Abstract
GPR68, an orphan G-protein coupled receptor, senses protons, couples to multiple G-proteins, and is also activated or inhibited by divalent metal ions. It has seven extracellular histidine residues, although it is not clear how these histidine residues play a role in both proton-sensing and metal ion modulation. Here we demonstrate that divalent metal ions are allosteric modulators that can activate or inhibit proton activity in a concentration- and pH-dependent manner. We then show that single histidine mutants have differential and varying degrees of effects on proton-sensing and metal ion modulation. Some histidine residues play dual roles in proton-sensing and metal ion modulation, while others are important in one or the other but not both. Two extracellular disulfide bonds are predicted to constrain histidine residues to be spatially close to each other. Combining histidine mutations leads to reduced proton activity and resistance to metal ion modulation, while breaking the less conserved disulfide bond results in a more severe reduction in proton-sensing over metal modulation. The small-molecule positive allosteric modulators (PAMs) ogerin and lorazepam are not affected by these mutations and remain active at mutants with severely reduced proton activity or are resistant to metal ion modulation. These results suggest GPR68 possesses two independent allosteric modulation systems, one through interaction with divalent metal ions at the extracellular surface and another through small-molecule PAMs in the transmembrane domains. A new GPR68 model is developed to accommodate the findings which could serve as a template for further studies and ligand discovery by virtual ligand docking.
Graphical Abstract
GPR68 is an understudied orphan G-protein coupled receptor (oGPCR). First cloned from an ovarian cancer cell line for ovarian cancer G-protein-couple receptor 1 (therefore also known as OGR1 in the literature),1 it is widely expressed in most tissues and cells1–3 and functions as a proton-sensing receptor.4,5 Together with two other orphan receptors, GPR4 and GPR65 (previously known as TDAG8), they form a subfamily of proton-sensing GPCRs.6–8 In responding to acidic extracellular conditions, GPR68 couples to multiple downstream signaling pathways via different families of G-proteins in different cells, including Gq/11, Gs, Gi/o, and G12/13,4,5,9,10 and has been implicated in a range of biological processes and pathological conditions, including pH homeostasis, insulin secretion, bone metabolism, learning and memory, blood flow and mechanosensing, inflammation, tumor metastasis and growth, and hematopoiesis.5,11–26 As one of the understudied GPCR targets of the NIH funded Illuminating the Druggable Genome (IDG) program (https://pharos.nih.gov/targets/Q15743), GPR68 has been receiving increasing attention in recent years and represents a potential therapeutic target for drug design and development.
As with other oGPCRs, small molecule ligands are highly desired for GPR68. For many years after it was cloned in 1996,1 there were no confirmed ligands that could activate or inhibit GPR68 except through protons (H+). In 2015, we had reported a small molecule ligand ogerin as the first novel positive allosteric modulator (PAM)5 and subsequently improved ogerin analogues,27 and three endogenous orphan peptides as PAMs for protons at GPR68,28 thus extending the number of available tool compounds for GPR68. The molecular mechanisms of allosteric modulation of proton activity by small-molecule ligands or peptides remain unknown. With H+ as an agonist, extracellular histidine (His, H) residues (Figures 1a, 1b) were identified as orthosteric binding sites,4 suggesting that proton-sensing receptors are a unique group of GPCRs with multiple extracellular orthosteric binding sites. A proposed mechanism of GPR68 activation by H+ is initiated upon protonation of extracellular His residues, which disrupts H-bond interactions between the His residues that keep the receptor in an inactive conformation.4 In the same study, the metal ions Zn2+ and Cu2+ were proposed to complex with unprotonated His residues to stabilize the receptor, and micromolar concentrations of Zn2+ and Cu2+ showed inhibitory activity at GPR684. Zn2+ and Cu2+ have since been used as GPR68 antagonists in pharmacological assays.24 Other groups have also reported on the metal ion activity at proton-sensing GPCRs. Abe-Ohya and colleagues identified divalent metal ions (Fe2+, Co2+, Mn2+, Ni2+, and Zn2+) as GPR68 agonists by measuring Gq-mediated IP accumulation under neutral pH conditions.29 Using SRE-Luciferase reporter assays, Negishi and colleagues later reported Co2+ and Mn2+, but not Fe2+ or Ni2+, at high micromolar concentrations activated zebrafish GPR68,30 while Mochimaru and colleagues showed that GPR68 extracellular regions are critical for the Mn2+ activity and that activity at 100 μM is dependent on pH conditions.39 However, these studies did not characterize metal ion activity under multiple pH conditions or at a sufficiently broad pH range to determine if metal ions were still active at high pH conditions, i.e., if metal ions act as allosteric modulators for H+. While we were designing and developing allosteric modulators for H+ at GPR68,5,27 we also tested and observed that divalent metal ions can activate or inhibit GPR68 activity but in a concentration-and pH-dependent manner. GPR68 has a total of 7 extracellular His residues, but it is not clear how individual extracellular His residues coordinate both proton-sensing and metal ion binding. We hypothesize that (i) divalent metal ions are allosteric modulators but not agonists, (ii) individual extracellular His residues may play different roles in proton-sensing and allosteric modulation of metal ions, and that (iii) His protonation (proton-sensing) and metal ion coordination are two linked and pH-dependent dynamic processes occurring at the extracellular regions. By characterizing functional activity of His mutants, we reveal differential roles of extracellular His residues in both proton-sensing and divalent metal ion modulation.
Figure 1.
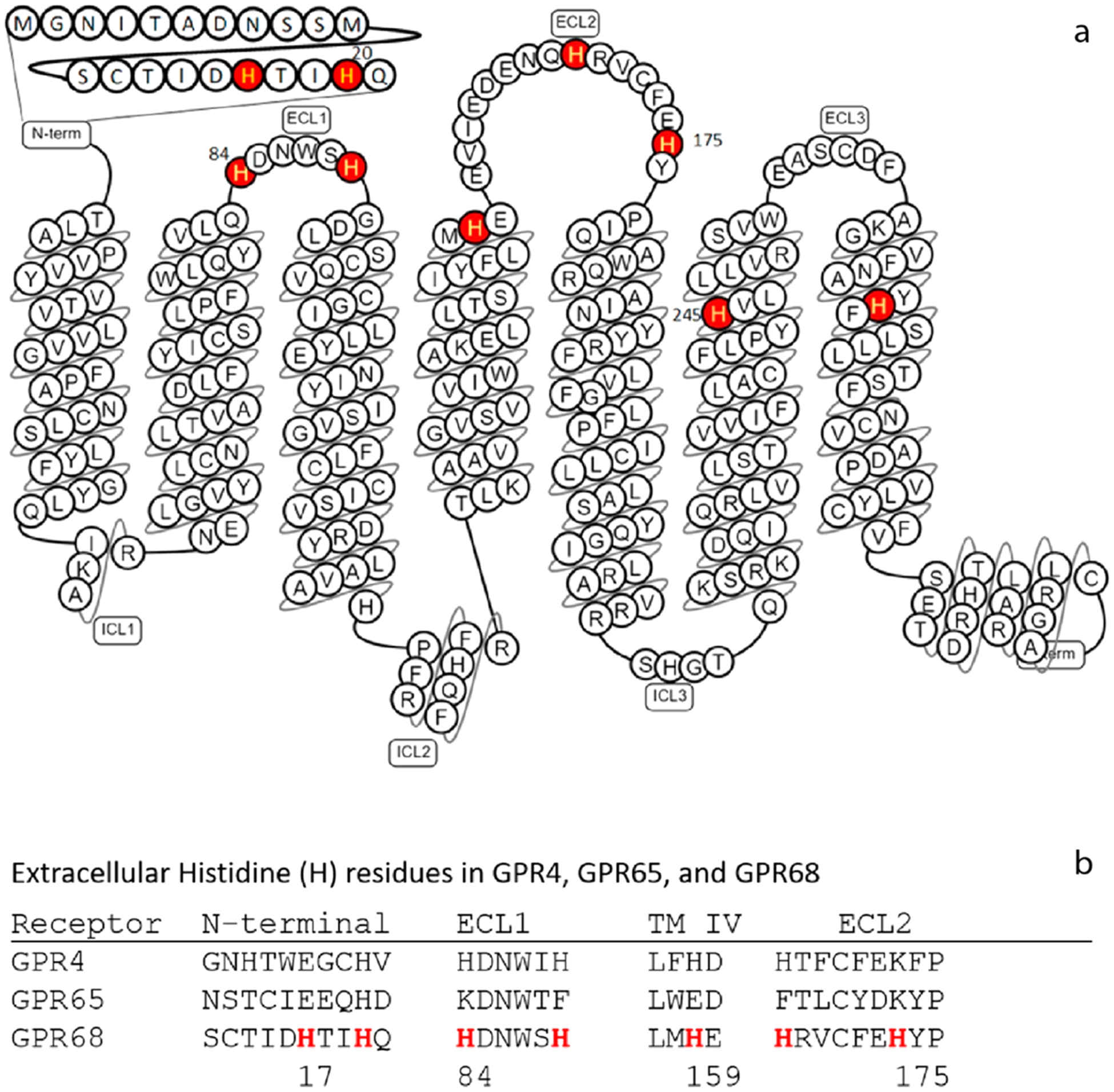
GPR68 snake model and partial sequence alignment to highlight the Histidine (H) in this study. (a) Histidine (H) residues are highlighted and mutated in this project, H17 and H20 in the N-terminal, H84 and H89 in the first extracellular loop (ECL1), H159 at the top of transmembrane helix 4 (TM IV), H169 and H175 in ECL2, H245 in TM VI, and H269 in TM VII. The model is modified based on the GPR68 model from gpcrdb.org. (b) Partial sequence alignment illustrates the extracellular Histidine (H) residues in GPR4, GPR65, and GPR68.
MATERIALS AND METHODS
Materials and Chemicals.
Human GPR68 as well as GPR4 and GPR65 cDNA was obtained from the cDNA Resource Center (cdna.org). GloSensor cAMP reporter plasmid was from Promega. Ogerin and lorazepam were purchased from Tocris or Sigma. Poly-L-Lysine (PLL) was from Sigma. Phosphodiesterase (PDE) inhibitory Ro 20–1724 was purchased from Tocris or Cayman Chemical. Luciferase substrate luciferin sodium salt was ordered from Golden Bio. 20x regular Hank’s balanced salt solutions (HBSS) or 20x Calcium- and Magnesium-free HBSS were purchased from Invitrogen. Organic buffer reagents, 2-(N-morpholino)ethanesulfonic acid (MES), 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid (HEPES), and (tris-(hydroxymethyl)methylamino)propanesulfonic acid (TAPS), metal salts, and other general chemicals were purchased from Sigma or Fisher Scientific.
Site-Directed Mutagenesis.
Mutations were created with standard site-directed mutagenesis procedure using PrimeSTAR Max Polymerase mix from TAKARA. Primers were synthesized by Eton Bioscience (RTP, NC), and mutations were confirmed with a commercial sequencing service provided by GENEWIZ (RTP, NC).
GloSensor cAMP Assays.
GPR68-mediated Gs activation and cAMP production were determined using the GloSensor cAMP assays. The following method was adopted from a previously published procedure27 with minor modifications. In detail, HEK293T cells were maintained and cotransfected with receptor DNA and GloSensor cAMP reporter plasmids in DMEM containing 10% FBS. Overnight transfected cells were plated in PLL coated 384-well white clear-bottom plates in DMEM supplemented with 1% dialyzed fetal bovine serum (dFBS), about 15,000 cells in 40 μL per well, for a minimum of 6 h up to 24 h. To prepare for the assays, cells were removed of culture medium (gently shaking off) and incubated for 1 h at 37 °C with 20 μL per well of loading buffer (1x Calcium- and Magnesium-free HBSS, 20 mM TAPS, pH 8.40) containing 2 mM luciferin and 10 μM PDE inhibitor Ro 20–1724. Drug buffers were prepared in 1x Calcium- and Magnesium-free HBSS supplemented with different organic buffer agents for different pH ranges, 20 mM MES for pH 5.00–6.60, 20 mM HEPES for pH 6.70–8.20, and 20 mM TAPS for pH 8.30–8.60. Fresh buffers were prepared each day to use within 24 h, and pH was adjusted with KOH at room temperature. Ligand stock solutions were prepared in DMSO at 10 or 20 mM and diluted with pH adjusted drug buffers. PDE inhibitor Ro 20–1724 was added to working solutions to maintain the final concentration of 10 μM. Metal ion stock solutions were made at 100 mM with water and diluted to desired concentrations. Fresh FeCl2 stock was prepared whenever Fe2+ is tested. To stimulate cells with desired pH solutions, cells were first removed of luciferin loading solutions (gently shaking off) and added with 25 μL per well of premixed drug solutions. The cell plate was incubated at room temperature for 20 min before luminescence was counted.
Molecular Modeling.
The GPR68 model was built based on a previous model with modifications.5 Two disulfide bonds, C13-C258 and C94-C172, were kept, and two parts of the structure (residues 12–22 and 82–90) were remodeled using MODELER 9.15.31 From the 1,000 generated models, one was selected with a tetrahedron conformation of four His residues (H17, H20, H84, and H169) coordinating with a Zn2+ ion. At reduced pH conditions, H20 and H169 are protonated, thus modeled with an alternative rotamer, not coordinated with the Zn2+.
Data Analysis.
Receptor-mediated Gs activation and cAMP production were quantified as relative luminescence unit (RLU), normalized, and analyzed in Prism. To obtain proton potency (pEC50), efficacy (Emax), and Hill slope (n) with GPR68 wild-type and mutant receptors, proton concentration-response curves were fitted to the Prism built-in four-parameter logistic function. To compare and quantify the overall effect of mutations on proton activity, we applied index Log(Max/EC50) to represent overall proton activity and ΔLog(Max/EC50) to represent the net change of mutations,32 independent of assay systems and receptor expression levels. For allosteric modulation of proton activity, proton concentration-responses in the absence and presence of increasing concentrations of test modulators and metal ions were fitted to the standard allosteric operational model33–35 to extract key allosteric parameters (allosteric modulator binding affinity KB, affinity cooperativity α, efficacy cooperativity β). In detail, affinity cooperativity α defines the reciprocal effects on the binding affinity between orthosteric agonists (protons in this case) and allosteric ligand; while efficacy cooperativity β defines the effects of the allosteric modulator on orthosteric agonist efficacy. For both α and β, >1 indicates increased binding affinity and efficacy, correspondingly; < 1 but > 0 indicates reduced binding affinity and efficacy, correspondingly. In accordance with the Black/Leff operational model used to analyze responses,32 results were normalized to the relative percentage of activity; therefore, basal activity can be constrained to 0. The orthosteric agonist binding affinity, KA, defined as protons binding affinity in the absence of modulator, is constrained to equal to the potency of protons (EC50) in the absence of modulator. Since modulators in this study have no agonist activity by themselves, the allosteric efficacy parameter τB is therefore constrained to 0 as well. All the other allosteric parameters (system Emax, α, β, KB, orthosteric agonist efficacy τA, and Hill slope n) are all shared globally for each group of data sets as required by the allosteric operational model. The system Emax can also be estimated from fitting curves with the Prism built-in logistic function and can also be constrained to a fixed value. Since the orthosteric agonist in this case is protons which always are present in the assay system and receptor compartment, the KB has no corresponding biochemical meaning as it represents the affinity of the modulator in the absence of the cobinding ligand in the model. We adopted the comprehensive allosteric index Log(αβ/KB) to quantify the overall allosteric activity of an allosteric modulator.32 Correspondingly, the difference of the index between reference ligand or receptor, ΔLog(αβ/KB), can be used to quantify the net change of allosteric activity,32 independent of assay systems and receptor expression levels.
RESULTS
Proton-Sensing GPCRs Have Unique and Different Proton Response Profiles.
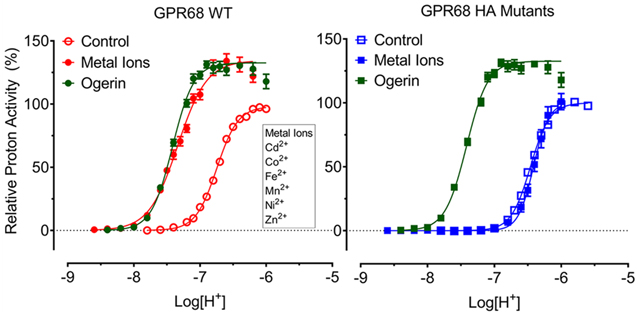
Experiments on proton-mediated cAMP production with GPR68 (as well as GPR4 and GPR65) in transiently transfected HEK293 T cells initially were conducted in a Hank’s balanced salt solution (HBSS)-based buffer5 which contains 1.3 mM Ca2+ and 0.9 mM Mg2+. To determine the effects of other divalent metal ions and reduce potential interference from existing Ca2+/Mg2+ in a regular HBSS-based buffer, we adopted the use of Ca2+/Mg2+ free HBSS-based buffers for all the assays as described.27 When required, divalent metal ions were supplemented in pH adjusted buffers as indicated. Ca2+/Mg2+ (1 mM each) increased proton efficacy by 15–30% but minimally affected proton potency at GPR68, GPR4, or GPR65 as compared to control conditions (Figure 2a and Table 1). As shown, proton-sensing GPCRs have different profiles and minimally overlap with each other in responding to proton stimulation-when GPR4 is partially active at pH 8.00, GPR65 or GPR68 is inactive; while GPR65 is fully activated at pH 7.20, GPR68 is only slightly active. However, they do share a common feature - all with steep curves as reflected by high Hill slopes. Because of the large Hill slope, receptor activity changes drastically within a narrow range of pH. Specifically, GPR68 activity goes from inactivity (0%) at pH 7.60 to full activation (100%) at pH 6.40 (Figure 2a), as proton concentration changes by less than 16-fold. Under normal physiological conditions (such as pH 7.40), GPR68 is maintained at a low activity (<10%); GPR65 is activated at about 50% capacity, while GPR4 is essentially fully activated, which could explain the high “constitutive activity” of GPR4 and GPR65 as reported in the literature.36–38
Figure 2.
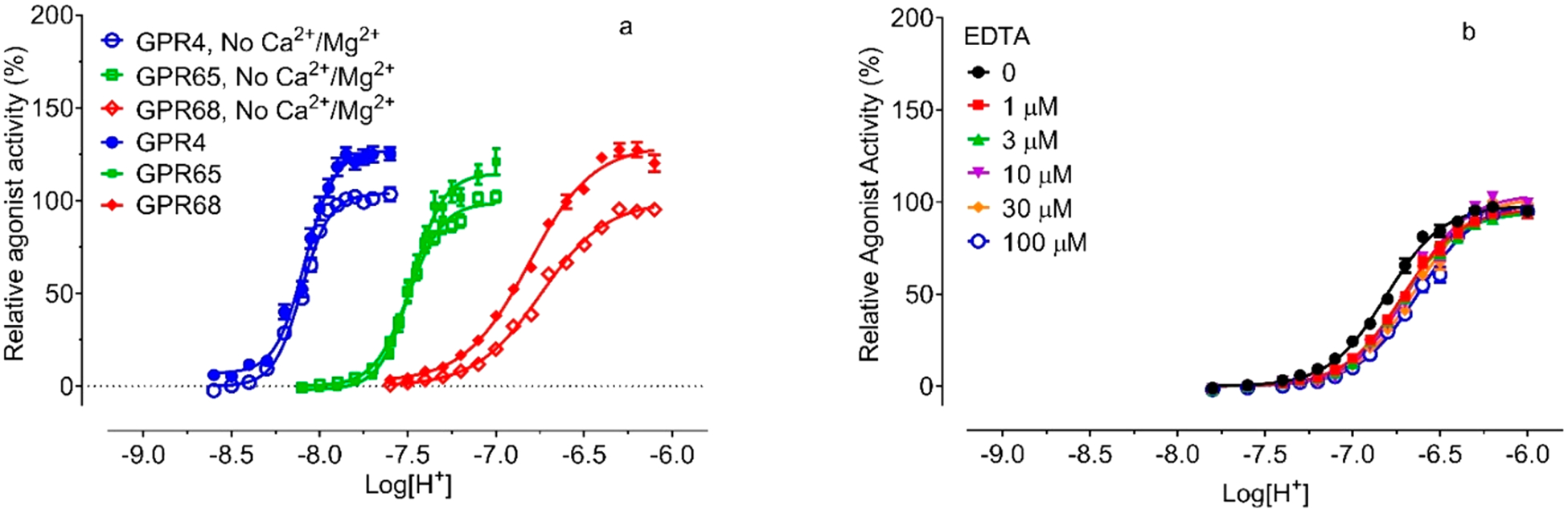
Proton concentration-responses at GPR4, GPR65, and GPR68 in the absence and presence of Ca2+/Mg2+ (a) and EDTA (b). Proton-mediated Gs-cAMP production was measured in transiently transfected HEK293T cells using the GloSensor cAMP assays. Results represented means ± SEM from a minimum of 3 independent assays, each in triplicate or quadruplicate, and were normalized to the proton activity in the absence of Ca2+/Mg2+ or EDTA for each receptor. Curves were analyzed using Prism 8.0 with the built-in four-parameter logistic function. Parameters (basal, Emax, potency, and Hill slope) were reported in Table 1.
Table 1.
Pharmacological Parameters for the Proton Concentration-Response Curves in the Absence and Presence of Ca2+ and Mg2+ and EDTAa
| receptor | condition | basal (%) | Emax (%) | potency (pH) | Hill slope |
|---|---|---|---|---|---|
| GPR4 | without Ca2+/Mg2+ | 0 | 100 | 8.11 ± 0.01 | 5.90 ± 0.31 |
| with 1 mM Ca2+/Mg2+ | 7.0 ± 2.5 | 127.2 ± 2.3 | 8.08 ± 0.01 | 5.14 ± 0.50 | |
| GPR65 | without Ca2+/Mg2+ | 0 | 100 | 7.49 ± 0.01 | 4.31 ± 0.23 |
| with 1 mM Ca2+/Mg2+ | 0.0 ± 2.5 | 114.8 ± 3.2 | 7.48 ± 0.01 | 4.85 ± 0.52 | |
| GPR68 | without Ca2+/Mg2+ | 0 | 100 | 6.74 ± 0.01 | 2.31 ± 0.06 |
| with 1 mM Ca2+/Mg2+ | 2.3 ± 1.9 | 129.8 ± 2.6 | 6.81 ± 0.01 | 2.37 ± 0.16 | |
| GPR68 | control | 0 (shared globally) | 98.2 ± 1.5 | 6.81 ± 0.01 | 2.66 ± 0.13 |
| 1 μM EDTA | 96.5 ± 1.6 | 6.72 ± 0.01 | 2.62 ± 0.13 | ||
| 3 μM EDTA | 94.8 ± 1.6 | 6.72 ± 0.01 | 2.64 ± 0.13 | ||
| 10 μM EDTA | 104.3 ± 1.9 | 6.68 ± 0.01 | 2.63 ± 0.13 | ||
| 30 μM EDTA | 103.1 ± 2.2 | 6.65 ± 0.01 | 2.56 ± 0.14 | ||
| 100 μM EDTA | 100.5 ± 2.5 | 6.63 ± 0.01 | 5.23 ± 0.15 |
Concentration-response curves are presented in Figure 2. Parameters were retrieved from fittings with pooled and normalized results using the Prism 8.4 built-in four-parameter logistic function.
As cell culture medium was removed before the cells were loaded with luciferase substrate (prepared in Ca2+/Mg2+ free HBSS-based buffers), which was also removed before stimulation with Ca2+/Mg2+ free HBSS-based buffers, therefore, residual Ca2+/Mg2+ levels are expected to be low. To ensure that residual metal ions were not interfering with other metal ions, we carried out proton concentration-responses in the presence of EDTA (1 to 100 μM) (Figure 2b). EDTA does not affect proton efficacy but reduced proton potency slightly from pH 6.81 to 6.63 at GPR68. This demonstrated that our assay system is not affected by potential residual divalent metal ions and is suitable for characterizing metal ion activity.
Functional Activity of Divalent Metal Ions Is Concentration- and pH-Dependent.
We next examined metal ion concentration responses under different pH conditions. For this study, we selected 9 divalent metal ions (Ca2+, Cd2+, Co2+, Cu2+, Fe2+, Mg2+, Mn2+, Ni2+, and Zn2+). Monovalent Li+ and trivalent Al3+ were also included for comparison in initial assays, but they showed little activity at proton-sensing receptors and were therefore excluded for further studies. Four pH points were chosen for each receptor to accommodate their different proton response profiles, so potential metal ion activity could be determined when the receptor was inactive, partially activated, or sub-maximally activated under different pH conditions. These pH conditions were set up to reveal if metal ions have potential agonist activity, allosteric activity (positive or negative), or antagonist activity.
As shown in Figure 3 and Figure S1 (concentration responses of metal ions at proton GPCRs under multiple pH conditions), different metal ions have diverse and pH-dependent concentration-response profiles at different proton-sensing receptors. Overall, GPR68 is more sensitive to metal ions than GPR4 or GPR65. Specifically, Ca2+ slightly activates receptors at high concentrations, while Mg2+ has minimal effect. Therefore, the observed small differences seen in Figure 2a were probably due to Ca2+ but not Mg2+. Compared with Ca2+ and Mg2+, other divalent metal ions have larger effects. Under mildly acidic conditions, Cd2+, Co2+, Fe2+, Mn2+, Ni2+, and Zn2+ can activate GPR68 to or over 100% of proton activity. The corresponding efficacy and potency values under different pH conditions are listed in Table 2. The efficacy values (fold of basal) of the metal ions trend lower and potency of metal ions trends higher when pH is reduced (Figure 3, Figure S1, and Table 2), indicating that metal ion activity is pH-dependent, characteristic evidence of allosteric modulation. In addition, Cd2+, Cu2+, and Zn2+ show bell-shaped responses-stimulation at low concentrations and inhibition at high concentrations, especially at GPR68 with peaks forming near 1 μM of Cd2+, Cu2+, and Zn2+. Similarly, relatively lower peak activity of Cd2+, Cu2+, and Zn2+ occurs at GPR65 near 0.3 μM.
Figure 3.
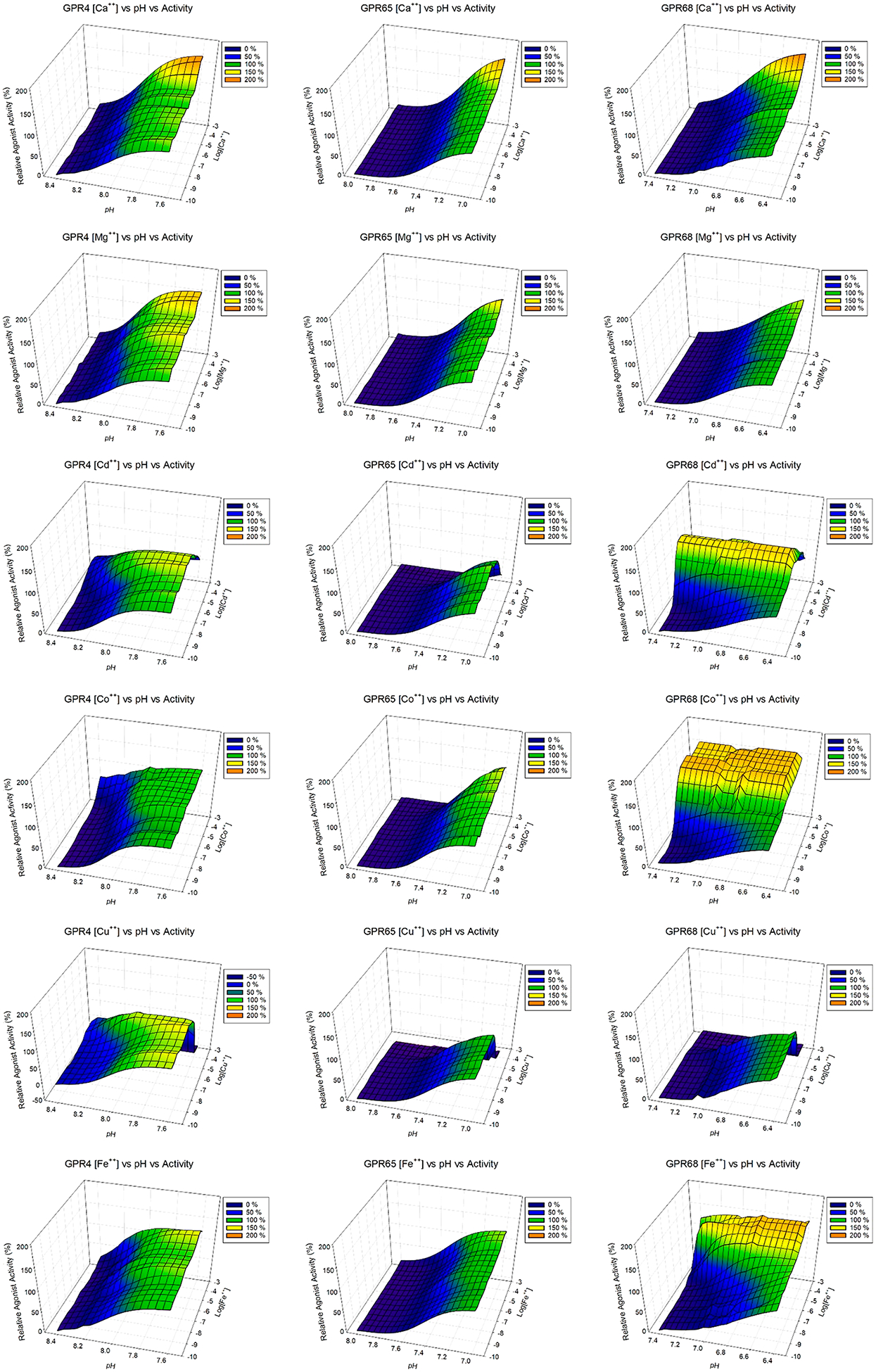
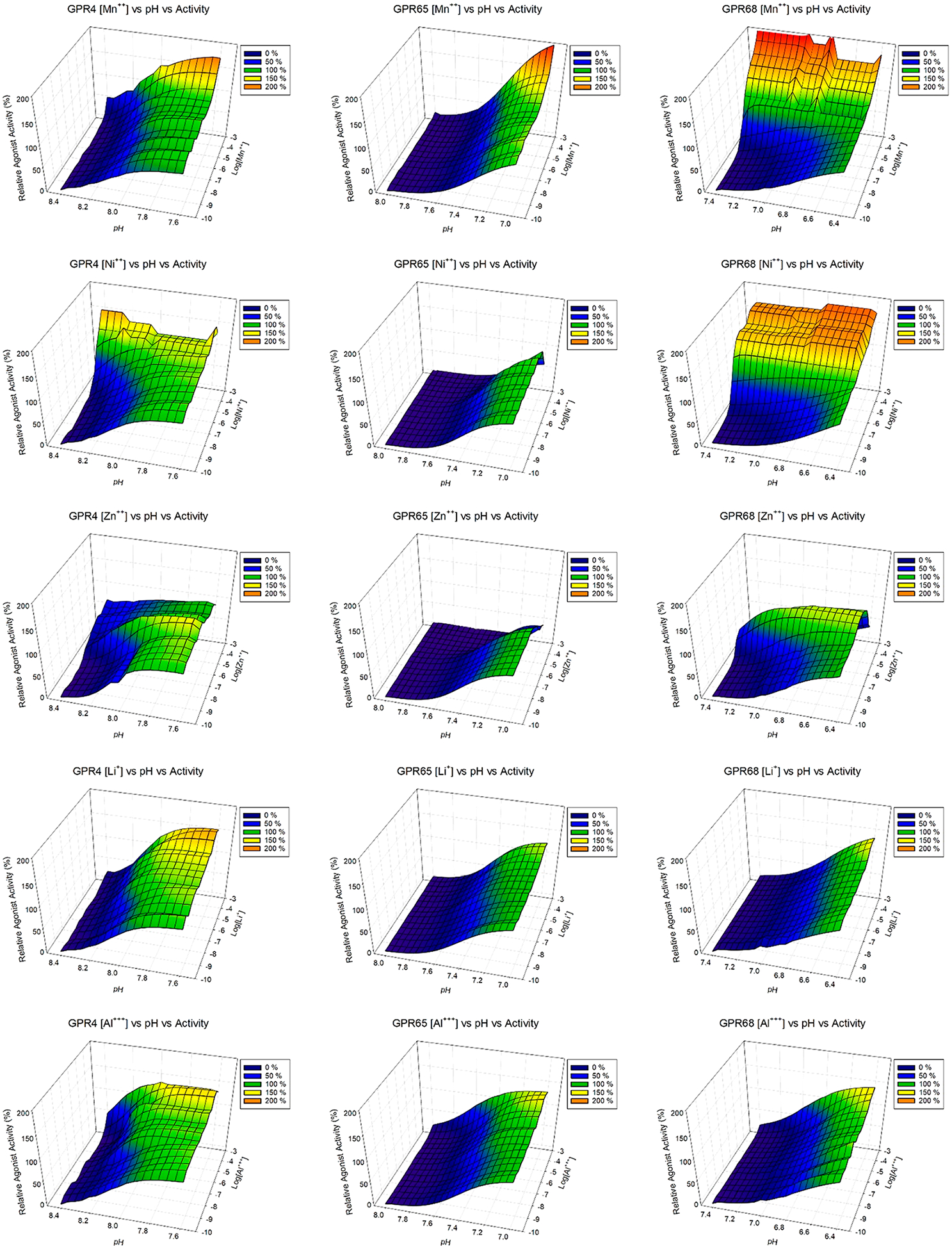
3D mesh presentations of GPR4 (left), GPR65 (middle), and GPR68 (right) activation by divalent metal ions under different pH conditions. GPR4 was tested at pH 8.40, 8.20, 8.00, and 7.60; GPR65 was tested at pH 8.00, 7.80, 7.40, and 7.00; and GPR68 was tested at pH 7.40, 7.00, 6.80, and 6.40. Results were normalized (0% at pH 8.40 and 100% at pH 7.60 for GPR4, 0% at pH 8.00 and 100% at pH 7.00 for GPR65, 0% at pH 7.40 and 100% at pH 6.40 for GPR68, all in the absence of metal ions) and represented means from a minimum of 3 independent assays, each in quadruplicate. Values at 4 pH conditions at each metal ion concentration (every half log between 1 mM to 0.1 nM) were fitted with the Prism built-in four-parameter logistic function to obtain potency and Hill slope values, which were used to carry out simulations in Prism to generate values at different pH points (every 0.05 pH interval between 8.40 and 7.60 for GPR4, 8.00 and 7.00 for GPR65, and 7.40 and 6.40 for GPR68). Simulated values were then used to produce 3D mesh plots in SigmaPlot. Values obtained from experiments at 4 pH points for each receptors were included in the 3D mesh plots. Metal ion dose-responses were also analyzed in Prism, and pharmacological parameters were listed in Table 2. Corresponding concentration response curves are presented in Figure S1.
Table 2.
Pharmacological Parameters of Divalent Metal Ion under Different pH Conditions at GPR68a
| efficacy and potency | Hill slope | |||||
|---|---|---|---|---|---|---|
| stimulation | inhibition | stimulation | inhibition | |||
| metal ions | pH | Emax at (μM) | pEC50 (nM) | pEC50 | Hill 1 | Hill 2 |
| Ca2+ | 7.40 | 1.9 ± 0.3 (1000) | 3.88 ± 0.23 | NA | 1.06 ± 0.49 | NA |
| 7.00 | 1.9 ± 0.3 (1000) | 3.78 ± 0.13 | 1.31 ± 0.42 | |||
| 6.80 | 2.1 ± 0.2 (1000) | 3.78 ± 0.11 | 1.19 ± 0.28 | |||
| 6.40 | 1.6 ± 0.1 (1000) | 3.81 ± 0.12 | 1.52 ± 0.55 | |||
| Cd2+ | 7.40 | 36.0 ± 6.8 (1) | 7.28 ± 0.05 (53) | 4.22 ± 0.20 | 1.41 ± 0.19 | 0.55 ± 0.19 |
| 7.00 | 5.1 ± 0.4 (1) | 7.69 ± 0.06 (20) | 4.21 ± 0.10 | 1.04 ± 0.11 | 0.80 ± 0.15 | |
| 6.80 | 3.0 ± 0.3 (1) | 7.75 ± 0.04 (18) | 3.81 ± 0.25 | 1.02 ± 0.10 | 0.58 ± 0.08 | |
| 6.40 | 1.3 ± 0.1 (1) | 7.62 ± 0.25 (24) | 3.31 ± 1.39 | 1.04 ± 0.50 | 0.47 ± 0.35 | |
| Co2+ | 7.40 | 33.5 ± 8.9 (30) | 7.14 ± 0.05 (72) | NA | 1.34 ±0.19 | NA |
| 7.00 | 4.4 ± 0.7 (30) | 7.45 ± 0.07 (35) | 0.99 ± 0.13 | |||
| 6.80 | 2.8 ± 0.4 (30) | 7.34 ± 0.08 (46) | 0.95 ± 0.14 | |||
| 6.40 | 1.4 ± 0.1 (30) | 6.39 ± 0.54 (407) | 0.34 ± 0.18 | |||
| Cu2+ | 7.40 | 2.7 ± 1.0 (1) | NA | NA | NA | NA |
| 7.00 | 1.6 ± 0.1 (1) | 6.09 ± 0.30 | 5.61 ± 0.17 | 1.18 ± 0.42 | 1.57 ± 0.44 | |
| 6.80 | 1.5 ± 0.1 (1) | 6.47 ± 0.18 | 5.43 ± 0.05 | 0.60 ± 0.15 | 2.22 ± 0.54 | |
| 6.40 | 1.0 ± 0.0 (1) | NA | 5.31 ± 0.03 | NA | 2.27 ± 0.26 | |
| Fe2+ | 7.40 | 10.2 ± 2.0 (30) | 4.13 ± 0.09 | NA | 0.64 ± 0.06 | NA |
| 7.00 | 3.7 ± 1.1 (30) | 5.91 ± 0.05 | 1.71 ± 0.29 | |||
| 6.80 | 3.0 ± 0.3 (30) | 6.26 ± 0.05 | 1.07 ± 0.11 | |||
| 6.40 | 1.3 ± 0.1 (30) | 5.33 ± 0.20 | 0.81 ± 0.27 | |||
| Mn2+ | 7.40 | 24.3 ± 5.3 (30) | 5.40 ± 0.05 | NA | 0.59 ± 0.04 | NA |
| 7.00 | 4.7 ± 0.2 (30) | 5.76 ± 0.07 | 0.52 ± 0.04 | |||
| 6.80 | 2.7 ± 0.2 (30) | 4.79 ± 0.27 | 0.32 ± 0.04 | |||
| 6.40 | 1.3 ± 0.1 (30) | 2.91 ± 1.07 | 0.42 ± 0.11 | |||
| Ni2+ | 7.40 | 20.2 ± 5.9 (30) | 6.91 ± 0.09 (123) | NA | 1.04 ± 0.19 | NA |
| 7.00 | 4.2 ± 0.7 (30) | 7.12 ± 0.09 (76) | 1.09 ± 0.22 | |||
| 6.80 | 2.9 ± 0.2 (30) | 7.00 ± 0.08 (100) | 0.95 ± 0.15 | |||
| 6.40 | 1.3 ± 0.1 (30) | 3.52 ± 1.61 | 0.27 ± 0.07 | |||
| Zn2+ | 7.40 | 9.1 ± 2.1 (1) | 6.68 ± 0.03 (209) | 5.33 ± 0.05 | 2.07 ± 0.20 | 1.55 ± 0.15 |
| 7.00 | 3.2 ±0.5 (1) | 6.88 ± 0.06 (132) | 4.86 ± 0.05 | 1.60 ± 0.25 | 1.17 ±0.15 | |
| 6.80 | 2.3 ± 0.2 (1) | 7.01 ± 0.08 (98) | 4.59 ± 0.05 | 1.09 ± 0.17 | 1.16 ±0.14 | |
| 6.40 | 1.3 ± 0.1 (1) | 6.92 ± 0.10 (120) | 3.89 ± 0.09 | 0.93 ± 0.19 | 0.84 ± 0.09 | |
Concentration-response curves are presented in Figure 3 and Figure S1. Emax (fold of basal) for the stimulation activity represented mean ± SEM from a minimum of 3 independent assays, each in triplicate or quadruplicate. The Emax for Ca2+ is determined at 1 mM, Cd2+, Cu2+, and Zn2+ at 1 μM, Co2+, Fe2+, Mn2+, and Ni2+ at 30 μM. Under each pH condition, buffer alone served as basal for normalization (with basal = 1.00). Potency and Hill slope values were retrieved from fittings with pooled and normalized results using the Prism 8.4 built-in four-parameter logistic function or bell-shaped concentration-response function.
However, the bell-shaped activity is different at GPR4. The peak shifts to higher concentrations at higher pH conditions for Cd2+, Cu2+, and Zn2+. Zn2+, at pH 8.40, displays only a stimulation phase with no inhibition phase at up to 1 mM. The efficacy of the stimulation phase by low concentrations of Cu2+ is much smaller (Figure 3, Figure S1, and Table 2) and is better displayed when the receptor is partially activated by protons, i.e., under pH 7.00 and 6.80 at GPR68, pH 8.20 and pH 8.00 at GPR4, and pH 7.40 at GPR65, but not at other higher or lower pH conditions. Other than Cd2+, Cu2+, and Zn2+, inhibitory activity is also observed with Co2+ and Ni2+ at GPR65 but not at GPR68 or GPR4, indicating that they may function as NAMs (negative allosteric modulators) at GPR65. Different from metal ions with bell-shaped responses, Co2+, Fe2+, Mn2+, and Ni2+ demonstrate pH-dependent agonist activity and with no inhibitory activity at up to 1 mM at GPR68 with higher efficacy than at GPR4 or GPR65. In summary, these results demonstrate that the activities of divalent metal ions are concentration- and pH-dependent, functioning as allosteric modulators at proton-sensing GPCRs. Co2+, Ni2+, Fe2+, and Mn2+ function as PAMs while others, especially Cd2+ and Zn2+, to a less degree Cu2+, function as PAMs at concentrations below 1 μM and as NAMs above 1 μM and at high micromolar concentrations. Under normal physiological conditions (pH 7.40) and especially mildly acidic conditions (such as pH 7.0–6.8), GPR68 activity is sensitively modulated by these divalent metal ions.
The pH-dependent activity profiles with metal ions indicate the activity is receptor-specific and concentration-dependent but may still be complicated with possible interference of metal ions on the luciferase activity. To determine if and how cAMP-dependent luciferase activity is affected by individual metal ions, we also tested them at parent HEK293T cells transfected to express the GloSensor reporter only, in the absence of Isoproterenol (ISO) for a potential stimulatory effect when basal activity is low and in the presence of 0.3 μM ISO for a potential inhibitory effect when basal activity is high (since ISO increases cAMP production through endogenous β2 adrenergic receptors) (Figure 4a). When basal luciferase activity is low, Ca2+/Mg2+ has minimal if any effect, and the presence of Cd2+, Cu2+, and Zn2+ has small inhibitory activity at high concentrations (over 30 μM), while the presence of Co2+, Fe2+, Mn2+, and Ni2+ has weak stimulatory activity at high concentrations (≥30 μM), much lower than the maximal stimulations of these metal ions at GPR68 at 7.40 (Figures 4b and 4c inserted bars and Table 2). With high concentrations of cAMP (high luciferase activity), Ca2+/Mg2+ and Fe2+ have no inhibitory effect, and Ni2+ has no activity until 3 mM. Mn2+ again showed small stimulatory activity at high concentrations. Clear inhibition is observed with Cd2+, Co2+, Cu2+, and Zn2+ starting at 100 μM. These results indicate that small levels of nonspecific stimulatory activity by some metal ions would not be expected to interfere with the characterizations of these metal ions at GPR68 for stimulatory activity. Nonspecific inhibitory activity by some metal ions occurs at concentrations higher than their inhibitory activity at GPR68 (Table 2); therefore, this would not be expected to be responsible for the observed inhibition at GPR68. Nevertheless, to avoid nonspecific inhibitory activity at high concentrations, we mainly focused on the stimulatory activity by low concentrations of metal ions at GPR68 WT and mutant receptors.
Figure 4.
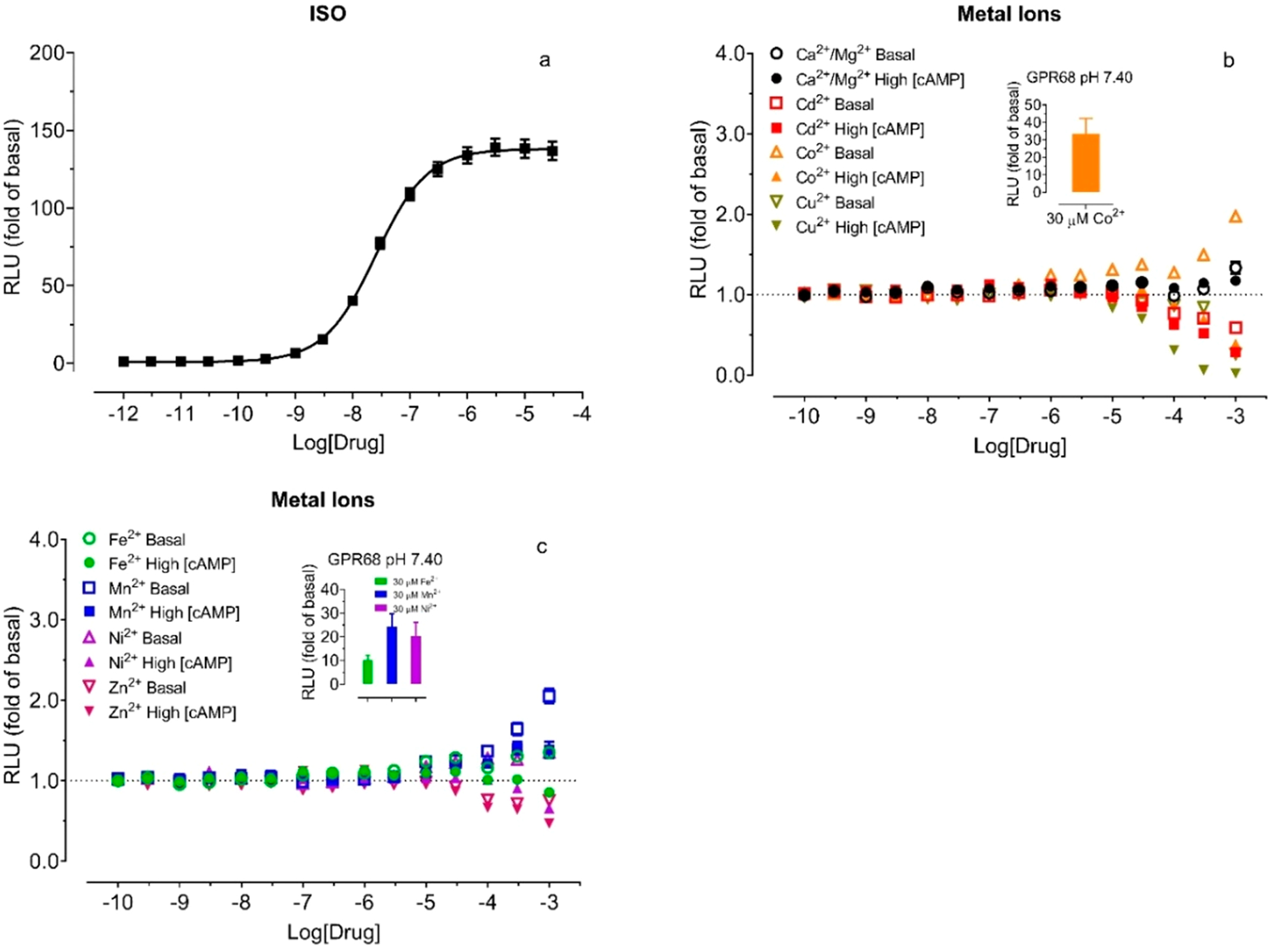
Effects of divalent metal ions on luciferase activity under basal and high concentrations of cAMP at control HEK293 T cells transiently transfected with the GloSensor cAMP reporter. The assays were conducted in the Ca2+/Mg2+-free HBSS-based buffer with 20 mM HEPES, 10 μM PDE inhibitor Ro 20–1724, pH 7.40, supplemented with indicated metal ions. (a) Isoproterenol (ISO) produced a concentration-dependent cAMP production through endogenous β2 adrenergic receptors, (b) Ca2+/Mg2+, Cd2+, Co2+, and Cu2+, (c) Fe2+, Mn2+, Ni2+, and Zn2+. Effects of metal ions were determined in the absence of ISO (basal) first and then in the presence of 0.3 μM ISO (high [cAMP]) in the same assay plate. Results were normalized to corresponding control (or basal) conditions in the form of the fold of basal and represented mean ± SEM from a minimum of 3 independent assays, each in a quadruplicate set. The curve was analyzed in Prism 8.4. For comparison, inserted bar graphs in panels b and c represented the activity of corresponding metal ions (fold of basal) at GPR68 at pH 7.40, with values from Table 2.
Divalent Metal Ions Are Allosteric Modulators at GPR68.
To quantify allosteric activity, we measured proton concentration responses in the absence and presence of increasing concentrations of metal ions at GPR68 wild-type receptors. Results were analyzed using the Prism built-in four-parameter logistic function to obtain proton potency, efficacy, and Hill slopes (Figure 5 and Table 3) and the standard allosteric operational model33–35 to retrieve key allosteric parameters-affinity cooperativity α, efficacy cooperativity β, and allosteric binding affinity KB (Table 4). Cu2+ was not included in these experiments due to its small potentiation only at pH 6.80 and 7.00, while Cd2+ (Figure 5c) and Zn2+ (Figure 5h) were tested at up to 1 μM to avoid inhibitory activity as indicated from Figure 3 and Figure S1. Consistent with the results in Figure 2, Ca2+ (Figure 5a) produces slight increases in proton efficacy and has little effect on proton potency, functioning as a weak β-PAM (β > 1 for increasing proton efficacy), while Mg2+ (Figure 5b) does not affect proton activity. The other metal ions potentiate proton activity, shifting proton concentration-response curves to the left with or without concomitant increases in maxima (Figures 5d, 5e, 5f, 5g). The estimated allosteric activity index Log(αβ/KB) (Table 4) follows the following order: Cd2+ > Co2+ > Ni2+ > Zn2+ > Mn2+ > Fe2+ > Ca2+ > Mg2+. At normal physiological conditions (pH 7.40), GPR68 is essentially inactive in the absence of metal ions but is at about 30% activity by 30 nM Cd2+ or Co2+ or Ni2+ or 1 μM Mn2+ and close to 100% activity by 100 nM Cd2+ or Co2+ or Ni2+ (Figures 5c, 5d, 5g). Fe2+ and Zn2+ (Figures 5e and 5h) are relatively weaker modulators but are also able to fully activate GPR68 under pH 7.00 at 300 nM for Zn2+ or 3 μM for Fe2+. With both α and β > 1, these metal ions are all αβ-PAMs for protons at GPR68.
Figure 5.
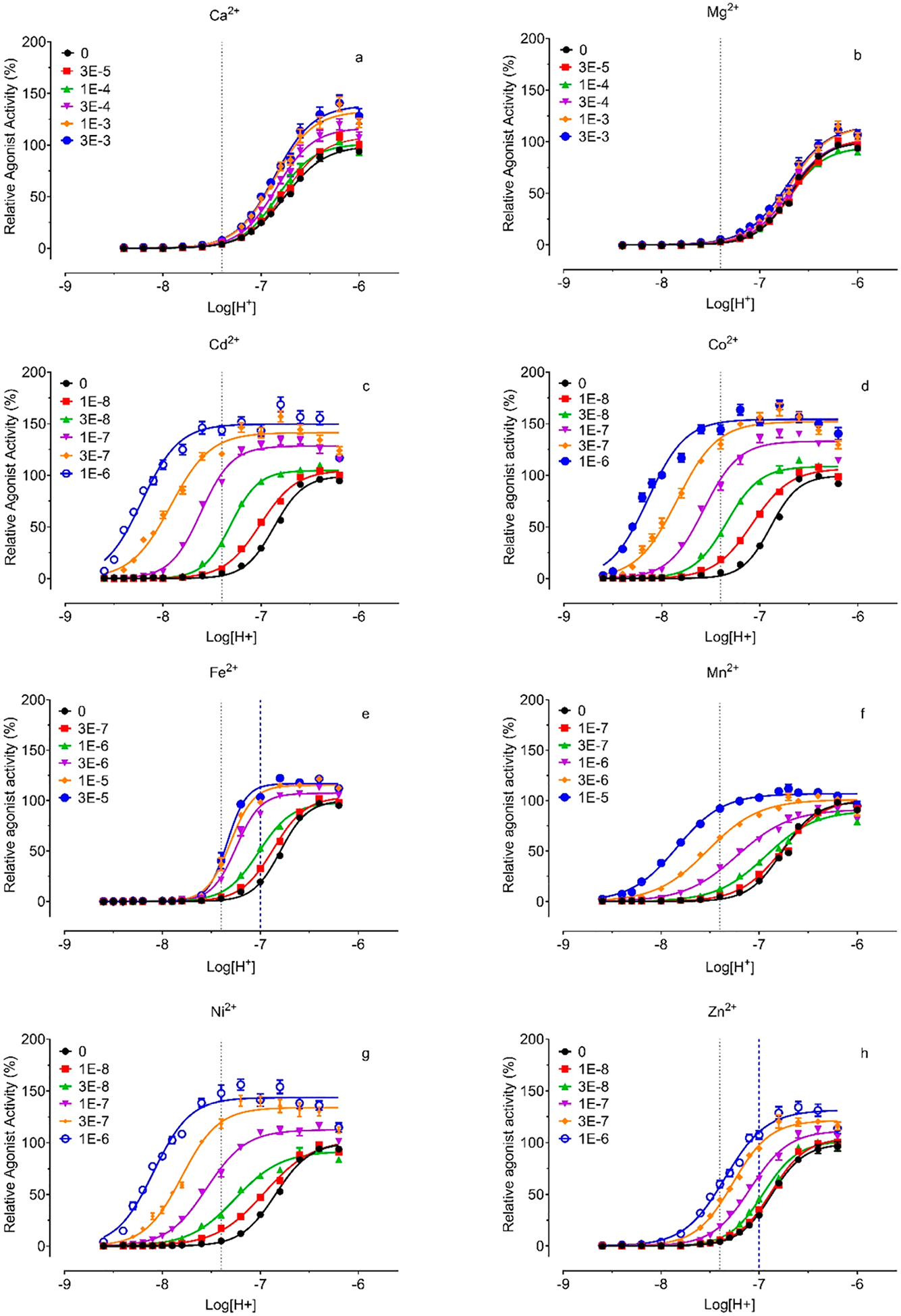
Proton concentration-responses in the absence and presence of increasing concentrations of Ca2+ (a), Mg2+ (b), Cd2+ (c), Co2+ (d), Fe2+ (e), Mn2+ (f), Ni2+ (g), and Zn2+ (h). Gs-cAMP production was measured in transiently transfected HEK293T cells using the GloSensor cAMP assays. Metal ion stock solutions were made in water and diluted in pH preadjusted Ca2+/Mg2+ free HBSS-based buffers. Results were normalized to proton responses (%) and represented means ± SEM from a minimum of 3 independent assays, each in quadruplicate. Curves were analyzed in Prism 8.4 using the built-in four-parameter logistic function. Allosteric parameters are listed in Table 4. Dashed lines indicate relative activity at pH 7.40 or pH 7.00.
Table 3.
Pharmacological Parameters of Protons at GPR68 in the Presence of Metal Ionsa
| metal and concentrations | pEC50 (pH) | Emax (%) | Hill slope | |
|---|---|---|---|---|
| Ca2+ | 0 | 6.76 ± 0.01 | 99.5 ± 1.7 | 2.11 ± 0.09 |
| 30 μM | 6.76 ± 0.01 | 108.3 ± 2.1 | 2.16 ± 0.11 | |
| 100 μM | 6.83 ± 0.01 | 101.2 ± 2.1 | 2.35 ± 0.14 | |
| 300 μM | 6.84 ± 0.01 | 116.6 ± 2.6 | 2.31 ± 0.15 | |
| 1 mM | 6.87 ± 0.02 | 133.1 ± 2.9 | 2.20 ± 0.14 | |
| 3 mM | 6.86 ± 0.02 | 138.6 ± 3.4 | 2.19 ± 0.16 | |
| Mg2+ | 0 | 6.69 ± 0.01 | 99.9 ± 1.5 | 2.48 ± 0.10 |
| 30 μM | 6.66 ± 0.01 | 103.2 ± 2.0 | 2.30 ± 0.11 | |
| 100 μM | 6.69 ± 0.01 | 94.7 ± 1.8 | 2.38 ± 0.12 | |
| 300 μM | 6.70 ± 0.02 | 102.3 ± 2.6 | 2.29 ± 0.15 | |
| 1 mM | 6.70 ± 0.02 | 115.1 ± 3.2 | 2.18 ± 0.15 | |
| 3 mM | 6.72 ± 0.02 | 115.7 ± 3.4 | 2.07 ± 0.15 | |
| Cd2+ | 0 | 6.87 ± 0.01 | 99.5 ± 1.1 | 2.55 ± 0.10 |
| 10 nM | 7.01 ± 0.01 | 105.0 ± 1.2 | 2.52 ± 0.11 | |
| 30 nM | 7.31 ± 0.01 | 104.7 ± 1.0 | 2.31 ± 0.11 | |
| 100 nM | 7.61 ± 0.01 | 128.4 ± 1.3 | 2.06 ± 0.12 | |
| 300 nM | 7.92 ± 0.02 | 141.4 ± 1.7 | 1.99 ± 0.13 | |
| 1000 nM | 8.22 ± 0.01 | 149.7 ± 1.9 | 1.54 ± 0.10 | |
| Co2+ | 0 | 6.89 ± 0.01 | 99.8 ± 1.2 | 3.03 ± 0.13 |
| 10 nM | 7.08 ± 0.01 | 106.5 ± 1.2 | 2.24 ± 0.09 | |
| 30 nM | 7.34 ± 0.01 | 108.6 ± 1.1 | 2.55 ± 0.12 | |
| 100 nM | 7.58 ± 0.01 | 133.0 ± 1.4 | 2.41 ± 0.13 | |
| 300 nM | 7.84 ± 0.01 | 152.0 ± 1.7 | 2.10 ± 0.11 | |
| 1000 nM | 8.14 ± 0.01 | 154.3 ± 1.7 | 2.27 ± 0.13 | |
| Fe2+ | 0 | 6.79 ± 0.01 | 100.2 ± 1.1 | 3.11 ± 0.12 |
| 0.3 μM | 6.87 ± 0.01 | 103.8 ± 1.0 | 2.63 ± 0.08 | |
| 1 μM | 7.02 ± 0.01 | 98.1 ± 0.8 | 2.58 ± 0.08 | |
| 3 μM | 7.25 ± 0.01 | 107.3 ± 1.1 | 3.49 ± 0.19 | |
| 10 μM | 7.31 ± 0.01 | 115.3 ± 1.2 | 3.60 ± 0.22 | |
| 30 μM | 7.34 ± 0.01 | 116.8 ± 1.3 | 4.42 ± 0.33 | |
| Mn2+ | 0 | 6.76 ± 0.01 | 99.2 ± 1.0 | 2.53 ± 0.08 |
| 0.1 μM | 6.77 ± 0.01 | 102.2 ± 1.2 | 2.00 ± 0.06 | |
| 0.3 μM | 6.93 ± 0.01 | 90.3 ± 1.2 | 1.73 ± 0.06 | |
| 1 μM | 7.22 ± 0.01 | 91.5 ± 1.3 | 1.61 ± 0.07 | |
| 3 μM | 7.53 ± 0.01 | 101.1 ± 1.2 | 1.66 ± 0.07 | |
| 10 μM | 7.84 ± 0.01 | 106.9 ± 1.1 | 1.85 ± 0.09 | |
| Ni2+ | 0 | 6.85 ± 0.01 | 100.2 ± 1.6 | 2.46 ± 0.10 |
| 10 nM | 6.96 ± 0.02 | 103.2 ± 2.3 | 1.75 ± 0.09 | |
| 30 nM | 7.26 ± 0.02 | 91.9 ± 1.5 | 1.85 ± 0.09 | |
| 100 nM | 7.55 ± 0.01 | 112.5 ± 1.3 | 2.16 ± 0.10 | |
| 300 nM | 7.80 ± 0.01 | 134.0 ± 1.8 | 2.31 ± 0.15 | |
| 1000 nM | 8.09 ± 0.01 | 143.6 ± 1.9 | 2.32 ± 0.15 | |
| Zn2+ | 0 | 6.78 ± 0.01 | 98.5 ± 1.3 | 2.46 ± 0.11 |
| 10 nM | 6.79 ± 0.01 | 98.6 ± 1.4 | 2.48 ± 0.12 | |
| 30 nM | 6.83 ± 0.01 | 91.1 ± 1.6 | 2.33 ± 0.14 | |
| 100 nM | 6.96 ± 0.02 | 96.4 ± 1.8 | 2.06 ± 0.14 | |
| 300 nM | 7.17 ± 0.02 | 98.8 ± 1.4 | 2.06 ± 0.12 | |
| 1000 mM | 7.27 ± 0.01 | 100.9 ± 1.3 | 2.10 ± 0.12 | |
Proton concentration response curves are presented in Figure 5. Parameters were extracted from curve fitting to the Prism 8.4 build-in four-parameter logistic function.
Table 4.
Allosteric Parameters at GPR68 Wild-Type (WT) and Mutant Receptorsa
| mutant | ligand | Log(α) | Log(β) | pKB | Log(αβ/KB) | Δ | τA | Hill |
|---|---|---|---|---|---|---|---|---|
| WT | Ca2+ | −0.31 ± 0.34 | 0.43 ± 0.28 | 3.23 ± 0.09 | 3.36 ± 0.07 | 1.68 ± 0.013 | 3.12 ± 0.38 | |
| WT | Mg2+ | NA | NA | NA | NA | NA | NA | |
| WT | Cd2+ | 1.32 ± 0.11 | 0.27 ± 0.07 | 6.39 ± 0.14 | 7.98 ± 0.14 | 1.71 ± 0.03 | 3.48 ± 0.15 | |
| WT | Co2+ | 1.12 ±0.11 | 0.37 ± 0.07 | 6.28 ± 0.11 | 7.77 ± 0.11 | 1.61 ± 0.05 | 2.90 ± 0.14 | |
| WT | Fe2+ | 0.38 ± 0.13 | 0.20 ± 0.06 | 5.26 ± 0.10 | 5.84 ± 0.08 | 1.80 ± 0.03 | 3.73 ± 0.31 | |
| WT | Mn2+ | 1.43 ± 0.09 | 0.17 ± 0.07 | 4.60 ± 0.07 | 6.20 ± 0.02 | 1.73 ± 0.06 | 3.08 ± 0.16 | |
| WT | Ni2+ | 1.42 ± 0.16 | 0.27 ± 0.08 | 5.92 ± 0.16 | 7.61 ± 0.09 | 1.68 ± 0.05 | 3.07 ± 0.20 | |
| WT | Zn2+ | 0.63 ±0.18 | 0.14 ± 0.04 | 6.34 ± 0.23 | 7.11 ± 0.11 | 1.82 ± 0.03 | 3.81 ± 0.18 | |
| C13A | Cd2+ | 1.02 ± 0.09 | 0.67 ± 0.13 | 5.31 ± 0.14 | 6.99 ± 0.10 | −0.99 | 1.31 ± 0.16 | 1.37 ± 0.17 |
| C258A | Cd2+ | 0.26 ± 0.11 | 0.94 ± 0.06 | 5.49 ± 0.05 | 6.69 ± 0.07 | −1.29 | 0.11 ± 0.15 | 1.15 ± 0.09 |
| C13A | Co2+ | 2.09 ± 0.38 | 0.56 ± 0.10 | 3.95 ± 0.46 | 6.60 ± 0.08 | −1.17 | 1.48 ± 0.13 | 1.61 ± 0.14 |
| C258A | Co2+ | 1.00 ± 0.12 | 0.74 ± 0.05 | 4.78 ± 0.16 | 6.52 ± 0.07 | −1.25 | 1.37 ± 0.06 | 1.31 ± 0.10 |
| C13A | Ni2+ | 0.76 ± 0.15 | 1.04 ± 0.16 | 4.86 ± 0.13 | 6.66 ± 0.09 | −0.95 | 0.82 ± 0.08 | 1.15 ± 0.11 |
| C258A | Ni2+ | 0.65 ± 0.12 | 0.80 ± 0.12 | 5.32 ± 0.09 | 6.77 ± 0.09 | −0.84 | 1.13 ± 0.16 | 1.24 ± 0.08 |
| C13A | Zn2+ | NA | NA | NA | NA | NA | NA | NA |
| C258A | Zn2+ | NA | NA | NA | NA | NA | NA | NA |
| WT | lorazepam | 0.30 ± 0.07 | 0.21 ± 0.06 | 4.87 ± 0.04 | 5.33 ± 0.03 | 0 | 1.88 ± 0.05 | 4.72 ± 0.24 |
| H17A | lorazepam | 0.21 ± 0.08 | 0.21 ± 0.06 | 5.11 ± 0.03 | 5.52 ± 0.02 | 0.19 | 1.74 ± 0.02 | 3.85 ± 0.18 |
| H20A | lorazepam | 0.25 ± 0.13 | 0.37 ± 0.06 | 4.76 ± 0.20 | 5.39 ± 0.10 | 0.06 | 1.62 ± 0.06 | 3.27 ± 0.16 |
| C13A | lorazepam | 0.16 ± 0.03 | 0.39 ± 0.10 | 5.13 ± 0.08 | 5.68 ± 0.13 | 0.35 | 1.26 ±0.14 | 2.27 ± 0.27 |
| C258A | lorazepam | −0.32 ± 0.10 | 0.77 ± 0.12 | 5.06 ± 0.12 | 5.51 ± 0.10 | 0.18 | 0.96 ± 0.17 | 1.46 ± 0.13 |
| 4HA | lorazepam | −0.08 ± 0.04 | 0.58 ± 0.06 | 4.83 ± 0.04 | 5.34 ± 0.06 | 0.01 | 1.41 ± 0.09 | 3.26 ± 0.23 |
| WT | ogerin | 0.67 ± 0.09 | 0.31 ± 0.05 | 4.32 ± 0.09 | 5.30 ± 0.04 | 0 | 1.64 ± 0.04 | 3.76 ± 0.07 |
| H17A | ogerin | 0.67 ± 0.17 | 0.18 ±0.12 | 4.47 ± 0.10 | 5.33 ± 0.04 | 0.03 | 1.78 ± 0.04 | 4.18 ± 0.40 |
| H20A | ogerin | 0.37 ± 0.10 | 0.33 ± 0.03 | 4.42 ± 0.08 | 5.12 ± 0.03 | −0.18 | 1.71 ± 0.05 | 3.29 ± 0.19 |
| C13A | ogerin | 0.15 ± 0.32 | 0.75 ± 0.22 | 4.28 ± 0.07 | 5.17 ± 0.02 | −0.13 | 1.55 ± 0.02 | 2.60 ± 0.03 |
| C258A | ogerin | 0.05 ±0.13 | 0.68 ± 0.07 | 4.49 ± 0.13 | 5.22 ± 0.03 | −0.08 | 1.42 ± 0.03 | 1.90 ± 0.12 |
| 4HA | ogerin | 0.27 ± 0.03 | 0.44 ± 0.10 | 4.20 ± 0.22 | 4.90 ±0.11 | −0.40 | 1.58 ± 0.11 | 3.73 ± 0.22 |
Concentration response curves are presented in Figures 5 and 10. Data was normalized to the control (0% for basal and 100% for proton Emax in the absence of modulator) and analyzed in Prism 8.4 using the standard allosteric operational model. Results represented mean ± SEM from a minimum of 3 assays, each in quadruplicate. Mutant 4HA = GPR68 H17A, H20A, H84A, and H169A. α stands for the affinity cooperativity, β stands for efficacy cooperativity, KB stands for allosteric ligand binding affinity, τA stands for orthosteric efficacy (in this case H+), and Hill is the slope factor. Log(αβ/KB) is the allosteric activity index, and Δ is the difference between mutant and wildtype for the same modulator. NA stands for not applicable.
Differential Roles of Histidine Residues in Proton-Mediated GPR68 Activity.
Previous studies, changing extracellular His residues to Phe and measuring Gq-mediated IP production in transiently transfected cells, found H17, H20, H84, H169, H245, and H269 were important for proton-mediated Gq activity, while H89, H159, and H175 were not.4 To systematically examine potential effects of these His residues, we mutated all 7 extracellular His and 2 TM His residues to Ala and Phe and compared with wild-type receptors in proton-mediated cAMP production in transiently transfected cells (Figures 6a, 6b and Table 5). The overall effects of mutations on proton activity are quantified by ΔLog(Max/EC50).32 In general, different single His mutations have varying degrees of effect on proton activity. For some mutants, moderate degrees of reduction in proton potency are compensated by small increases in proton efficacy (fold of basal), which results in little or no change in the ΔLog(Max/EC50) index value. The higher maximal responses in fold of basal are mainly due to a reduced basal activity as compared with wild-type receptors (results not shown). All single His mutant have high Hill values (comparable to WT); therefore, a small change in proton potency could mean a large change in overall activity (Figure 6). Consistently, reduced effects on proton potency are observed at H20 and H169 to either Ala or Phe mutation. Both H20A and H20F mutations reduce proton activity by 0.6 log unit, while H169A and H169F reduce proton activity by 0.14 log unit. Surprisingly, all the other single extracellular His to Ala or Phe mutations have little or no effect on proton potency. A discrepancy is observed between HA vs HF mutations. If protonation is important for receptor activation at these sites, both HA and HF mutations would be expected to reduce proton potency. This discrepancy may indicate that these His residues may be less important for protonation but more important for the receptor structure. In contrast to extracellular His mutations, however, mutations in the TM domains, H245F in the TM VI and H269F in the TM VII, result in a reduction of 0.9 and 0.6 log unit, respectively, while corresponding H245A and H269A mutations have smaller effects on proton activity. The discrepancy between HF vs HA substitution suggests that H245 or H269 may play structural roles rather than being protonated during the receptor activation process. It has been suggested that H245 points outward into the lipid membrane environment.4 In summary, these mutational results indicate that H20 and H169 are more important than other extracellular His residues for proton activity, while TM His residues may be complicated with their potential structural roles.
Figure 6.
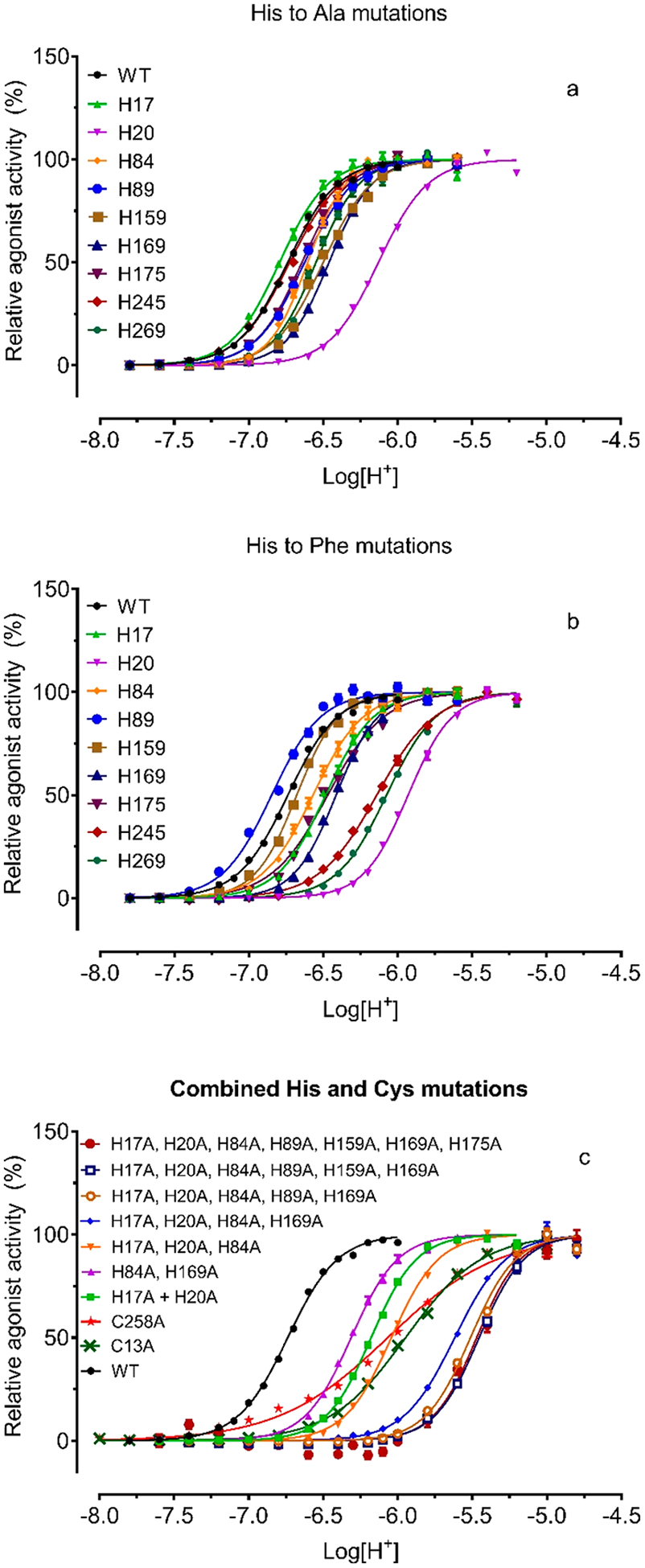
Effect of histidine mutations on proton activity. (a) Single His to Ala mutations; (b) single His to Phe mutations; (c) Cys mutation and combined His to Ala mutations. GloSensor-cAMP production was measured in transiently transfected HEK293T cells using the GloSensor cAMP assays. Results were independently normalized (0–100%) for potency comparison and represented means ± SEM from a minimum of 6 independent assays, each in quadruplicate. Curves were analyzed in Prism using the built-in four-parameter logistic function. Pharmacological parameters were reported in Table 5.
Table 5.
Pharmacological Parameters of GPR68 Wild-Type and Histidine Mutant Receptorsa
| receptor | efficacy (fold) | potency (pH) | ΔLog (Emax/EC50) | Hill slope (n) |
|---|---|---|---|---|
| WT | 58.5 ± 3.3 | 6.77 ± 0.01 | 0 | 2.77 ± 0.09 (68) |
| H17A | 71.6 ± 5.7 | 6.78 ± 0.01 | 0.10 | 2.60 ± 0.06 (26) |
| H17F | 81.1 ± 10.8 | 6.50 ± 0.02 | −0.12 | 2.89 ± 0.06 (8) |
| H20A | 80.4 ± 5.1 | 6.04 ± 0.04 | −0.59 | 2.48 ± 0.06 (28) |
| H20F | 93.1 ± 10.3 | 5.93 ± 0.02 | −0.63 | 2.98 ± 0.19 (6) |
| H84A | 87.7 ± 6.3 | 6.60 ± 0.01 | 0.01 | 3.33 ± 0.10 (32) |
| H84F | 61.4 ± 8.9 | 6.58 ± 0.04 | −0.16 | 2.93 ± 0.23 (6) |
| H89A | 63.0 ± 7.0 | 6.49 ± 0.05 | −0.24 | 2.84 ± 0.19 (20) |
| H89F | 47.9 ± 5.0 | 6.86 ± 0.01 | 0.01 | 2.72 ± 0.08 (23) |
| H159A | 91.0 ± 5.8 | 6.50 ± 0.02 | −0.07 | 2.67 ± 0.09 (44) |
| H159F | 73.3 ± 13.3 | 6.68 ± 0.01 | 0.01 | 3.15 ± 0.31 (12) |
| H169A | 89.1 ± 7.3 | 6.44 ± 0.02 | −0.14 | 3.12 ± 0.10 (31) |
| H169F | 92.7 ± 5.2 | 6.42 ± 0.01 | −0.15 | 3.16 ± 0.13 (8) |
| H175A | 72.5 ± 6.0 | 6.63 ± 0.01 | −0.04 | 3.02 ± 0.18 (30) |
| H175F | 78.5 ± 12.5 | 6.48 ± 0.03 | −0.16 | 2.41 ± 0.14 (8) |
| H245A | 52.5 ± 3.6 | 6.76 ± 0.01 | −0.05 | 2.61 ± 0.08 (32) |
| H245F | 31.9 ± 2.6 | 6.15 ± 0.02 | −0.88 | 2.07 ± 0.07 (15) |
| H269A | 62.9 ± 13.8 | 6.55 ± 0.01 | −0.18 | 3.03 ± 0.22 (6) |
| H269F | 65.9 ± 5.7 | 6.11 ±0.01 | −0.60 | 2.72 ± 0.07 (18) |
| H17A, H20A | 79.4 ± 8.0 | 6.13 ±0.01 | −0.50 | 2.81 ± 0.08 (32) |
| H84A, H169A | 100.6 ± 3.5 | 6.29 ± 0.03 | −0.24 | 3.04 ± 0.17 (23) |
| H17A, H20A, H84A (3HA) | 89.4 ± 6.8 | 6.03 ± 0.02 | −0.55 | 2.81 ± 0.11 (20) |
| H17A, H20A, H84A, H169A (4HA) | 64.6 ± 3.9 | 5.61 ± 0.03 | −1.11 | 2.86 ± 0.09 (37) |
| H17A, H20A, H84A, H89A, H169A (5HA) | 41.9 ± 3.7 | 5.51 ± 0.01 | −1.40 | 2.90 ± 0.15 (8) |
| H17A, H20A, H84A, H89A, H159A, H169A (6HA) | 25.9 ± 4.0 | 5.45 ± 0.02 | −1.67 | 2.92 ± 0.17 (8) |
| H17A, H20A, H84A, H89A, H159A, H169A, H175A (7HA) | 4.2 ± 1.1 | 5.40 ± 0.05 | −2.51 | 2.45 ± 0.41 (4) |
| C13A | 27.9 ± 1.9 | 5.93 ± 0.02 | −1.16 | 1.92 ± 0.05 (51) |
| C258A | 24.5 ± 1.5 | 6.08 ± 0.04 | −1.06 | 1.43 ± 0.08 (28) |
Proton concentration responses are presented in Figure 6, where proton efficacy was individually normalized for comparison of potency. Actual proton efficacy in the format of the fold of basal for each receptor is presented in this table. Results were analyzed in Prism 8.4 using the built-in four-parameter logistic function. Values represented mean ± SEM from a minimum of n independent assays, each in triplicate or quadruplicate. ΔLog(Emax/EC50) = mutant Log(Emax/EC50) − WT Log(Emax/EC50); negative values stand for reduction, and positive values stand for enhancement as compared with the GPR68 WT receptor.
It has initially been proposed that GPR68 activation involves protonation on multiple extracellular His residues;4 therefore, we created mutants with combined His to Ala mutations (Figure 6c and Table 5). The mutant with both H17A and H20A mutations is similar to H20A alone, consistent with the data showing that the H17A mutation has little effect. Although H84A alone has no effect, the mutant with triple His mutations (3HA for H17A + H20A + H84A) shifts the proton curve to the right of the mutant with double His mutations of H17A and H20A. Similarly, combining H84A with H169A mutation reduces proton activity more than H169A alone. The mutant with four His mutations (4HA = H17A + H20A + H84A + H169A) reduces proton potency further, by over 10-fold (1 log unit), to below pH 6.00. Further addition of H89A (5HA for H17A + H20A + H84A + H89A + H169A) and H159A (6HA for H17A + H20A + H84A + H89A + H159A + H169A) reduces proton activity by 30-fold (1.5 log unit). The 7HA mutant with all extracellular His residues replaced by Ala displays further reduced proton potency and has less than 10% of WT efficacy, in combination leading to over 300-fold (2.5 log unit) reduced proton activity. In summary, mutants with combined extracellular His mutations display gradually reduced proton activity. The 7HA mutant, with all extracellular His residues replaced by Ala, remains active but with severely reduced potency and efficacy. These results are consistent with the notion that extracellular His residues are important for proton activity, presumably through coordinated protonation at multiple His residues.
Differential Roles of Histidine Residues in Allosteric Modulation by Divalent Metal Ions.
To determine if and how allosteric modulation by metal ions is affected at His mutants, we tested all HA mutants with metal ion concentration responses. Since metal ion activity is pH-dependent and each mutant has a slightly different potency for protons, proper control of pH conditions is critical for the functional assays. We chose a pH condition at or close to the pEC50 point of each receptor, under which allosteric modulation of proton activity would be most sensitive for detection. Specifically, H17A, H245A, and wildtype (WT) were tested at pH 6.80; H20A was tested at pH 6.10; H84A and H175A were tested at pH 6.60; and H89A, H159A, H169A, and H269A were tested at pH 6.40. As shown in Figures 7a–7g, single HA mutations have diverse and complex activity profiles with metal ions. To facilitate comparison, we quantified the effects of mutations with ΔLog(Max/EC50)32 as compared with corresponding values at WT, and the results are presented in a heat map (Figure 8). Cu2+ (Figure 7c) shows small PAM activity at GPR68 WT but displays inhibitory activity at all mutant receptors in a similar pattern and within the same narrow range, 300 nM to 10 μM. Although the inhibitory activity at mutant GPR68 occurs before nonspecific inhibitory activity in control assays (Figure 4c), the steep inhibition profiles make characterization and comparison difficult; therefore, Cu2+ was excluded from further assays. It is clear that ECL2 H169 is the most important His residue for metal ions, and mutation H169A abolishes the PAM activity for both Cd2+ (Figure 7a) and Zn2+ (Figure 7g), greatly reducing the PAM activity for other metal ions. Compared with H169, H20 also abolishes the PAM activity for Zn2+ and greatly reduces the PAM activity for other metal ions. Slightly different from H20 or H169, H17 demonstrates interesting and differential effects among the tested metal ions, as it eliminates the PAM activity for Zn2+, severely diminishes the PAM activity for both Cd2+ (Figure 7a) and Co2+ (Figure 7b), has moderate reductions for Fe2+ (Figure 7d) and Ni2+ (Figure 7f), but barely affects Mn2+ (Figure 7e). For Cd2+, H17A right shifts the bell-shaped curve with peaking concentration at 10 μM (vs 1 μM at WT) (Figure 7a). H20A right shifts the curve further with a peaking concentration of over 100 μM. However, H169A precludes PAM activity and only displays inhibitory activity. The estimated inhibitory potency of Cd2+ is similar at HA mutants, 182 μM at H17A, 398 μM at H20A, and 123 μM at H169A vs 71 μM at WT. For Zn2+, H17A and H20A show a little PAM activity at 0.3 to 3 μM (too small to quantify) but demonstrate clear inhibition as H169A with similar efficacy. A wider range of inhibitory potency of Zn2+ is observed, 34 μM at WT, 30 μM at H17A, 145 μM at H20A, and 2.1 μM at H169A. These results indicate that H17, H20, and H169 are critical for PAM activity while remaining His or other residues are important for NAM activity for Zn2+. Compared with H17, H20, and H169, a different pattern is observed for H84 and H89. These have very similar profiles and reduce activity the most for Co2+ and Ni2+ ions while having relatively moderate reductions for other metal ions. Different from other extracellular His residues, H159 and H175 have small and similar reducing activity for all metal ions (except for Mn2+ at H159), suggesting H159 and H175 are not critical residues for metal ions. Similar to H159 and H169 and different from other extracellular His residues, two TM His residues, H245 and H269, also have smaller effects on metal ions. H245A mutation essentially has no effect on Cd2+, Ni2+, or Zn2+, while H269A mutation results in the same degree of reduction for all tested metal ions, except for Mn2+, which is compensated by increased efficacy (Figure 7e), similar to their nonessential role in proton sensing and consistent with the notion that they, individually, play more structural roles than directly involving metal ion modulation. In summary, these results indicate that extracellular H17, H20, H84, H89, and H169 are important for PAM activity of divalent metal ions.
Figure 7.
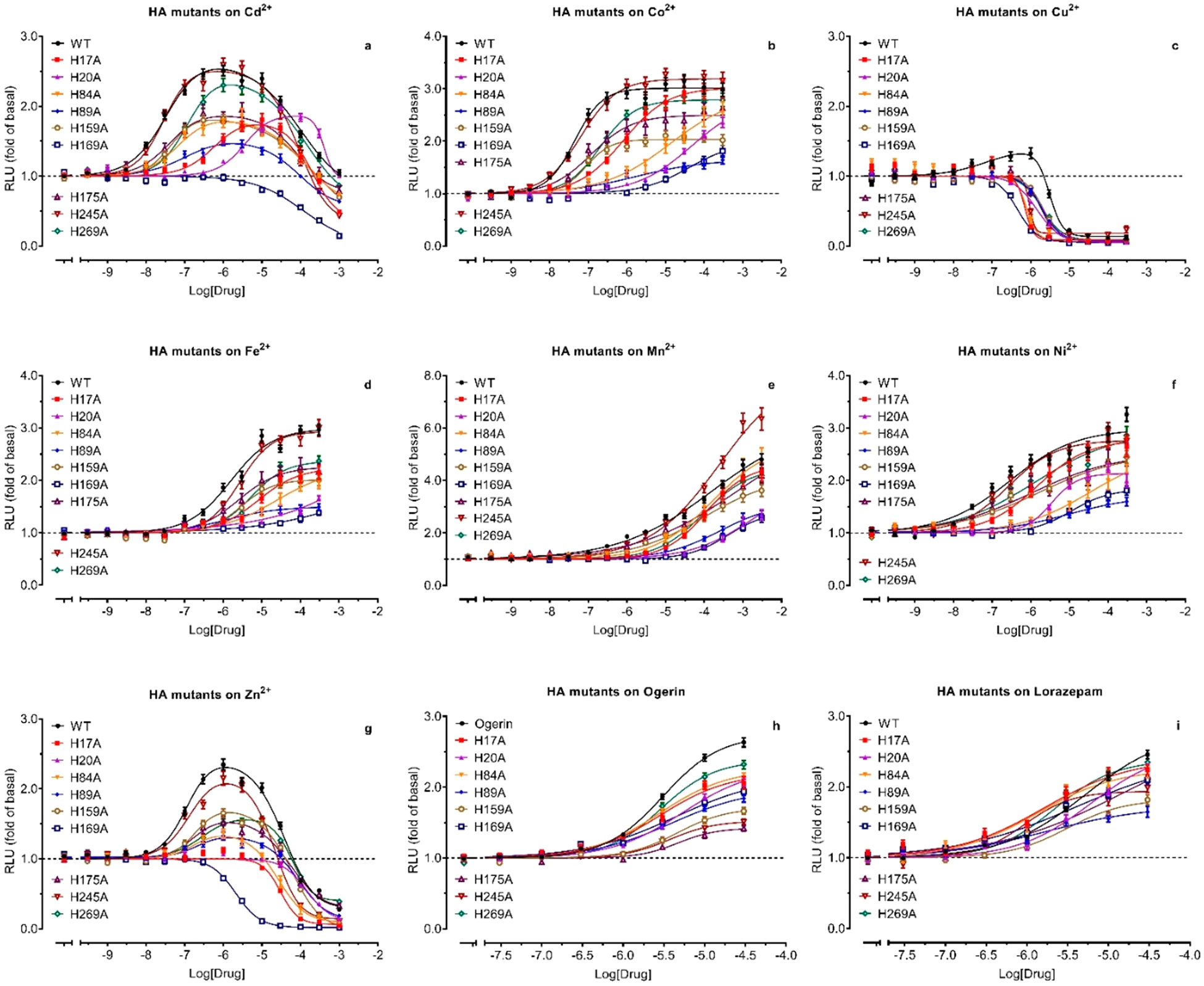
Effect of Histidine to Alanine mutations on allosteric modulation of protons by metal ions (a–g) and ogerin (h) and lorazepam (i). Functional assays were conducted with HEK293T cells transiently transfected with the receptor and GloSensor cAMP reporter. Concentration-responses were determined under a specific pH condition close to the proton potency at a corresponding receptor, specifically, pH 6.80 for WT, H17A, and H245A; pH 6.10 for H20A; pH 6.60 for H84A and H175A; and pH 6.40 for H89A, H159A, H169A, and H269A. Results were normalized to corresponding basal (1.0, in the absence of modulators) and represented mean (fold of basal) ± SEM from a minimum of 3 assays, each in a quadruplicate set. Curves were analyzed in Prism 8 using the built-in four-parameter logistic or Bell-shaped function. Pharmacological parameters were listed in Table S1.
Figure 8.
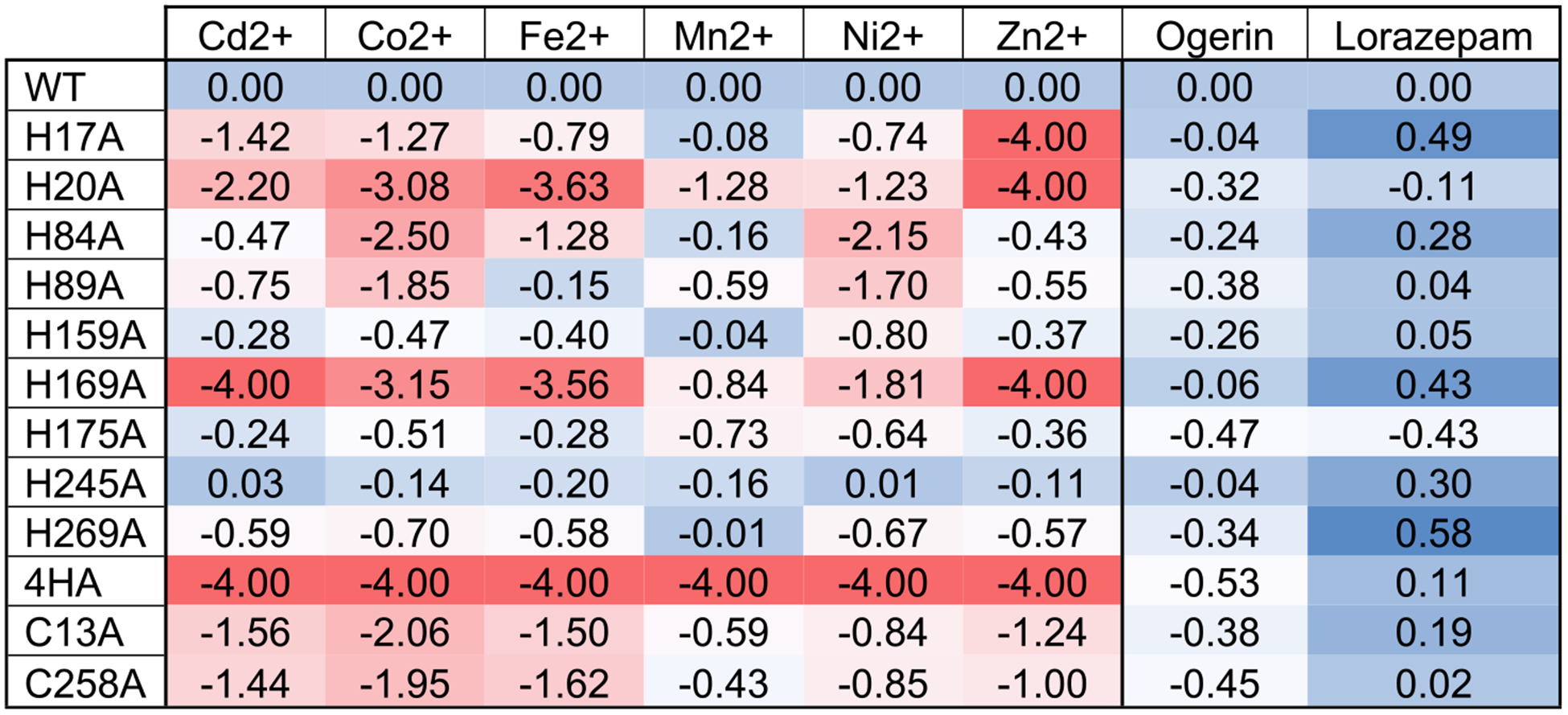
Heat map presentation of effects of Histidine and Cysteine mutations on metal ions and ogerin and lorazepam. For each ligand, the ΔLog(Max/EC50) value between mutant and wild-type (WT) receptors was calculated from the parameters retrieved from the curves in Figure 7. Negative values stand for reduced activity, and positive values stand for enhanced activity. Results were analyzed in an Excel sheet using the built-in conditional formatting function. For the mutant and ligand pair with no observed PAM activity, Cd2+ at H169A, Zn2+ at H17A, H20A, and H169A mutants, and metals at 4HA (H17A + H20A + H84A + H169A), ΔLog(Max/EC50) values could not be calculated. We set the value at the lowest (−4.00) for presentation purposes. Detail parameters were listed in Table S1.
Small-Molecule PAMs Are Minimally Affected by Single Histidine Mutation.
Compared with metal ion modulations, the small molecule PAMs ogerin and lorazepam5 are affected by His mutations to a much smaller extent (Figures 7h, 7i, Figure 8). Furthermore, profiles for ogerin are also different from those for lorazepam. In general, for most His mutations, small reductions are observed for ogerin, but small degrees of enhancement are observed for lorazepam. The His residues (such as H17 and H169) critical for metal ions have very little effect on ogerin (ΔLog(Max/EC50 is −0.04 and −0.06, respectively) but do enhance lorazepam activity by 3.0-and 2.7-fold (ΔLog(Max/EC50 is 0.49 and 0.43, respectively). H20 and H175 are the only mutations with reducing effects on both ogerin and lorazepam. The largest difference between ogerin and lorazepam is observed at TM VII H269A, which reduces ogerin activity by about over 2.2-fold and enhances lorazepam activity by 3.8-fold, leading to an over 8-fold difference between ogerin and lorazepam. In summary, when compared with metal ions, ogerin and lorazepam are less but differently affected by single HA mutations, suggesting extracellular His residues are not directly involved in binding interactions with ogerin or lorazepam.
4HA Mutations Abolish Metal Ion Modulation but Minimally Affect Allosteic Modulation by Ogerin and Lorazepam.
We have shown that H20 and H169 are important for proton sensing, while H17, H20, H84, and H169 are important for metal ion binding and modulation. We next measured metal ion dose-response relationships at the 4HA mutant (H17, H20, H84, H169) under pH 5.70 (pEC50 is 5.61, Table 5). The 4HA mutant becomes resistant to the positive allosteric modulation by any tested metal ions but remains sensitive to the negative modulation by Cd2+ and Zn2+ (Figure 8 and Figures 9a vs 9b). The inhibitory potency of Cd2+ is 204 μM, slightly higher than 71 μM at WT receptors. However, the inhibitory potency of Zn2+ is 8.9 μM, higher than 2.1 μM at a single H169A mutation but lower than H17A (30 μM) or H20A (145 μM) or WT (34 μM) receptors. The results indicate that these His residues are critical for the PAM activity but not responsible for the NAM modulation for Cd2+ or Zn2+. As with the individual HA mutant, the 4HA mutant is still sensitive to ogerin and lorazepam modulation (Figure 8, Figure 9e), displaying slightly reduced activity for ogerin and slightly enhanced activity for lorazepam. This demonstrates that the 4HA mutant is not a dead mutant but defective only to the positive modulation by metal ions. Full concentration-response curves in the absence and presence of increasing concentrations of metal ions (Figure 10) further confirm that the mutant is resistant to metal ion modulation under all assay conditions but does remain active with ogerin and lorazepam with the allosteric activity index ΔLog(αβ/KB) of 4.91 for ogerin and 5.33 for lorazepam (Table 4). These values are slightly reduced for ogerin and not changed for lorazepam as compared with corresponding index values at the GPR68 WT receptor.
Figure 9.
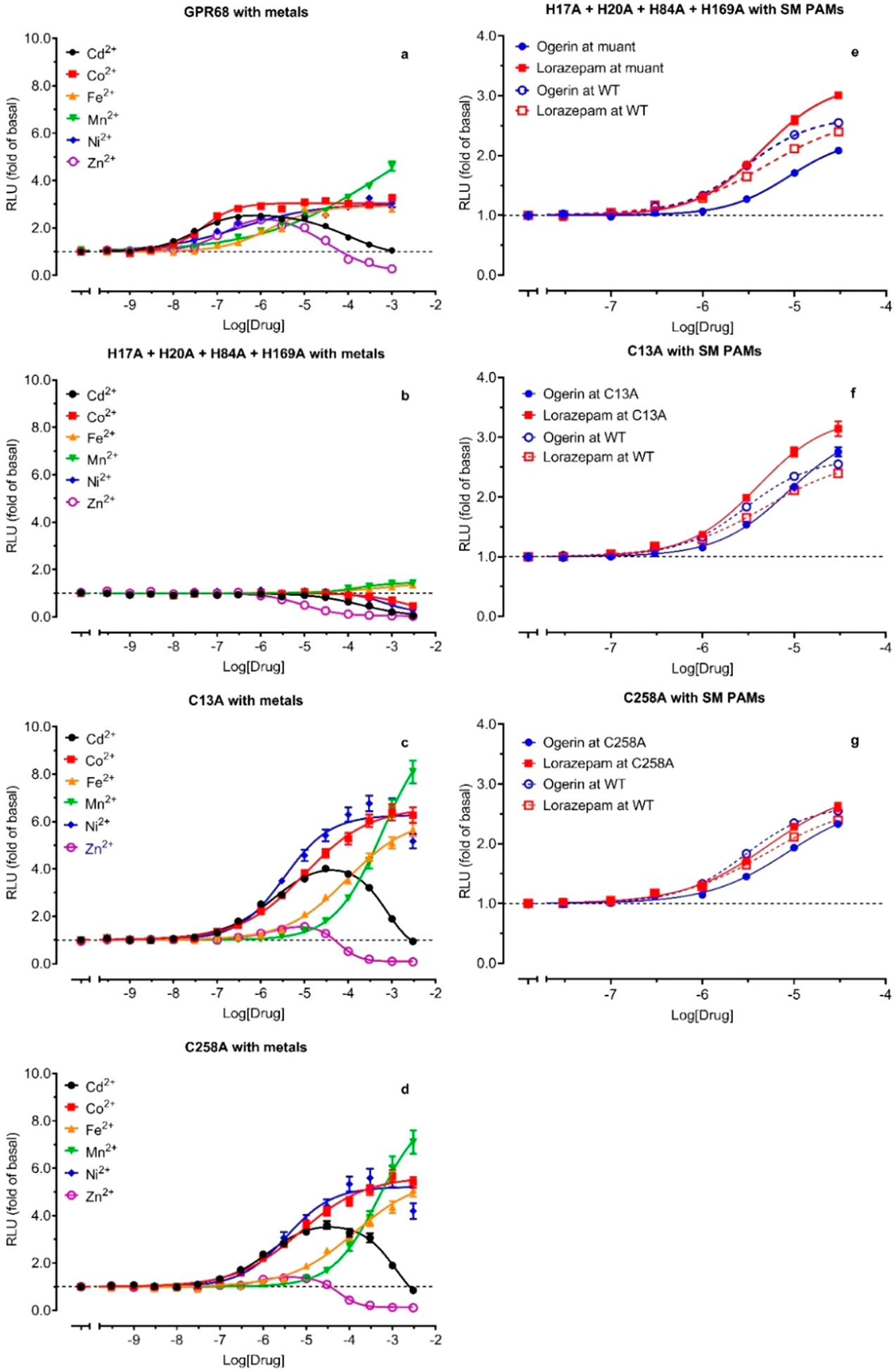
Effects of combined Histidine mutations and single Cysteine mutations on allosteric modulations by metal ions (b, c, d) and ogerin and loraepam (e, f, g). Functional assays were conducted with HEK293T cells transiently transfected with the receptor and GloSensor cAMP reporter. Concentration-responses of metal ions and PAMs were determined under a specific pH condition close to the proton potency at the corresponding receptor, specifically, pH 6.80 for WT, pH 5.70 for 4HA mutant (H17A, H20A, H84A, H169A), and pH 6.00 for C13A and C258A. Results were normalized to corresponding basal (1.0, in the absence of modulators) and represented mean (fold of basal) ± SEM from a minimum of 3 assays, each in a quadruplicate set. Curves were analyzed in Prism 8.4 using the built-in four-parameter logistic or Bell-shaped function. GPR68 wild-type modulation by metal ions (a) were from Figure 7. Pharmacological parameters were listed in Table S1.
Figure 10.
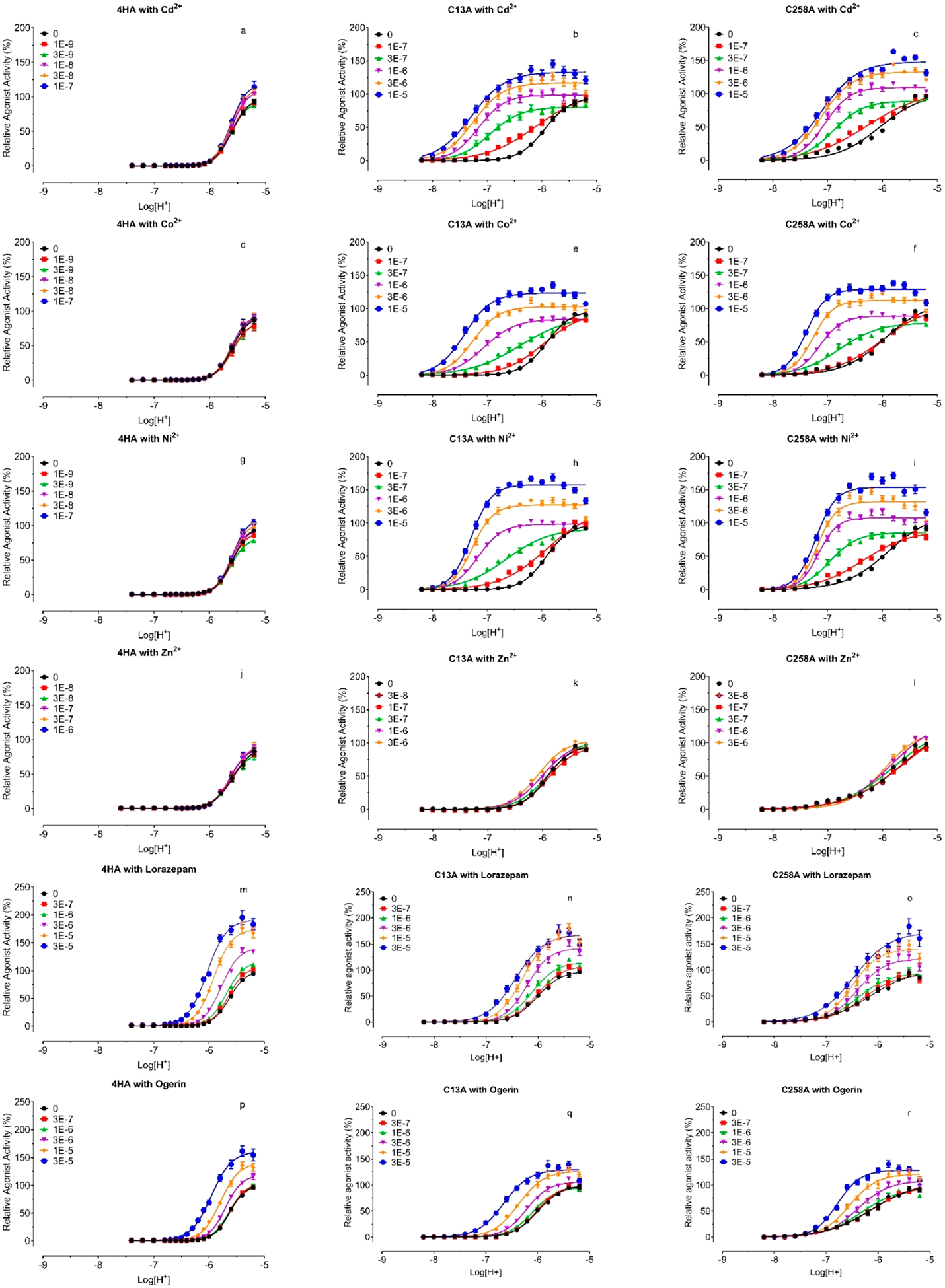
Proton concentration-responses at GPR68 4HA (a, d, g, j, m, and p), C13A (b, e, h, k, n, and q), and C258A (c, f, i, l, o, and r) mutants in the absence and presence of increasing concentrations of Cd2+ (a, d, and g), Co2+ (b, e, and h), Ni2+ (c, f, and I), Zn2+ (j, k, and l), lorazepam (m, n, and o), and ogerin (p, q, and r). Gs-cAMP production was measured in transiently transfected HEK293T cells using the GloSensor cAMP assays. Metal ion stock solutions were made in water. Lorazepam and ogerin stock solutions were made in DMSO and diluted in pH preadjusted Ca2+/Mg2+ free HBSS-based buffers. Results were normalized to proton responses (%) and represented means ± SEM from a minimum of 3 independent assays, each in quadruplicate. Curves were analyzed in Prism 8 using the built-in four-parameter logistic function. The data were also analyzed using the standard allosteric operation model in Prism, and allosteric parameters were listed in Table 3.
Structural Roles of the Disulfide Bond between C13 and C258.
The above results identified extracellular His residues important for proton sensing and metal ion modulation. The 4HA mutant remains to be fully activated by protons with reduced potency (Table 5 and Figure 6) but becomes resistant to positive allosteric modulation by any tested metal ions (Figures 8 and 9), implying that these His residues may form a binding pocket for metal ions. This requires them to be physically close to each other. H84 and H169 could be constrained within proximity through the highly conserved disulfide bond between C94 in TM III and C172 in ECL2. H17 and H20, however, being close to each other in the N-terminus, would need a structural constraint to bring them to be near H84 and/or H169. One possibility is a disulfide bond between C13 (N-Terminal) and C258 (ECL3) (Figure 1a). If this is the case, breaking the disulfide bond would be expected to release the structural constraint and reduce or eliminate metal ion modulation. We therefore first mutated C13 and C258 to Ala and tested mutant receptors for proton-mediated receptor activation. Mutant C13A or C258A has a similar effect on proton activity and reduces proton potency by about 15-fold and proton efficacy by about 50% (Figure 6c and Table 5), a larger effect than any single His mutation. In addition, both C13A and C258A have reduced steepness of the proton response curves as compared with WT or other mutants as indicated by lower Hill slopes (Table 5). These results indicate that C13 and C258 form a disulfide bond which is critical to constrain the N-terminal and ECL3 for potentially coordinated proton binding and receptor activation. Without the disulfide bond, the homotropic cooperativity of proton binding (high Hill values) is reduced. We then examined C13A and C258A for allosteric modulation by metal ions, carried out at a pH of 6.00, close to corresponding proton potency when allosteric activity could be easily observed. As shown in Figures 9c and 9d, C13A and C258A display similar response profiles to metal ions. Compared with GPR68 WT (Figure 9a), C13A and C258A curves are right-shifted by over 16-fold (average of 1.29 log unit for C13A and 1.21 log unit for C258A, Table 4) but with increased efficacy, except for Zn2+, which displays residual PAM activity at C13A (Figure 9c) or C258A (Figure 9d). The bell-shaped concentration-response profile for Cd2+ and Zn2+ remains but with a plateau at 30 μM for Cd2+ and 10 μM for Zn2+ at C13A and C258A vs a plateau at 1 μM at WT (Figures 9c, 9d vs 9a). These results indicate that the C13A or C258A mutant retains the metal ion binding pocket but also that conformational configuration is probably less optimized for metal ion binding. To fully characterize the 4HA (Figures 10a, 10d, 10g, and 10j) and Cys (Figures 10b, 10c, 10e, 10f, 10h, 10i, 10k, and 10l) mutants with metal ions under a wider pH range, we also measured proton concentration-responses in the absence and presence of increasing concentrations of metal ions. We used higher metal ion concentrations for C13A and C258A mutants to compensate for the reduced affinity. As displayed in Figure 10 vs WT results in Figure 5 and quantified in Table 4, the allosteric activity index ΔLog(αβ/KB) is reduced approximately 10-fold as compared with corresponding values at GPR68 WT. Low PAM activity by Zn2+ is visible at C13A or C258A, but the extent of allosteric potentiation is too small to quantify. In addition, Zn2+ turns out to have inhibitory activity at above 1 μM at GPR68 WT, and at above 10 μM at C13A or C258A receptors, measurable PAM signal windows are reduced at mutant receptors when the inhibitory activity starts to dominate between 1 and 10 μM.
As a comparison, we also tested ogerin and lorazepam to determine if allosteric modulation is affected by 4HA mutations (Figures 10m and 10p) or at C13A (Figures 10n and 10q) or C258A (Figures 10o and 10r). Not surprisingly, C13A or C258A remains sensitive to ogerin and lorazepam with small changes in allosteric activity (Figure 8, Figures 9f and 9g), an effect very similar to those seen with single HA mutants (Figure 8). Detailed characterization of allosteric modulation of proton activity by ogerin and lorazepam at the Cys mutants (Figure 10) confirms that ogerin or lorazepam activity is largely unchanged, with a small reduction of Log(αβ/KB) for ogerin and a small increase for lorazepam. These results are consistent with those from single HA or 4HA mutant and indicate that the allosteric binding pocket for ogerin and or lorazepam is probably not altered by C13A or C258A mutation.
A Model Explains GPR68’s Histidine Moieties.
Based on the above results, we developed a model to highlight important and differential roles of extracellular His residues of GPR68 in both proton binding and metal ion allosteric modulation (Figure 11). Two disulfide bonds, one highly conserved between C94 in TM III and C172 in ECL2 and another relatively less conserved between C13 in the N-terminus and C258 in ECL3, bring and constrain the 4 extracellular residues (H17, H20, H84, and H169) close to each other in space and form a coordinated complex with metal ions (Zn2+ in this case as an example). Complexing with Zn2+ could constrain these His residues in a tetradentate configuration and stabilize local receptor conformations. Depending on the surrounding microenvironment, His residues may have changed pKa. Since H17 and H84 are not important for proton activity, we propose that their corresponding pKa values are reduced and may not be protonated even at reduced pH; therefore, they will maintain interactions with Zn2+ ion. While H20 and H169 are both important for proton activity, they may have increased pKa values and therefore would facilitate proton binding and receptor activation under high pH ranges. Mutation of one or two of these 4 His residues would destabilize the tetradentate complex with Zn2+, resulting in a mutant insensitive to metal ion modulation. Protonated H20 and H169 residues break away from complexing with Zn2+ (Figure 11, right). The positively charged H169 forms interactions with a nearby D85. These regional changes, triggered by protonation on H20 and H169, initiate receptor conformational changes and the activation process. In the absence of metal ions (such as Ca2+/Mg2+-free HBSS buffers), these His residues are still close to each in space but may not be in the same configuration as in the presence of a metal ion. Therefore, proton binding (His protonation) could not occur until pH reaches lower points (such as below pH 6.8). Further mutational and functional studies are needed to examine D85, as well as other charged residues and aromatic residues in extracellular regions of the GPR68 in proton activity and metal ion modulation.
Figure 11.
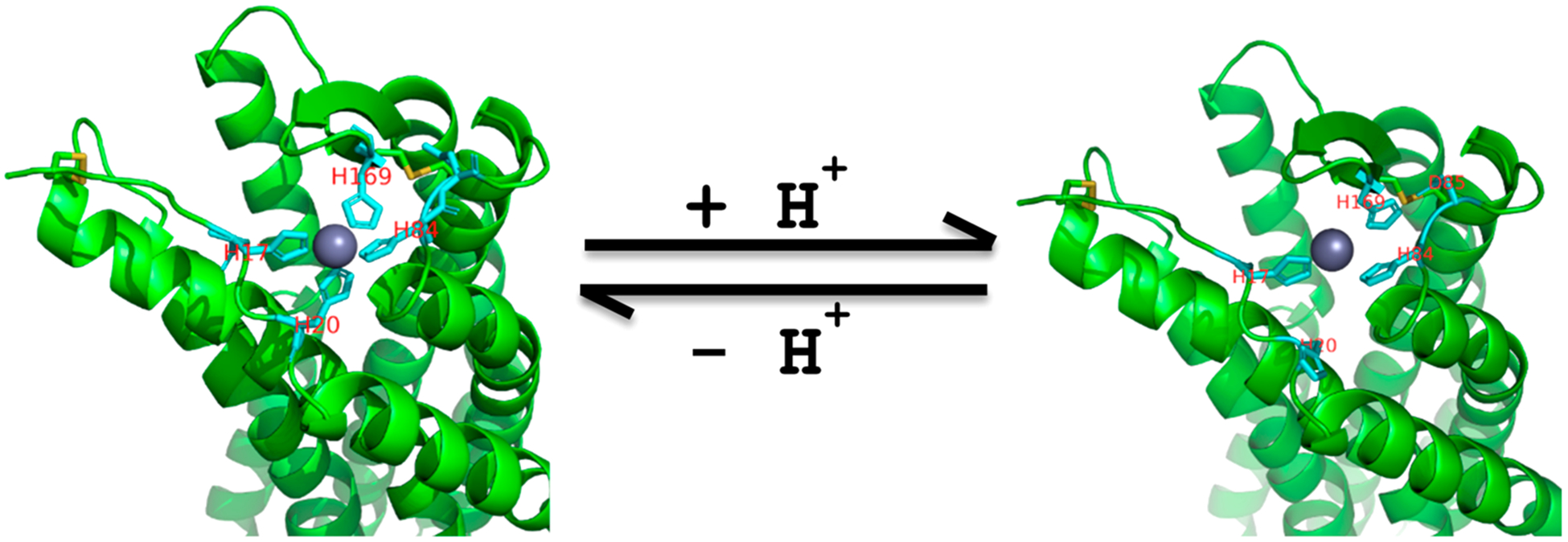
Extracellular divalent metal ion binding models. Left, H17, H20, H84, and H169 of GPR68 form tetradentate coordination with Zn2+ under high pH conditions. Right, protonated H20 breaks away, and protonated H169 breaks away and forms interactions with nearby D85. Two disulfide bonds (in yellow) help constrain the Zn2+ binding site.
DISCUSSION
In this paper we show that GPR68 extracellular His residues play differential roles in interconnected proton-sensing and metal ion modulation. GPR68 contains seven extracellular His residues and two TM His residues, and its activation by protons is allosterically modulated by divalent metal ions. We systematically mutated all these His residues to either Ala or Phe to eliminate their capacity to become protonated under acidic conditions, individually and/or in combination, and performed functional assays to study proton activity and the allosteric modulation by divalent metal ions. Our results revealed that proton activity and metal ion modulation are affected by individual His mutations in differential ways and to varying degrees. In combination, multiple H → A mutations are able to block proton activity or render resistance to allosteric modulation by any tested metal ions. We also revealed a second disulfide bond between C13 and C258. Together with the highly conserved disulfide bond of C94 and C172, they could play an important structural role for both proton activity and metal ion modulation. Breaking the C13-C258 disulfide bond severely reduced proton activity, proton homotropic cooperativity (Hill slope), and allosteric activity of metal ions. In contrast, small molecule PAMs (ogerin or lorazepam) are minimally affected by either single or combined mutations. GPR68 appears to be allosterically modulated by two independent mechanisms: one by divalent metal ions in extracellular regions and another by small-molecule modulators in TM domains.
We showed that GPR68 has steep proton concentration-response curves. This feature is consistent with the fact that GPR68 plays an important role in pH homeostasis, which requires the receptor to respond quickly and efficiently to small extracellular pH changes. Mutational assays indicate that N-terminal H20 and ECL2 H169, individually, are more important than the other His residues in proton activity. While mutants with combined His mutations displayed gradually reduced proton activity, a mutant with all 7 extracellular His residues mutated to Ala displayed over 300-fold reduced proton activity, indicating that GPR68 requires multiple His residues to be protonated in a coordinated manner for full activation. This is consistent with the findings by Ludwig and colleagues,4 although we measured a different signaling activity (cAMP production). Comparison with the previous study4 revealed more similarities than differences. H17F mutation right-shifted proton potency in IP accumulation, while H17A or H17F in our assays had little effect on proton-mediated cAMP production. H20F mutation resulted in a big reduction in IP accumulation, consistent with our results with H20A or H20F mutation for cAMP production. TM VI H245F or TM VII H269F mutation led to a severe reduction in IP accumulation, also consistent with our observation with H245F or H269F on cAMP production. However, we found the H245A or H269A mutation resulted in little or a moderate change in proton activity. The discrepancy (H245F vs H245A or H269F vs H269A) was not observed with any single extracellular His mutation. Therefore, the different functional results with HF vs HA, depending on where the residue is located (extracellular regions or TM domains), suggest that TM His residues are probably not the sites for protonation but may play a more important structural role.
Metal ions are well-known allosteric modulators for many GPCRs.39,40 Melanocortin-4 receptor (MC4),41 adrenergic α1A,42 GPR83,43 and 5-HT744 are sensitive to Zn2+ and or Cu2+, while GPR39 is modulated by Zn2+ and Ni2+ ions45–47 and is referred to as a Zn2+-receptor.48,49 At GPR68, divalent metal ions (Cd2+, Co2+, Cu2+, Fe2+, Mn2+, Ni2+, and Zn2+) potentiate proton activity, especially under mildly acidic conditions. GPR68 is a rare GPCR, to our knowledge, that can be allosterically modulated by these aforementioned divalent metal ions. Therefore, it is important to ensure that assay buffers do not contain residual divalent metal ions.
Abe-Ohya and colleagues29 reported that metal ions (Co2+, Fe2+, Mn2+, Ni2+, and Zn2+) activate GPR68-mediated Gq-IP accumulation. These assays were carried out at “neutral” pH conditions (probably pH 7.4). If compared with our results at a single pH 7.40 point and by measuring cAMP production, their results are consistent with ours, namely that these metal ions activate GPR68 in a concentration-dependent manner. Other reports30,50 using SRE-luciferase reporter assays failed to confirm metal ions, such as Ni2+, Fe2+, and Zn2+, as “agonists” at GPR68 under multiple pH conditions. Several factors could be responsible for the discrepancy. Specifically, SRE-Luciferase reporter activity is mainly downstream of G12/13-protein activation, needs a longer incubation time than Gq-IP accumulation or cAMP production, and is less sensitive than cAMP assays. In addition, the pH stability control in the presence of CO2 and at 37 °C for a long period of incubation could be challenging. To determine if extracellular His residues are important for metal ion activity, previous studies used mutant GPR68 receptors that might not be activated by proton stimulation. Specifically, H245F and a mutant with 5 HF mutations (H17F + H20F + H84F + H169F + H269F) showed no activity to metal ions, but their responses to reduced pH conditions were not examined.29,30 If it is assumed that metal ions are agonists independent of protons, this supports the conclusion that extracellular His residues are critical for metal ions (since they failed to activate the His mutant receptors). However, these metal ions are allosteric modulators for protons. Thus, their activity is dependent on proton activity, and the characterization of metal ion allosteric activity requires mutants that are still responsive to protons.
The metal ions Co2+, Cu2+, Fe2+, Mn2+, Ni2+, and Zn2+ are all essential metals playing critical roles in a wide range of physiological processes, and deficiency or overdose of these essential metal ions is implicated in numerous disorders, especially neurological conditions.51–53 Among them, Zn2+ is probably the most important essential metal ion, cosecreted with glutamate into synapses and with insulin from β-cells, and functions as a neuromodulator and a signaling messenger.54–58 High micromolar Co2+ has been widely used as a chemical inducer of hypoxia in vitro models.59 Cd2+, a major environmental contaminant, is a neurotoxic, and carcinogenic heavy metal and accumulation in the human body may be related to a variety of different diseases.60,61 Our study revealed that all these metal ions could potentiate proton activity at GPR68 at normal physiological pH conditions at concentrations as low as 30 nM, an extracellular level that may be attained in the synapses of the brain. More importantly, since they are PAMs at low concentrations, their activity could be dramatically enhanced under mildly acidic conditions, which are usually associated with neurological disorders or neuroinflammatory conditions. At high micromolar concentrations, Cd2+ and Zn2+ also have NAM activity. Bell-shaped biphasic responses of Zn2+ were previously observed at 5-HT1A binding assays with peak stimulation at about 10 μM62 as well as Glycine receptors with peak responses at 1 to 10 μM and may be associated with high and low-affinity Zn2+ sites.63,64 Compared with GPR39, a so-called Zn2+-sensing GPCR, GPR68 is much more sensitive with a potency of 209 nM under pH 7.40 vs 81 μM for the Gq-IP pathway and 380 μM for the Gs-cAMP pathway at GPR39.46 GPR68 is also a sensitive GPCR sensor for other divalent metal ions, especially Cd2+, Co2+, and Ni2+ with a potency of 50, 72, and 123 nM, respectively, under pH 7.40. These ions become more potent when pH drops to 7.00–6.80. GPR68 is highly expressed in the brain and pancreas; therefore, PAM or NAM activity of divalent metal ions at GPR68 under neutral or mildly acid conditions is physiologically and pathophysiologically relevant.
Our functional results support the existence of an additional but less conserved disulfide bond between C13 and C258 of GPR68. This less conserved disulfide bond in the GPCR family also exists in structures of CXCR4,65 CCR5,66 AT1,67,68 AT2,69,70 P2Y1,71 P2Y12,72,73 CysLT1,74 and CysLT275 receptors. Together with the highly conserved disulfide bond in the GPCR family between TM III and ECL2 (C94 and C172, respectively, in this case), the two disulfide bonds bring N-terminal H17 and H20, ECL2 H84, and ECL3 H169 within proximity to coordinate tetradentate interactions with a metal ion (Zn2+ in this case) under high pH conditions. H20 and H169 (but not H17 or H84) are protonated at above neutral pH conditions. Protonated His residues break away from interactions with Zn2+ and form new interactions with nearby residues. These dynamic interactions, initiated by protonation on H20 and H169, trigger conformational changes and receptor activation processes. Without a Zn2+ forming complex with these His residues, H20 and H169 will not be protonated until well below neutral pH conditions. It is worth pointing out that Zn2+ binding is observed in the extracellular regions of several crystal structures of 7TM receptors,76 especially in the PAF receptor crystal structure (PDB code 5ZKQ) where tight tetradentate interactions of Zn2+ with H8, E259, H268, and N272 were well-defined.77 In addition, in a β2 structure (PDB code 6WXT), E107 and H172 form an extracellular binding site for a Ni2+ ion.78 Our current model may not be applicable to all metal ions or explain all observed activity in this study but could serve as a starting template for further optimizations. Further mutational and pharmacological studies are needed to identify (i) potential interaction partners for His residues (such as H20 and H169) after protonation, (ii) other residues that may coordinate with metal ions, and (iii) residues for the putative second metal-binding site and their potential roles in NAM activity. Before GPR68 structures become available, homology models are essential tools for docking and virtual screening. Revealing molecular mechanisms by which GPR68 is activated by protons and allosterically modulated by divalent metal ions would facilitate designing mechanism-based selective ligands for GPR68, especially when available small molecule ligands are limited and none of them produces antagonism.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Rodger Zuo and Violetta Weinreb for plasmid preparation.
Funding
The project was supported by NIMH R21 MH120422-02 (X.P.H. and T.P.K.), The National Institute of Mental Health Psychoactive Drug Screening Program (NIMH PDSP) (X.P.H., T.P.K., and B.L.R.), and NIH R35 GM122481 (S.G. and B.K.S.). The NIMH PDSP is directed by B.L.R., MD, PhD, at the University of North Carolina at Chapel Hill, Chapel Hill, NC, and Project Officer Jamie Driscoll at NIMH, Bethesda, MD, USA.
ABBREVIATIONS
- GPCR
G-protein coupled receptor
- PAM
positive allosteric modulator
- NAM
negative allosteric modulator
- HBSS
Hanks’ balanced salt solution
- ECL1
the first extracellular loop
- ECL2
the second extracellular loop
- ECL3
the third extracellular loop
- TM
transmembrane
Footnotes
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.0c00576
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.0c00576.
Supplementary Table 1, pharmacological parameters of metal ions and small molecule PAMs at GPR68 WT and mutant receptors; Supplementary Figure 1, metal ion concentration response curves at proton-sensing GPCRs under different pH conditions (PDF)
Accession Codes
GPR68: Q15743 (OGR1_Human)
REFERENCES
- (1).Xu Y, and Casey G (1996) Identification of human OGR1, a novel G protein-coupled receptor that maps to chromosome 14. Genomics 35, 397–402. [DOI] [PubMed] [Google Scholar]
- (2).Regard JB, Sato IT, and Coughlin SR (2008) Anatomical profiling of G protein-coupled receptor expression. Cell 135, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Herzig M, Dasgupta P, Kaemmerer D, Sänger J, Evert K, Schulz S, and Lupp A (2019) Comprehensive assessment of GPR68 expression in normal and neoplastic human tissues using a novel rabbit monoclonal antibody. Int. J. Mol. Sci 20 (21), 5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ludwig M-G, Vanek M, Guerini D, Gasser JA, Jones CE, Junker U, Hofstetter H, Wolf RM, and Seuwen K (2003) Proton-sensing G-protein-coupled receptors. Nature 425, 93–98. [DOI] [PubMed] [Google Scholar]
- (5).Huang X-P, Karpiak J, Kroeze WK, Zhu H, Chen X, Moy SS, Saddoris KA, Nikolova VD, Farrell MS, Wang S, Mangano TJ, Deshpande DA, Jiang A, Penn RB, Jin J, Koller BH, Kenakin T, Shoichet BK, and Roth BL (2015) Allosteric ligands for the pharmacologically dark receptors GPR68 and GPR65. Nature 527, 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Seuwen K, Ludwig M-G, and Wolf RM (2006) Receptors for protons or lipid messengers or both? J. Recept. Signal Transduction Res 26, 599–610. [DOI] [PubMed] [Google Scholar]
- (7).Yang L, Sanderlin E, Justus C, and Krewson E (2015) Emerging roles for the pH-sensing G protein-coupled receptors in response to acidotic stress. Cell Health Cytoskeleton 7, 99–109. [Google Scholar]
- (8).Alexander SPH, Christopoulos A, Davenport AP, Kelly E, Mathie A, Peters JA, Veale EL, Armstrong JF, Faccenda E, Harding SD, Pawson AJ, Sharman JL, Southan C, Davies JA, and CGTP Collaborators (2019) THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: G protein-coupled receptors. Br. J. Pharmacol 176 (Suppl1), S21–S141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Li J, Guo B, Wang J, Cheng X, Xu Y, and Sang J (2013) Ovarian cancer G protein coupled receptor 1 suppresses cell migration of MCF7 breast cancer cells via a Gα12/13-Rho-Rac1 pathway. J. Mol. Signaling 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Singh LS, Berk M, Oates R, Zhao Z, Tan H, Jiang Y, Zhou A, Kirmani K, Steinmetz R, Lindner D, and Xu Y (2007) Ovarian cancer G protein-coupled receptor 1, a new metastasis suppressor gene in prostate cancer. J. Natl. Cancer Inst 99, 1313–1327. [DOI] [PubMed] [Google Scholar]
- (11).Wiley SZ, Sriram K, Salmerón C, and Insel PA (2019) GPR68: an emerging drug target in cancer. Int. J. Mol. Sci 20 (3), 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Saxena H, Deshpande DA, Tiegs BC, Yan H, Battafarano RJ, Burrows WM, Damera G, Panettieri RA, Dubose TD, An SS, and Penn RB (2012) The GPCR OGR1 (GPR68) mediates diverse signalling and contraction of airway smooth muscle in response to small reductions in extracellular pH. Br. J. Pharmacol 166, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wiley SZ, Sriram K, Liang W, Chang SE, French R, McCann T, Sicklick J, Nishihara H, Lowy AM, and Insel PA (2018) GPR68, a proton-sensing GPCR, mediates interaction of cancer-associated fibroblasts and cancer cells. FASEB J. 32, 1170–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ichimonji I, Tomura H, Mogi C, Sato K, Aoki H, Hisada T, Dobashi K, Ishizuka T, Mori M, and Okajima F (2010) Extracellular acidification stimulates IL-6 production and Ca(2+) mobilization through proton-sensing OGR1 receptors in human airway smooth muscle cells. Am. J. Physiol. Lung Cell Mol. Physiol 299, L567–77. [DOI] [PubMed] [Google Scholar]
- (15).Xu J, Mathur J, Vessières E, Hammack S, Nonomura K, Favre J, Grimaud L, Petrus M, Francisco A, Li J, Lee V, Xiang F-L, Mainquist JK, Cahalan SM, Orth AP, Walker JR, Ma S, Lukacs V, Bordone L, Bandell M, Laffitte B, Xu Y, Chien S, Henrion D, and Patapoutian A (2018) GPR68 senses flow and is essential for vascular physiology. Cell 173, 762–775.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Nakakura T, Mogi C, Tobo M, Tomura H, Sato K, Kobayashi M, Ohnishi H, Tanaka S, Wayama M, Sugiyama T, Kitamura T, Harada A, and Okajima F (2012) Deficiency of proton-sensing ovarian cancer G protein-coupled receptor 1 attenuates glucose-stimulated insulin secretion. Endocrinology 153, 4171–4180. [DOI] [PubMed] [Google Scholar]
- (17).Mogi C, Nakakura T, and Okajima F (2014) Role of extracellular proton-sensing OGR1 in regulation of insulin secretion and pancreatic β-cell functions. Endocr. J 61, 101–110. [DOI] [PubMed] [Google Scholar]
- (18).de Vallière C, Wang Y, Eloranta JJ, Vidal S, Clay I, Spalinger MR, Tcymbarevich I, Terhalle A, Ludwig M-G, Suply T, Fried M, Kullak-Ublick GA, Frey-Wagner I, Scharl M, Seuwen K, Wagner CA, and Rogler G (2015) G Protein-coupled pH-sensing Receptor OGR1 Is a Regulator of Intestinal Inflammation. Inflamm. Bowel Dis 21, 1269–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Okajima F (2013) Regulation of inflammation by extracellular acidification and proton-sensing GPCRs. Cell. Signalling 25, 2263–2271. [DOI] [PubMed] [Google Scholar]
- (20).Chandra V, Karamitri A, Richards P, Cormier F, Ramond C, Jockers R, Armanet M, Albagli-Curiel O, and Scharfmann R (2016) Extracellular acidification stimulates GPR68 mediated IL-8 production in human pancreatic β cells. Sci. Rep 6, 25765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Krieger NS, Yao Z, Kyker-Snowman K, Kim MH, Boyce BF, and Bushinsky DA (2016) Increased bone density in mice lacking the proton receptor OGR1. Kidney Int. 89, 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Parry DA, Smith CEL, El-Sayed W, Poulter JA, Shore RC, Logan CV, Mogi C, Sato K, Okajima F, Harada A, Zhang H, Koruyucu M, Seymen F, Hu JC-C, Simmer JP, Ahmed M, Jafri H, Johnson CA, Inglehearn CF, and Mighell AJ (2016) Mutations in the pH-Sensing G-protein-Coupled Receptor GPR68 Cause Amelogenesis Imperfecta. Am. J. Hum. Genet 99, 984–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Schneider JW, Goetsch SC, Leng X, Ludwig SM, Russell JL, Yang C-P, and Zhang Q-J (2012) Coupling hippocampal neurogenesis to brain pH through proneurogenic small molecules that regulate proton sensing G protein-coupled receptors. ACS Chem. Neurosci 3, 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Mohebbi N, Benabbas C, Vidal S, Daryadel A, Bourgeois S, Velic A, Ludwig M-G, Seuwen K, and Wagner CA (2012) The proton-activated G protein coupled receptor OGR1 acutely regulates the activity of epithelial proton transport proteins. Cell. Physiol. Biochem 29, 313–324. [DOI] [PubMed] [Google Scholar]
- (25).Imenez Silva PH, Katamesh-Benabbas C, Chan K, Pastor Arroyo EM, Knöpfel T, Bettoni C, Ludwig M-G, Gasser JA, Brandao-Burch A, Arnett TR, Bonny O, Seuwen K, and Wagner CA (2020) The proton-activated ovarian cancer G protein-coupled receptor 1 (OGR1) is responsible for renal calcium loss during acidosis. Kidney Int. 97 (5), 920–933. [DOI] [PubMed] [Google Scholar]
- (26).He X, Feng S, Hawkins C, Lawley L, Fan W, Xu Y, Zha X-M, and Fang J (2020) G protein-coupled receptor 68 increases the number of B lymphocytes. Am. J. Blood Res 10, 15–21. [PMC free article] [PubMed] [Google Scholar]
- (27).Yu X, Huang X-P, Kenakin TP, Slocum ST, Chen X, Martini ML, Liu J, and Jin J (2019) Design, Synthesis, and Characterization of Ogerin-Based Positive Allosteric Modulators for G Protein-Coupled Receptor 68 (GPR68). J. Med. Chem 62, 7557–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Foster SR, Hauser AS, Vedel L, Strachan RT, Huang X-P, Gavin AC, Shah SD, Nayak AP, Haugaard-Kedström LM, Penn RB, Roth BL, Bräuner-Osborne H, and Gloriam DE (2019) Discovery of Human Signaling Systems: Pairing Peptides to G Protein-Coupled Receptors. Cell 179, 895–908.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Abe-Ohya R, Ishikawa T, Shiozawa H, Suda K, and Nara F (2015) Identification of metals from osteoblastic ST-2 cell supernatants as novel OGR1 agonists. J. Recept. Signal Transduction Res 35, 485–492. [DOI] [PubMed] [Google Scholar]
- (30).Negishi J, Omori Y, Shindo M, Takanashi H, Musha S, Nagayama S, Hirayama J, Nishina H, Nakakura T, Mogi C, Sato K, Okajima F, Mochimaru Y, and Tomura H (2017) Manganese and cobalt activate zebrafish ovarian cancer G-protein-coupled receptor 1 but not GPR4. J. Recept. Signal Transduction Res 37, 401–408. [DOI] [PubMed] [Google Scholar]
- (31).Webb B, and Sali A (2016) Comparative protein structure modeling using MODELLER. Curr. Protoc Bioinformatics 54, 5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Kenakin T (2017) A scale of agonism and allosteric modulation for assessment of selectivity, bias, and receptor mutation. Mol. Pharmacol 92, 414–424. [DOI] [PubMed] [Google Scholar]
- (33).Ehlert FJ (2005) Analysis of allosterism in functional assays. J. Pharmacol. Exp. Ther 315, 740–754. [DOI] [PubMed] [Google Scholar]
- (34).Price MR, Baillie GL, Thomas A, Stevenson LA, Easson M, Goodwin R, McLean A, McIntosh L, Goodwin G, Walker G, Westwood P, Marrs J, Thomson F, Cowley P, Christopoulos A, Pertwee RG, and Ross RA (2005) Allosteric modulation of the cannabinoid CB1 receptor. Mol. Pharmacol 68, 1484–1495. [DOI] [PubMed] [Google Scholar]
- (35).Kenakin T (2005) New concepts in drug discovery: collateral efficacy and permissive antagonism. Nat. Rev. Drug Discovery 4, 919–927. [DOI] [PubMed] [Google Scholar]
- (36).Bektas M, Barak LS, Jolly PS, Liu H, Lynch KR, Lacana E, Suhr K-B, Milstien S, and Spiegel S (2003) The G protein-coupled receptor GPR4 suppresses ERK activation in a ligand-independent manner. Biochemistry 42, 12181–12191. [DOI] [PubMed] [Google Scholar]
- (37).Tobo M, Tomura H, Mogi C, Wang J-Q, Liu J-P, Komachi M, Damirin A, Kimura T, Murata N, Kurose H, Sato K, and Okajima F (2007) Previously postulated “ligand-independent” signaling of GPR4 is mediated through proton-sensing mechanisms. Cell. Signalling 19, 1745–1753. [DOI] [PubMed] [Google Scholar]
- (38).Martin AL, Steurer MA, and Aronstam RS (2015) Constitutive Activity among Orphan Class-A G Protein Coupled Receptors. PLoS One 10 (9), No. e0138463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).van der Westhuizen ET, Valant C, Sexton PM, and Christopoulos A (2015) Endogenous allosteric modulators of G protein-coupled receptors. J. Pharmacol. Exp. Ther 353, 246–260. [DOI] [PubMed] [Google Scholar]
- (40).Ananthanarayanan VS, and Kerman A (2006) Role of metal ions in ligand-receptor interaction: insights from structural studies. Mol. Cell. Endocrinol 246, 53–59. [DOI] [PubMed] [Google Scholar]
- (41).Link R, Veiksina S, Tahk M-J, Laasfeld T, Paiste P, Kopanchuk S, and Rinken A (2020) The constitutive activity of melanocortin-4 receptors in cAMP pathway is allosterically modulated by zinc and copper ions. J. Neurochem 153 (3), 346–361. [DOI] [PubMed] [Google Scholar]
- (42).Ciolek J, Maïga A, Marcon E, Servent D, and Gilles N (2011) Pharmacological characterization of zinc and copper interaction with the human alpha(1A)-adrenoceptor. Eur. J. Pharmacol 655, 1–8. [DOI] [PubMed] [Google Scholar]
- (43).Müller A, Kleinau G, Piechowski CL, Müller TD, Finan B, Pratzka J, Grüters A, Krude H, Tschöp M, and Biebermann H (2013) G-protein coupled receptor 83 (GPR83) signaling determined by constitutive and zinc(II)-induced activity. PLoS One 8, No. e53347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Satała G, Duszyńska B, Lenda T, Nowak G, and Bojarski AJ (2018) Allosteric Inhibition of Serotonin 5-HT7 Receptors by Zinc Ions. Mol. Neurobiol 55, 2897–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Holst B, Egerod KL, Schild E, Vickers SP, Cheetham S, Gerlach L-O, Storjohann L, Stidsen CE, Jones R, Beck-Sickinger AG, and Schwartz TW (2007) GPR39 signaling is stimulated by zinc ions but not by obestatin. Endocrinology 148, 13–20. [DOI] [PubMed] [Google Scholar]
- (46).Sato S, Huang X-P, Kroeze WK, and Roth BL (2016) Discovery and characterization of novel GPR39 agonists allosterically modulated by zinc. Mol. Pharmacol 90, 726–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Storjohann L, Holst B, and Schwartz TW (2008) Molecular mechanism of Zn2+ agonism in the extracellular domain of GPR39. FEBS Lett. 582, 2583–2588. [DOI] [PubMed] [Google Scholar]
- (48).Sunuwar L, Gilad D, and Hershfinkel M (2017) The zinc sensing receptor, ZnR/GPR39, in health and disease. Front. Biosci., Landmark Ed 22, 1469–1492. [DOI] [PubMed] [Google Scholar]
- (49).Młyniec K, Singewald N, Holst B, and Nowak G (2015) GPR39 Zn(2+)-sensing receptor: a new target in antidepressant development? J. Affective Disord 174, 89–100. [DOI] [PubMed] [Google Scholar]
- (50).Mochimaru Y, Negishi J, Murakami S, Musha S, Sato K, Okajima F, and Tomura H (2018) Metals Differentially Activate Ovarian Cancer G Protein-Coupled Receptor 1 in Various Species. Zool. Sci 35, 109–114. [DOI] [PubMed] [Google Scholar]
- (51).Maret W (2016) The metals in the biological periodic system of the elements: concepts and conjectures. Int. J. Mol. Sci 17 (1), 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Zoroddu MA, Aaseth J, Crisponi G, Medici S, Peana M, and Nurchi VM (2019) The essential metals for humans: a brief overview. J. Inorg. Biochem 195, 120–129. [DOI] [PubMed] [Google Scholar]
- (53).Mezzaroba L, Alfieri DF, Colado Simão AN, and Vissoci Reiche EM (2019) The role of zinc, copper, manganese and iron in neurodegenerative diseases. NeuroToxicology 74, 230–241. [DOI] [PubMed] [Google Scholar]
- (54).Blakemore LJ, and Trombley PQ (2017) Zinc as a Neuromodulator in the Central Nervous System with a Focus on the Olfactory Bulb. Front. Cell. Neurosci 11 (297), 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Sensi SL, Paoletti P, Koh J-Y, Aizenman E, Bush AI, and Hershfinkel M (2011) The neurophysiology and pathology of brain zinc. J. Neurosci 31, 16076–16085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Maret W (2017) Zinc in cellular regulation: the nature and significance of “zinc signals. Int. J. Mol. Sci 18, 2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Maret W (2017) Zinc in pancreatic islet biology, insulin sensitivity, and diabetes. Prev. Nutr. Food Sci 22, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Besser L, Chorin E, Sekler I, Silverman WF, Atkin S, Russell JT, and Hershfinkel M (2009) Synaptically released zinc triggers metabotropic signaling via a zinc-sensing receptor in the hippocampus. J. Neurosci 29, 2890–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Muñoz-Sánchez J, and Chánez-Cárdenas ME (2019) The use of cobalt chloride as a chemical hypoxia model. J. Appl. Toxicol 39, 556–570. [DOI] [PubMed] [Google Scholar]
- (60).Rani A, Kumar A, Lal A, and Pant M (2014) Cellular mechanisms of cadmium-induced toxicity: a review. Int. J. Environ. Health Res 24, 378–399. [DOI] [PubMed] [Google Scholar]
- (61).Wang B, and Du Y (2013) Cadmium and its neurotoxic effects. Oxid. Med. Cell. Longevity 2013, 898034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Satała G, Duszyńska B, Stachowicz K, Rafalo A, Pochwat B, Luckhart C, Albert PR, Daigle M, Tanaka KF, Hen R, Lenda T, Nowak G, Bojarski AJ, and Szewczyk B (2016) Concentration-Dependent Dual Mode of Zn Action at Serotonin 5-HT1A Receptors: In Vitro and In Vivo Studies. Mol. Neurobiol 53, 6869–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Bloomenthal AB, Goldwater E, Pritchett DB, and Harrison NL (1994) Biphasic modulation of the strychnine-sensitive glycine receptor by Zn2+. Mol. Pharmacol 46, 1156–1159. [PubMed] [Google Scholar]
- (64).Laube B, Kuhse J, Rundström N, Kirsch J, Schmieden V, and Betz H (1995) Modulation by zinc ions of native rat and recombinant human inhibitory glycine receptors. J. Physiol. (Oxford, U. K.) 483 (Pt 3), 613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Wu B, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, and Stevens RC (2010) Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, Li T, Ma L, Fenalti G, Li J, Zhang W, Xie X, Yang H, Jiang H, Cherezov V, Liu H, Stevens RC, Zhao Q, and Wu B (2013) Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science 341, 1387–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Zhang H, Unal H, Gati C, Han GW, Liu W, Zatsepin NA, James D, Wang D, Nelson G, Weierstall U, Sawaya MR, Xu Q, Messerschmidt M, Williams GJ, Boutet S, Yefanov OM, White TA, Wang C, Ishchenko A, Tirupula KC, Desnoyer R, Coe J, Conrad CE, Fromme P, Stevens RC, Katritch V, Karnik SS, and Cherezov V (2015) Structure of the Angiotensin receptor revealed by serial femtosecond crystallography. Cell 161, 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Wingler LM, McMahon C, Staus DP, Lefkowitz RJ, and Kruse AC (2019) Distinctive activation mechanism for angiotensin receptor revealed by a synthetic nanobody. Cell 176, 479–490.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Zhang H, Han GW, Batyuk A, Ishchenko A, White KL, Patel N, Sadybekov A, Zamlynny B, Rudd MT, Hollenstein K, Tolstikova A, White TA, Hunter MS, Weierstall U, Liu W, Babaoglu K, Moore EL, Katz RD, Shipman JM, Garcia-Calvo M, Sharma S, Sheth P, Soisson SM, Stevens RC, Katritch V, and Cherezov V (2017) Structural basis for selectivity and diversity in angiotensin II receptors. Nature 544, 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Asada H, Horita S, Hirata K, Shiroishi M, Shiimura Y, Iwanari H, Hamakubo T, Shimamura T, Nomura N, Kusano-Arai O, Uemura T, Suno C, Kobayashi T, and Iwata S (2018) Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog. Nat. Struct. Mol. Biol 25, 570–576. [DOI] [PubMed] [Google Scholar]
- (71).Zhang D, Gao Z-G, Zhang K, Kiselev E, Crane S, Wang J, Paoletta S, Yi C, Ma L, Zhang W, Han GW, Liu H, Cherezov V, Katritch V, Jiang H, Stevens RC, Jacobson KA, Zhao Q, and Wu B (2015) Two disparate ligand-binding sites in the human P2Y1 receptor. Nature 520, 317–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Zhang J, Zhang K, Gao Z-G, Paoletta S, Zhang D, Han GW, Li T, Ma L, Zhang W, Müller CE, Yang H, Jiang H, Cherezov V, Katritch V, Jacobson KA, Stevens RC, Wu B, and Zhao Q (2014) Agonist-bound structure of the human P2Y12 receptor. Nature 509, 119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Zhang K, Zhang J, Gao Z-G, Zhang D, Zhu L, Han GW, Moss SM, Paoletta S, Kiselev E, Lu W, Fenalti G, Zhang W, Müller CE, Yang H, Jiang H, Cherezov V, Katritch V, Jacobson KA, Stevens RC, Wu B, and Zhao Q (2014) Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature 509, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Luginina A, Gusach A, Marin E, Mishin A, Brouillette R, Popov P, Shiriaeva A, Besserer-Offroy É, Longpré J-M, Lyapina E, Ishchenko A, Patel N, Polovinkin V, Safronova N, Bogorodskiy A, Edelweiss E, Hu H, Weierstall U, Liu W, Batyuk A, Gordeliy V, Han GW, Sarret P, Katritch V, Borshchevskiy V, and Cherezov V (2019) Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs. Sci. Adv 5, No. eaax2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Gusach A, Luginina A, Marin E, Brouillette RL, Besserer-Offroy É, Longpré J-M, Ishchenko A, Popov P, Patel N, Fujimoto T, Maruyama T, Stauch B, Ergasheva M, Romanovskaia D, Stepko A, Kovalev K, Shevtsov M, Gordeliy V, Han GW, Katritch V, Borshchevskiy V, Sarret P, Mishin A, and Cherezov V (2019) Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors. Nat. Commun 10, 5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Zarzycka B, Zaidi SA, Roth BL, and Katritch V (2019) Harnessing Ion-Binding Sites for GPCR Pharmacology. Pharmacol. Rev 71 (4), 571–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Cao C, Tan Q, Xu C, He L, Yang L, Zhou Y, Zhou Y, Qiao A, Lu M, Yi C, Han GW, Wang X, Li X, Yang H, Rao Z, Jiang H, Zhao Y, Liu J, Stevens RC, Zhao Q, Zhang XC, and Wu B (2018) Structural basis for signal recognition and transduction by platelet-activating-factor receptor. Nat. Struct. Mol. Biol 25, 488–495. [DOI] [PubMed] [Google Scholar]
- (78).Masureel M, Zou Y, Picard L-P, van der Westhuizen E, Mahoney JP, Rodrigues JPGLM, Mildorf TJ, Dror RO, Shaw DE, Bouvier M, Pardon E, Steyaert J, Sunahara RK, Weis WI, Zhang C, and Kobilka BK (2018) Structural insights into binding specificity, efficacy and bias of a β2AR partial agonist. Nat. Chem. Biol 14, 1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


