Abstract
Background
Celastrol is a potential anti-tumor agent in hepatocellular carcinoma (HCC). Identifying the molecular determinants of the anti-HCC effect of celastrol is still challenging. In this study, we undertook to associate circular RNAs (circRNAs) with the anti-HCC molecular determinants of celastrol.
Methods
Cell colony formation, proliferation, migration, invasion and apoptosis were determined using the colony formation, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-2H-tetrazolium bromide (MTS), transwell and flow cytometry assays, respectively. The levels of circRNA slit guidance ligand 3 (circ_SLIT3), miR-223-3p and C-X-C motif chemokine receptor 4 (CXCR4) were gauged by quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot. Ribonuclease R (RNase R) and actinomycin D assays were performed to assess the stability of circ_SLIT3. Targeted relationships among circ_SLIT3, miR-223-3p and CXCR4 were confirmed by the dual-luciferase reporter assay. In vivo assays were performed to detect the roles of celastrol and circ_SLIT3 on tumor growth in vivo.
Results
Celastrol repressed HCC cell proliferation, migration, invasion, and enhanced apoptosis in vitro and suppressed tumor growth in vivo. Celastrol down-regulated circ_SLIT3 expression in HCC cells, and celastrol exerted an anti-tumor effect on HCC in vitro and in vivo by down-regulating circ_SLIT3. Mechanistically, circ_SLIT3 directly interacted with miR-223-3p, and circ_SLIT3 controlled CXCR4 expression by sponging miR-223-3p. Moreover, miR-223-3p was involved in the celastrol/circ_SLIT3-mediated regulation on HCC progression. Furthermore, celastrol exerted the anti-HCC effect in vitro through the miR-223-3p/CXCR4 axis.
Conclusion
Our present work first identified the circ_SLIT3/miR-223-3p/CXCR4 axis as a novel mechanism of the anti-HCC effect of celastrol, providing a new insight into the involvement of circRNAs in the anti-tumor molecular determinants of celastrol.
Keywords: hepatocellular carcinoma, celastrol, circ_SLIT3, miR-223-3p, CXCR4
Introduction
Hepatocellular carcinoma (HCC) accounts for ~85% of primary liver cancer cases and is one of the most prevalent malignancies worldwide.1 Although advanced therapeutic approaches have significantly improved the outcome of early-stage HCC, the overall survival of advanced disease is still poor.2 Therefore, developing more efficacious treatments has always been challenging.
Celastrol, a pentacyclic triterpene, is one of the natural active components isolated from the medicinal plant Tripterygium wilfordii Hook F.3 Emerging evidence is pointing towards celastrol as a potential anti-tumor agent in various cancers, including HCC.4,5 Moreover, recent studies have indicated the involvement of noncoding RNAs (ncRNAs), such as microRNAs (miRNAs), in the molecular mechanism underlying the anti-HCC activity of celastrol.6 For example, Li et al reported that celastrol suppressed HCC cell metastasis by interfering with microRNA (miRNA)-224 activity.7 Although the anti-tumor property of celastrol has been widely accepted, our understanding of its anti-HCC molecular action is still limited.
Circular RNAs (circRNAs), a novel class of noncoding RNAs, are covalently closed RNA circles that are attractive candidates as key players of HCC carcinogenesis by functioning as sponges of available miRNAs.8 For example, Yu et al unraveled that hsa_circ_0001445 weakened the aggressive progression of HCC through sequestering miR-17-3p.9 Li et al reported that hsa_circ_101368 contributed to HCC cell malignant behaviors by working as a miR-200a sponge.10 As for circRNA slit guidance ligand 3 (circ_SLIT3, hsa_circ_0074903), produced by the back-splicing of SLIT3, it has been discovered to be overexpressed in HCC.11 Nevertheless, no insight has been gained into the precise action of circ_SLIT3 in HCC progression. Furthermore, it is still unclear whether circ_SLIT3 was a downstream mediator of celastrol in repressing HCC progression.
For these reasons, we undertook to associate circ_SLIT3 with the anti-HCC molecular determinants of celastrol, with the hope that such association might provide a novel insight into the molecular basis of the anti-tumor property of celastrol.
Materials and Methods
Specimen Collection
Ethics approval of this study was obtained from the Ethics Committee of Affiliated Hospital of the Shaanxi University of Traditional Chinese Medicine, and all 50 volunteers signed written informed consent. These patients were pathologically confirmed with primary HCC at Affiliated Hospital of the Shaanxi University of Traditional Chinese Medicine from October 2017 to May 2019. Tumor tissues and adjacent normal hepatic tissues were collected after hepatectomy. Histological analysis of tumor tissues and normal tissues was confirmed by our hospital using haematoxylin and eosin (H&E) stain as reported.12 The study was carried out in accordance with the World Medical Association Declaration of Helsinki.
Cell Culture and Treatment
Huh7 and Hep3B HCC cells (Bnbio, Beijing, China) were used in this study. An immortalized THLE-2 cell line (Bnbio) was used as a control of noncancerous cells. Huh7 and Hep3B cells were propagated at 37°C, 5% CO2 in complete growth medium (Dulbecco’s modified Eagle’s medium + 10% fetal bovine serum + 2 mM L-glutamine) provided by Thermo Fisher Scientific (Waltham, VA, USA). The cultivation of THLE-2 was performed using BEGM Bullet Kit as recommended by the manufacturers (Lonza, Basel, Switzerland). Huh7 and Hep3B cells of ~70% confluence were treated with 0.5 or 1 µM of celastrol (Acme, Shanghai, China) for 48 h to assess the anti-HCC effect of celastrol in vitro.
Generation of Gene, miRNA Overexpressing and Knockdown Cells
The sequence of circ_SLIT3 was synthesized by Genewiz (Suzhou, China), digested by EcoR I and BamH I enzymes (TaKaRa, Dalian, China) and then cloned into pCD25-ciR or pLC5-ciR vector (Geneseed, Guangzhou, China) to produce circ_SLIT3 overexpressing plasmid (circ_SLIT3) or circ_SLIT3 lentiviral vector (lenti-circ_SLIT3). Nontarget corresponding vector (Vector or lenti-NC) was used as negative controls. To produce lentivirus for circ_SLIT3 overexpression in cells, lenti-circ_SLIT3 was transfected into HEK293T packing cells (Bnbio) together with lentiviral packaging mix (Addgene, Cambridge, MA, USA). Virus particles were collected at 72 h after transfection and then used to transduce Hep3B cells in media containing 8 µg/mL of polybrene (Yesen, Shanghai, China). Vector-transduced cells were selected using 2.5µg/mL of puromycin over 96 h. For in vitro circ_SLIT3 overexpression studies, Huh7 and Hep3B cells (1.0 × 105) were transiently transfected with 100 ng of circ_SLIT3 overexpressing plasmid using Lipofectamine 3000 (Thermo Fisher Scientific). For in vitro alteration of miR-223-3p studies, Huh7 and Hep3B cells (1.0 × 105) were introduced with 30 nM of miR-223-3p mimic (5ʹ-ACCCCAUAAACUGUUUGACUGU-3ʹ, Ribobio, Guangzhou, China), inhibitor to conceal miR-223-3p (anti-miR-223-3p, 5ʹ-ACAGUCAAACAGUUUAUGGGGU-3ʹ, Ribobio) or corresponding scrambled sequences (miR-NC mimic, 5ʹ-ACGUGACACGUUCGGAGAATT-3ʹ; anti-miR-223-3p, 5ʹ-CUAACGCAUGCACAGUCGUACG-3ʹ) using Lipofectamine 3000. The knockdown of C-X-C motif chemokine receptor 4 (CXCR4) was carried out using 50 nM of CXCR4-siRNA (si-CXCR4, 5ʹ-UUAUCUGAAGUGUAUAUACUG-3ʹ), with NON-siRNA (si-NC, 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ) as the control group. Twenty-four hours post-transfection, cells were plated or harvested for further analyses.
Cell Colony Formation, Proliferation and Apoptosis Assays
For colony formation assays, about 120 cells after various treatments were plated in triplicate into 6-well plates in complete medium and cultured under standard protocols for 14 days. After being stained with 1% crystal violet, the number of colonies (over 50 cells) formed on each well was determined using a microscope (Leica, Wetzlar, Germany) at 10× magnification. For proliferation assays, about 1000 treated cells were plated into 96-well plates in triplicate and cultured under standard protocols. After 1, 2 and 3 days, 10 µL of 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-2H-tetrazolium bromide (MTS, Promega, Madison, WI, USA) reagent was used to monitor the number of viable cells based on the absorbance at OD570 nm. For apoptosis assays, a total of 10,000 events were analyzed for each sample using a FACScan flow cytometer (BD Biosciences, Cowley, UK) after being double-stained with 10 µL of fluorescein isothiocyanate (FITC)-labeled Annexin V and 5 µL of propidium iodide (PI) as recommended by the manufacturers.
Transwell Migration and Invasion Assays
Transwell migration and invasion assays were conducted using 24-well Boyden chambers (Corning, Tewksbury, MA, USA) with 8-µm pore inserts and Matrigel-precoated membrane inserts (BD Biosciences), respectively. Treated cells were seeded onto the upper surface of the chamber at 10,000 cells per well for migration assays and 50,000 cells for invasion assays in serum starvation media. The lower chamber was filled with a complete growth medium. The penetrated cells through the membrane inserts were stained with 1% crystal violet after 24 h incubation and counted with the Leica microscope at 100× magnification to get an average in five random fields.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA including small RNA extracted from tissues and cells with the RNeasy Mini Kit (Qiagen, Hilden, Germany) averaged 120 ± 16 ng/µL when was quantified by a spectrophotometer. For circ_SLIT3 and mRNA quantification, reverse transcription (RT) from 50 ng of RNA was conducted using GoScript RT System (Promega) and qRT-PCR was done using SYBR Green Master (Thermo Fisher Scientific) with gene-specific primers (Table 1). For miR-223-3p quantification, miScript RT Kit for RT and SYBR Green mix for qRT-PCR were used as recommended by the manufacturers (Qiagen). Human β-actin or U6 was used for normalization with specific primers in Table 1. Fold changes of circ_SLIT3, miR-223-3p and CXCR4 gene were calculated using the equation 2−ΔΔCt.13
Table 1.
Primers Sequences Used for PCR
| Primers for PCR (5ʹ-3ʹ) | ||
|---|---|---|
| Circ_SLIT3 | Forward | GCTGGTCTATGACAGCCTGA |
| Reverse | TTCTGGCACTCGTACTGGTC | |
| SLIT3 linear mRNA | Forward | TCGTGGGCAAAGACTCCTAC |
| Reverse | GGGGAACTCAGGCTGTCATA | |
| CXCR4 | Forward | GAGGGGATCAGTATATACAC |
| Reverse | GGGAAGCGTGATGACAAAG | |
| miR-223-3p | Forward | GCCGAGACCCCAUAAACUG |
| Reverse | CAGTGCGTGTCGTGGAGT | |
| β-actin | Forward | CTCGCCTTTGCCGATCC |
| Reverse | GGGGTACTTCAGGGTGAGGA | |
| U6 | Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT | |
Ribonuclease R (RNase R) and Actinomycin D Assays
In RNase R assays, 2.5 µg of total RNA was incubated with 10 U of RNase R (Geneseed) at 37°C for 20 min. In actinomycin D assays, cells were incubated with actinomycin D (Leagene, Beijing, China) in a final concentration of 5 µM for 8, 16 and 24 h.
Western Blot
Protein extracts from tissues and cells were analyzed by Western blot.14 Proteins (100 µg) were resolved on 5–12% polyacrylamide gels and transferred onto Immobilon-P membranes (Millipore, Shanghai, China). Membranes were probed with the following antibodies: primary antibodies anti-Ki67 (ab197547; dilution 1:1000), anti-B cell lymphoma-2 (anti-Bcl-2, ab238042; dilution 1:1000), anti-CXCR4 (ab124824; dilution 1:100), anti-Bcl2 associated X (anti-Bax, ab104156; dilution 1:1000), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH, ab22555; dilution 1:3000), and anti-rabbit IgG secondary antibody labeled by horseradish peroxidase (HRP) (ab20518). All antibodies were obtained from Abcam (Cambridge, UK) and used as per the manufacturing protocols, and chemiluminescence reagents were purchased from Thermo Fisher Scientific for visualization. Protein bands were analyzed by Image J software (Rawak Software, Stuttgart, Germany).
Dual-Luciferase Reporter Assay
CircInteractome online database was used to predict the miRNAs that potentially bind to circ_SLIT3. Analysis of miR-223-3p targets was done using the starBase v.3 online tool. The fragments of circ_SLIT3 and CXCR4 3ʹUTR harboring the putative miR-223-3p-binding sites or mutated target sites were individually cloned into the pmirGLO vector (Promega) to create corresponding wild-type or mutant-type luciferase reporter constructs. Each reporter construct (200 ng) was cotransfected with 30 nM of miR-223-3p mimic or negative mimic control into Huh7 and Hep3B cells (1.0 × 105) in 24-well plates. Forty-eight hours post-transfection, 20 µL of cell lysates was used to detect the luciferase activity with Dual-Luciferase Assay (Promega).
In vivo Studies
Female BALB/c nude mice (6-week-old, n = 24) from Yisonbio (Shanghai, China) were used in this study and divided into four groups (n = 6 each group): PBS control, celastrol, celastrol+lenti-NC and celastrol+lenti-circ_SLIT3. Briefly, un-transduced, lenti-NC- or lenti-circ_SLIT3-transduced Hep3B cells (5.0 × 106) were injected under the subcutaneous layer into the left flank of the nude mice. The administration of celastrol (2 mg/kg) or PBS was performed after 5-day implantation by intraperitoneal injection every 5 days. Tumor growth was monitored by calculating tumor volume with the formula: volume = (length × width2)/2. At the end of the experiments, all mice were euthanized and tumor tissues were analyzed. All protocols for mouse experiments were approved by the Animal Care and Use Committee of Affiliated Hospital of the Shaanxi University of Traditional Chinese Medicine. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Immunohistochemistry
Mice tumor tissues were fixed with 4% neutral-buffered formalin, dehydrated, embedded in paraffin, and then sectioned at 4 µm intervals. The sections were dewaxed and then heated in 0.01 M of sodium citrate buffer for antigen retrieval. After the incubation of methanolic H2O2, tissue sections were incubated with anti-CXCR4 (ab227767; dilution 1:50), anti-Ki67 (ab197547; dilution 1:100), anti-Bcl-2 (ab238042; dilution 1:100) and anti-Bax (ab32503; dilution 1:250) antibodies, followed by the incubation with HRP-labeled secondary antibody (ab6721, all from Abcam). Band signals were visualized by 3,3ʹ-diaminobenzidine substrate (Thermo Fisher Scientific) and then were counterstained with hematoxylin and mounted.
Statistical Analysis
Data were expressed as the mean ± standard deviation (SD). The Student’s t-test and one-way analysis of variance (ANOVA) were used to compare the measured parameters, using Prism v.7 software (GraphPad Software, La Jolla, CA, USA). Mann–Whitney U-test was used to compare the difference in gene expression between tumor tissues and matched normal tissues. Independent relationships between gene expression levels in tumor specimens were determined by the Spearmen correlation analysis. A two-tailed p-value < 0.05 was considered significant, with 3 biological × 3 technical replicates.
Results
Celastrol Inhibited HCC Cell Proliferation, Migration, Invasion and Enhanced Apoptosis in vitro
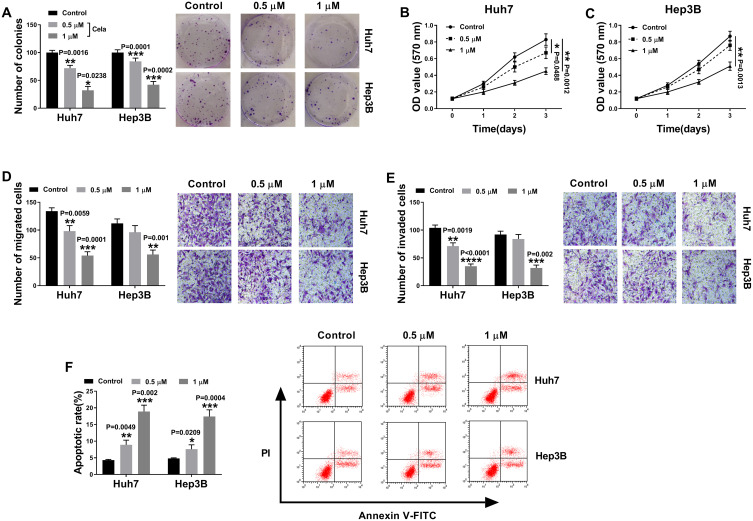
We firstly examined the anti-tumor effect of celastrol on HCC in vitro. The data of colony formation and MTS assays showed that in contrast to the negative control, celastrol induced a striking inhibition in cell colony formation (Figure 1A) and proliferation (Figure 1B and C). Transwell assays revealed that celastrol resulted in reduced abilities of cell migration (Figure 1D) and invasion (Figure 1E). Conversely, cell apoptosis was promoted by celastrol in both Huh7 and Hep3B cells (Figure 1F).
Figure 1.
Celastrol performed a potential anti-tumor effect on HCC in vitro. Huh7 and Hep3B cells were treated with mock, 0.5 or 1 µM of celastrol (Cela) for 48 h. (A) Colony formation assay for the colony formation capacity of treated cells. (B and C) MTS assay for cell proliferation. (D and E) Transwell assay for cell migration and invasion. (F) Flow cytometry for cell apoptosis. n = 3; data were presented as mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001 or ****P < 0.0001 vs Control.
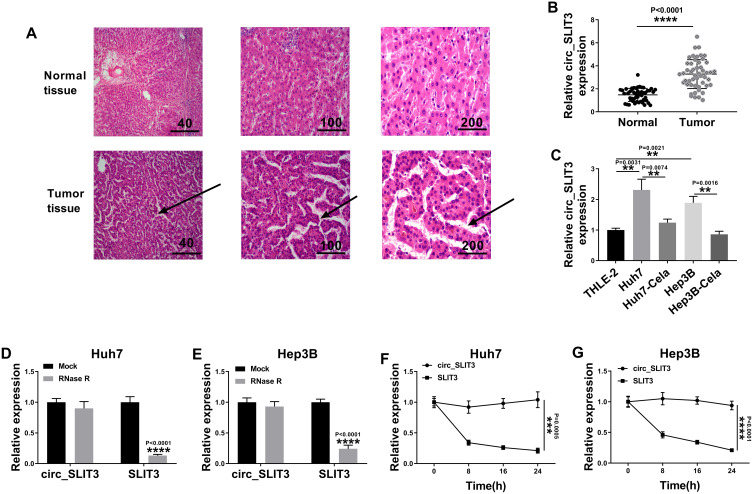
Circ_SLIT3 Was Overexpressed in HCC Tissues and Cell Lines
To evaluate the involvement of circ_SLIT3 in HCC progression, we collected 50 pairs of HCC tissues and adjacent normal tissues confirmed by H&E stain. As an example, the properties of the cells of HCC tissues (patients: 56 years old, TNM stage I–II, no lymph node metastasis) were disordered arrangement and variable size (Figure 2A). As demonstrated by qRT-PCR, circ_SLIT3 level was up-regulated in HCC tissues and cells (Figure 2B and C). Of interest, celastrol led to a significant reduction in the expression of circ_SLIT3 in the two HCC cells (Figure 2C). We then performed RNase R and actinomycin D assays to assess the stability of circ_SLIT3. By contrast, RNase R caused a significant decrease in the level of the corresponding linear transcript but barely affected circ_SLIT3 level (Figure 2D and E). The incubation of cells with actinomycin D resulted in reduced expression of SLIT3 linear mRNA, and circ_SLIT3 level was not affected in the assayed time frame (Figure 2F and G).
Figure 2.
Celastrol down-regulated circ_SLIT3 expression in HCC cells. (A) Confirmation of HCC tissues and matched normal tissues by H&E stain. Arrow represented disordered arrangement of tumor tissues compared with normal tissues. Relative circ_SLIT3 expression by qRT-PCR in 50 pairs of HCC tissues and matched normal tissues (B), THLE-2, Huh7, Hep3B cells and 1 µM of celastrol (Cela)-treated HCC cells (C). The levels of circ_SLIT3 and SLIT3 linear transcript by qRT-PCR in total RNA incubated with RNase R (D and E), Huh7 and Hep3B cells incubated with actinomycin D (F and G). n = 3; data were presented as mean ± SD; **P < 0.01, ***P < 0.001 or ****P < 0.0001 vs Normal, THLE-2 cells, Huh7 cells, Hep3B cells, Mock or circ_SLIT3.
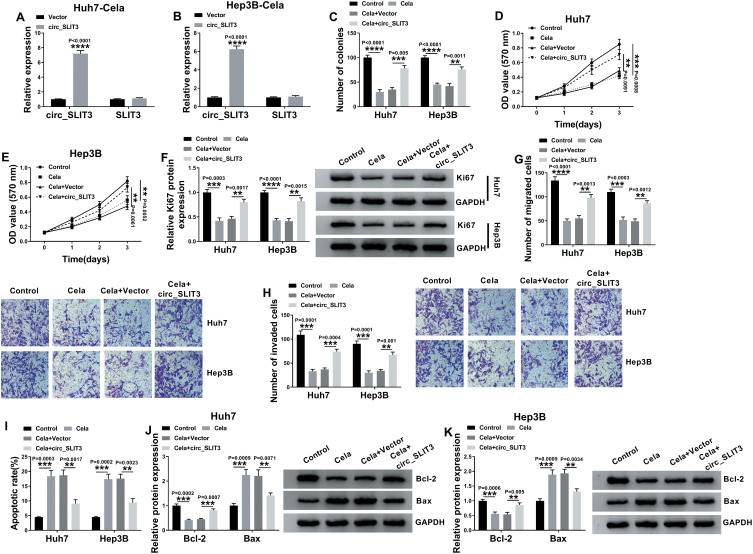
Celastrol Exerted Anti-HCC Effect in vitro by Down-Regulating circ_SLIT3
To directly evaluate whether circ_SLIT3 was a downstream effector of celastrol function, we performed a rescue experiment by up-regulating its level with circ_SLIT3 overexpressing plasmid. In comparison to the control group, transient transfection of circ_SLIT3 overexpressing plasmid led to a striking up-regulation of circ_SLIT3 expression in the two celastrol-treated HCC cell lines but barely affected the expression of the corresponding linear transcript (Figure 3A and B). Function analyses showed that circ_SLIT3 overexpression remarkably abolished celastrol-mediated anti-colony formation (Figure 3C) and anti-proliferation (Figure 3D and E) effects. Western blot data revealed that celastrol induced a prominent decrease in the expression of proliferating marker Ki67, and this effect was dramatically reversed by circ_SLIT3 overexpression (Figure 3F). Moreover, the increased circ_SLIT3 level notably abrogated celastrol-mediated anti-migration (Figure 3G), anti-invasion (Figure 3H) and pro-apoptosis (Figure 3I) effects. Importantly, celastrol triggered a distinct decrease in anti-apoptotic protein Bcl-2 and a strong increase in pro-apoptotic protein Bax in the two HCC cells, and these effects were reversed by the overexpression of circ_SLIT3 (Figure 3J and K).
Figure 3.
Celastrol performed an anti-tumor effect on HCC in vitro through down-regulating circ_SLIT3. (A and B) The levels of circ_SLIT3 and SLIT3 linear transcript by qRT-PCR in celastrol (Cela)-treated Huh7 and Hep3B cells transfected with Vector or circ_SLIT3. Huh7 and Hep3B cells were transfected with Vector or circ_SLIT3 for 24 h and then treated with 1 µM of celastrol (Cela) for 48 h, followed by the detection of cell colony formation using a colony formation assay (C), cell proliferation by MTS assay (D and E), Ki67 level by Western blot (F), cell migration and invasion by transwell assay (G and H), cell apoptosis by flow cytometry (I), the levels of Bcl-2 and Bax by Western blot (J and K). Vector: negative control plasmid, circ_SLIT3: circ_SLIT3 overexpressing plasmid. n = 3; data were presented as mean ± SD; **P < 0.01, ***P < 0.001 or ****P < 0.0001 vs Vector, Control or Cela+Vector.
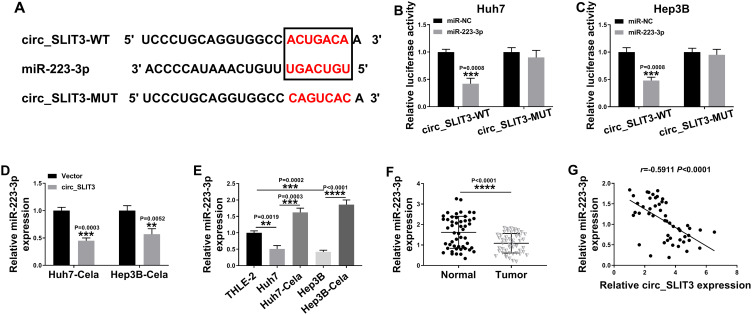
Circ_SLIT3 Directly Interacted with miR-223-3p
To further understand the role of circ_SLIT3, we used online prediction tools to help identify its targeted miRNAs. Of interest, CircInteractome online database revealed a putative miR-223-3p binding sequence (ACUGACA) within circ_SLIT3 (Figure 4A). To verify this, we cloned the circ_SLIT3 fragment harboring the putative target sites into a luciferase plasmid and mutated the seed region. With wild-type luciferase reporter (circ_SLIT3-WT) and miR-223-3p mimic induced a striking down-regulation in luciferase activity (Figure 4B and C). When the miR-223-3p-binding sites were mutated (circ_SLIT3-MUT), no reduction in luciferase was discovered in the presence of miR-223-3p mimic (Figure 4B and C). Moreover, miR-223-3p level was significantly reduced by circ_SLIT3 overexpression in the two celastrol-treated HCC cell lines (Figure 4D). Additionally, miR-223-3p level was remarkably underexpressed in HCC cells and tissues (Figure 4E and F). In contrast, celastrol caused a prominent augmentation in miR-223-3p expression in the two HCC cell lines (Figure 4E). More importantly, in HCC tissues, we observed a strong inverse correlation between miR-223-3p expression and circ_SLIT3 level (Figure 4G).
Figure 4.
Circ_SLIT3 interacted with miR-223-3p by binding to miR-223-3p. (A) Schematic of the putative miR-223-3p binding sequence with circ_SLIT3 identified by CircInteractome tool and the mutation of the seed region. (B and C) Dual-luciferase assays in Huh7 and Hep3B cells. The expression of miR-223-3p by qRT-PCR in celastrol (Cela)-treated Huh7 and Hep3B cells transfected with Vector or circ_SLIT3 (D), THLE-2, Huh7, Hep3B cells and 1 µM of celastrol (Cela)-treated HCC cells (E), and 50 pairs of HCC tissues and matched normal tissues (F). Vector: negative control plasmid, circ_SLIT3: circ_SLIT3 overexpressing plasmid. (G) Correlation between miR-223-3p expression and circ_SLIT3 level in HCC tissues using the Spearman test. n = 3; data were presented as mean ± SD; **P < 0.01, ***P < 0.001 or ****P < 0.0001 vs miR-NC mimic, Vector, THLE-2 cells, Huh7 cells, Hep3B cells or Normal.
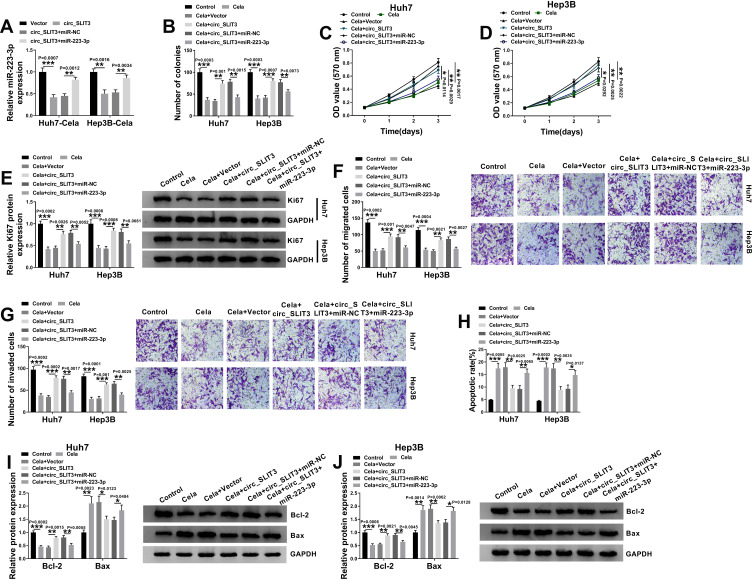
miR-223-3p Was Involved in the Regulation of the Celastrol/circ_SLIT3 Axis on HCC Progression in vitro
To examine whether miR-223-3p represented a functionally important mediator of circ_SLIT3 in celastrol-mediated anti-HCC effect, we cotransfected miR-223-3p mimic and circ_SLIT3 overexpressing plasmid into the two HCC cells before celastrol treatment. As shown by qRT-PCR, the reduction of circ_SLIT3 overexpression on miR-223-3p level was prominently abolished by miR-223-3p mimic (Figure 5A). “Rescue” experiments wherein miR-223-3p was up-regulated significantly reversed circ_SLIT3 overexpression-mediated pro-colony formation (Figure 5B), pro-proliferation (Figure 5C–E), pro-migration (Figure 5F), pro-invasion (Figure 5G) and anti-apoptosis (Figure 5H–J) effects in the two celastrol-treated HCC cell lines.
Figure 5.
The celastrol/circ_SLIT3 axis regulated HCC progression in vitro by miR-223-3p. (A) miR-223-3p expression by qRT-PCR in celastrol (Cela)-treated Huh7 and Hep3B cells transfected with Vector, circ_SLIT3, circ_SLIT3+miR-NC mimic or circ_SLIT3+miR-223-3p mimic. Huh7 and Hep3B cells were transfected with Vector, circ_SLIT3, circ_SLIT3+miR-NC mimic or circ_SLIT3+miR-223-3p mimic for 24 h and then treated with 1 µM of celastrol (Cela) for 48 h, followed by the determination of cell colony formation by colony formation assay (B), cell proliferation by MTS assay (C and D), Ki67 level by Western blot (E), cell migration and invasion by transwell assay (F and G), cell apoptosis by flow cytometry (H), the levels of Bcl-2 and Bax by Western blot (I and J). Vector: negative control plasmid, circ_SLIT3: circ_SLIT3 overexpressing plasmid. n = 3; data were presented as mean ± SD; *P < 0.05, **P < 0.01 or ***P < 0.001 vs Vector, circ_SLIT3+miR-NC, Control, Cela+Vector or Cela+circ_SLIT3+miR-NC.
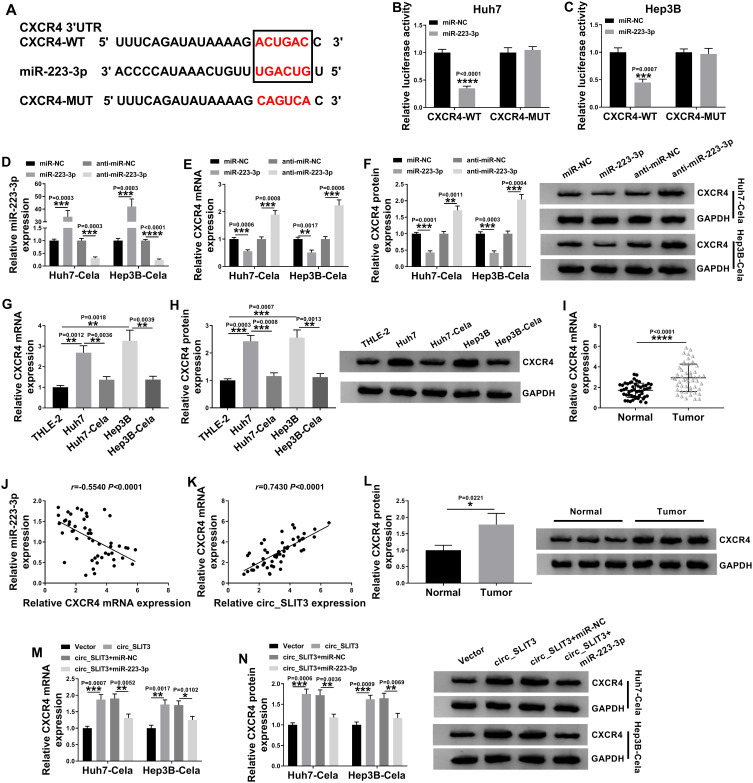
Circ_SLIT3 Controlled CXCR4 Expression by Working as a miR-223-3p Sponge
Using the starBase v.3 online tool, the predicted data showed that CXCR4 harbored a potential target sequence (ACUGAC) for miR-223-3p within its 3ʹUTR (Figure 6A). To confirm this, we used CXCR4 3ʹUTR wild-type (CXCR4-WT) or mutant-type (CXCR4-MUT) luciferase reporter in dual-luciferase assays. Transient transfection of CXCR4-WT in the presence of miR-223-3p mimic triggered a remarkable down-regulation of relative luciferase activity, and this effect was completely abrogated by the mutation of the miR-223-3p putative target sites (Figure 6B and C). To determine whether miR-223-3p influenced CXCR4 expression, we manipulated miR-223-3p level in the two HCC cells. The transfection efficiencies of miR-223-3p mimic and anti-miR-223-3p were gauged by qRT-PCR (Figure 6D). As expected, CXCR4 mRNA and protein levels were significantly reduced by miR-223-3p overexpression, while they were remarkably elevated by miR-223-3p silencing (Figure 6E and F). Furthermore, in HCC cell lines, CXCR4 expression was up-regulated and was prominently down-regulated by celastrol at both mRNA and protein levels (Figure 6G and H). Interestingly, in HCC tissues, CXCR4 mRNA level was dramatically up-regulated, and it was inversely correlated with miR-223-3p expression and positively correlated with circ_SLIT3 expression (Figure 6I–K). Strikingly, CXCR4 protein level was augmented in HCC tissues (Figure 6L). These data together indicated that CXCR4 was directly targeted and inhibited by miR-223-3p.
Figure 6.
Circ_SLIT3 worked as a miR-223-3p sponge to control CXCR4 expression. (A) Schematic of the miR-223-3p putative target sites within CXCR4 3ʹUTR and mutated the seed region. (B and C) Dual-luciferase experiments in Huh7 and Hep3B cells. miR-223-3p (D) and CXCR4 mRNA (E) levels by qRT-PCR, and CXCR4 protein expression by Western blot (F) in celastrol (Cela)-treated Huh7 and Hep3B cells transfected with miR-NC mimic, miR-223-3p mimic, anti-miR-NC or anti-miR-223-3p. (G and H) CXCR4 mRNA and protein levels in THLE-2, Huh7, Hep3B cells and celastrol (Cela)-treated HCC cells. (I) CXCR4 mRNA expression in 50 pairs of HCC tissues and matched normal tissues. (J and K) Correlations among circ_SLIT3, miR-223-3p and CXCR4 mRNA levels in HCC tissues using the Spearman test. (L) CXCR4 protein expression in HCC tissues and matched normal tissues. (M and N) CXCR4 mRNA and protein levels in celastrol (Cela)-treated Huh7 and Hep3B cells transfected with Vector, circ_SLIT3, circ_SLIT3+miR-NC mimic or circ_SLIT3+miR-223-3p. Vector: negative control plasmid, circ_SLIT3: circ_SLIT3 overexpressing plasmid. n = 3; data were presented as mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001 or ****P < 0.0001 vs miR-NC mimic, anti-miR-NC, THLE-2 cells, Huh7 cells, Hep3B cells, Normal, Vector or circ_SLIT3+miR-NC mimic.
Subsequently, we asked whether circ_SLIT3 influenced CXCR4 expression in the two celastrol-treated HCC cell lines. By contrast, CXCR4 mRNA and protein levels were significantly increased by the elevated level of circ_SLIT3, and these effects were strikingly abolished by miR-223-3p mimic (Figure 6M and N).
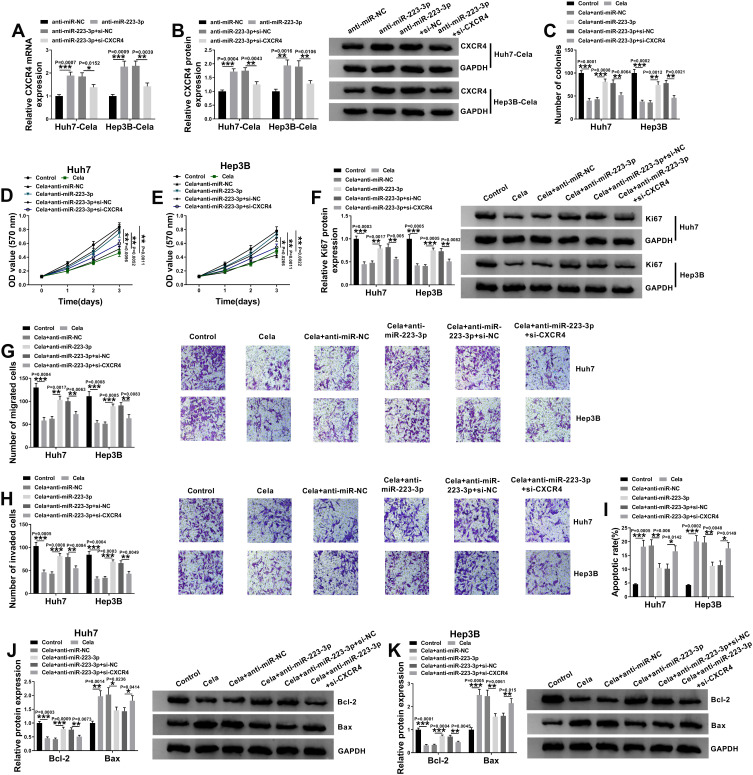
Celastrol Exerted Anti-HCC Effect in vitro by the miR-223-3p/CXCR4 Axis
We next determined the relationship between miR-223-3p and CXCR4 in celastrol-mediated anti-HCC effect in vitro. In addition to the increased impact on CXCR4 expression (Figure 7A and B), miR-223-3p knockdown led to a significant enhancement in cell colony formation (Figure 7C), proliferation (Figure 7D–F), migration (Figure 7G), invasion (Figure 7H), as well as a striking suppression in cell apoptosis (Figure 7I–K) in the two celastrol-treated HCC cell lines. Further analyses revealed that these effects of miR-223-3p knockdown were remarkably reversed by CXCR4 silencing with si-CXCR4 (Figure 7A–K).
Figure 7.
miR-223-3p depletion reversed the anti-tumor effect of celastrol on HCC in vitro by up-regulating CXCR4. (A and B) The levels of CXCR4 mRNA and protein levels in celastrol (Cela)-treated Huh7 and Hep3B cells transfected with anti-miR-NC, anti-miR-223-3p, anti-miR-223-3p+si-NC or anti-miR-223-3p+si-CXCR4. Huh7 and Hep3B cells were transfected with anti-miR-NC, anti-miR-223-3p, anti-miR-223-3p+si-NC or anti-miR-223-3p+si-CXCR4 for 24 h and then treated with 1 µM of celastrol (Cela) for 48 h, followed by the measurement of cell colony formation by colony formation assay (C), cell proliferation by MTS assay (D and E), Ki67 level by Western blot (F), cell migration and invasion by transwell assay (G and H), cell apoptosis by flow cytometry (I), the levels of Bcl-2 and Bax by Western blot (J and K). n = 3; data were presented as mean ± SD; *P < 0.05, **P < 0.01 or ***P < 0.001 vs anti-miR-NC, anti-miR-223-3p+si-NC, Control, Cela+anti-miR-NC or Cela+anti-miR-223-3p+si-CXCR4.
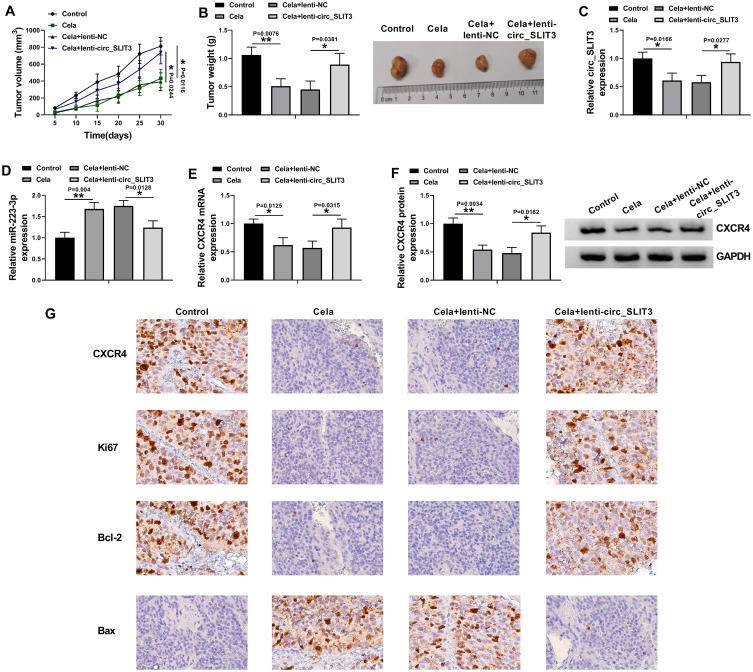
Celastrol Suppressed Tumor Growth in vivo Through Down-Regulating circ_SLIT3
Given our data that circ_SLIT3 inhibition was crucial for celastrol-mediated anti-HCC activity in vitro, we also asked whether celastrol affected tumor growth by circ_SLIT3 in vivo. The administration of celastrol led to a striking reduction in tumor growth (Figure 8A and B). Remarkably, when Hep3B cells were transduced with lenti-circ_SLIT3, celastrol-mediated tumor growth inhibition was significantly abolished (Figure 8A and B). Moreover, celastrol triggered a remarkable decrease in circ_SLIT3 and CXCR4 levels, as well as a striking increase in miR-223-3p expression in the xenograft tumors, and these effects were dramatically reversed by lenti-circ_SLIT3 transduction (Figure 8C–G). Furthermore, immunohistochemistry results showed that celastrol elevated Bax expression and inhibited Ki67 and Bcl-2 levels in the tumor tissues, and these effects were abolished by circ_SLIT3 overexpression (Figure 8G).
Figure 8.
Celastrol exerted anti-tumor effect on tumor growth in vivo by circ_SLIT3. Un-transduced, lenti-NC-infected or lenti-circ_SLIT3-transduced Hep3B cells were implanted under the subcutaneous layer into the nude mice (n = 6 each group). The administration of celastrol (2 mg/kg) was performed after 5 days implantation by intraperitoneal injection every 5 days. (A) Growth curves of the xenograft tumors. (B) The average weight of the excised tumors. The levels of circ_SLIT3 (C), miR-223-3p (D) and CXCR4 mRNA (E) by qRT-PCR and CXCR4 protein expression by Western blot (F) in the xenograft tumors. (G) The expression of CXCR4, Ki67, Bcl-2 and Bax by immunohistochemistry in the xenograft tumors. n = 6; data were presented as mean ± SD; *P < 0.05 or **P < 0.01 vs Control or Cela+lenti-NC.
Discussion
In recent years, many studies have highlighted the anti-tumor function of Chinese herbal medicine.15 As an active product extracted from a medicinal plant, celastrol has been identified as a potential anti-tumor agent in numerous cancers, such as prostate cancer, myeloma and colorectal cancer.16–18 Moreover, work in a number of laboratories has demonstrated the anti-HCC property of celastrol.4,19 However, the molecular determinants underlying the anti-tumor activity of celastrol are still not fully understood. Previous work has illuminated the importance of noncoding RNAs, such as miRNAs, in the anti-tumor mechanisms of celastrol.7,20 Considered the up-regulated expression of circ_SLIT3 in HCC,11 we undertook to associate circ_SLIT3 with the mechanism of the anti-HCC activity of celastrol. Our work expanded on the molecular targets of celastrol in inhibiting HCC progression.
In this report, we firstly validated the anti-HCC activity of celastrol in vitro and in vivo. Given our data that celastrol down-regulated the expression of circ_SLIT3 in HCC cells, we hypothesized that circ_SLIT3 was crucial for celastrol-mediated anti-HCC effect. By a series of function experiments, we first demonstrated that celastrol exerted the anti-HCC effect by down-regulating circ_SLIT3. As has been reported for other circRNAs,21,22 circ_SLIT3 was also intrinsically stable and resistant to RNase R due to the lack of both 3ʹ and 5ʹ ends.23
Some of circRNAs are posited as miRNA sponges to liberate mRNAs targeted by miRNAs during tumorigenic processes.8 For the first time, we identified the role of circ_SLIT3 as a miR-223-3p molecular sponge. MiR-223-3p was a strong candidate in the current work, considering its pivotal role in multiple cancers, including penile cancer and bladder cancer.24,25 Moreover, miR-223-3p was reported to be implicated in the pathogenesis of HCC.26,27 Here, our data first uncovered the involvement of miR-223-3p in the anti-HCC activity of celastrol. More importantly, we highlighted that celastrol exerted anti-HCC activity through targeting circ_SLIT3/miR-223-3p axis.
Using the starBase online tool, we found that miR-223-3p potentially targeted CXCR4. CXCR4, a chemokine receptor, functions as a potent oncogenic driver in tumorigenesis.28 Recent studies have shown the association between the anti-tumor mechanism of celastrol and CXCR4 expression in various cancers, including HCC.29,30 Here, we were first to demonstrate that CXCR4 was a direct and functional target of miR-223-3p in celastrol-treated HCC cells. More intriguingly, we also first pointed out the function of circ_SLIT3 to protect against CXCR4 repression by sequestering miR-223-3p. Additionally, in vivo assays were hampered at present by the lack of direct evidence for the novel mechanism of celastrol-mediated anti-HCC action, which would be performed in further work.
In summary, our current work outlined a novel regulatory mechanism, the circ_SLIT3/miR-223-3p/CXCR4 axis, for the anti-tumor function of celastrol in HCC. Our findings provided a new insight into the involvement of circRNAs in the anti-tumor molecular determinants of celastrol, reinforcing celastrol as a promising therapeutic opportunity for HCC treatment.
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. 81704066), Subject Innovation Team of Shaanxi University of Chinese Medicine (2019-YL06).
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, et al. Hepatocellular carcinoma: from diagnosis to treatment. Surg Oncol. 2016;25(2):74–85. doi: 10.1016/j.suronc.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 3.Chen SR, Dai Y, Zhao J, et al. A mechanistic overview of triptolide and celastrol, natural products from Tripterygium wilfordii Hook F. Front Pharmacol. 2018;9:104. doi: 10.3389/fphar.2018.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang W, He W, Li PP, et al. Protective effects of Celastrol on diethylnitrosamine-induced hepatocellular carcinoma in rats and its mechanisms. Eur J Pharmacol. 2016;784:173–180. doi: 10.1016/j.ejphar.2016.04.045 [DOI] [PubMed] [Google Scholar]
- 5.Li PP, He W, Yuan PF, et al. Celastrol induces mitochondria-mediated apoptosis in hepatocellular carcinoma Bel-7402 cells. Am J Chin Med. 2015;43(1):137–148. doi: 10.1142/S0192415X15500093 [DOI] [PubMed] [Google Scholar]
- 6.Li CW, Chang PY, Chen BS. Investigating the mechanism of hepatocellular carcinoma progression by constructing genetic and epigenetic networks using NGS data identification and big database mining method. Oncotarget. 2016;7(48):79453–79473. doi: 10.18632/oncotarget.13100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Li Y, Liu D, Sun H, Liu J. miR-224 is critical for celastrol-induced inhibition of migration and invasion of hepatocellular carcinoma cells. Cell Physiol Biochem. 2013;32(2):448–458. doi: 10.1159/000354450 [DOI] [PubMed] [Google Scholar]
- 8.Xiong DD, Dang YW, Lin P, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16(1):220. doi: 10.1186/s12967-018-1593-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. doi: 10.1016/j.jhep.2018.01.012 [DOI] [PubMed] [Google Scholar]
- 10.Li S, Gu H, Huang Y, et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle. 2018;17(19–20):2349–2359. doi: 10.1080/15384101.2018.1526599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu L, Wang T, Ge Q, et al. Circular RNA signature in hepatocellular carcinoma. J Cancer. 2019;10(15):3361–3372. doi: 10.7150/jca.31243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ning TZ, Kin WW, Mustafa S, et al. Detection of Entamoeba histolytica in experimentally induced amoebic liver abscess: comparison of three staining methods. Asian Pac J Trop Biomed. 2012;2(1):61–65. doi: 10.1016/S2221-1691(11)60191-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng EK, Tsang WP, Ng SS, et al. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer. 2009;101(4):699–706. doi: 10.1038/sj.bjc.6605195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y, Xie Z, Chen J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer. 2019;18(1):73. doi: 10.1186/s12943-019-1007-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo H, Vong CT, Chen H, et al. Naturally occurring anti-cancer compounds: shining from Chinese herbal medicine. Chin Med. 2019;14:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuchta K, Xiang Y, Huang S, et al. Celastrol, an active constituent of the TCM plant Tripterygium wilfordii Hook.f., inhibits prostate cancer bone metastasis. Prostate Cancer Prostatic Dis. 2017;20(2):156–164. doi: 10.1038/pcan.2016.61 [DOI] [PubMed] [Google Scholar]
- 17.Tozawa K, Sagawa M, Kizaki M. Quinone methide tripterine, celastrol, induces apoptosis in human myeloma cells via NF-κB pathway. Int J Oncol. 2011;39(5):1117–1122. doi: 10.3892/ijo.2011.1161 [DOI] [PubMed] [Google Scholar]
- 18.Barker EC, Kim BG, Yoon JH, et al. Potent suppression of both spontaneous and carcinogen-induced colitis-associated colorectal cancer in mice by dietary celastrol supplementation. Carcinogenesis. 2018;39(1):36–46. doi: 10.1093/carcin/bgx115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajendran P, Li F, Shanmugam MK, et al. Celastrol suppresses growth and induces apoptosis of human hepatocellular carcinoma through the modulation of STAT3/JAK2 signaling cascade in vitro and in vivo. Cancer Prev Res (Phila). 2012;5(4):631–643. doi: 10.1158/1940-6207.CAPR-11-0420 [DOI] [PubMed] [Google Scholar]
- 20.Yao SS, Han L, Tian ZB, et al. Celastrol inhibits growth and metastasis of human gastric cancer cell MKN45 by down-regulating microRNA-21. Phytother Res. 2019;33(6):1706–1716. doi: 10.1002/ptr.6359 [DOI] [PubMed] [Google Scholar]
- 21.Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi: 10.18632/oncotarget.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Li X, Lu L, et al. Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J Clin Lab Anal. 2018;32(5):e22379. doi: 10.1002/jcla.22379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi: 10.1038/nrm.2015.32 [DOI] [PubMed] [Google Scholar]
- 24.Pinho JD, Silva GEB, Teixeira Júnior AAL, et al. MIR-107, MIR-223-3P and MIR-21-5P reveals potential biomarkers in penile cancer. Asian Pac J Cancer Prev. 2020;21(2):391–397. doi: 10.31557/APJCP.2020.21.2.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugawara S, Yamada Y, Arai T, et al. Dual strands of the miR-223 duplex (miR-223-5p and miR-223-3p) inhibit cancer cell aggressiveness: targeted genes are involved in bladder cancer pathogenesis. J Hum Genet. 2018;63(5):657–668. doi: 10.1038/s10038-018-0437-8 [DOI] [PubMed] [Google Scholar]
- 26.Oksuz Z, Serin MS, Kaplan E, et al. Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p could be used as novel non-invasive biomarkers for HCV-positive cirrhosis and hepatocellular carcinoma. Mol Biol Rep. 2015;42(3):713–720. doi: 10.1007/s11033-014-3819-9 [DOI] [PubMed] [Google Scholar]
- 27.Pratedrat P, Chuaypen N, Nimsamer P, et al. Diagnostic and prognostic roles of circulating miRNA-223-3p in hepatitis B virus-related hepatocellular carcinoma. PLoS One. 2020;15(4):e0232211. doi: 10.1371/journal.pone.0232211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatterjee S, Behnam Azad B, Nimmagadda S. The intricate role of CXCR4 in cancer. Adv Cancer Res. 2014;124:31–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yadav VR, Sung B, Prasad S, et al. Celastrol suppresses invasion of colon and pancreatic cancer cells through the downregulation of expression of CXCR4 chemokine receptor. J Mol Med (Berl). 2010;88(12):1243–1253. doi: 10.1007/s00109-010-0669-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kun-Ming C, Chih-Hsien C, Chen-Fang L, et al. Potential anticancer effect of celastrol on hepatocellular carcinoma by suppressing CXCR4-related signal and impeding tumor growth in vivo. Arch Med Res. 2020;51(4):297–302. doi: 10.1016/j.arcmed.2020.03.001 [DOI] [PubMed] [Google Scholar]