Abstract
The ongoing COVID-19 pandemic has highlighted the immediate need for the development of antiviral therapeutics targeting different stages of the SARS-CoV-2 life cycle. We developed a bioluminescence-based bioreporter to interrogate the interaction between the SARS-CoV-2 viral spike (S) protein and its host entry receptor, angiotensin-converting enzyme 2 (ACE2). The bioreporter assay is based on a nanoluciferase complementation reporter, composed of two subunits, large BiT and small BiT, fused to the S receptor-binding domain (RBD) of the SARS-CoV-2 S protein and ACE2 ectodomain, respectively. Using this bioreporter, we uncovered critical host and viral determinants of the interaction, including a role for glycosylation of asparagine residues within the RBD in mediating successful viral entry. We also demonstrate the importance of N-linked glycosylation to the RBD’s antigenicity and immunogenicity. Our study demonstrates the versatility of our bioreporter in mapping key residues mediating viral entry as well as screening inhibitors of the ACE2-RBD interaction. Our findings point toward targeting RBD glycosylation for therapeutic and vaccine strategies against SARS-CoV-2.
Keywords: coronavirus, bioluminescence, bioreporter, viral entry, high-throughput screening, 2019-nCoV, SARS-CoV-2, COVID-19
Graphical abstract
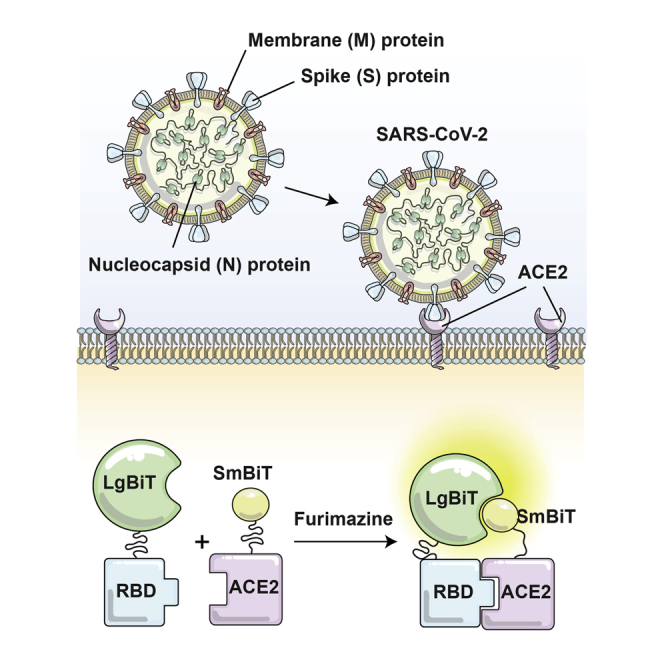
In this study, Azad et al. developed a nanoluciferase complementation reporter to interrogate the interaction between the SARS-CoV-2 viral spike protein and its host entry receptor. Using this bioreporter, they uncovered critical host and viral determinants of the interaction, including a role for glycosylation of asparagine residues within the RBD in mediating successful viral entry.
Introduction
As of December 22, 2020, there were globally more than 75 million confirmed SARS-CoV-21 infections resulting in nearly 1.7 million deaths,2 and with no signs of the pandemic ebbing in the near future, effective therapeutics and vaccines are desperately needed. Entry of the enveloped SARS-CoV-2 virus into mammalian cells is mediated by the viral spike (S) protein that binds to the angiotensin-converting enzyme 2 (ACE2) cell receptor and initiates fusion of the viral and cell membranes.3, 4, 5 This critical role in the virus infection cycle has made the S protein the focus of therapeutic development, including the identification of neutralizing antibodies,6 peptide-based S protein binders,7 and small molecule inhibitors of proteases involved in S protein maturation.5 As for many enveloped virus surface proteins, S is heavily glycosylated, and it has been speculated that these post-translational modifications could facilitate immune evasion or perhaps play a fundamental role in the determination of virus tropism.7 Interestingly, two N-linked glycan modifications occur within the conserved receptor-binding domain (RBD) of the S protein. The RBD mediates the binding of the S protein to ACE2,6,8 and while there have been a number of documented polymorphisms in the amino acid sequence of the RBD from clinical isolates around the world,9 these two glycosylation sites are uniformly conserved. This suggested to us the possibility that glycosylation of the RBD is important for its binding to the cellular ACE2 receptor or, as suggested earlier, inhibits immune recognition. To test these ideas, we constructed a bioreporter to rapidly assess the interactions between RBD variants and the ACE2 receptor. We took advantage of the recently developed nanoluciferase (NanoLuc) binary technology (NanoBiT)10, 11, 12 to create a surrogate assay for virus-host cell interactions. Our bioreporter provides a simple and rapid system to carry out a structure-function analysis of critical amino acids in the RBD that modulate its interaction with ACE2, as well as screen potential inhibitors of this host-virus interaction. We demonstrate that the two conserved N-glycan modifications in the RBD are required for efficient binding to ACE2 and infection with S pseudotyped viruses.
Results
SARS-CoV-2 NanoBiT bioreporter for detecting ACE2-RBD interactions
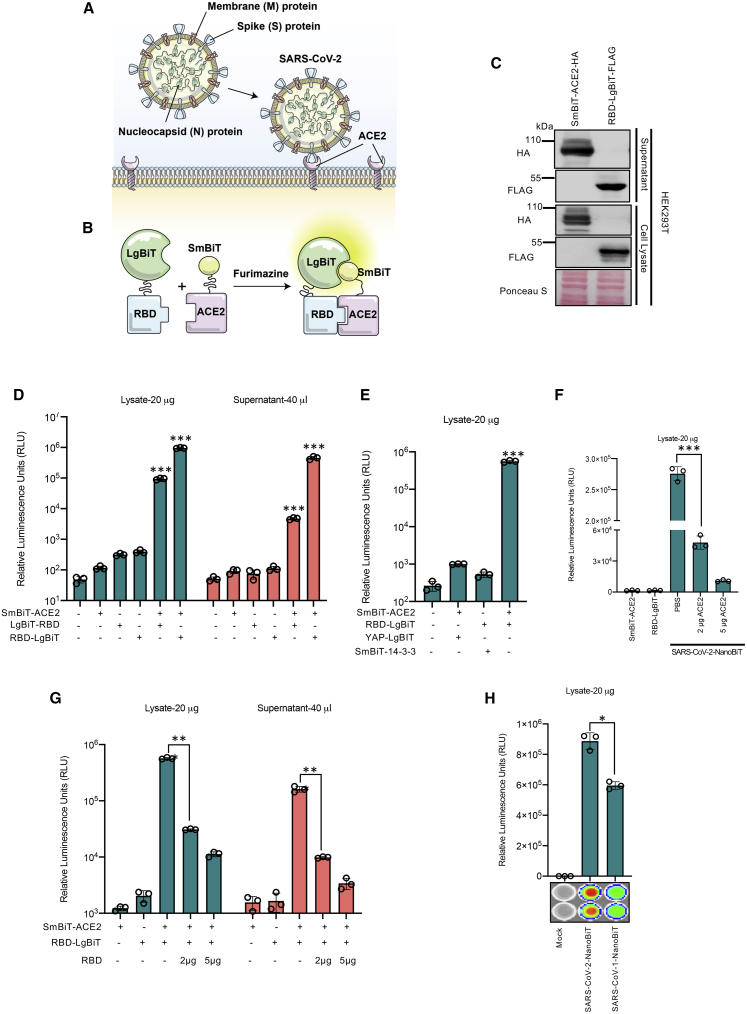
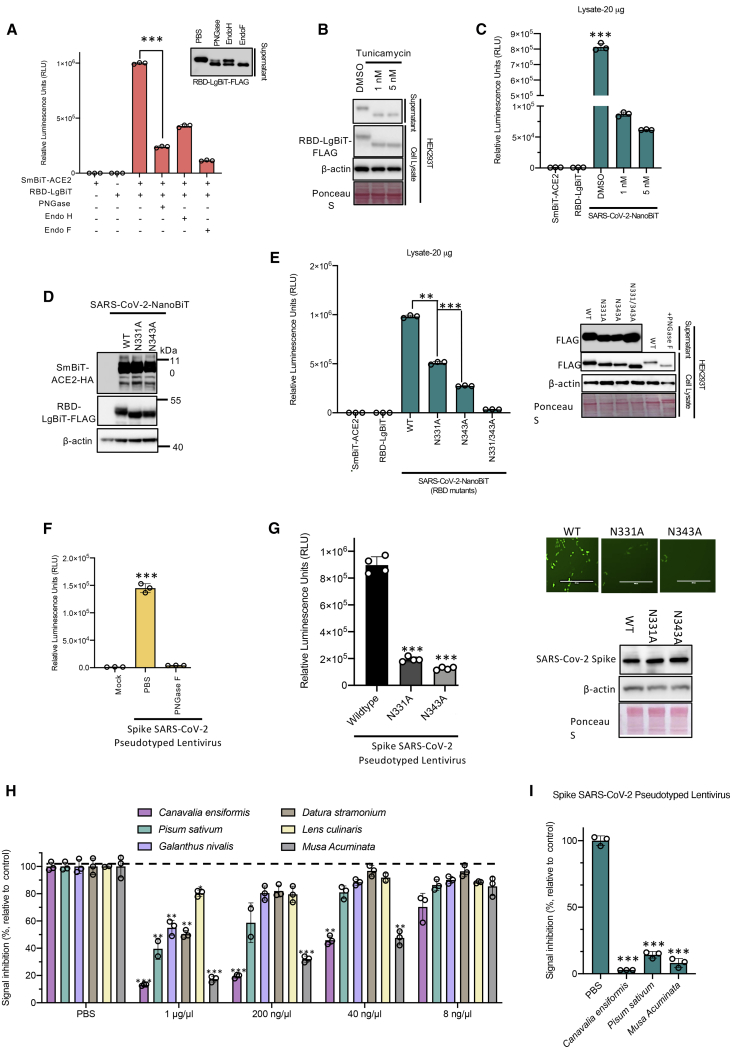
Several different reporter fragment complementation-based strategies have been used to interrogate protein-protein interactions,13 including split-luciferase schemes.14, 15, 16, 17 Conventional split-luciferase bioreporters can be limited in their application due to their relatively large sizes, poor stability, and the short half-lives of their catalyzed luminescent reactions. A recently reported NanoLuc (from Oplophorus gracilirostris)18 does not possess these limitations, and a NanoLuc-based fragment complementation system has been reported.10, 11, 12 Our bioreporter uses NanoLuc fragments linked to the RBD and ACE2, creating a bioreporter that can rapidly and sensitively serve as a surrogate for virus-host cell interactions (Figure 1A). Using published sequences and structural homology analysis,6,8,19,20 we designed a SARS-CoV-2 RBD sequence spanning residues 331–524 of the S protein (194 aa; Figure S1) for one component of the bioreporter. For the other component we used the soluble ectodomain of ACE2 (residues 1–740), as this has been shown to be sufficient to interact with the RBD.21 Since the RBD is the smaller protein of the two partners of interest,11 we linked the RBD with the larger fragment of the split luciferase (LgBiT) while ACE2 was fused to the small fragment (SmBiT, Figure 1B). A glycine-serine linker was inserted between ACE2 or the RBD and its respective NanoBiT component11 to enhance folding and flexibility of the fusion proteins. To facilitate the production of the interacting partners as secreted molecules, we carried out a series of codon optimization studies and tested different secretion signals. We found good production and complementation with an interleukin-12 secretion signal linked to the SmBiT-ACE2 fusion protein while an immunoglobulin κ (IgK) secretory leader sequence worked best at the N terminus of the RBD-LgBiT protein. Transfection of these constructs into 293T cells showed strong expression in cell lysates and secretion into supernatants, as confirmed by immunoblot analyses (Figure 1C). We then carried out luciferase assays using either cell lysates or supernatants from 293T cells co-transfected with the RBD and ACE2 NanoBiT fusion constructs in both orientations (Figure 1D). As can be seen, SmBiT-ACE2 and either RBD-LgBiT or LgBiT-RBD (>105 relative light units [RLU] versus ~104 RLU in Figure 1D) produced strong luminescent signals compared to transfection of individual components. Supernatants from the co-transfected cells also produced strong luminescence in the assay mirroring our findings with the cell lysates (Figure 1D). The large dynamic range of the assay makes it amenable to high-throughput screening, and indeed bioluminescent signals could even be observed using an in vivo imaging system (IVIS) (Figure S2A) or with the naked eye (Figure S2B). We validated the specificity of the interactions we were detecting in two ways. First, we co-transfected SmBiT-ACE2 or LgBiT-RBD with LgBiT-YAP15 or SmBiT-14-3-3, respectively. The YAP15 and 14-3-3 constructs have been previously demonstrated to interact with each other in a phosphorylation-dependent manner and rescue the NanoBiT structure;11 however, these proteins are not known to interact with ACE2 and the RBD. We used these constructs as negative controls to illustrate that the SARS-CoV-2-NanoBiT interaction is specific to the SARS-CoV-2 S RBD and ACE2 interactions. As predicted, LgBiT-Yap15 and SmBiT-14-3-3 proteins did not complement SmBiT-ACE2 or LgBit-RBD, respectively (Figure 1E).
Figure 1.
Establishment of a NanoLuc complementation-based bioreporter for measuring ACE2 interaction with the SARS-CoV-2 spike (S) RBD
(A) SARS-CoV-2 uses ACE2 as a viral entry receptor. (B) Schematic depicting mechanism of action for the bioreporter. (C and D) 293T cell were transfected with codon-optimized ACE2 and RBD constructs. 48 h post-transfection, cell lysates and supernatants were harvested. 10 μg of protein or 10 μL of supernatant was resolved by SDS-PAGE. (C) Immunoblot analysis of HA-tagged SmBiT-ACE2 and FLAG-tagged RBD-LgBiT expression. (D) Luminescence was quantified by a luciferase assay using FMZ as substrate (n = 3 biological replicates, mean ± SD; one-way ANOVA, ∗∗∗p < 0.005 relative to RBD-LgBiT alone, Dunnett’s correction for multiple comparisons). Lysates and supernatants were analyzed independently. (E) LgBiT-YAP-15 and SmBiT-14-3-3 were co-transfected with SmBiT-ACE2 and RBD-LgBiT, respectively, to demonstrate the specificity of the bioreporter (n = 3 biological replicates, mean ± SD; one-way ANOVA, ∗∗∗p < 0.005 relative to mock, Dunnett’s correction for multiple comparisons). (F) Recombinant ACE2 purified from 293T cells was incubated for 15 min with cell lysate containing RBD-LgBiT at room temperature. Equal amounts of lysates containing SmBiT-ACE2 were added (total 20 μg) and incubated for 5 minutes. Luciferase assay was performed using CTZ as substrate. (n = 3 biological replicates, mean ± SD; one-way ANOVA, ∗∗∗p < 0.005, Tukey’s correction for multiple comparisons). (G) Recombinant RBD purified from 293T cells was incubated for 15 min with cell lysate or supernatant containing SmBiT-ACE2 at room temperature. Equal amounts of lysates or supernatants containing RBD-LgBiT were added and incubated for 5 min. Luciferase assay was performed using CTZ as substrate (n = 3 biological replicates, mean ± SD; one-way ANOVA, ∗∗∗p < 0.005, Tukey’s correction for multiple comparisons). (H) Bioreporter assay was performed on lysates of 293T cells co-transfected with SmBiT-ACE2 and either SARS-CoV or SARS-CoV-2 RBD-LgBiT constructs, respectively. IVIS imaging of bioluminescence is shown below.
NanoLuc can utilize coelenterazine (CTZ) or its synthetic optimized derivative, furimazine (FMZ), as substrates to produce glow-type luminescence. CTZ represents a widely accessible and cost-effective alternative to FMZ with well-documented and stable formulations. We repeated the SARS-CoV-2-NanoBiT assay to compare FMZ and CTZ at a final concentration of 3.33 μM (Figure S3). Our assay results from transfected lysates (Figure S3A) and supernatants (Figure S3B) demonstrate comparable luminescence regardless of the substrate used. For the purpose of our newly developed biosensor, CTZ is a comparable alternative to FMZ, and it can be used as a cost-effective substrate.
We then evaluated the impact of recombinant soluble ACE2 protein (rACE2), a SARS-CoV-2 entry inhibitor under clinical investigation as an antiviral,22 and recombinant RBD (rRBD) on the SARS-CoV-2 NanoBiT. For these experiments, we transfected 293T cells independently with either the RBD-LgBiT or SmBiT-ACE2 constructs to obtain lysates or culture supernatants containing either RBD-LgBiT or SmBiT-ACE2. In the absence of added recombinant protein, the combined lysates or supernatants produced robust signals (Figure 1F). However, upon pre-incubation of RBD-LgBiT with rACE2 prior to adding the SmBiT-ACE2 containing lysate, we observed a dose-dependent reduction in luminescent signals (Figure 1F). Similarly, pre-incubation of SmBiT-ACE2 with recombinant RBD resulted in the loss of luminescence in the bioreporter reporter assay (Figure 1G). These results serve as a proof of principle that our bioreporter can identify molecules that disrupt the ACE2-RBD interaction. We also constructed a SARS-CoV-1 NanoBiT bioreporter. Earlier work suggested that the SARS-CoV-2 RBD binds with higher affinity to ACE2 than does the SARS-CoV-1 RBD, potentially contributing to enhanced transmissibility of the virus,8 while others have shown that the RBDs from these two viruses have comparable affinities.6,23 In our hands, the SARS-CoV-1 NanoBiT produced only a modestly reduced signal relative to the SARS-CoV-2 bioreporter (Figure 1H; Figure S4A), supporting the idea that two virus RBDs have comparable affinity for ACE2.
Characterization of critical determinants of ACE2-RBD interaction
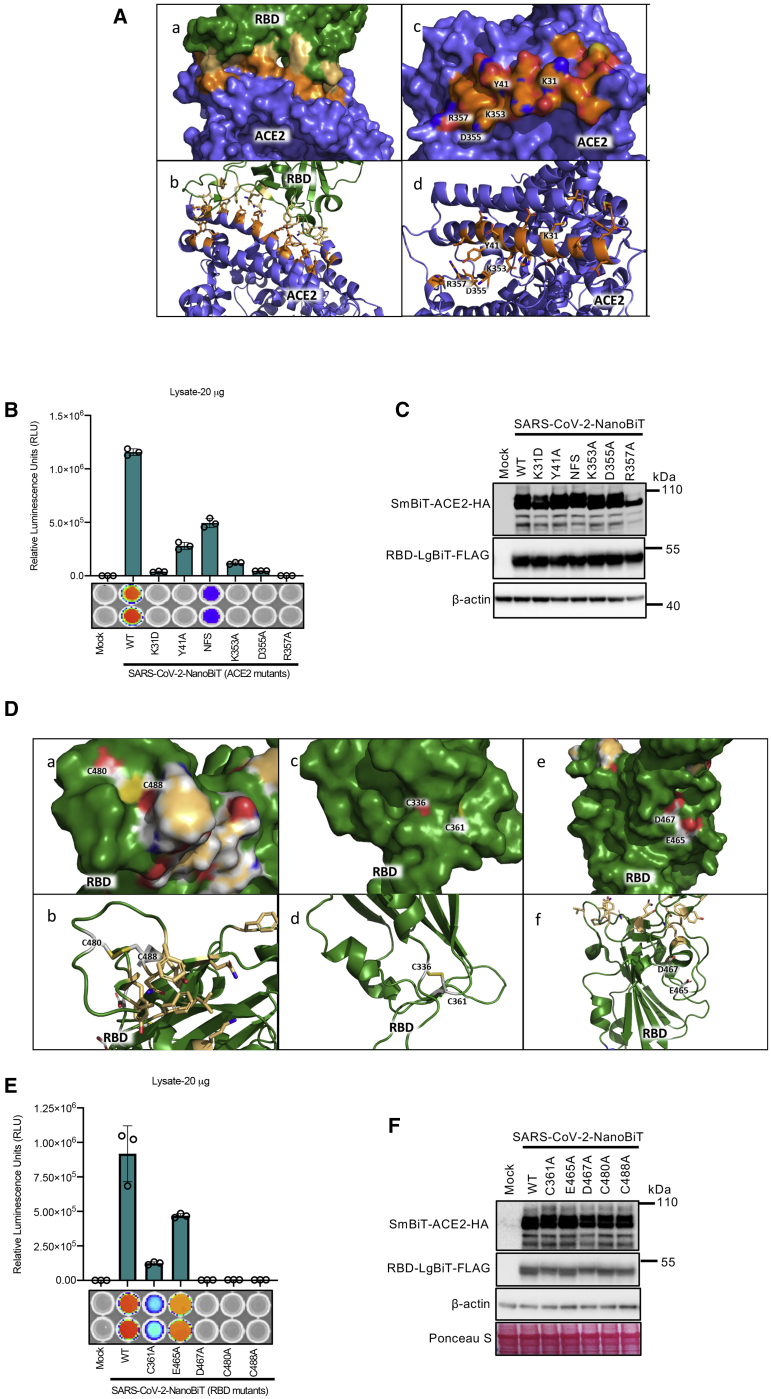
While the interaction of ACE2 with the SARS-CoV S protein has been well characterized, the critical host and viral determinants of ACE2-SARS-CoV-2 S interaction remain under investigation. We next utilized the bioreporter assay to characterize key residues of both ACE2 and the SARS-CoV-2 RBD that mediate their interaction. Previous mutational analyses of the ACE2 catalytic domain identified critical residues interacting with the SARS-CoV S S1 domain.3,24 These studies identified K31, Y41, K353, D355, and R357 as key amino acids mediating ACE2’s interaction with the SARS-CoV RBD (Figure 2A). We mutated these residues individually in SmBiT-ACE2 to examine whether these amino acids were similarly important for ACE2’s interaction with the SARS-CoV-2 RBD (Figures 2B and 2C; Figure S4B). When tested in the bioreporter, we observed that all of these mutations significantly impaired the ACE2-RBD interaction, with the K31D, D355A, and R357A mutations completely abolishing the interaction. Rat cells are non-permissive to both SARS-CoV and SARS-CoV-2 infection. The incorporation of rat ACE2 residues 82–84 (amino acids NFS) into the human ACE2 (hACE2) sequence was previously shown to strongly impair the hACE2-RBD interaction with SARS-CoV.3 Introduction of the NFS residues into SmBiT-ACE2 similarly impaired the SARS-CoV-2 ACE2-RBD bioreporter (Figures 2B and 2C; Figure S4B), suggesting that these ACE2 residues contribute to the species tropism of SARS-CoV-2.
Figure 2.
Mutational analyses reveal critical host and viral determinants of ACE-SARS-CoV-2 RBD interaction
(A) Binding interface of the RBD and ACE2 with potential critical ACE2 residues of interest highlighted. (B and C) A bioreporter assay (B) and immunoblot analysis (C) were performed on cells co-transfected with RBD-LgBiT and SmBiT-ACE2 mutant constructs. IVIS imaging of bioreporter assay results are shown at the bottom of (B). (D) Binding interface of the RBD and ACE2 with potential critical RBD residues of interest highlighted. (E and F) Biosensor (BS) assay (E) and immunoblot analysis (F) were performed on cells co-transfected with RBD mutant-LgBiT constructs and SmBiT-ACE2 constructs. IVIS imaging of bioluminescence is shown at bottom of (E). Immunoblot analyses were performed to confirm proper expression of the constructs.
We next utilized previous mutational analyses with the SARS-CoV RBD to guide our study of the critical SARS-CoV-2 RBD residues mediating the ACE2 interaction.21 SARS-CoV RBD cysteine residues 348, 467, and 474, as well as the acidic residues E452 and D454, were shown to be critical to this domain’s interaction with ACE221 (Figure 2D). The homologous residues in the SARS-CoV-2 RBD (corresponding to C361, E465, D467, C480, and C488) were mutated to alanine in the RBD-LgBiT construct in order to evaluate their role in ACE2 association. Four of the mutations caused a major loss (>80%) of luminescent signal produced by the bioreporter assay (C361A, D467A, C480A, and C488A), suggesting that these residues in the SARS-CoV2 RBD are critical for ACE2 interaction (Figures 2E and 2F; Figure S4C). Alternatively, the E465A mutation caused a modest drop in signal, suggesting that this residue is less critical for SARS-CoV-2 in mediating an interaction with ACE2 in comparison to SARS-CoV.
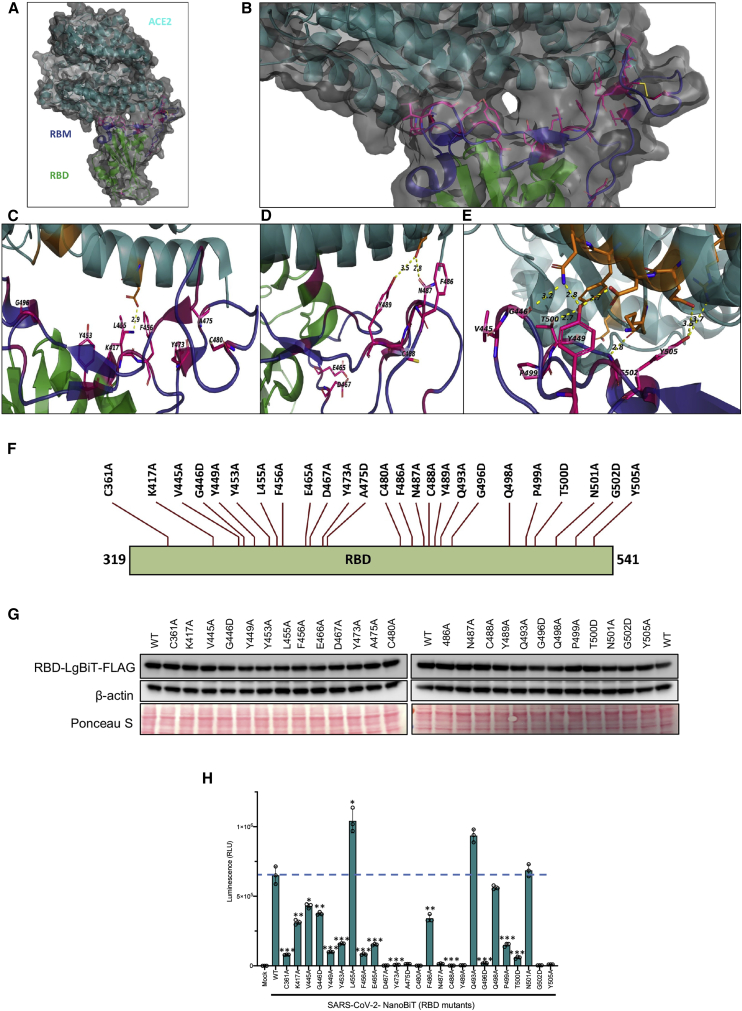
Since our bioreporter is sensitive to the initial point mutations we examined, we expanded our mutational analyses to include other potential critical residues, based on analysis of the 3D crystal structure of the ACE2-RBD binding interface (Figures 3A–3E). We used site-directed mutagenesis to create an additional alanine mutation in the RBD (Figure 3F) and analyze their impact on ACE2-RBD interactions (Figures 3G and 3H). We demonstrated that 21 out of the 25 tested SARS-CoV-2 RBD mutations significantly reduced binding to ACE2 (Figure 3H). To further illustrate the potential of the assay in a high-throughput screen, we analyzed its reproducibility in a 384-well plate assay (Figure S6) and found minimal variability. Collectively, these mutational analyses, along with high reproducibility of the assay, demonstrate that the bioreporter is a useful tool for high-throughput structure-function analysis of viral and host determinants of the ACE2-RBD interaction.
Figure 3.
RBD amino acids in 3D structure of the bound ACE2 and schematic representation of mutations in the RBD
(A) Overall 3D structure of the RBD-ACE2 interaction. The RBD is colored in green, the receptor-binding motif (RBM) of the RBD is in dark blue, and ACE2 is in dark cyan. C361 is in the bottom middle of the overall structure in the RBD core section and is not depicted in the following illustrations. (B) Enlarged view of the overall structure depicting the RBD target mutation sites in stick representation, colored in magenta. (C–E) Stick representation of target mutation sites at the contact site of two molecules. Dotted lines connect the mutant amino acids (aa) to their contacting amino acids in ACE2. The structure is from PDB: 6M0J. (F) Illustration of the amino acid changes used to examine the RBD in this study. (G) Immunoblot of LgBiT-RBD mutant expression from the cell lysates of transfected HEK293T cells. β-actin and total protein loading are shown as controls. (H) Bioreporter assay with LgBiT-RBD mutants demonstrating altered binding affinity of various mutants.
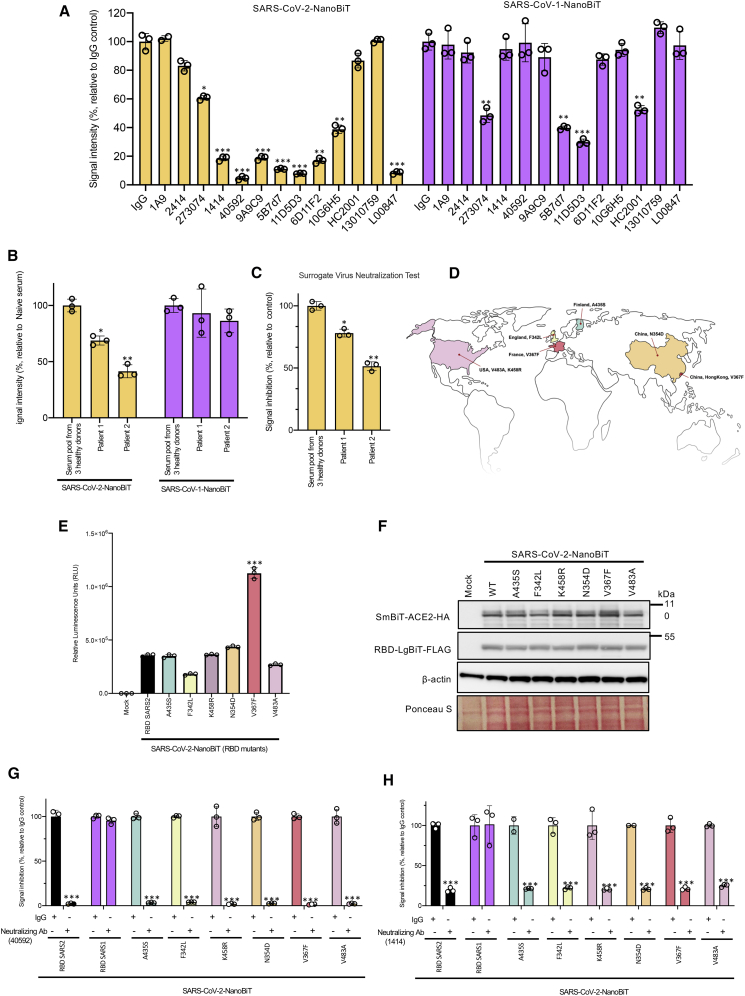
The SARS-CoV-2 bioreporter is sensitive to neutralizing antibodies
Monoclonal antibodies targeting the RBD are under consideration as SARS-CoV-2 therapeutics.25 We screened 13 different commercially available monoclonal SARS-CoV-2 S RBD antibodies with the SARS-CoV-2 bioreporter (Figure 4A). Seven of these antibodies (1414, 40592, 9A9C9, 5B7d7, 11D5D3, 6D11F2, and 10G6H5) are reported to not only bind the RBD but also to neutralize infection of cells with an S pseudotyped lentivirus. Interestingly, these seven monoclonal antibodies were the most effective at blocking RBD-ACE2 interactions measured with the SARS-CoV-2 bioreporter. We applied the antibody collection to the SARS-CoV-1 bioreporter and found that while most SARS-CoV-2 antibodies did not cross-react, antibodies 5B7d7 and11D5D3 showed some ability to disrupt RBD-ACE2 interactions for both virus strains. Non-specific mouse IgG and monoclonal antibody 1A9, which binds to the S2 subdomain of the S protein, did not disrupt the signal generated by the SARS-CoV-2 bioreporter, supporting that the specificities of the signals were observed. We tested our bioreporters with serum from two patients recovered from SARS-CoV-2 infections at the Ottawa Hospital and pooled serum from three healthy volunteers (Figure 4B). In these experiments, we compared our SARS-CoV-2 bioreporter to a widely used, commercially available ELISA kit that is designed to act as surrogate for virus neutralization (Figure 4C).26 For the bioreporter experiments, SARS-CoV-2 RBD-LgBiT was co-incubated with sera for 25 min, followed by the addition of SmBiT-ACE2 for an additional 5 min. At this point, substrate was added, and luminescence measured. The bioreporter was able to distinguish seroconverters from healthy donors, as both convalescent patients’ sera significantly reduced the bioreporter signal (Figure 4B, left panel). Interestingly, sera from convalescent SARS-CoV-2 patients failed to disrupt the SARS-CoV-1 RBD-ACE2 interaction (Figure 4B, right panel), suggesting a lack of cross-reactivity in these patients’ neutralizing antibody response. The signal from our bioreporter compared well with the results produced by a receptor-ligand binding ELISA (Figure 4C).26 Note, however, the NanoBiT assay is more rapid (25 min compared to >1.5 h for the ELISA-based assay) and more accessible in terms of cost and technical feasibility.
Figure 4.
SARS-CoV-2-NanoBiT enables facile detection of SARS-CoV-2 seroconversion
(A) SARS-CoV-2 (left) or SARS-CoV (right) bioreporter assays were performed with an array of anti-RBD monoclonal antibodies or control IgG. (B) SARS-CoV-2 BS (left) or SARS-CoV bioreporter (right) assays were performed with serum pooled from three healthy donors or two recovered SARS-CoV-2 patients for 25 min. A luciferase assay was performed 5 min after SmBiT-ACE2 addition. (C) SARS-CoV-2 RBD-HRP was incubated for 15 min with the serum pool from three healthy donors or recovered patients from SARS-CoV2 for 25 min. Then, samples were added to a plate coated with ACE2 for 15 min. After washing, tetramethylbenzidine (TMB) solution was added and the amount of RBD-HRP binding was evaluated by measuring the optical density (OD) at 450 nm. (D) Geographic distribution of the SARS-CoV-2 strains with the indicated RBD mutations. (E) BS assays were performed on cells co-transfected with the indicated RBD mutant-LgBiT constructs and SmBiT-ACE2 constructs. (F) Immunoblot analyses were performed to confirm the proper expression of the constructs. (G and H) Cell lysates transfected with different RBD-LgBiT mutants were incubated for 25 min with anti-RBD monoclonal antibodies 40592 in (G) and 1414 in (H). Luciferase assays were performed 5 min after SmBiT-ACE2 addition.
SARS-CoV-2 genome sequencing has revealed the emergence of RBD mutations in global strains. We investigated the influence of six emerging RBD mutations found in SARS-CoV-2 genome sequences worldwide on the ACE2-RBD interaction: V367F (France and Hong Kong/China), N354D (China), A435S (Finland), F342L (England), and K458R and V483A (USA)27 (Figure 4D). The bioreporter assay revealed that these SARS-CoV-2 variants displayed variable binding to ACE2 (Figures 4E and 4F). Interestingly, the V367F mutant displayed a >3-fold enhanced interaction with ACE2, while the F342L mutation decreased reporter activity 2-fold. The enhanced affinity of V367F RBD mutation to ACE2 is consistent with a recent study describing enhanced viral entry in HEK293T-ACE2/TMPRSS2 cells with lentivirus pseudotyped with V367F S compared to wild-type (WT) S.9 Similarly, these mutations also have the potential to impact the efficacy of RBD-targeted monoclonal antibodies and vaccination strategies. We analyzed the cross-reactivity of two SARS-CoV-2 RBD-targeted monoclonal antibodies toward the different RBD variants using a bioreporter assay (Figures 4G and 4H). Our results demonstrated that both monoclonal antibodies tested could effectively block all the mutants’ interactions with ACE2, highlighting that specific monoclonal RBD antibodies have the potential to work effectively against multiple circulating SARS-CoV-2 strains encoding different RBD variants.
N-linked glycosylation of the RBD is required for ACE2-RBD interaction
We found that bacterially produced recombinant SARS-CoV-2 RBD was not able to block SmBiT-ACE2’s interaction with SARS-CoV-2 RBD-LgBiT (Figure S5A), in contrast to our results with recombinant RBD produced in mammalian cells (Figure 1G). An important distinction with bacterial expression systems is their inability to produce mammalian-type glycosylation, suggesting a potential role for protein glycosylation in the ACE2-RBD interaction. As discussed earlier, a recent study demonstrated that the S protein contains 22 N-linked glycosylation sites,7 including 2 in the RBD at asparagine residues 331 and 343. To evaluate the relevance of N-linked glycosylation of the RBD on the ACE2-RBD interaction, we pre-treated RBD-LgBiT- or SmBiT-ACE2-containing lysates with peptide:N-glycosidase F (PNGase F), endoglycosidase H (Endo H), or Endo F enzymes, which cleave N-linked oligosaccharides, and subsequently interrogated their ability to interact with their complementary partner using the bioreporter assay. Interestingly, RBD-LgBiT’s treatment with either enzyme significantly impaired its interaction with the ACE2 ectodomain (Figure 5A). Our results suggest that Endo H’s cleavage of S N-linked glycans occurs with lower efficiency, consistent with previous studies reporting a high level of complex N-linked glycans, a poor substrate for Endo H, at N331 and N343.28 We next performed a bioreporter assay using RBD-LgBiT cell lysates derived from 293T cells treated with tunicamycin, an inhibitor of N-linked glycosylation in eukaryotic cells. Immunoblot analyses demonstrated that RBD-LgBiT-transfected cells, which were treated with tunicamycin, produced RBD-LgBiT with an apparent lower molecular weight, suggesting a loss of glycosylation (Figure 5B). Tunicamycin treatment resulted in significantly reduced bioreporter activity in lysates and supernatants from cells co-transfected with RBD-LgBiT and SmBiT-ACE2 (Figure 5C; Figures S5B and S5C). Taken together, these data suggest that N-linked glycosylation of the SARS-CoV-2 S protein RBD is necessary for its interaction with ACE2.
Figure 5.
N-linked glycosylation of SARS-CoV-2 S RBD is critical to its interaction with ACE2
(A) 293T cells transfected with either SmBiT-ACE2 or RBD-LgBiT were harvested. 25 μL of supernatants was pre-treated for 1 h at 37°C with Endo H, Endo F, PNGase F, or untreated, as indicated. Lysates were subsequently mixed and incubated for 15 min at room temperature, then assessed by luciferase assay using CTZ as a substrate (n = 3 biological replicates, mean ± SD; one-way ANOVA, ∗∗∗p < 0.005, Tukey’s correction for multiple comparisons). (B) 293T cells were transfected with RBD-LgBiT and 24 h after transfection were treated with tunicamycin for 16 h. Lysates and supernatants were prepared and analyzed by immunoblotting. (C) RBD-LgBiT cell lysates from (B) were incubated at room temperature with cell lysates from SmBiT-ACE2-transfected cells. After 15 min, the biosensor assay was performed. (D and E) Immunoblot analysis (D) and a bioreporter assay (E) were performed on cells co-transfected with the indicated RBD-glycosylation site mutants of the LgBiT-RBD constructs and SmBiT-ACE2 constructs. Immunoblot confirming equaling expression of Lg-RBD mutants is shown in right panel. (F) SARS-CoV-2 S pseudotyped lentivirus encoding ZsGreen and luciferase reporters was incubated with PBS ot or PNGase F for 1 h and then used to infect HEK293T-ACE2 cells. 48 h post-transduction, cells were evaluated for measuring luciferase activity. (G) SARS-CoV-2 S mutant pseudotyped lentiviruses infectivity assay as in (F). (H) Plant lectins were screened for the ability to disrupt CoV-NanoBiT. Cell lysates transfected with RBD-LgBiT were incubated for 1 h with different lectins from shown species. Luciferase assays were performed 5 min after SmBiT-ACE2 addition. (I) Plant lectins from Canavalia ensiformis (jack bean), Pisum sativum (pea), and Musa acuminata (banana) were evaluated for inhibitory effects against SARS-CoV-2 S pseudotyped lentivirus infection in HEK293T-ACE2 cells.
To more directly determine the importance of RBD glycosylation, we substituted alanine residues for aspargines at positions 331 and 343. Immunoblot analyses of the point mutations in RBD-LgBiT revealed that N331A and N343A mutations resulted in decreased molecular weights, consistent with the two sites being functional glycosylation sites (Figure 5D), and paralleled the changes in mobility of RBD produced from tunicamycin-treated cells. We evaluated the impact of mutating these two glycosylation sites on ACE2-RBD interactions using our bioreporter assay (Figures 5D and 5E; Figures S5D and S5E). Both of the N331A and N343A mutations significantly impacted complementation between ACE2 and RBD fusion proteins. To further validate the importance of N-linked glycosylation to SARS-CoV-2 infectivity using an orthogonal approach, we also used Endo H and PNGase F treatment on S protein pseudotyped lentiviruses.29 Consistent with the bioreporter data (Figures 5A–5E), enzymatic removal of N-linked glycosylation abrogated the infectivity of the S pseudotyped lentivirus (Figure 5F). We then used site-directed mutagenesis to create full-length S mutants (N331A and N343A) and used these to create S pseudotyped lentiviruses. Consistent with our SARS-CoV-2-NanoBiT data (Figure 5E), both mutations produced significant decreases in S pseudotyped lentivirus infectivity (Figure 5G; Figures S5F and S5G). Overall, these data provide direct evidence that SARS-CoV-2 S depends on N-linked glycosylation of RBD to mediate its interaction with the ACE2 ectodomain.
SARS-CoV-2-NanoBiT identifies lectins as antiviral therapeutic candidates
SARS-CoV-2 S is glycosylated with oligomannose- and complex-type glycans.7 We sought to examine the therapeutic potential of targeting these N-linked glycans by testing mannose-binding plant lectins for anti-viral effects. We screened lectins from Canavalia ensiformis (jack bean), Pisum sativum (pea), Galanthus nivalis (snow drop), Datura stramonium (jimson weed/thorn apple), Musa acuminata (banana), and Lens culinaris (lentil) for their ability to disrupt the SARS-CoV-2 RBD-ACE2 interaction using our bioreporter (Figure 5H; Figure S5H). Our results demonstrate a diverse range of antiviral effects. While the lentil lectin displayed no significant inhibition of the interaction across the tested concentration range (8–1,000 ng/μL), other lectins showed some efficacy, with the jack bean (Canavalia ensiformis) lectin demonstrating the strongest impact. The antiviral effects for the top three lectin candidates (Pisum sativum, Musa acuminata, and Canavalia ensiformis) were validated for their ability to inhibit S pseudotyped lentivirus at 100 ng/μL (Figure 5I). Consistent with the CoV-NanoBiT data, both lectins inhibited pseudovirion entry, with Canavalia ensiformis lectin showing a >900-fold decrease while the pea lectin showed only an 8-fold decrease. Canavalia ensiformis lectin’s antiviral effects are consistent with previous work suggesting that mannose-binding lectins can inhibit authentic SARS-CoV infection.30 Collectively, our work suggests targeting the glycosylation of S represents a viable therapeutic target that warrants further investigation.
N-linked glycosylation of the SARS-CoV-2 S RBD is critical to its immunogenicity and antigenicity
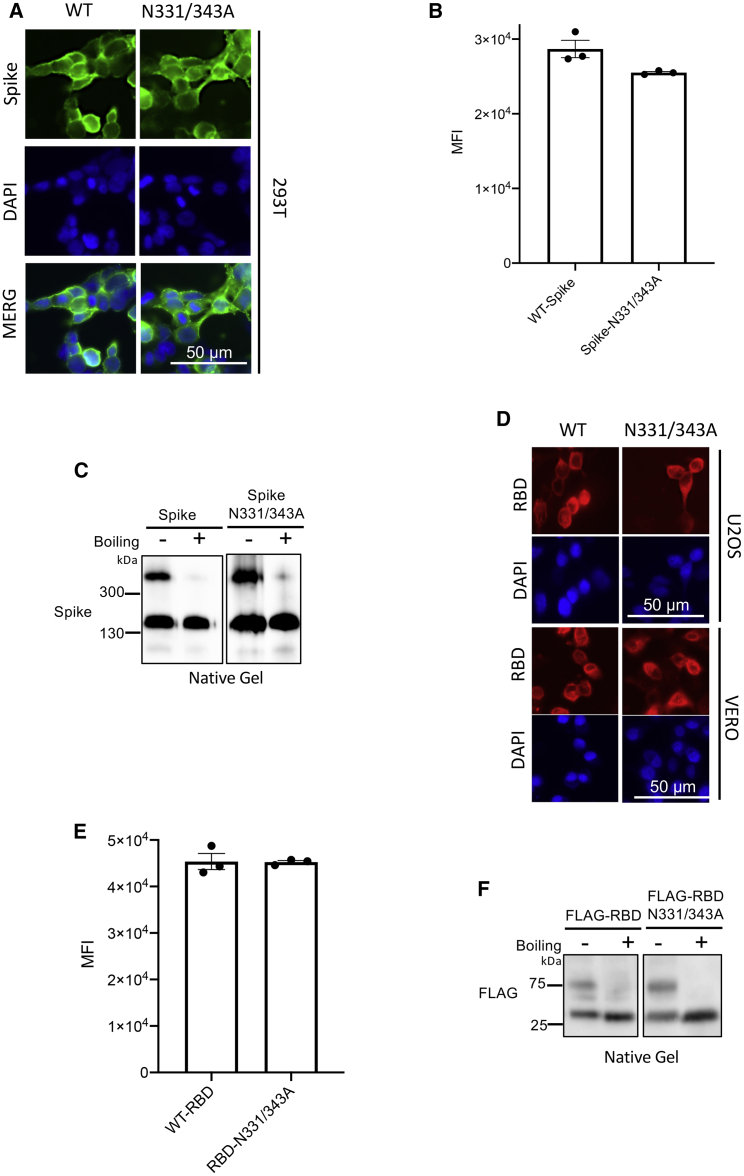
We sought to examine the possibility that deglycosylation of S/RBD was disrupting protein conformation. To investigate this, we also analyzed cell surface RBD expression of the S glycosite mutant (N331A/N343A). In our models, we observed no major differences in cell surface expression of the S N331A/N343A double mutant relative to the WT protein, using either immunofluorescence or flow cytometric analyses (Figures 6A and 6B). Furthermore, native PAGE analyses revealed that the S mutant retained its ability to trimerize (Figure 6C). Taken together, these data suggest the glycosite mutations do not cause major structural changes. Our data cannot exclude, however, that minor conformation changes in the proximity of the RBD glycosites result in a reduced ACE2 binding capacity. Similar to the WT construct, we demonstrated that the N331A/N343A RBD-TMD construct maintained its ability to trimerize and localize to the cell surface, as demonstrated by immunofluorescence (Figure 6D), flow cytometry (Figure 6E), and native PAGE analysis (Figure 6F), suggesting that there were no major conformational changes resulting from the mutations.
Figure 6.
N-linked glycosylation does not influence S localization and trimerization
(A–C) HEK293T cells were transfected with wild-type (WT) or N331A/N343A mutant S expression constructs and (A) immunofluorescence and (B) flow cytometric analyses were performed to visualize cell surface S levels. (C) Native-PAGE analysis of S trimerization. Immunoblotting was performed to visualize S complexes. (D–F) U2OS/VERO cells were transfected with WT or N331A/N343A mutant RBD-TMD expression constructs. (D and E) Immunofluorescence (D) and flow cytometric (E) analyses were performed to visualize cell surface RBD levels. (F) Immunoblotting of RBD-TMD complexes on native PAGE gel.
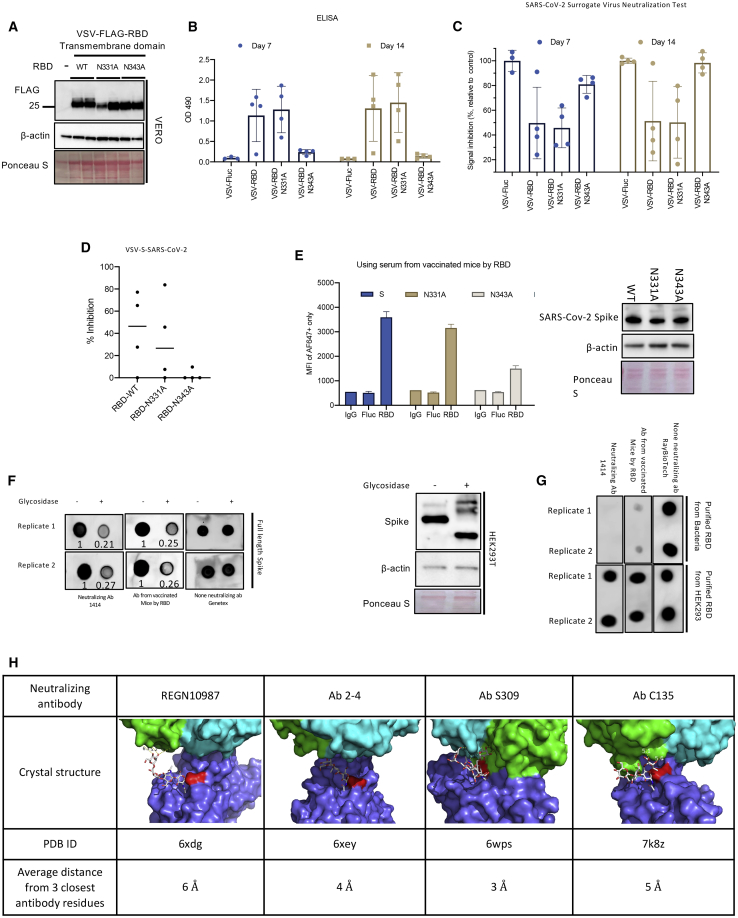
We also examined the influence of N-linked glycosylation on the RBD’s immunogenicity and antigenicity. There is a large global effort to generate effective SARS-CoV-2 vaccines.31 Several of these vaccine strategies are focused on using S or RBD as the central immunogen. This is in line with a recent study demonstrating that RBD is an immunodominant antigen that is targeted by most neutralizing antibodies in COVID-19 patients.32,33 As the glycosylation status of the RBD in these different vaccine strategies can vary depending on the models used for vaccine generation, we wished to examine the role of N-linked glycosylation on the immunogenicity of an RBD-based vaccine. Therefore, we engineered vesicular stomatitis virus (VSV)Δ51 virus expressing the S transmembrane domain (TMD) fused with either WT RBD (RBD-TMD) or glycosite mutant derivatives (Figure 7A). We focused our vaccine approach on delivering RBD as an antigen because RBD is the immunodominant target of the humoral response,32 with >90% of neutralizing antibodies in patients targeting this domain.33 We immunized mice with these different VSVΔ51 vaccine strains and analyzed the RBD-specific IgG levels (Figure 7B) and neutralizing antibody response, using two surrogate neutralization assays, in mouse sera samples (Figures 7C and 7D). Both VSVΔ51-RBD-TMD WT and N331A induced a significant IgG response against the RBD, while VSVΔ51-RBD-TMD N343A-immunized mice sera had significantly less anti-RBD IgG. Analogously, the N343A mutation impaired neutralizing antibody generation, as measured in parallel by an ELISA-based surrogate neutralization assay (Figure 7C)26 and a pseudovirus assay (Figure 7D)34. Our observations point to an important role for N-linked glycosylation of the N343 glycosite in the immunogenicity of RBD.
Figure 7.
N-linked glycosylation of RBD influences antigenicity and immunogenicity
(A) Immunoblot analysis of VSVΔ51-RBD-TMD-infected cell lysates. (B) Serum anti-RBD IgG levels of mice vaccinated with VSVΔ51-RBD-TMD WT and mutants were measured using ELISA. (C–E) Neutralizing antibody response was also measured using an ELISA-based surrogate neutralization assay (C) or a VSV-S pseudovirus-based assay (D) using 64-fold diluted serum. (E). HEK293T cells were transfected with expression constructs for WT, N331A, or N343A S and analyzed by flow cytometric analysis for S cell surface expression using sera from VSVΔ51-FLuc or VSVΔ51-RBD-TMD-WT vaccinated mice as a primary antibody. Immunoblot analysis showing total S expression in transfected cells is shown on the right panel. (F) Dot blot analyses of Endo F1/F2/F3-treated cell lysates probing with different RBD-targeted antibodies or sera from VSVΔ51-RBD-TMD-WT vaccinated mice. Immunoblot analyses showed the effect of glycosidase treatment on S protein migration (right panel). (G) Dot blot analyses comparing recognition of recombinant RBD purified from bacteria or HEK293 to different RBD-targeted antibodies or sera from VSVΔ51-RBD-TMD-WT-vaccinated mice. (H) Crystal structure of S complexed with four known neutralizing antibodies reveals the proximity of the binding interface near the RBD glycosites.
We investigated the role of N-linked glycosylation in the immunogenicity of RBD. 293T cells were transfected with full-length S constructs expressing either the full-length S protein, the N331A mutant, or the N343A mutant (Figure 7E, right panel). Flow cytometry analysis was performed on the transfected cells using serum from VSVΔ51-RBD-TMD (WT)-vaccinated mice. This serum bound WT and N331A S equally; however, there was decreased binding to the N343A mutant (Figure 7E, left panel). This suggests the N343 glycosylation is critical to recognition by RBD antibodies. In order to further probe the role of S glycosylation on the glycoprotein’s antigenicity, we treated S-transfected cell lysates with Endo F1, F2, and F3 (glycosidases less sensitive to the native conformation of proteins). Effective cleavage of glycans from RBD by Endo F was also confirmed by a lack of binding to a biotinylated concanavalin A (Con A) lectin, via dot blot (Figures S7A and S7B), as well as measuring the levels of the released glycans by a total carbohydrate assay kit (Figures S7C and S7D). We then probed the lysates with different RBD-targeted antibodies. Immunoblot analyses revealed that the S-transfected lysates were recognized by serum from VSVΔ51-RBD-TMD-vaccinated mice, a neutralizing antibody targeting RBD (1414), and a non-neutralizing antibody (GeneTex) (Figure 7F). However, the glycosidase-treated lysate was only recognized by the non-neutralizing antibody. This pointed toward the importance of N-linked glycosylation to RBD antigenicity. Similar results were observed using sera from convalescent COVID-19 patients to perform dot blot analysis on N331A and N343A S mutants. While the N331A mutation had minimal impact on sera binding, the N343A caused a major drop in patient sera binding, suggesting an important role for N-linked glycosylation of S N343 in the protein’s antigenicity (Figure S7E). This was further supported by the fact that recombinant RBD from bacteria, which are incapable of matching mammalian glycosylation profile, was poorly recognized by 1414 and the mouse sera (Figure 7G). An examination of the crystal structures of the S RBD complexed with neutralizing antibodies (REGN10987, Ab2-4, C135, and Ab S309) revealed the epitope recognized by all of these antibodies was in close proximity to N343 (<6 Å; Figure 7H).35 Collectively, these data suggest that N343 glycosylation and its effects on proximal protein conformation are integral to the antigenicity of RBD.
Discussion
Previous studies using split-luciferase reporters have examined viral protein interactions;36,37 however, we think that the data presented herein are the first report of a NanoLuc complementation reporter-based assay to probe virus binding to host receptor ACE2. While the RBD in our bioreporter may not capture trimerization-related and full-length S epitopes, we have validated the system’s ability to successfully test potential therapeutics, including monoclonal antibodies and receptor decoys. The bioreporter also enabled the evaluation of emerging RBD mutations on SARS-COV-2 infectivity and monoclonal antibody efficacy. This represents a valuable application as we begin to identify the novel emerging SARS-CoV-2 S mutants in the global population.
Our observation that monoclonal RBD antibodies have conserved efficacy against various RBD variants suggests that vaccines capable of inducing a strong neutralizing antibody response against the SARS-COV-2 RBD should display strong cross-reactive efficacy in the global population. Although it has been speculated that glycan clusters on the S protein could impede immune recognition or antibody activity, our bioreporter data suggest that, for SARS-CoV-2 S RBD, this may not be the primary role of glycosylation. Indeed, given the strong conservation of these glycosylation sites in clinical isolates around the world, we think that appropriate glycosylation at N331 and N343 could provide a conserved target for vaccine development.
We think that our data provide direct evidence demonstrating that N-linked glycosylation of the SARS-CoV-2 RBD is an important mediator of ACE2 binding. This is consistent with the findings of a pair of studies published during the revision of our manuscript.38,39 We utilized plant lectins to target this post-translational modification and inhibit the ACE2-RBD interaction and demonstrated its potential as an antiviral using a pseudovirion system. The most potent antiviral lectins identified in this study were jack bean lectin, banana lectin, and Pisa sativum lectin, all of which have a binding specificity toward high mannose or hybrid glycans. Therefore, their antiviral effects are in line with previous reports characterizing the RBD N-linked glycans as a heterogeneous mixture of oligomannose and hybrid type glycosylation.40,41 A recent study demonstrated that it was viable to engineer a banana lectin to inhibit influenza A virus infection in mice while minimizing mitogenic effects associated with lectins.42 Lectins or another carbohydrate binding agent may similarly act as a lead candidate to enable the development of a SARS-CoV-2 S glycan-targeted lectin. Alternatively, our finding that glycosylation is essential for RBD binding to ACE2 suggests that it may be possible to use specific glycosylation inhibitors as an antiviral approach to blunt SARS-CoV-2 infections, especially if given acutely in a locoregional fashion. For example, iminosugars that disrupt appropriate processing of N-linked glycan groups have been shown to act as broad-spectrum antivirals against viruses that are dependent on one N-linked glycan on a glycoprotein for infectivity.43 Our results are consistent with a recent study describing an important role for the N331 and N343 glycosites in viral entry.39
Previous studies have highlighted the importance of glycosylation on viral antigenicity. In fact, several HIV neutralizing antibodies have been shown to be glycan-dependent, with viral escape associated with deletion of a glycan.44,45 Our study establishes a similar role for N-linked glycosylation in the antigenicity of RBD (Figures 7E and 7G). Specifically, our data suggest that the N343 glycan regulates epitope recognition of several RBD-targeted neutralizing antibodies. This is also consistent with a recent report describing potent neutralizing antibodies forming directed interactions with the N343 glycan.46
Whereas several other SARS-CoV-2 regions are less conserved, the RBD glycopeptide is highly conserved and represents a prime immunogen to drive neutralizing antibody responses. Our study illustrates that the immunogenicity of RBD is strongly influenced by N-linked glycosylation (Figures 7A–7D). Mutational analyses revealed that the N343 glycan was critical for RBD’s recognition by neutralizing antibodies and, in the context of an RBD-targeted vaccine, the N343A mutation significantly decreased the immunogenicity as measured by blunted induction of anti-RBD IgG and neutralizing antibodies (Figures 7B–7D). These results suggest that a crucial consideration for vaccines using the RBD as an immunogen is the utilization of a production system that will generate the proper glycosylation of the RBD, as this influences the efficacy of the neutralizing antibody response.
SARS-CoV-2-NanoBiT represents a versatile and rapid tool for the identification of novel inhibitors and molecular determinants of coronavirus infection by probing the interaction of monomeric RBD with ACE2. It can be easily adapted for use in high-throughput screening of drug libraries or phage display libraries. Our work further highlights the untapped potential of the NanoLuc complementation-based reporter strategy in identifying antiviral drugs targeting other host-virus interactions, which will undoubtedly be critical to our global response against future pandemics.
Materials and methods
Plasmid construction
Inserts outlined in Table S1 were ordered from GenScript. Bioreporter subunits were cloned into the BamHI/NotI sites of pcDNA3.1 to generate mammalian expression constructs. SARS-CoV-2 RBD-TMD WT and mutant constructs were cloned into the BamHI/NotI sites of pcDNA3.1 or the XhoI/NheI sites of VSV backbone plasmid.
Cell culture
293T (ATCC CRL-3216), HEK293 (ATCC CRL-1573), and HEK293T-ACE2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma) containing 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin (Invitrogen). HEK293T-ACE2 cells were previously described. These 293T cells stably overexpress full-length ACE2 via lentiviral transduction.29
In vitro NanoLuc assay
293T cells (3 × 105 cells) were plated in 12-well plates in triplicate 24 h before transfection. Five hundred nanograms of the bioreporter constructs was transfected using PolyJet transfection reagent (SignaGen Laboratories). After 48 h, supernatant or cells lysates were collected. Cells were lysed using passive lysis buffer (Promega). NanoLuc luciferase assays were performed using one of two substrates: FMZ/ (Nano-Glo cell reagent, Promega) or native CTZ (3.33 μM final concentration; Nanolight Technologies-Prolume, Pinetop, AZ, USA). A Synergy microplate reader (BioTek, Winooski, VT, USA) was used to measure luminescence. Results are presented as RLU normalized to control. The data presented are the mean of three independent experiments.
Bioluminescence imaging
Lysates or supernatants from 293T cells transfected with the bioreporter construct were imaged in a 96-well plate. Nano-Glo cell reagent was added to the lysate in a 96-well plate as per the manufacturer’s protocols for the NanoLuc luciferase assay kit (Promega). Plates were imaged with the IVIS 200 series (Xenogen). Data acquisition and analysis were performed using the Living Image v2.5 procedure in Igor Pro 4.09 software.
Structure analysis
Protein sequence alignments were performed using ClustalW.47 X-ray crystal structures for the RBDs of SARS-CoV and SARS-CoV-2 in complex with ACE2 were obtained from the Protein Data Bank (PDB: 2AJF and 6M0J, respectively) and visualized with PyMOL (v2.0; Schrödinger)19,20. The RBD alignment and root-mean-square deviation (RMSD) calculations were also performed in PyMOL.
Bioreporter-based neutralization assay
For the neutralization assay with monoclonal antibodies and patient sera, 5 μg of RBD-LgBiT containing cell lysates was incubated at 37°C for 25 min with candidate antibodies or serum. Then, 50 μg of SmBiT-ACE2-transfected cell lysate was added and then incubated for an additional 5 min at room temperature. The amount of mentioned protein was determined using a bicinchoninic acid (BCA) assay, and it refers to total protein. Subsequently, a luciferase assay was performed. The following monoclonal RBD antibodies were tested in the bioreporter-based neutralization assay: 1A9 (GeneTex, GTX632604); 2414 (Active Motif, 91349), 1414 (Active Motif, 91361), 273074 (Abcam, ab273074), 40592 (Sino Biological, 40592-MM57), 9ACA (GenScript, 5B7D7), 11D11F2 (GenScript), 10G6H5 (Genscript), HC2001 (GenScript), and L00847 (Genscript Biotech).
Pseudotyped lentivirus assay
SARS-CoV-2 S pseudotyped lentivirus was produced as previously described using plasmids kindly provided by Dr. Jesse Bloom (Fred Hutchinson Cancer Research Center, Seattle, WA, USA).29 For glycosite mutants, HDm-IDTSpike-fixK was mutated using a QuikChange SDM kit (Stratagene) using the primers listed in Table S1, as per the manufacturer’s protocols. Briefly, HEK293 cells were co-transfected with HDM-IDTSpike-fixK, pHAGE-CMV-Luc2-IRES-ZsGreen-W, and pSPAX2. 48 h post-transfection, cell supernatants containing virus were collected and treated with either PNGase F or Endo H for 1 h. HEK293T-ACE2 cells were subsequently transduced and transduction efficiency was assessed by a luciferase assay using the Bright-Glo luciferase assay system (Promega) or fluorescence microscopy (EVOS cell imaging system, Thermo Fisher Scientific). Where indicated, lentivirus titers were measured using Lenti-X GoStix Plus (Takara) as per the manufacturer’s protocols. For lectin inhibition assays, S pseudotyped lentivirus was co-incubated with lectins for 1 h, and then the virus/lectin mixture was applied to HEK293-ACE2 cells as described above.
Surrogate SARS-CoV-2 neutralization assay
Receptor-ligand binding ELISA was performed as per the manufacturer’s protocols (GenScript, L00847).
Statistical analysis
All graphs and statistical analyses were generated using Excel or GraphPad Prism v.8. Means of two groups were compared using a two-tailed unpaired Student’s t test. Means of more than two groups were compared by one-way ANOVA with a Dunnett’s or Tukey’s multiple comparison correction. Alpha levels for all tests were 0.05, with a 95% confidence interval. Error was calculated as the standard deviation (SD). Measurements were taken from distinct samples. For all analyses, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; n.s., not significant. Data were reproduced by two different operators.
Other materials
PNGase F (P0704S) and Endo H (P0702S) were purchased from NEB. A native deglycosylation kit containing Endo F1, F2, and F3 was purchased from Sigma-Aldrich (NDEGLY-1KT). RBD (230-30162-100) and ACE2 (00707-01-05B) recombinant protein was purchased from RayBiotech. Lectins used in this study were purchased from Sigma-Aldrich (L2766, L5380, L8275, and L7647) or MilliporeSigma (L1277). Biotinylated Con A was purchased from Vector Laboratories (B1055).
VSVΔ51 virus rescue
Inserts (see Table S1 for detailed sequences) were ordered from GenScript (Piscataway, NJ, USA). SARS-CoV-2 RBD-TMD constructs were cloned into the XhoI/NheI sites of VSV backbone plasmid. Recombinant VSVΔ51 viruses expressing SARS-CoV-2 WT or mutant RBD-TMD were rescued as previously described.48
In vivo vaccination studies
Female 6-week-old BALB/C mice (Charles River Laboratories, Malvern, PA, USA) were vaccinated intravenously with 1E7 plaque-forming units (PFU) of VSVΔ51-expressing RBD-TMD WT or mutants (N331A or N343A). Sera were collected from mice using saphenous vein bleeds at days 7 and 14 post-inoculation using sera collection tubes. Blood was incubated on ice for 30 min and then centrifuged to separate sera.
Flow cytometry staining
Cells were stained first with anti-FLAG (M2, Sigma, F3165) for 30 min at 4°C. Cells then washed twice with fluorescence-activated cell sorting (FACS) buffer (0.5% bovine serum albumin [BSA]/PBS), then resuspended in 1% paraformaldehyde (PFA) buffer for analysis by flow cytometry on a BD LSRFortessa. Data were analyzed with FlowJo software.
Immunofluorescence
Cells were seeded in a 24-well plate with poly-l-lysine in an appropriate number to achieve 80% confluency at the time of immunostaining. Cells were fixed in 5% formaldehyde for 10 min. Cells were subsequently incubated with primary and secondary antibodies diluted in PBS with 1% BSA and 10% goat serum.
Native PAGE analysis
Whole-cell lysates were harvested from transfected cells using NativePAGE sample kit (Invitrogen, BN2008). Samples were homogenized in buffer containing 2% digitonin on ice and centrifuged to clarify the lysates. 10 μg of total proteins was loaded onto a pre-cast NativePAGE Bis-Tris gel (Invitrogen, BN1002BOX). The upper cathode chamber was filled with 200 mL of 1× NativePAGE dark blue cathode buffer (BN2002), and the lower anode chamber was filled with 550 mL of NativePAGE anode (running) buffer (Invitrogen, BN2001). Once the dye front reached one-third the length of the gel, the dark blue cathode buffer was replaced with light blue cathode buffer. The gel was run for 1.5 h at a constant voltage of 150 V.
Patient sera and ethics approval
All animal studies were approved by the Institutional Animal Care Committee of the University of Ottawa and carried out in accordance with guidelines of the National Institutes of Health and the Canadian Council on animal care. All sera samples were collected with informed consent from individuals being treated at the Ottawa Hospital General Campus under a protocol approved by the Institutional Ethics Board.
SDS-PAGE electrophoresis and immunoblotting
Whole-cell lysates were obtained by lysing cells in radioimmunoprecipitation assay (RIPA) buffer (pH 7.4; 25 mM Tris-HCl, 150 mM NaCl, 1% Nonidet P-40 [NP-40], 0.5% sodium deoxycholate, SDS) and 1× protease inhibitor cocktail (Roche) on ice. Protein concentration was measured by a Pierce BCA assay (Thermo Scientific), and 10 μg of cell extract was mixed into DTT-Laemmli buffer and boiled for 5 min. Samples were resolved using the NuPAGE SDS-PAGE system (Invitrogen) and transferred to a nitrocellulose membrane. Blots were probed with primary antibodies, including anti-FLAG (1:1,000, MilliporeSigma, F3165), anti-β-actin (1:10,000, Sigma, A5441), anti-hemagglutinin (HA) (1:1,000, Sigma, H6908), anti-RBD (1:1,000, RayBiotech, 130-10759), and anti-S (1:1,000, GeneTex, 1A9), and then washed and probed with the appropriate secondary antibodies, including anti-mouse, anti-rabbit (MilliporeSigma, A9169) or anti-goat (Abcam, Cambridge, UK, ab97110). Blots were imaged using the ChemiDoc MP imaging system (Bio-Rad Laboratories, Mississauga, ON, Canada). Clarity western enhanced chemiluminescence (ECL) substrate (Bio-Rad) was used to visualize the blot.
Dot blot
Cell lysates were harvested using RIPA buffer supplemented with a protease/phosphatase inhibitor cocktail on ice. Protein concentration was determined by a BCA assay, and 5–10 μg of whole-cell lysate was loaded directly onto nitrocellulose membrane. After 15 min of incubation at room temperature (RT), membranes were blocked in 5% milk in Tris-buffered saline with Tween 20 (TBST), then sequentially incubated with primary and secondary antibodies. For detection of RBD glycosylation, Carbo-Free blocking solution (Vector Laboratories) was used for blocking, then, after sample loading, the membrane was incubated in biotinylated lectin for 30 min at RT. Blots were incubated with streptavidin horseradish peroxidase (HRP) conjugate and developed as described above.
VSV-S neutralization assay
VSV pseudotyped with S (VSV-S) was a kind gift from Dr. Sean Whelan (Washington University, St. Louis, MO, USA).34 Vero E6 cells were seeded in 96-well plates such that 4E5 cells were in each well at the time of infection. Serial dilutions of mouse sera were performed and then co-incubated with an equal volume of VSV-S (2,000 PFU per well) and incubated for 1 h at 37°C. After 1 h, media on the cell were replaced with 60 μL of the virus/serum and incubated for 1 h at 37°C. Wells were then topped up with carboxymethylcellulose (CMC) in DMEM supplemented with 10% FBS for a final concentration of 3% CMC and incubated 24 h at 34°C. GFP foci were imaged and counted using a Cellomics ArrayScan VTI HCS reader.
Acknowledgments
LgBiT-YAP15 and 14-3-3-SmBiT plasmids were gifts from Dr. Yang (Queen’s University, Canada). Schematic illustrations were designed by Dr. Mina Ghahremani (Designs that Cell). T.A. acknowledges the Banting Fellowship for funding support. R.S. is supported by post-doctoral funding from CIHR. T.A., R.S., N.T.M., M.J.F.C., and J.P. are recipients of funding support from MITACS Accelerate. J.C.B., J.-S.D., and C.S.I. are supported by grants from the CCSRI, CIHR, and TFFRI. Z.T. acknowledges NSERC for funding support in the form of a graduate scholarship. We thank Dr. Sean Whelan for sharing VSV pseudotyped with spike protein. Thanks to This Week in Virology for inspiration, specifically Dr. Vincent Racaniello. Thanks to Dr. Hamid Mahdizadeh for his valuable feedbacks.
Author contributions
Conceptualization, T.A., R.S., M.G., K.N., P.A.G., and J.C.B.; methodology, T.A. and R.S.; investigation, T.A., R.S., Z.T., A.P., N.T.M., N.A., M.C., T.J., E.E.F.B., J.P., M.G., R.S., R.R., and R.A.; formal analysis, T.A. D.W.C., and S.G.; visualization, T.A.; illustration,T.J., E.E.F.B., M.G.; writing – original draft, T.A., R.S., Z.T., and J.C.B.; writing – reviewing & editing, T.A., R.S., Z.T., S.B., A.C.G.,M.C., R.R., R.A., P.A.G., C.S.I., J.-S.D., and J.C.B.; supervision; funding acquisition, J.C.B.; program administration, J.C.B.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.02.007.
Supplemental information
References
- 1.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . 2020. Coronavirus disease 2019 (COVID-19) weekly epidemiology update.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200324-sitrep-64-covid-19.pdf?sfvrsn=703b2c40_2 [Google Scholar]
- 3.Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun C., Chen L., Yang J., Luo C., Zhang Y., Li J., Yang J., Zhang J., Xie L. SARS-CoV-2 and SARS-CoV spike-RBD structure and receptor binding comparison and potential implications on neutralizing antibody and vaccine development. bioRxiv. 2020 doi: 10.1101/2020.02.16.951723. [DOI] [Google Scholar]
- 7.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozono S., Zhang Y., Ode H., Tan T.S., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., Suzuki T., Tokunaga K. Naturally mutated spike proteins of SARS-CoV-2 variants show differential levels of cell entry. bioRxiv. 2020 doi: 10.1101/2020.06.15.151779. [DOI] [Google Scholar]
- 10.Dixon A.S., Schwinn M.K., Hall M.P., Zimmerman K., Otto P., Lubben T.H., Butler B.L., Binkowski B.F., Machleidt T., Kirkland T.A. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 2016;11:400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- 11.Nouri K., Azad T., Lightbody E., Khanal P., Nicol C.J., Yang X. A kinome-wide screen using a NanoLuc LATS luminescent biosensor identifies ALK as a novel regulator of the Hippo pathway in tumorigenesis and immune evasion. FASEB J. 2019;33:12487–12499. doi: 10.1096/fj.201901343R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nouri K., Azad T., Ling M., Janse van Rensburg H.J., Pipchuk A., Shen H., Hao Y., Zhang J., Yang X. Identification of celastrol as a novel YAP-TEAD inhibitor for cancer therapy by high throughput screening with ultrasensitive YAP/TAZ-TEAD biosensors. Cancers (Basel) 2019;11:1596. doi: 10.3390/cancers11101596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azad T., Tashakor A., Hosseinkhani S. Split-luciferase complementary assay: applications, recent developments, and future perspectives. Anal. Bioanal. Chem. 2014;406:5541–5560. doi: 10.1007/s00216-014-7980-8. [DOI] [PubMed] [Google Scholar]
- 14.Remy I., Michnick S.W. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 15.Paulmurugan R., Gambhir S.S. Monitoring protein-protein interactions using split synthetic Renilla luciferase protein-fragment-assisted complementation. Anal. Chem. 2003;75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azad T., Janse van Rensburg H.J., Lightbody E.D., Neveu B., Champagne A., Ghaffari A., Kay V.R., Hao Y., Shen H., Yeung B. A LATS biosensor screen identifies VEGFR as a regulator of the Hippo pathway in angiogenesis. Nat. Commun. 2018;9:1061. doi: 10.1038/s41467-018-03278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ataei F., Torkzadeh-Mahani M., Hosseinkhani S. A novel luminescent biosensor for rapid monitoring of IP3 by split-luciferase complementary assay. Biosens. Bioelectron. 2013;41:642–648. doi: 10.1016/j.bios.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 18.Hall M.P., Unch J., Binkowski B.F., Valley M.P., Butler B.L., Wood M.G., Otto P., Zimmerman K., Vidugiris G., Machleidt T. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Crystal structure of the 2019-nCoV spike receptor-binding domain bound with the ACE2 receptor. bioRxiv. 2020 doi: 10.1101/2020.02.19.956235. [DOI] [PubMed] [Google Scholar]
- 20.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 21.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han D.P., Penn-Nicholson A., Cho M.W. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350:15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.C., Tiu C., Hu Z., Chen V.C., Young B.E., Sia W.R. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 27.Ou J., Zhou Z., Dai R., Zhang J., Lan W., Zhao S., Wu J., Seto D., Cui L., Zhang G., Zhang Q. Emergence of RBD mutations in circulating SARS-CoV-2 strains enhancing the structural stability and human ACE2 receptor affinity of the spike protein. bioRxiv. 2020 doi: 10.1101/2020.03.15.991844. [DOI] [Google Scholar]
- 28.Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., Aoki K., Kellman B.P., Bridger R., Barouch D.H. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28:586–601.e6. doi: 10.1016/j.chom.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12:12. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keyaerts E., Vijgen L., Pannecouque C., Van Damme E., Peumans W., Egberink H., Balzarini J., Van Ranst M. Plant lectins are potent inhibitors of coronaviruses by interfering with two targets in the viral replication cycle. Antiviral Res. 2007;75:179–187. doi: 10.1016/j.antiviral.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 32.Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.M., Zeng Q., Tahan S., Droit L. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 36.Wei X.-F., Gan C.-Y., Cui J., Luo Y.-Y., Cai X.-F., Yuan Y., Shen J., Li Z.Y., Zhang W.L., Long Q.X. Identification of compounds targeting hepatitis B virus core protein dimerization through a split luciferase complementation assay. Antimicrob. Agents Chemother. 2018;62:e01302–e01318. doi: 10.1128/AAC.01302-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng Q., Wang D., Xiang X., Gao X., Hardwidge P.R., Kaushik R.S., Wolff T., Chakravarty S., Li F. Application of a split luciferase complementation assay for the detection of viral protein-protein interactions. J. Virol. Methods. 2011;176:108–111. doi: 10.1016/j.jviromet.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shajahan A., Supekar N.T., Gleinich A.S., Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30:981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., Aoki K., Kellman B.P., Bridger R., Barouch D.H. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28:586–601.e6. doi: 10.1016/j.chom.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Covés-Datson E.M., King S.R., Legendre M., Gupta A., Chan S.M., Gitlin E., Kulkarni V.V., Pantaleón García J., Smee D.F., Lipka E. A molecularly engineered antiviral banana lectin inhibits fusion and is efficacious against influenza virus infection in vivo. Proc. Natl. Acad. Sci. USA. 2020;117:2122–2132. doi: 10.1073/pnas.1915152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller J.L., Tyrrell B.E., Zitzmann N. Mechanisms of antiviral activity of iminosugars against dengue virus. In: Hilgenfeld R., Vasudevan S.G., editors. Dengue and Zika: Control and Antiviral Treatment Strategies. Springer Singapore; 2018. pp. 277–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker L.M., Phogat S.K., Chan-Hui P.-Y., Wagner D., Phung P., Goss J.L., Wrin T., Simek M.D., Fling S., Mitcham J.L., Protocol G Principal Investigators Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wibmer C.K., Bhiman J.N., Gray E.S., Tumba N., Abdool Karim S.S., Williamson C., Morris L., Moore P.L. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog. 2013;9:e1003738. doi: 10.1371/journal.ppat.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azad T., Singaravelu R., Crupi M.J.F., Jamieson T., Dave J., Brown E.E.F., Rezaei R., Taha Z., Boulton S., Martin N.T. Implications for SARS-CoV-2 vaccine design: fusion of spike glycoprotein transmembrane domain to receptor-binding domain induces trimerization. Membranes (Basel) 2020;10:215. doi: 10.3390/membranes10090215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.