Abstract
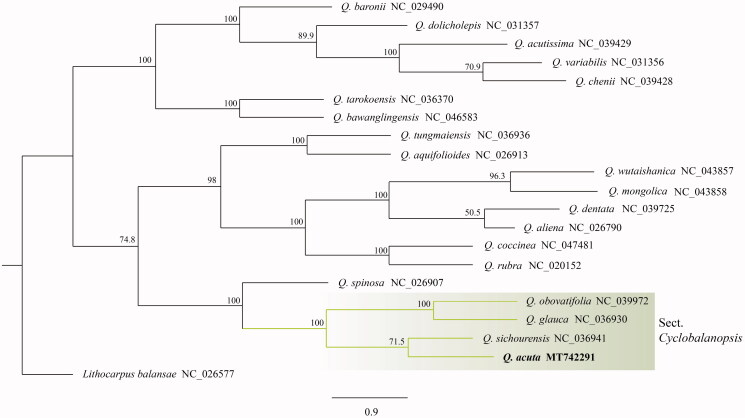
We are reporting the complete plastid genome (plastome) of Quercus acuta, an evergreen broad-leaved oak endemic to East Asia. This species is important for maintaining the warm-temperate evergreen forest biome in East Asia. The Q. acuta plastome is 160,522 base pairs (bp) long, with two inverted repeat (IR) regions (25,839 bp each) that separate a large single copy (LSC) region (90,199 bp) and a small single copy (SSC) region (18,645 bp). The phylogenetic tree shows that Quercus acuta is closely related to Quercus sichourensis with strong bootstrap support.
Keywords: Complete plastid genome, Cyclobalanopsis, evergreen broad-leaved oak, phylogenetic analysis, Quercus acuta
The genus Quercus L. is one of the most abundant and economically important woody plant genera in the Northern Hemisphere (Manos et al. 1999). Its evolutionary success is attributed to the species diversity, which varies among geographic regions and follows ecological divergence within each region (Han et al. 2020; Hipp et al. 2020). Quercus species can be locally endemic or widespread, extending from the equator to the boreal regions of Europe and from sea level to 4000 m in China (Kremer and Hipp 2020). Quercus section Cyclobalanopsis species dominate in the subtropical and warm-temperate evergreen broad-leaved forests of East Asia (Deng et al. 2018) where species such as Quercus acuta Thunb. and Quercus hondae Makino maintain a high degree of endemism (Qian and Ricklefs 2000).
To date, many plastid genomes (plastomes) have been sequenced for Quercus and have provided useful phylogenetic information to solve the taxonomical problems attributed to hybridization (Yang et al. 2016). However, Cyclobalanopsis plastome sequence availability is scarce, especially when considering their high level of endemism in East Asia. Quercus acuta is known as an evergreen broad-leaved oak tree endemic to the warm-temperate regions of East Asia, such as China, Korea and Japan. This species is an important resource for maintaining regional forest biomes (Lee and Choi 2010). However, their distribution in China, along with taxonomic problems, still remains controversial (Ohashi et al. 2006; Deng et al. 2008). Therefore, the plastom sequence for this species will be an important basis for improving our understanding of the evolutionary process and solving taxonomic problems in inter/intra species within especially section Cyclobalanopsis.
Quercus acuta samples were collected from Jeju Island, South Korea (N33°19′11″, E126°37′25″). The voucher specimen (MFDS-C-7840) was stored in the herbarium at the Korea Institute of Oriental Medicine (KIOM: sgyang81@kiom.re.kr). The DNA library was constructed and sequenced on the MGI-seq 2000 platform (LAS, Seoul, Korea) and generated 71,993,390 raw reads (150 bp paired-end). The Q. acuta plastid genome was constructed using NOVOPlasty 4.1 (Dierckxsens et al. 2017), with the Q. gilva rbcL gene sequence (Zeng et al. 2019; MK986651) as the seed. The sequence was checked by reference mapping using Geneious 10.2.3 (Kearse et al. 2012), on which 86,571 reads were assembled with an 80X coverage. The annotation was separately performed in Geneious 10.2.3 (Kearse et al. 2012), and was manually corrected for the start and stop codons and for the intron/exon boundaries. The annotated plastome sequence was deposited in GenBank (accession number: MT742291). The phylogenetic tree was constructed by downloading the complete plastome sequences of 20 related species (Quercus: 19 species, Lithocarpus: 1 species) from the NCBI database and alignments were performed with MAFFT (Katoh and Toh 2010). The maximum likelihood (ML) analysis was performed with RAxML v.8.0 (Stamatakis 2014) using default parameters and 1000 bootstrap replicates. For RAxML tree, the general time-reversible (GTR) model of nucleotide substation was used with the Gamma model of rate heterogeneity.
The Q. acuta plastome is 160,522 bp long, with two inverted repeat (IR) regions (25,839 bp each) that separate a large single copy (LSC) region (90,199 bp) and a small single copy (SSC) region (18,645 bp). This is the second shortest reported Quercus plastome. It contains 130 genes that encode 85 proteins, eight ribosomal RNAs, and 37 transfer RNAs, and the G + C content is 36.9% overall, 34.8% in the LSC region, and 42.8% in the IR regions. The gene content and order were comparable to other published Quercus plastome (Yang et al. 2016; Zeng et al. 2019). The phylogenetic tree (Figure 1) shows that Q. acuta is closely related to Q. sichourensis, with strong bootstrap support. Section Cyclobalanopsis, comprised of Q. acuta, Q. sichourensis, Q. glauca, and Q. obovatifolia, form a monophyletic sister clade to Q. spinosa. These data are useful for the phylogenetic and evolutionary studies of Quercus and Fagaceae.
Figure 1.
A phylogenetic tree (RAxML) using the plastome sequences of 21 species, with Lithocarpus balansae as the outgroup. The numbers above each node indicate the bootstrap value with 1000 replicates.
Funding Statement
This research was supported by the National Research Foundation of Korea (NRF) [no. 2019R1F1A1057589].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI (accession no. MT742291) at https://www.ncbi.nlm.nih.gov. All high-throughput sequencing data files are available from the GenBank Sequence Read Archive (SRA) accession number: SRR13068430.
References
- Deng M, Cao LM, Cao M.. 2008. A new distribution record of Quercus acuta Thunberg in Hongkong and Guangdong Province. Guihaia. 28:731–734. [Google Scholar]
- Deng M, Jiang XL, Hipp AL, Manos PS, Hahn M.. 2018. Phylogeny and biogeography of East Asian evergreen oaks (Quercus section Cyclobalanopsis; Fagaceae): insights into the Cenozoic history of evergreen broad-leaved forests in subtropical Asia. Mol Phylogenet Evol. 119:170–181. [DOI] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han EK, , Cho WB, , Park JS, , Choi IS, , Kwak M, , Kim BY, , Lee JH. 2020. A Disjunctive marginal edge of evergreen broad-Leaved oak (Quercus gilva) in East Asia: the high genetic distinctiveness and unusual diversity of Jeju Island populations and insight into a massive, independent postglacial colonization. Genes. 11(10):1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp AL, Manos PS, Hahn M, Avishai M, Bodénès C, Cavender ‐Bares J, Crowl AA, Deng M, Denk T, Fitz‐Gibbon S, et al. 2020. Genomic landscape of the global oak phylogeny. New Phytol. 226(4):1198–1212. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H.. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 26(15):1899–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer A, Hipp AL.. 2020. Oaks: an evolutionary success story. New Phytol. 226(4):987–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Choi BH.. 2010. Distribution and northernmost limit on the Korean Peninsula of three evergreen trees. Korean J Pl Taxon. 40(4):267–273. [Google Scholar]
- Manos PS, Doyle JJ, Nixon KC.. 1999. Phylogeny, biogeography, and processes of molecular differentiation in Quercus subgenus Quercus (Fagaceae). Mol Phylogenet Evol. 12(3):333–349. [DOI] [PubMed] [Google Scholar]
- Ohashi H, Ohashi K, Takahashi K.. 2006. Identity of Quercus acuta Thunb. (Fagaceae) recorded from Taiwan and China. J Japanese Bot. 81:268–274. [Google Scholar]
- Qian H, Ricklefs RE.. 2000. Large-scale processes and the Asian bias in species diversity of temperate plants. Nature. 407(6801):180–182. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhou T, Duan D, Yang J, Feng L, Zhao G.. 2016. Comparative analysis of the complete chloroplast genomes of five Quercus species. Front Plant Sci. 7:959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng QM, Liu B, Lin RQ, Jiang YT, Liu ZJ, Chen SP.. 2019. The complete chloroplast genome sequence of Quercus gilva (Fagaceae). Mitochondrial DNA Part B. 4(2):2493–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank of NCBI (accession no. MT742291) at https://www.ncbi.nlm.nih.gov. All high-throughput sequencing data files are available from the GenBank Sequence Read Archive (SRA) accession number: SRR13068430.