 | rs34053895 | 0.81 | A | C | 0.024 | 0.014 | 1.718 | 1.42–2.06 | 3.51×10−9 | 8.7×10−3 | LINC00348 | Intergenic | 0.39 |
| 13 | rs35502061 | 0.72 | G | A | 0.024 | 0.014 | 1.703 | 1.41–2.05 | 5.97×10−9 | .011 | SOGA2P1 | Intergenic | 2.69 |
| 13 | rs66828456 | 0.74 | A | C | 0.027 | 0.015 | 1.707 | 1.42–2.04 | 8.63×10−9 | .013 | LINC00348 | Intergenic | 4.24 |
| 15 | rs35256367 | 0.50 | G | A | 0.023 | 0.014 | 1.605 | 1.43–2.07 | 1.10×10−8 | .014 | ATP10A | Intronic | 4.41 |
Note: Chr = chromosome, SNP = single nucleotide polymorphism, A1, A2 = alleles 1 (minor) and 2, AF = allele 1 frequency, OR = odds ratio, SE = standard error, CADD61,62 = combined annotation dependent depletion score. For scale, note that a CADD score of 20 means that a variant is among the top 1% of deleterious variants in the human genome. All variants presented in the table were imputed.
Gene-based Results and Pathway Functional Enrichment Tests
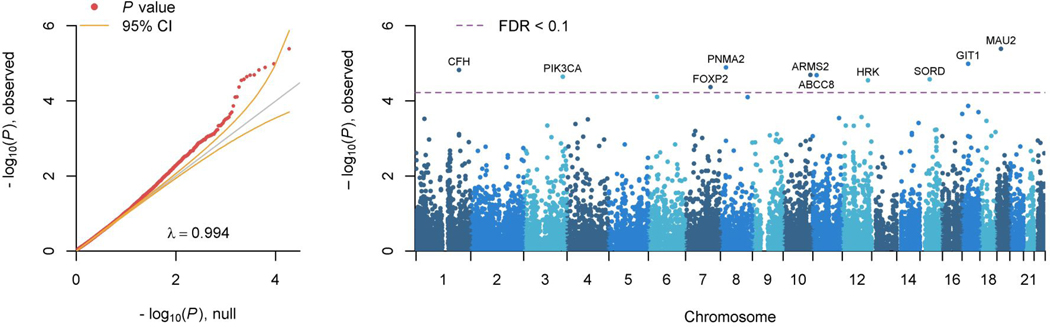
Mapping top positional SNP hits to genes suggested 22 gene associations, including chromosome 13 genes, Daschund family transcription factor 1 (DACH1), Ubiquitin-protein ligase protein (UBE3A), and Kelch-like family member 1 (KLHL1) on chromosome 15 (Supplementary Table S1). Gene-based analysis using MAGMA (FUMA1) identified an additional ten genes significantly associated (q < 0.1) with suicide death (Figure 2; Supplementary Table S1). GO pathway results included enrichment of histone modification sites SETD6, COPR5, GATAD2A. Full gene and Gene Ontology (GO, http://www.geneontology.org/) pathway enrichment results are presented in Supplementary Tables S2-S3. Genes showing psychiatric associations are in green (Supplementary Table S2).
Figure 2. GWAS Gene-Based Results.
FUMA gene-based test results: qq plot and Manhattan plot of >18,000 genes. Y-axes for both plots are identical and reflect observed p-values. The x-axis on the qq-plot is the number of significant p-values expected under H0, and the x-axis on the Manhattan plot maps each chromosome. The purple dashed line indicates threshold for FDR-corrected nominal statistical significance; 10 genes met this threshold for nominal significance.
In addition to functional pathways, FUMA analyses indicated significant enrichment for schizophrenia (p = 1×10−11) and bipolar disorder (p = 1×10−17) in the GWAS Catalog (https://www.ebi.ac.uk/gwas/; Table S1). IW-scoring in SNP-Nexus41 suggested regulatory functional significance for one SNP (chr13:71553748:C/T). Ten of the implicated genes from positional or gene-based testing have evidenced genome-wide significant differential gene expression in postmortem brain analyses in schizophrenia, autism, or bipolar disorder (FDR<0.1; PsychENCODE Consortium, Supplementary Table S4).42
Polygenic Scores, SNP Heritability, and Genetic Correlations
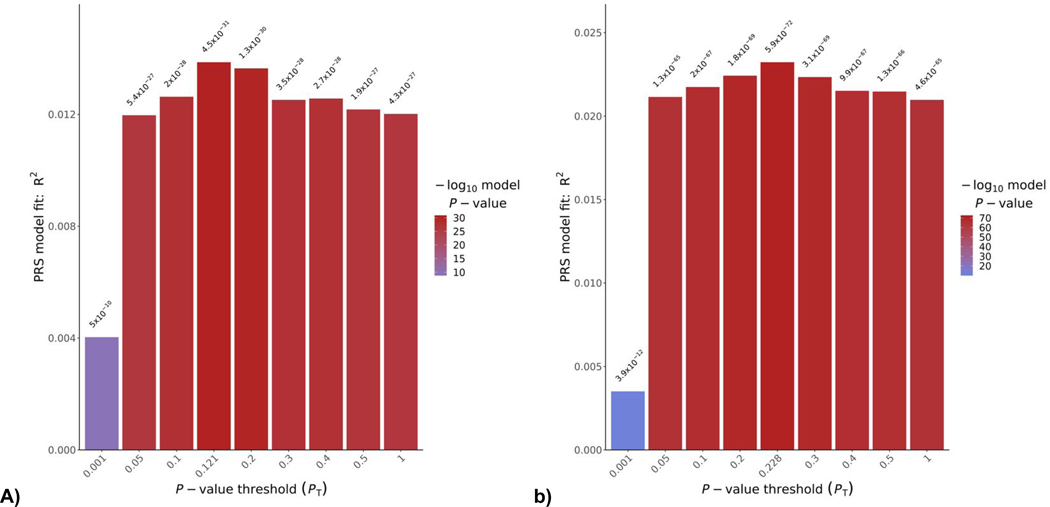
In European ancestry training and test samples comprising independent case and control cohorts, and accounting for 20 ancestry covariates, suicide PS significantly predicted suicide death case status. Suicide waves 1 and 2 comprise 1,321 and 2,092 suicide cases, respectively, and were pruned for relatedness within waves prior to training and testing analyses. These case-control predictions are plotted across p-value thresholds in Figure 3.
Figure 3. Cross Validation of Suicide Polygenic Case-Control Prediction.
Polygenic prediction of suicide death case status across two independent cohorts of cases and controls. Training GWAS summary statistics are used to score the test set for suicide polygenic risk. P-value thresholds are plotted on the x-axis from 0.1–1.0, reflecting the top 10% to 100% of the common variants from the training GWAS. Plots present model fit when a) predicting case-control status with wave 1 suicide vs. Generation Scotland summary statistics and b) predicting with wave 2 suicide vs. UK10K summary statistics. Scores are adjusted for 20 PCs. Summary data for the best fitting and 1.0 p-value threshold models are presented in Table S29.
The LDSC common variant h2 estimate, based on the summary statistics from a logistic GWAS with five ancestry covariates and pruning to remove related samples, was 0.2463, SE = 0.0356. The h2 estimate on the liability scale = 0.1567, SE = 0.0218 when considering the .2% reported population prevalence of completed suicide in Utah. λ in the model was 1.20, λ1000 = 1.04. These λs remained stable with follow-up testing of 20 PCs and with increased pruning. The suicide death cases differed significantly from the two UK control groups on PS of phenotypes relevant to suicide death. These differences were in the expected directions. Original discovery GWAS for all PS phenotypes were filtered to exclude any that used these control cohorts (Supplementary Table S5).
Consistent with hypotheses, significant PS elevations included alcohol use, autism spectrum disorder, child IQ, depressive symptoms, disinhibition, major depression, loneliness, and schizophrenia. The largest effect sizes were observed for PS for behavioral disinhibition, major depressive disorder, and schizophrenia (Figure 4). LD Hub37 provided estimates of SNP-based shared genetic covariance for several phenotypes (Supplementary Table S6), though none of these SNP-based genetic correlations reached statistical significance. Additionally, we disaggregated suicide by mode of death into four categories (see Supplementary Methods S2): gun, overdose, asphyxiation, and violent trauma. We epidemiologically characterized these groups by testing associations with 30 ICD-10 derived clinical antecedents (Supplementary Figure S9; Supplementary Tables S7-S9). Additionally, we conducted PS association testing with mode of death in all cases. No associations met multiple testing adjusted significance criteria (q < 0.1; Supplementary Figure S10; Supplementary Tables S10-S12).
Figure 4. Notable Elevations of Psychiatric Polygenic Risk in Suicide Cases.
P-value thresholds are plotted on the x-axis. Scores are adjusted for 20 PCs. Largest effect sizes and significance levels were observed for major depressive disorder, schizophrenia, and behavioral disinhibition. Summary data for the best fitting and 1.0 p-value threshold models for all eight phenotypes—also including alcohol use, autism spectrum disorder, child IQ, depressive symptoms, and loneliness—are presented in Table S29.
Sex Differences
Epidemiological analyses of sex differences in electronic health records data indicated that suicide cases of both sexes evidenced clinical diagnostic clusters of 1) internalizing-trauma-cluster B psychiatric disorders and 2) metabolic-cardiovascular-obesity medical disorders (Supplementary Tables S13-S18). Female cases were observed to have a higher overall number of diagnoses relative to males, which could reflect increased severity in females, decreased severity in males, decreased likelihood of males receiving diagnosis, and/or decreased help-seeking in males. This is broadly consistent with observed higher relative prevalence rates of gun-related death in males and overdose death in females. All associations of ICD and PS with mode of death are presented for females and males separately in Supplementary Figures S11-S14. All PS analyses include ancestry covariates. All corresponding statistics are reported in Supplementary Tables S19-S24. No sex-stratified polygenic scores findings met multiple testing adjusted significance criteria (q < 0.1).
DISCUSSION
Results from this analysis, the first adequately powered comprehensive genomic study of suicide death, yield several insights and suggest important applications for future extension. The GWAS of suicide death identified two genome-wide significant loci on chromosomes 13 and 15. The significant SNP-based heritability estimate of suicide death in this GWAS (25%, or 16% on the liability scale) is greater than those previously reported for suicide ideation/attempt (5%)14,15 and attempt within psychiatric diagnosis (3–10%)43 and is closer to previous estimates reported for suicide death (35–48%).13 Fully half of the genes implicated by our results overlap with schizophrenia results from the GWAS Catalog (p = 1×10−11), and two of these 11 genes have prior associations with risk of suicidal behavior (HS3ST3B, NCAN; for relevant literature see Supplementary Table S25; allele frequencies and effect sizes by cohort in our study are presented in Table S26).
All but four of these schizophrenia-associated genes also drove a significant GWAS Catalog association with bipolar disorder (p=1×10–17), supporting a hypothesis of genetic liability to more non-specific psychiatric illness. Of particular interest, the region on chromosome 19p13.11 in our study has evidence from the most recent, well-powered studies of both bipolar disorder44 and schizophrenia.45 In addition, our region on chromosome 16q21 has evidence not only from several large GWAS of schizophrenia, but also from a large meta-analysis of autism spectrum disorder.45–50
Genome-wide significant SNPs on chromosome 13 (Table 1) are all intergenic and vary with respect to combined annotation dependent depletion (CADD) score, an index for scoring the deleteriousness of single nucleotide variants as well as insertion/deletions variants in the human genome. Notably the variant with the strongest GWAS effect size, SNV rs34399104, reaches a CADD score of almost 20, indicating that it is in the top 1% of deleterious variants on the genome.
Direct comparison of this GWAS with 110 effect sizes reported in previous literature12–15,43,51 identified just one nominally significant variant in the largest GWAS of suicidal behavior,43 rs4870888, nearest to FER1L6 and FER1l6-AS2. Table S27 provides direct comparison of the previously reported effect sizes with those in this GWAS. SNV rs7989250 (chr 13) was reported in the UK Biobank GWAS for ordinal suicidality and reached a p <.05 threshold for replication in the Otsuka et al. suicide GWAS, but did not reach threshold in this GWAS (Table S27).
We were also able to cross-validate prediction of suicide death case-control status with polygenic scores for suicide death, accounting for population stratification. While within-case PS validation relies on stronger assumptions than other PS case-control comparisons, the considerable overlapping polygenic signal across p-value thresholds in this study (n = 3,413 deaths) is consistent with the two existing smaller polygenic analyses predicting suicidality. In one, suicidal behavior PRS predicted increased odds of suicide death with n = 127 deaths14 and in the other, a significant SNP-based genetic correlation of suicide death was observed between two independent cohorts (n = 746 deaths).13
Perhaps most compellingly, suicide death was strongly associated with polygenic scores for multiple psychiatric and psychological traits. As noted above, these included alcohol use, autism spectrum disorder, child IQ, depression, disinhibition, schizophrenia, and loneliness (Figure S15 presents additional PRS plots). These results were consistent with expectations, as strong associations in cases relative to both control cohorts were with behavioral disinhibition and major depressive disorder—arguably the two most critical antecedents of suicide death.52 More generally, these results are consistent with the emerging consensus that genetic correlations across psychiatric diagnoses and traits are substantial.53 Future research with an even larger suicide death cohort will be needed to determine the ultimate cause of these PS associations with suicide death, with leading hypotheses arguing for the presence of a “p-factor” capturing general liability to psychopathology,54 contrasting those arguing for pleiotropic effects across a small set of psychiatric disorder clusters.55
Limitations
Several limitations should be noted. First, it is possible that population stratification or platform confounds stemming from convenience controls could lead to false positives. To address this risk, we employed very conservative filters of both loci and subjects to minimize false positives. Values of λGC<1.05 are generally considered benign, although inflation in λGC is proportional to sample size.56 Both Generation Scotland and UK10k founder controls were extensively assessed for ancestral comparability to both 1000 Genomes Utah CEU25 and Utah European ancestry suicides, both matching extremely well. Incorporating two separate control cohorts (and formal analytical comparison of these cohorts to the corresponding 1KG populations) also allowed us to detect and filter out loci related to platform-specific technical artifacts, and subtle variation in population structure, prior to case-control analyses. Table S28 presents results of tests for differences in loadings for up to 200 PCs by cohort and by case-control status. Before and after imputation, we excluded any and all variants generating q-values <.1 in a control-control GWAS. Additionally, employing ancestral PC Mahalanobis (i.e., centroid) distance pruning, we have leveraged the fact the CEU 1KG subjects are largely ancestrally homogenous with our case samples to restrict our controls (and cases) to a highly ancestrally homogenous group.
It is also possible that the use of mixed healthy controls and controls with common medical conditions, as in UK10K, could exert a mild bias on effect estimates toward the null, which we do not view as highly problematic, given that the direction of the bias effectively reduces the risk of false positive findings. In the suicide case cohort, rates of common medical conditions are much higher than population base rates. However, in stratified control analyses, effect sizes specific to cases vs. UK10K were elevated relative to Generation Scotland, which is inconsistent with potential confounding of signal due to other disorders. Furthermore, effect sizes for PS for other disorders, across 1000 p-value thresholds, were far greater between the cases and controls than between the control groups, wherein differences were negligible, and distributions of PS often overlapped. None of the implicated variants were significantly different in AF between control groups in the control-control GWAS. However, it should be noted that AF is relatively low for the top variants across all cohorts.
Another limitation relates to electronic medical record data, in which prevalence for disorders is always confounded with age, and in a number of ways. For example, missing diagnosis could be more or less likely in a given decade during which a case was assessed. Younger ages are more likely to have complete electronic medical record information yet be less likely to be diagnosed with co-morbid medical conditions, schizophrenia, or personality disorders.
Importantly, in this study there was insufficient diversity to examine effect sizes for non-European ancestries. We expect a sufficient number of cases for a Mexican-American ancestry GWAS within five years, and we will prioritize diverse ancestry groups in order to minimize potential health disparities stemming from homogeneous summary statistics.57
Future Directions
A recent review of suicide prediction models has indicated that positive predictive values for suicide attempt are quite high (.9), but positive predictive values for suicide death continue to hover near zero.58,59 In this study, we have discovered multiple genome-wide significant loci/genes, strong polygenic signal, and significantly increased genetic risks for behavioral disinhibition, major depression, depressive symptoms, autism spectrum disorder, psychosis, and alcohol use disorder in cases. Future modeling of multiple polygenic risks and phenotypic risk factors may help isolate important moderators of risk and improve objective risk measures of suicide death. Importantly, ethical challenges associated with predictive models of suicide death are significant, and must be addressed proactively across psychiatric and medical domains.6,60
Supplementary Material
Acknowledgements:
This work was supported by the National Institute of Mental Health (H.C., grant number R01MH099134; A.D., grant number K01MH093731), a research contract from Janssen Research, LLC (H.C. & Q.L.); the American Foundation for Suicide Prevention (A.D. & A.B.), the Brain & Behavior Research Foundation (A.D. & A.S., NARSAD Young Investigator Awards); the University of Utah EDGE Scholar Program (A.D.); the Clark Tanner Foundation (HC, TD, AVB), and a donation from the Sharon Kae Lehr Endowed Research Fund in Memory of James Raymond Crump (DG). Partial support for all datasets housed within the Utah Population Data Base is provided by the Huntsman Cancer Institute (HCI), http://www.huntsmancancer.org/, and the HCI Cancer Center Support grant, P30CA-42014 from the NIH. Generation Scotland received core support from the Chief Scientist Office of the Scottish Government Health Directorates (CZD/16/6) and the Scottish Funding Council (HR03006). Genotyping of the GS samples was carried out by the Genetics Core Laboratory at the Wellcome Trust Clinical Research Facility, Edinburgh, Scotland and was funded by the Medical Research Council UK and the Wellcome Trust (Wellcome Trust Strategic Award “Stratifying Resilience and Depression Longitudinally” (STRADL) Reference 104036/Z/14/Z). Genotyping was performed by U of Utah Genomic Core and by Illumina, Inc with support from Janssen Research & Development, LLC. This work was also supported by research funding from Janssen Research & Development, LLC to University of Utah.
Previous presentation: World Congress of Psychiatric Genetics, Oct 26–31, 2019, Anaheim, CA, USA.
Footnotes
Disclosures: Dr. Qingqin Li, a co-author, is employed by Janssen Pharmaceuticals. All other authors have no financial relationships with commercial interests.
REFERENCES
- 1.Turecki G. & Brent DA Suicide and suicidal behaviour. Lancet (London, England) 387, 1227–1239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hedegaard HCS, Warner M. Suicide mortality in the United States, 1999–2017 in National Center for Health Statistics Data Brief Vol. 330 (CDC/NCHS, Hyattsville, MD, 2018). [Google Scholar]
- 3.Kochanek K, Murphy S, Xu J. & Tejada-Vera B. Deaths: Final Data for 2014. National Vital Statistics Reports 65, 1–121 (2016). [PubMed] [Google Scholar]
- 4.Murphy SL, Mathews TJ, Martin JA, Minkovitz CS & Strobino DM Annual Summary of Vital Statistics: 2013–2014. Pediatrics 139(2017). [DOI] [PubMed] [Google Scholar]
- 5.Case A. & Deaton A. Rising morbidity and mortality in midlife among white non-Hispanic Americans in the 21st century. Proceedings of the National Academy of Sciences 112, 15078 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barak-Corren Y. et al. Predicting Suicidal Behavior From Longitudinal Electronic Health Records. American Journal of Psychiatry 174, 154–162 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Franklin JC et al. Risk Factors for Suicidal Thoughts and Behaviors: A Meta-Analysis of 50 Years of Research. Psychological Bulletin 143, 187-U121 (2017). [DOI] [PubMed] [Google Scholar]
- 8.McGuffin P, Marusic A. & Farmer A. What can psychiatric genetics offer suicidology? Crisis 22, 61–5 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Mullins N. et al. Genetic relationships between suicide attempts, suicidal ideation and major psychiatric disorders: a genome-wide association and polygenic scoring study. Am J Med Genet B Neuropsychiatr Genet 165b, 428–37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turecki G. Suicidal behavior: is there a genetic predisposition? Bipolar Disord 3, 335–49 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Coon H. et al. Genome-wide significant regions in 43 Utah high-risk families implicate multiple genes involved in risk for completed suicide. Mol Psychiatry (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galfalvy H. et al. A genome-wide association study of suicidal behavior. Am J Med Genet B Neuropsychiatr Genet 168, 557–63 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Otsuka I. et al. Genome-wide association studies identify polygenic effects for completed suicide in the Japanese population. Neuropsychopharmacology 44, 2119–2124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strawbridge RJ et al. Identification of novel genome-wide associations for suicidality in UK Biobank, genetic correlation with psychiatric disorders and polygenic association with completed suicide. EBioMedicine 41, 517–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erlangsen A. et al. Genetics of suicide attempts in individuals with and without mental disorders: a population-based genome-wide association study. Mol Psychiatry (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Brien E. et al. Familial mortality in the Utah population database: characterizing a human aging phenotype. 62, 803–812 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Kerber R, O’Brien E, Smith KR & Mineau GP Familial aggregation of elderly cause-specific mortality: analysis of extended pedigrees in Utah, 1904–2002 in Kinship and demographic behavior in the past 243–258 (Springer, 2008). [Google Scholar]
- 18.World Health, O. ICD-10 : international statistical classification of diseases and related health problems : tenth revision. 2nd ed edn (World Health Organization, Geneva, 2004). [Google Scholar]
- 19.Schrijvers DL, Bollen J. & Sabbe B.G.J.J. o.a.d. The gender paradox in suicidal behavior and its impact on the suicidal process. 138, 19–26 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Skogman K, Alsen M. & Ojehagen A. Sex differences in risk factors for suicide after attempted suicide - A follow-up study of 1052 suicide attempters. Social Psychiatry and Psychiatric Epidemiology 39, 113–120 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Utah Medical Examiner Act. in 26–4 (https://le.utah.gov/xcode/Title26/Chapter4/C26-4_1800010118000101.pdf, United States, 2016).
- 22.Jorde LB Inbreeding in the Utah Mormons: an evaluation of estimates based on pedigrees, isonymy, and migration matrices. Ann Hum Genet 53, 339–55 (1989). [DOI] [PubMed] [Google Scholar]
- 23.Smith BH et al. Generation Scotland: the Scottish Family Health Study; a new resource for researching genes and heritability. BMC Medical Genetics 7, 74 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter K. et al. The UK10K project identifies rare variants in health and disease. Nature 526, 82–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shabalin AA et al. RaMWAS: fast methylome-wide association study pipeline for enrichment platforms. Bioinformatics 34, 2283–2285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shabalin A, Clark S, Hattab M, Aberg K. & van den Oord E. RaMWAS: Fast Methylome-Wide Association Study Pipeline for Enrichment Platforms. R package version 1.2.0 edn (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das S. et al. Next-generation genotype imputation service and methods. Nat Genet 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh PR et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet 48, 1443–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell S, Chang C, Sinai N.-N.L. o.B.M.P.L.a.M. & School of Medicine. PLINK 1.9 beta. (2017). [Google Scholar]
- 32.Zhou X. & Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet 44, 821–4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe K, Taskesen E, van Bochoven A. & Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nature Communications 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLean CY et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Euesden J, Lewis CM & O’Reilly PF PRSice: polygenic risk score software. Bioinformatics 31, 1466–1468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hastie T, Tibshirani R. & Friedman JH The Elements of Statistical Learning: Data Mining, Inference, and Prediction, (Springer, 2009). [Google Scholar]
- 37.Zheng J. et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature genetics 47, 291 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narum SR Beyond Bonferroni: less conservative analyses for conservation genetics. Conservation genetics 7, 783–787 (2006). [Google Scholar]
- 40.Hochberg Y. & Benjamini Y. More powerful procedures for multiple significance testing. Stat Med 9, 811–8 (1990). [DOI] [PubMed] [Google Scholar]
- 41.Dayem Ullah AZ et al. SNPnexus: assessing the functional relevance of genetic variation to facilitate the promise of precision medicine. Nucleic acids research 46, W109-W113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gandal MJ et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mullins N. et al. GWAS of Suicide Attempt in Psychiatric Disorders and Association With Major Depression Polygenic Risk Scores. Am J Psychiatry 176, 651–660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stahl EA et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet 51, 793–803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pardinas AF et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet 50, 381–389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Autism Spectrum Disorders Working Group of The Psychiatric Genomics, C. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol Autism 8, 21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ikeda M. et al. Genome-Wide Association Study Detected Novel Susceptibility Genes for Schizophrenia and Shared Trans-Populations/Diseases Genetic Effect. Schizophr Bull 45, 824–834 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Periyasamy S. et al. Association of Schizophrenia Risk With Disordered Niacin Metabolism in an Indian Genome-wide Association Study. JAMA Psychiatry (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goes FS et al. Genome-wide association study of schizophrenia in Ashkenazi Jews. Am J Med Genet B Neuropsychiatr Genet 168, 649–59 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Ruderfer DM et al. Significant shared heritability underlies suicide attempt and clinically predicted probability of attempting suicide. Mol Psychiatry (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nock MK et al. Prevalence, correlates, and treatment of lifetime suicidal behavior among adolescents: results from the National Comorbidity Survey Replication Adolescent Supplement. JAMA Psychiatry 70, 300–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anttila V. et al. Analysis of shared heritability in common disorders of the brain. 360, eaap8757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caspi A. et al. The p factor: one general psychopathology factor in the structure of psychiatric disorders? 2, 119–137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee PH et al. Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell 179, 1469–1482.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Price AL, Zaitlen NA, Reich D. & Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet 11, 459–63 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin AR et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nature Genetics 51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belsher BE, Smolenski DJ & Pruitt LD Positive Predictive Values and Potential Success of Suicide Prediction Models-Reply. JAMA Psychiatry (2019). [DOI] [PubMed] [Google Scholar]
- 59.Belsher BE et al. Prediction Models for Suicide Attempts and Deaths: A Systematic Review and Simulation. JAMA Psychiatry 76, 642–651 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Adkins DE Machine Learning and Electronic Health Records: A Paradigm Shift. American Journal of Psychiatry 174, 93–94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kircher M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nature genetics 46, 310 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rentzsch P, Witten D, Cooper GM, Shendure J. & Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic acids research 47, D886–D894 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.