Abstract
The availability and repartition of fucosylated glycans within the gastrointestinal tract contributes to the adaptation of gut bacteria species to ecological niches. To access this source of nutrients, gut bacteria encode α-l-fucosidases (fucosidases) which catalyze the hydrolysis of terminal α-l-fucosidic linkages. We determined the substrate and linkage specificities of fucosidases from the human gut symbiont Ruminococcus gnavus. Sequence similarity network identified strain-specific fucosidases in R. gnavus ATCC 29149 and E1 strains that were further validated enzymatically against a range of defined oligosaccharides and glycoconjugates. Using a combination of glycan microarrays, mass spectrometry, isothermal titration calorimetry, crystallographic and saturation transfer difference NMR approaches, we identified a fucosidase with the capacity to recognize sialic acid-terminated fucosylated glycans (sialyl Lewis X/A epitopes) and hydrolyze α1–3/4 fucosyl linkages in these substrates without the need to remove sialic acid. Molecular dynamics simulation and docking showed that 3′-Sialyl Lewis X (sLeX) could be accommodated within the binding site of the enzyme. This specificity may contribute to the adaptation of R. gnavus strains to the infant and adult gut and has potential applications in diagnostic glycomic assays for diabetes and certain cancers.
Electronic supplementary material
The online version of this article (10.1007/s00018-020-03514-x) contains supplementary material, which is available to authorized users.
Keywords: Gut microbiota, Glycoside hydrolase, Mucus, Mucin glycosylation, Lewis epitopes, Antennary fucose
Introduction
The microbial community inhabiting the human gut (gut microbiota) exerts a profound effect on human health through, e.g. polysaccharide digestion, metabolite and vitamin production, maturation of the immune system and protection against pathogens [1]. The adult gut microbiota is dominated by members of Firmicutes and Bacteroidetes phyla whereas the infant gut microbiota is dominated by Bifidobacterium that are adapted to utilize human milk oligosaccharides (HMOs), which are one of the major glycans found in breast milk. HMOs are composed of a linear or branched backbone containing galactose (Gal), N-acetylglucosamine (GlcNAc) and glucose (Glc), which can be decorated with fucose (Fuc) and/or sialic acid (Sia) residues, depending on the mother’s secretory status [2, 3]. In the adult colon, gut bacteria have not only access to non-digestible polysaccharides from the diet, but also to complex oligosaccharides from host mucins [4–6]. Mucins are large glycoproteins with a high carbohydrate content of up to 80%. Mucin-type O-glycans consists of N-acetylgalactosamine (GalNAc), Gal and GlcNAc, containing glycan chains modified by fucosylation, sialylation and sulfation [7–9]. The main source of glycan diversity is provided by the peripheral terminal epitopes that show considerable variation. The H1 structure (α1,2-fucose) is found in populations carrying the secretor gene [10], and individuals may also express the Lewis gene and the Lewis B (LeB) histoblood group antigen if they are secretors, while non-secretors express Lewis A (LeA) [11]. Another phenotype (SeW—weak secretor) is characterized by the expression of both LeA and LeB antigens [12]. The presentation of the major mucin glycan epitopes, sialic acid and fucose, varies along the GI tract with a decreasing gradient of fucose and ABH blood group expression and an increasing gradient of sialic acid from the ileum to the colon [7]. These gradients are reversed in mice, where the small intestine is dominated by sialylated structures and the colon with those terminating in fucose [13]. These glycans provide a potential source of nutrients to members of the gut microbiota [5]. In particular, α-l-fucosidases (α-fucosidases) are key enzymes for the degradation and metabolism of intestinal mucin glycans or HMOs by gut microbes and therefore, contribute to shaping the composition of the gut microbiota by favoring different bacterial species and influencing health and disease. Currently, α-fucosidases which catalyze the release of α-1–2, α-1–3, α-1–4 and α-1–6 linked fucose are classified into glycoside hydrolase (GH) families 29 and 95 (CAZy, www.cazy.org). All GH95 enzymes functionally characterized so far show strict substrate specificity to the terminal Fuc α1-2Gal linkage and hydrolyze the linkage via an inverting mechanism whereas GH29 enzymes show relatively relaxed substrate specificities with hydrolysis proceeding via a retaining mechanism (www.cazy.org). It was suggested that GH29 can be divided into two subfamilies. One contains fucosidases with relaxed substrate specificities that can act on 4-nitrophenyl α-l-fucopyranoside (pNP-Fuc) (referred to as GH29-A) (EC 3.2.1.51), whereas the members of the other subfamily show strict specificity for terminal α-(1–3/4)-fucosidic linkages with little/no activity on pNP-Fuc (GH29-B) (EC 3.2.1.111) as shown for fucosidases from Streptomyces and Bacteroidetes thetaiotaomicron [14, 15]. The GH29-A subfamily includes fucosidases from Thermotoga maritima [16], soil metagenome [17] or bacterial pathogens [18, 19] whereas the GH29-B subfamily includes fucosidases from Bifidobacterium bifidum (BbAfcB) [20] Clostridium perfringens (CpAfc2) [21] and Streptococcus pneumoniae (SpGH29c) [22]. Despite the importance of fucose in regulating bacterial intestinal colonization in adults and infants, only a limited number of fucosidases have been studied at a biochemical level from human gut symbionts.
Ruminococcus gnavus is a prevalent member of the gut microbial community belonging to the Firmicutes division [23, 24]. R. gnavus is an early colonizer of the human gut [25] but persists in healthy adults where it belongs to the 57 species detected in more than 90% of human faecal samples by metagenomic sequencing [23]. In the past few years, an increasing number of studies are reporting a disproportionate representation of R. gnavus in diseases, such as inflammatory bowel disease [26]. In our previous work, we showed that R. gnavus ability to grow on HMOs or mucins was strain dependent [27, 28], underscoring the importance of analysing glycan utilization by members of the human gut microbiota at the strain level. These differences are reflected by the distribution of GH families between R. gnavus strains [27]. For example, R. gnavus E1 genome lacks a sialidase encoding gene whereas R. gnavus ATCC 29149 encodes a GH33 enzyme which has been functionally characterized as an intramolecular trans-sialidase [29] and is associated with a unique sialic acid metabolism pathway which forms the basis of R. gnavus ATCC 29149 adaptation to mucus [30]. In contrast, both R. gnavus E1 and R. gnavus ATCC 29149 genomes harbor fucosidase encoding genes belonging to GH29 or GH95 families [27], but their functional characterization has not been reported. To gain further biochemical and structural insights into R. gnavus strategy to utilize mucin glycans, we determined the substrate and linkage specificities of a range of fucosidases belonging to GH29 and 95 family from R. gnavus ATTC 29149 and E1 strains. We identified and characterized a fucosidase from R. gnavus E1 with the capacity to recognize fucosylated glycans capped with sialic acid and to hydrolyze α1–3/4 fucosyl linkages in these substrates without the need to remove sialic acid. This unique specificity may contribute to the adaptation of R. gnavus strains to distinct nutritional niches. Since changes in abundances of sialyl fucosylated epitopes on human glycans have been associated in several diseases, such as diabetes and certain cancers, these novel fucosidases may have potential in diagnostic glycomic assays.
Materials and methods
Materials
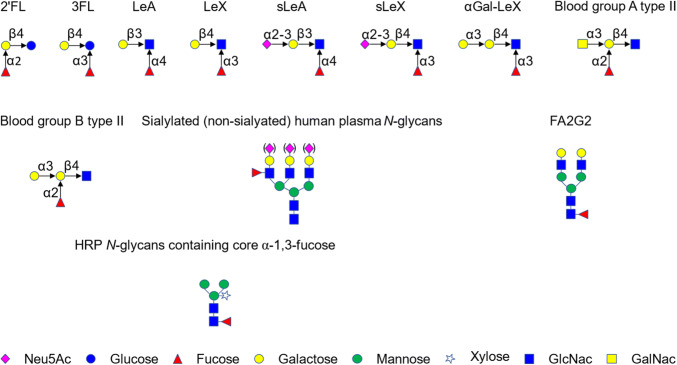
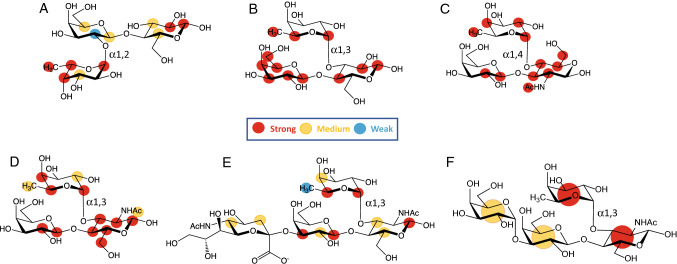
All chemicals were obtained from Sigma (St Louis, MO, USA) unless otherwise stated. The structure of the oligosaccharides used in this work is shown in Fig. 1. 3′-Sialyl Lewis X (sLeX) was purchased from Carbosynth Limited (Campton, UK), Lewis A (LeA), α1–3Gal-Lewis X (αGal-LeX), Blood group A/B tetrasaccharide type II (Blood group A/B type II) were from Elicityl (Crolles, France), Lewis X (LeX) used for activity assay was from Dextra Laboratories (Reading, UK), LeX/LeA/N-Acetylneuraminic acid (Neu5Ac) used for ITC were from Carbosynth Limited (Campton, UK), LeX used for STD NMR was from the Consortium for Functional Glycomics (CFG). 3′-Sialyl Lewis A (sLeA), sialylated and desialylated human plasma N-glycans and FA2G2 N-glycans were from Ludger (Oxford, UK). Horseradish peroxidase (HRP) treated with BM03341 plant specific PNGase was a kind gift from Dr Lucy Crouch (Newcastle University). E. coli strain (Tuner DE3 pLacI) was from Merck (Darmstadt, Germany).
Fig. 1.
Fucosylated oligosaccharides used in this study. Monosaccharide symbols follow the Symbol Nomenclature for Glycans system [98]
Cloning, expression, mutagenesis and purification of GH29 and GH95 fucosidases
Ruminococcus gnavus ATCC 29149 or E1 genomic DNA (gDNA) was purified from the cell pellet of a bacterial overnight culture (1 ml) following centrifugation (5000g, 5 min) using the GeneJET Genomic DNA Purification Kit (ThermoFisher, UK), according to the manufacturer’s instructions. The full-length sequence of E1_10125 and E1_10180, excluding the signal sequence were cloned into the pOPINF expression system [31], introducing an His-tag at the N terminus. The D221A mutant of E1_10125 was produced by NZYTech (Lisbon, Portugal). The E1_10125G260M mutant was generated using the NZY Mutagenesis kit (Lisbon, Portugal). The ATCC_03833 and ATCC_00842 sequences exempt of the signal sequence were cloned into pET28a with N terminal His tag by Prozomix (Haltwhistle, UK). The E1_10587 was synthesized by NZYTech (Lisbon, Portugal) into pHTP1 with N terminal His tag.
The primers used are listed in Table S1. DNA manipulation was carried out in E. coli DH5α cells. Sequences were verified by DNA sequencing at Eurofins MWG (Ebersberg, Germany) or Earlham Institute (formerly TGAC, Norwich, UK). E. coli TunerDE3pLacI cells were transformed with the recombinant plasmids according to manufacturer's instructions. The expression was carried out in 1000 ml LB media growing cells at 37 °C until OD600 nm reached 0.4 to 0.6 and then induced at 16 °C for 48 h. The cells were harvested by centrifugation at 7000g for 10 min. The His-tagged proteins were purified by immobilized metal affinity chromatography (IMAC) and further purified by gel filtration (Superdex 75 and 200 columns) on an Akta system (GE Health Care Life Sciences, Little Chalfont, UK). Protein purification was assessed by standard SDS–polyacrylamide gel electrophoresis using the NuPAGE Novex 4–12% Bis–Tris (Life Technologies, Paisley, UK). Protein concentration was measured with a NanoDrop (Thermo Scientific, Wilmington, USA) and using the extinction coefficient calculated by ProtParam (ExPASy-Artimo, 2012) from the peptide sequence.
Glycan microarrays
Three concentrations (5, 50 and 200 µg/ml) of recombinant His6-tagged E1_10125 D221A mutant were screened for binding to Core H glycan microarray glycans at the Consortium for Functional Glycomics (CFG).
STD NMR experiments
An Amicon centrifuge filter unit with a 10 kDa MW cutoff was used to exchange the protein in 25 mM d19-2,2-bis(hydroxymethyl)-2,2′,2″-nitrilotriethanol pH* 7.4 (uncorrected for the deuterium isotope effect on the pH glass electrode) D2O buffer and 50 mM NaCl. All the ligands were dissolved in 25 mM d19-2,2-bis(hydroxymethyl)-2,2′,2″-nitrilotriethanol pH* 7.4, 50 mM NaCl. A concentration of 50 µM was used for the enzyme and 2 mM for the ligands. The STD NMR spectra were performed on a Bruker Avance 800.23 MHz at 278 K. The on- and off-resonance spectra were acquired using a train of 50 ms Gaussian selective saturation pulses using a variable saturation time from 0.5 to 5 s, for binding epitope mapping determination (STD build-up curves). Residual protein resonances were filtered out using a T2 filter of 40 ms. All the spectra were performed with a spectral width of 10 kHz and 32768 data points using 256 or 512 scans. Binding epitope mappings were obtained by determining the initial slopes () calculated by performing a least-squares fitting of the following mono-exponential curve:
where is the STD intensity for a saturation time, , is the maximum STD intensity and is the rate constant for saturation transfer. In the limit, :
Importantly, gives a value that is independent of any relaxation or rebinding effects, allowing for a more accurate binding epitope. The value of was then normalized against the proton with the largest intensity to give values in the range of 0–100%, which were then mapped onto the ligand structure to give the corresponding binding epitope mapping.
X-ray crystallography
The E1_10125 fucosidase His tag was removed using 3C-protease overnight at 4 °C at a mass ratio of 20:1, the His tag and 3C-protease was then removed by passing the sample over a nickel sepharose column. The final crystallization condition was 0.2 M magnesium chloride, 25% PEG 3350, 0.1 M bis–tris pH 5.5, 10 mM 2-Fucosyllactose (2′FL). Sitting drop vapour diffusion crystallization experiments of E1_10125 or E1_10125 D221A were set up at a concentration of 20 mg/ml and monitored using the VMXi beamline at Diamond Light Source [32]. Crystals were cryoprotected using the crystallization condition with the addition of 15% ethylene glycol. Wild-Type and D221A E1_10125 mutant diffraction experiments were performed at Diamond Light Source on beamlines I04 (wavelength 0.9795 Å) and I03 (wavelength 0.9763 Å), respectively. The data were processed with Xia2 making use of aimless, dials and pointless [33–36]. The data were phased using PHASER using pdb 4OUE as a molecular replacement model and refined using REFMAC [37] and Coot [38] within the CCP4 software environment [39]. The PDB REDO server [40] was used to optimize the refinement parameters and the model was validated using the Molprobity server [41].
Molecular dynamics (MD) simulation and docking of sLeX into E1_10125 D221A
Docking calculations were run using the crystal structure of E1_10125 D221A mutant as it showed the highest resolution. First, a MD simulation was carried out to explore the flexibility of the side chains surrounding the fucose binding subsite and the whole binding site. The input coordinates for the simulation were produced by loading the X-ray coordinates into Schrödinger Maestro [42] and processed using the protein preparation wizard [43]. Protons were added to the structure and then all buffer ions, buffer and structurally non-essential water molecules were removed. PROPKA [44] was then used to predict the ionization state of polar residues at pH 7. The OPLS force field was then used to minimize the protein structure, converging heavy atoms to a threshold of 0.3 Å.
The system was then simulated using the Amber PMEMD software [45]. The system was solvated using TIP3P water, placed within a truncated octahedron buffered to 10 Å, to a net charge of zero using Na+ ions. The parameter set for the protein atoms and structural ions used was taken from the ff14SB and gaff force fields. The system was initially minimized with constraints of 20 kcal mol−1 Å−2 placed on solute atoms and then minimized a second time with no constraints placed on solute atoms. The system was then heated to a temperature of 300 K and raised to a pressure of 1 atm in two separate 500 ps steps under constrains of 20 kcal mol−1 Å−2 placed on solute atoms. Over the course of four 500 ps steps constrains were then released in 5 kcal mol−1 Å−2 increments. The system was then simulated over the course of 500 ns with a 2 fs time step, with frame sampling every 0.5 ns. The SHAKE algorithm [46] was used to constrain bonds involving hydrogen atoms. A Berendsen barostat and a Langevin thermostat with a 5 ps−1 collision frequency were used to maintain constant pressure and temperature, respectively. Non-bonding atom bond cutoff was set to 8 Å.
The trajectory file from the MD simulation was then clustered using cpptraj [47] to produce 20 average structures. The kmeans clustering algorithm within cpptraj was used with a random set of initial points. The clustering was calculated for every tenth frame and based on the distance between atoms measured using root-mean-square deviation of atomic positions (RMSD) without fitting structures to each other prior to calculating RMSD.
The 20 average structures were then imported into Schrödinger Maestro and processed using the protein preparation wizard (see above). Protons were replaced in the structure then all buffer ions and structurally non-essential water molecules introduced during MD simulation were removed. The OPLS force field was then used to minimize the protein structure, converging heavy atoms to a threshold of 0.3 Å. Protein structures showing a wide-open binding site were selected from the 20 average structures to be used for docking of the tetrasaccharide ligand. In order to perform the docking calculations, a cubic grid with a 10 Å × 10 Å × 10 Å inner box and a 30 Å × 30 Å × 30 Å outer box with the centroid placed at the middle of the binding site was generated. sLeX was built within Maestro and prepared using LigPrep [48] and low-energy conformers generated using MacroModel [49]. sLeX was then docked using Glide [50] with standard precision enhanced by 2 times without canonicalization and without sampling ring conformations.
Isothermal titration calorimetry (ITC)
ITC experiments were performed using the PEAQ-ITC system (Malvern, Malvern, UK) with a cell volume of 200 µl. Prior to titration, protein samples (E1_10125D221A) were exhaustively dialyzed into 50 mM citrate buffer pH 6. The ligand was dissolved in the dialysis buffer. The cell protein concentration was 100 µM and the syringe ligand concentration was 2 mM for all ligands tested except 20 mM for Neu5Ac. Three controls with titrant (sugar) injected into the buffer, buffer injected to protein, buffer injected into buffer, were subtracted from the data. The analysis was performed using the Malvern software, using a single-binding site model. Experiments were carried out in triplicate.
Activity assays and kinetics
The optimum pH of fucosidases was determined with 0.5 mM pNP-Fuc for all fucosidase tested apart for E1_10587 where 5 mM pNP-Fuc was used instead, in 50 mM citrate buffer (pH 3, 4, 5 and 6), 50 mM sodium phosphate buffer (pH 6, 7, 7.5 and 8) and 50 mM Tris buffer (pH 8.5 and 9.3). The final concentration of enzyme was 1 µM for ATCC_00842, 2 µM for E1_10180, 20 µM for E1_10125, 0.015 µM for ATCC_03833 and 10 µM for E1_10587. The reaction duration was optimized to measure the reaction rates under initial conditions. After incubation at 37 °C, the reaction was stopped by adding 50 µl of 1 M of sodium carbonate into 200 µl of reaction (200 µl of 1 M of sodium carbonate into 40 µl of reaction for E1_10587). The amount of fucose released was determined using a 96-well plate reader (BMG Labtech, Ortenberg, Germany) by measuring absorbance at 405 nm.
Kinetic studies against pNP-Fuc were performed in 50 mM citrate buffer at optimal pH (pH 6 for ATCC_00842, E1_10180, E1_10125, ATCC_03833; pH 5 for E1_10587) with increasing amounts of pNP-Fuc and a constant enzyme concentration at 37 °C. The series of pNP-Fuc concentrations were chosen to ensure that there were at least three points below and above the Km value. The amount of enzyme was determined to fulfil free-ligand approximation, i.e. the enzyme concentration was linear with product formation. The reaction duration was optimized to measure the reaction rates under initial conditions. A standard curve was made with a range of pNP-Fuc from 0 to 140 µM and in the same experimental condition as the enzymatic reactions. Kinetic parameters were calculated based on the Michaelis–Menten equation using a non-linear regression analysis program (Prism 6, GraphPad, San Diego, USA). Kinetic parameters for E1_10587 were calculated based on the Michaelis–Menten equation:
To determine the substrate specificity of the enzymes against fucosylated oligosaccharides (2′FL, 3FL, LeA and LeX), each enzyme was incubated with the substrate (0.1 mM) at 37 °C in 50 mM citrate buffer at optimal pH. For kinetic assays with E1_10125, 50 µM to 350 µM of LeX, 50 µM to 1400 µM of sLeX and 20 µM to 400 µM αGal-LeX was used. Higher concentrations of LeX or αGal-LeX were not used because they contained significant amount of l-fucose, which affected the accuracy of quantification. The enzyme in the reaction was 0.01 µM for LeX and sLeX, 1 nM for αGal-LeX. The time course of reaction used for LeX, sLeX and αGal-LeX was 9 min, 30 min and 20 min, respectively. The fucose released was quantified using the k-fucose kit (Megazyme, Wicklow, Ireland) combined with the diaphorase/resazurin assay [51]. Briefly, 40 µl of reaction mixed with 97 µl of mixed reagent (50 µl of dH2O, 20 µl of reaction buffer pH9.5, 5 µl of NADP + , 2 µl of l-fucose dehydrogenase suspension, 10 µl of 1 mM resazurin solution, 10 µl of 10U/ml solution of Diaphorase) and incubated at room temperature for 20 min before measuring the fluorescence of resorufin using a 96-wells plate reader (BMG Labtech, Ortenberg, Germany) with an excitation at 550 nm and an emission at 584 nm. One unit of activity was defined as the amount of enzyme needed to release 1 µmol of product per min under the conditions described above. The kinetic parameters of E1_10125 fucosidase against sLeX were calculated based on Michaelis–Menten equation using a non-linear regression analysis program (Prism 6, GraphPad, San Diego, USA). The curve of initial rate against substrate concentration for LeX and αGal-LeX was linear, which indicated that the substrate concentration was far below Km, therefore their kcat/Km was estimated from the slope of the curve [52], k = (kcat/Km)[E]0 where k is the slope and [E]0 represents the enzyme concentration.
LC–MS/MS analysis
10 µM of enzyme was incubated with a range of substrates including sLeA (5 µM), sLeX (5 µM), sialylated and de-sialylated human plasma N-glycans released from 0.5 µl human plasma, HRP (0.01 µg/µl), FA2G2 (5 ng/ µl), blood group A type II (5 µM) or blood group B type II (5 µM). Reactions (20 µl) were performed in 50 mM citrate buffer at pH 6 and 37 °C for 24 h. Reactions were then dried down using Savant SpeedVac centrifugal evaporator (Thermo Fisher, Wilmington, USA), labelled at the reducing end with procainamide using the glycan labelling kit with sodium cyanoborohydride reductant (Ludger, Oxford, UK) and purified using S-cartridges (Ludger, Oxford, UK) to remove the excess dye. The samples were dried by speed vacuum and resuspended in 50 µl of acetonitrile:water solvent. Then the reactions were injected onto a Waters ACQUITY UPLC Glycan BEH amide column (2.1 × 150 mm, 1.7 µm particle size, 130 Å pore size) at 40 °C on a Dionex Ultimate 3000 UHPLC instrument with a fluorescence detector (λex = 310 nm, λem = 370 nm) coupled to a Bruker Amazon Speed ETD. A 50 mM ammonium formate solution pH 4.4 (Ludger, Oxford, UK) was used as mobile phase A and acetonitrile (Romil, UK) was used as mobile phase B. For the plasma samples, a 70 min gradient was used with mobile phase B from 76 to 51% from 0 to 53.5 min at a flow rate of 0.4 ml/min followed by mobile phase B from 51 to 0% from 53.5 min to 55.5 min at flow rate of 0. 2 ml/min, and 2 min stabilization, mobile phase B from 0 to 76% from 57.5 min to 59.5 min at a flow rate 0.2 ml/min, and then last for 6 min, from 65.5 min to 66.5 min, the flow rate was changed back to 0.4 ml/min and then equilibrated for 3.5 min. HRP samples used a 75 min gradient starting from 80 to 62% mobile B. A 70–62% gradient was used for FA2G2 glycan. For the shorter fucosyl-oligosaccharides, an 85 min gradient was used from 85 to 65% mobile B. The Amazon Speed was operated in the positive ion mode using the following settings: source temperature 180 °C; gas flow 4 L/min; capillary voltage, 4500 V; ICC target, 200,000; maximum accumulation time, 50 ms; rolling average, 2; number of precursor ions selected, 3; scan mode, enhanced resolution; mass range scanned, 400 to 1700. Singly charged ions were excluded for CID except for HRP and fucosyl-oligosaccharide samples.
Bioinformatics analyses
For sequence similarity networks (SSN) analysis, the sequences encoding GH29 and GH95 fucosidases were extracted from the Interpro database 66.0 (https://www.ebi.ac.uk/interpro/) after removing redundant sequences by CD-HIT Suite [53] (https://weizhong-lab.ucsd.edu/cdhit_suite/cgi-bin/index.cgi?cmd=cd-hit). Additional sequences included those corresponding to functionally characterized GH29 and GH95 from the CAZy database (www.cazy.org) as well as the R. gnavus E1 and ATCC29149 fucosidases (6 GH29 and 7 GH95). The amino acid sequences were then used to create SSN using the Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST) [54]. The network setting for GH29 and GH95 was made to combine proteins in one node sharing over 70% and 45% identity, respectively and nodes were linked by edges when their sequences shared over 40% (the e-value threshold was 1094) and 35% identity (the e-value threshold was 10130) for GH29 and GH95, respectively. The SSN data were visualized using Cytoscape 3.6 [55].
ProtParam (ExPASy) [56] was used to determine the length, molecular weight and theoretical pI of the fucosidases under study. LALIGN was used to do pairwise sequence alignment and obtain sequence similarities [57]. SignalP 5.0 Server was used to predict the presence and nature of signal peptides as well as cleavage sites [58]. The TMHMM Server v. 2.0 was used to predict the presence of transmembrane helices [59]. CW-PRED was used for the detection of LPXTG and LPXTG-like motif and thus the prediction of cell-wall proteins in Gram-positive bacteria [60]. PSORTb v3.0.2 was used for bacterial protein subcellular localization prediction [61].
Results
Sequence similarity network identified strain-specific fucosidases in R. gnavus ATCC 29149 and E1
The genome of R. gnavus E1 encodes 4 predicted GH29 fucosidases (named E1_10125, E1_10180, E1_10623 and E1_11127) and 4 GH95 fucosidases (named E1_10181, E1_10587, E1_30029, E1_40027), whereas R. gnavus ATCC 29149 encodes 2 GH29 (named ATCC_03411, ATCC_03833) and 3 GH95 fucosidases (named ATCC_00842, ATCC_01058, ATCC_03121) (for detailed information, see Table S2). A sequence similarity network (SSN) analysis was conducted to identify putative functional relationships between GH29 or GH95 fucosidases from R. gnavus strains and related protein sequences (Fig. 2). The SSN analysis covered 6736 amino acid sequences from the GH29 family from Interpro database 66.0 and CAZy (www.cazy.org/GH29_characterized.html) and 825 GH95 sequences from Interpro IPR027414 (including 8 sequences from functionally characterized GH95 www.cazy.org/GH95_characterized.html).
Fig. 2.
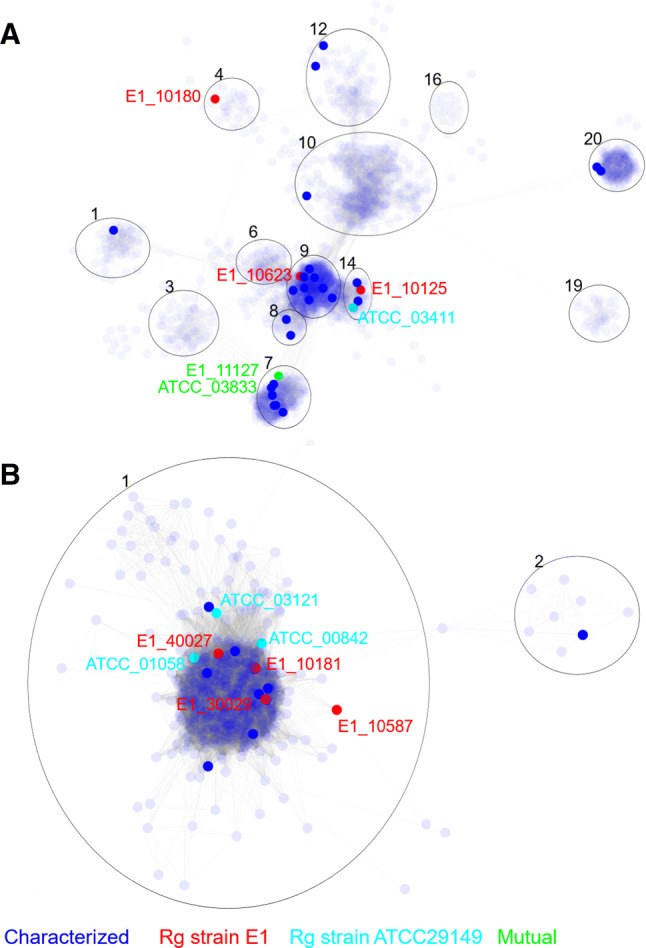
The distribution of R. gnavus GH29 and GH95 fucosidases based on SSN analysis. a Partial representation of SSN analysis of GH29 family containing fucosidases from R. gnavus E1 and ATCC29149 strains. b Representation of the SSN central cluster of GH95 family containing all GH95 from R. gnavus E1 and ATCC29149 strains. Blue node: sequences extracted from the CAZy database encoding functionally characterized enzymes. Red nodes sequences from R. gnavus E1 strain. Cyan nodes, sequences from R. gnavus ATCC29149 strain. Green nodes, sequences common to both R. gnavus E1 and ATCC29149 strains
For GH29, 5318 representative nodes in the SSN analysis were separated into 44 clusters according to their sequence similarity which may indicate a similar biochemical function and ligand specificity (Fig. S1A). GH29 fucosidases from R. gnavus E1 and ATCC 29149 strains were found in cluster 4 (E1_10180), cluster 7 (ATCC_03833 and E1_11127), cluster 9 (E1_10623) and cluster 14 (E1_10125 and ATCC_03411) (Fig. 2a). E1_10125 shares 94.5% sequence similarity with ATCC_03411, whereas ATCC_03833 is 99.8% similar to E1_11127. The catalytic domain of E1_10623 was 62.9% similar to Blon_2336 from Bifidobacterium longum subsp. infantis ATCC 15697 [62]. Based on this analysis, E1_10125, E1_10180 and ATCC_03833 were chosen as representatives of R. gnavus GH29 fucosidases for functional characterization.
The GH95 family includes fewer sequences and functionally characterized proteins (www.cazy.org). The SSN analysis of GH95 fucosidases led to the identification of 825 nodes, 627 of which were found in the cluster 1 (Fig. 2b). All GH95 fucosidases from R. gnavus E1 and ATCC 29149 strains fall within the same cluster. Based on the pre-screening for expression (not shown), ATCC_00842 and E1_10587, sharing 60% sequence similarity, were selected as representative GH95 fucosidases of R. gnavus ATCC 29149 and E1, respectively for further characterization.
R. gnavus fucosidases from GH29 and GH95 families display novel substrate specificities
The genes encoding the selected GH29 and GH95 fucosidases from R. gnavus ATCC 29149 and E1 strains were heterologously expressed in E. coli and the His6-tag recombinant proteins purified by immobilized metal ion affinity chromatography and gel filtration (see Materials and Methods for details). E. coli Tuner DE3 pLacI strain was chosen as heterologous host as it does not display any endogenous β-galactosidase activity (due to the deletion of the LacZ gene) that may interfere with the enzymatic characterization of the recombinant enzymes. The activity of the purified enzymes was first screened against the synthetic substrate pNP-Fuc. The optimum pH of all fucosidases tested, determined using pNP-Fuc, was found to be pH 6 apart for E1_10587, which was pH 5 (Fig. S2).
The kinetic parameters were determined at the optimum pH by calculating the initial rate of reaction with increasing pNP-Fuc concentrations. Fucosidase ATCC_03833 showed the highest catalytic efficiency with a kcat of 83.6 s−1 and a Km of 179.1 µM (Table 1). These values are consistent with fucosidases belonging to GH29 subfamily A [15, 17, 63, 64]. Fucosidases ATCC_00842 and E1_10180 also showed activity against pNP-Fuc, but their high Km suggest that pNP-Fuc is not a good ligand for these enzymes. Fucosidase E1_10125 displays the lowest activity against pNP-Fuc of the characterized GH29 subfamily fucosidases as shown by its low kcat of 1.8 10–3 s−1 [15, 20–22, 65, 66]. E1_10587 was merely active on pNP-Fuc as also reported for other GH95 fucosidases [21].
Table 1.
Kinetic parameters of R. gnavus fucosidases towards pNP-Fuc
| E1_10125 | E1_10180 | ATCC_03833 | ATCC_00842 | E1_10587a | |
|---|---|---|---|---|---|
| Catalytic efficiency (s−1 M−1) | 7.61 | 16.91 | 4.67 ×105 | 28.14 | 0.72 |
| Km (µM) | 237.9 ± 39.69 | ND | 179.1 ± 28.77 | 2.96 × 103 ± 3.63 × 102 | 1.50 × 104 |
| kcat (s−1) | 1.8 10–3 ± 8.8 10–5 | ND | 83.6 ± 2.97 | 0.0832 ± 0.0032 | 0.0108 |
ND could not be determined (using concentrations up to 20 mM pNP-Fuc)
aKinetic parameters were determined from the progress curve
Next, the substrate specificity of the recombinant fucosidases was tested on a range of fucosylated oligosaccharides. The specific activity was determined based on fucose release against 2′FL (Fucα1,2Galβ1,4Glc), 3FL (Galβ1-4[Fucα1-3]Glc), Lewis A (LeA, Galβl-3[Fucα1-4]GlcNAc) and Lewis X (LeX, Galβ1-4[Fucα1-3]GlcNAc) (Table 2). From this analysis, fucosidases E1_10125 and E1_10180 showed substrate specificity towards α1,3/4 fucosylated linkages while fucosidases ATCC_00842 and ATCC_03833 showed preference for α1,2 linkages (Table 2). E1_10587 showed lower activity against all tested substrates, therefore, it was not possible to assess its substrate specificity.
Table 2.
Specific activity of R. gnavus fucosidases towards fucosylated oligosaccharides
| Specific activity (U/µmol) | ||||
|---|---|---|---|---|
| 2′FL | 3FL | LeA | LeX | |
| E1_10125 | NS | 63.23 ± 2.03 | 83.88 ± 2.56 | 114.72 ± 1.76 |
| E1_10180 | NS | 0.119 ± 0.001 | 0.45 ± 0.02 | 0.36 ± 0.01 |
| ATCC_03833 | 5.47 ± 0.28 | 4.82 10–2 ± 7.09 10–4 | 0.069 ± 0.003 | ND |
| ATCC_00842 | 1.13 104 ± 3.06 102 | 15.96 ± 0.67 | NS | NS |
| E1_10587 | NS | NS | NS | NS |
ND not detected under experimental conditions, NS not significant, less than 0.01 U/µmol
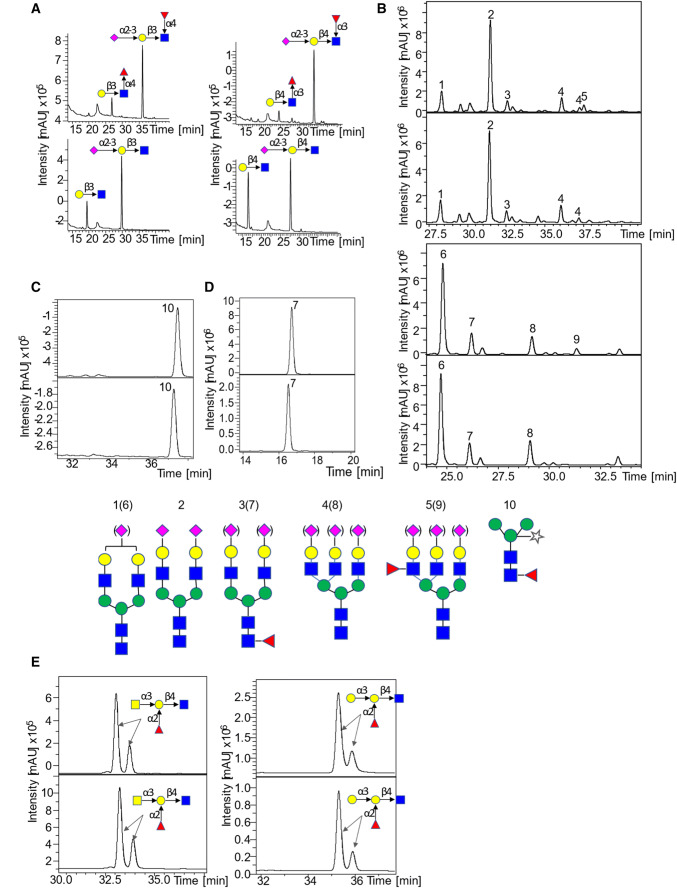
The activity of the recombinant fucosidases was further tested on more complex oligosaccharides and glycoproteins including sialyl Lewis X (Neu5Acα2-3Galβ1-4[Fucα1-3]GlcNAc, sLeX), sialyl Lewis A (Neu5Acα2-3Galβl-3[Fucα1-4]GlcNAc, sLeA), sialylated or desialylated human plasma N-glycans, horseradish peroxidase N-glycans containing core α-1,3-fucose (HRP), blood group A type II, blood group B type II and α-1,6-fucosylated biantennary N-glycan (FA2G2) and the products of the reactions analyzed by LC–MS/MS (Table S3). Interestingly, this screening revealed that E1_10125 was active against α-1,3- and α-1,4-fucosylated substrates presenting a terminal sialic acid modification. The chromatograms clearly showed the appearance of peaks corresponding to Neu5Acα2-3Galβl-3GlcNAc and Neu5Acα2-3Galβ1-4GlcNAc and the disappearance of the peaks corresponding to sLeA and sLeX (Fig. 3a). The use of sialylated and de-sialylated N-glycans from human plasma confirmed the ability of E1_10125 to accommodate sialyl residues in terminal location of fucosylated N-glycans (Fig. 3b), as shown by the disappearance of the peak corresponding to fucosylated antennary N-glycan upon incubation with E1_10125. In contrast, no reaction product was detected when HRP glycans (core α1,3-fucose), FA2G2 (α1,6-fucose) or blood group antigens (α1,2-fucose) were used as substrates as shown by LC–MS traces (Fig. 3c–e).
Fig. 3.
LC–MS/MS analysis of R. gnavus GH29 fucosidase E1-10125 towards various fucosylated substrates. a LC–MS/MS analysis of the products released from the enzymatic reaction of E1-10125 with LeA (left) and LeX (right), the upper graph is the negative control and the lower one corresponds to the enzymatic reaction. B LC–MS analysis of the products released from the enzymatic reaction of E1-10125 with sialylated (upper) and desialylated (lower) human plasma. The negative controls are showed on top of the enzymatic reactions. c LC–MS analysis of the products released from the enzymatic reaction of E1-10125 with HRP (core α1,3-fucose) (lower). The negative control is shown in the upper panel. D. LC–MS analysis of the products released from the enzymatic reaction of E1-10125 with FA2G2 (α1,6-fucose) (lower). The negative control is shown in the upper panel. e LC–MS analysis of the products released from the enzymatic reaction of E1-10125 with blood group A type II (left) and blood group B type II (right) (upper). The negative control is shown in the upper panel
R. gnavus GH29 E1_10125 fucosidase can accommodate terminal sialic acid moieties in α1,3/4 antennary fucosylated substrates
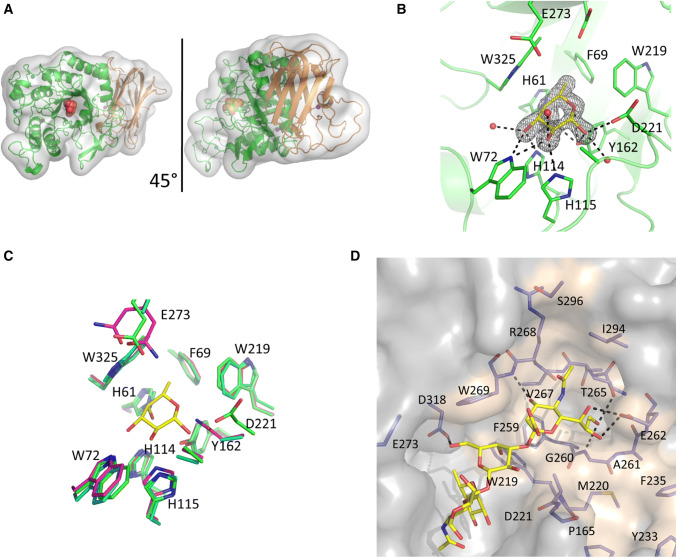
To further investigate E1_10125 ligand specificity, the enzyme was crystallized in the presence of 2′FL, providing the crystal structure of the complex showing the β-fucose anomer bound in the active site (Fig. 4). Data collection and refinement statistics are detailed in Table 3. Electron density maps allowed modelling of residues 23–527. The enzyme consists of two distinct domains, a catalytic domain (PF001120) domain comprising residues 46–366 in N-terminal and a F5/8 Type C domain (PF00754) covering residues 385–526 in C-terminal (Fig. 4a). The catalytic domain displays a (α/β)8 which is typical of GH29 enzymes (www.cazy.org) whereas the type C-domains shows structural homology with carbohydrate binding module (CBM) belonging to CBM32 family (www.cazy.org). The macromolecular architecture is conserved with the recently solved GH29 fucosidase enzymes from S. pneumoniae (RMSD: 0.969 Å) [22] and Bifidobacterium longum subsp. infantis (RMSD: 1.128 Å) [67]. Residues 23–45 wrap around the C-terminal β-sandwich domain. The catalytic machinery sits in a cleft in the center of the N-terminal domain (Fig. 4b). By proximity to the bound fucose residue and homology to other fucosidases, Asp221 was identified as the catalytic nucleophile and Glu273 as the acid/base (amino acid numbering based on recombinant protein sequence) (Fig. 4b). Trp325 creates a CH–π interaction with the bound ligand and His61, Trp72, His114, His115 and Tyr162 provides additional hydrogen bonding interactions. Phe59 and Trp219 provide a hydrophobic pocket to accommodate the fucose C6 methyl group (Fig. 4b). The fucose binding site is conserved with the S. pneumoniae [22] and B. longum GH29 fucosidases [67] and is henceforth referred to as the − 1 subsite (Fig. 4c). In an attempt to obtain crystal structures in complex with the substrate, an active site mutant (E1_10125 D221A) was generated. However, following co-crystallization experiments with 2′FL, fucose was found bound in the active site, indicating residual fucosidase activity. Clear electron density was present for both α- and β-fucose anomers (Fig. S3A and S3B). It was not possible to obtain crystals of E1_10125 D221A in the absence of 2′FL. Attempts were made to displace the fucose molecule with other fucosylated ligands; however, these were unsuccessful, perhaps due to the substrate binding site being present at a tightly packed interface between two symmetry related protein molecules. No crystal structures could be obtained with other substrates tested including l-fucose, Neu5Ac, 3FL, LeA, LeX and sLeX.
Fig. 4.
Crystal structure of R. gnavus GH29 fucosidase E1_10125. a Cartoon representation of E1_10125 fucosidase, the catalytic domain is coloured green and the proposed CBM is coloured orange. A fucose residue in a sphere representation indicates the location of the active site. The views are related by a 45° rotation around the y axis. b The E1_10125 fucose binding site. The β-anomer of fucose is shown in yellow with nearby active site residues shown in green. Black dashed lines indicate hydrogen-bonding interactions. Fo–Fc difference map density for the fucose residue is displayed as a black mesh, contoured at 2σ. c The fucose binding sites of E1_10125 (green), S. pneumoniae GH29 fucosidase (magenta), and B. longum subsp. infantis GH29 fucosidase (cyan) are aligned. Residue numbers refer to E1_10125. The binding site residues are conserved across the three structures and differences present at the D221 and E273 positions are catalytic mutants. Fucose bound in the E1_10125 is show in yellow for reference. d Model of the orientation and conformation of sLeX bound to R. gnavus E1_10125 proposed by MD simulations
Table 3.
Data collection and refinement statistics
| Data set | WT—Fucose | D221A—Fucose |
|---|---|---|
| PDB identifier | 6TR3 | 6TR4 |
| Data collection | ||
| Space group | C2 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 164.2, 48.8, 132.1 | 49.8, 74.1, 76.5 |
| α, β, γ (°) | 90.0, 151.1, 90.0 | 82.5, 80.4, 70.4 |
| Resolution (Å) | 63.90–1.70 (1.73–1.70) | 69.54–1.45 (1.47–1.45) |
| Rmerge | 0.08 (0.59) | 0.04 (0.11) |
| Rmeas | 0.10 (0.72) | 0.06 (0.16) |
| I/σI | 6.9 (1.1) | 12.7 (3.4) |
| CC half | 0.99 (0.58) | 0.99 (0.96) |
| Completeness | 99.1 (97.5) | 92.7 (56.9) |
| Redundancy | 3.1 (2.9) | 2.0 (1.8) |
| Refinement | ||
| Resolution (Å) | 63.86–1.70 | 69.54–1.45 |
| No. of reflections | 55,403 | 166,349 |
| Rwork/Rfree | 0.174/0.227 | 0.141/0.161 |
| No. of atoms | ||
| Protein | 4022 | 8134 |
| Ligand/ion | 30 | 48 |
| Water | 282 | 1507 |
| B-factors | ||
| Protein | 25.2 | 11.5 |
| Ligand/ion | 39.0 | 6.7 |
| Water | 26.9 | 23.6 |
| r.m.s.d | ||
| Bond lengths (Å) | 0.03 | 0.01 |
| Bond angle (°) | 2.8 | 1.4 |
| Ramachandran statistics (%) | ||
| Favoured | 96 | 97 |
| Outliers | 0 | 0 |
Numbers in parenthesis refer to the highest resolution shell
Superimposition of the E1_10125 crystal structure with that of α-1,3/4-fucosidase from B. longum subsp. infantis D172A/E217A mutant complexed with lacto-N-fucopentaose II (pdb 3UET) [67] (Fig. S4C) or with S. pneumoniae SpGH29CT D171N/E215Q in complex with LeA (pdb 6ORF) (Fig. S4D) or with LeX (pdb 6OR4) [62] (Fig. S3E) indicate that there are unlikely to be E1_10125 interactions that form a distinct + 1 site (GlcNAc in LeA and LeX trisaccharide antigens). E1_10125 Trp269 is conserved with S. pneumoniae and B. longum fucosidases, maintaining the + 2 site. More specifically, Trp269 is likely to form a CH/π stacking interaction with the galactose ring, and Asp318 to form a hydrogen bond with the galactose C6 hydroxyl. In the E1_10125 crystal structure, adjacent to the proposed + 2 site is an open platform comprises primarily neutral and hydrophobic residues, which would accommodate Neu5Ac (Fig. S3F). This is in marked contrast to the S. pneumoniae and B. longum homologue structures where this region is partially occluded by incoming loops. Molecular modelling calculations were carried out to further support this hypothesis and to provide a model for the orientation of the sialic acid ring of sLeX when bound to E1_10125 fucosidase (Fig. 4d). Following 500 ns MD simulations of the E1_10125 D221A mutant and docking of the sLeX ligand, the molecular model showed that sialic acid ring sits in the neighboring subsite. Polar contacts are established between nearby residues, Glu262 and Trp269; in particular, and hydroxyl groups present at sialic acid C4, C8 and C9 positions. This analysis confirmed that the E1_10125 fucosidase enzyme shows an open binding site able to accommodate the sLeX ligand.
In the absence of a complex structure of E1_10125 with a fucosylated oligosaccharide and in order to test the hypothesis that the cavity could accommodate the sialic acid moiety, E1_10125 R268W and E1_10125 G260M mutants were produced in which these introduced side-chains are expected to block access to the cavity. The E1_10125 R268W mutant showed a complete loss of activity towards all substrates tested including pNP-Fuc, 2′FL, 3FL, blood group A type II, blood group B type II, LeX and sLeX (data not shown) whereas the E1_10125 G260M mutant showed a significant decrease in activity towards sLeX down to 28% activity while 76% activity remained towards LeX (Table S4), suggesting that the cavity is important to accommodate terminal modifications of the fucosylated substrates.
Glycan arrays were then used to further define the ligand and linkage specificity of E1_10125 (Fig. S5). The purified recombinant His6-tagged E1_10125 D221A inactive mutant was screened at three protein concentrations against the Core H glycan microarray glycans at the Consortium for Functional Glycomics (CFG). Among the 585 glycans screened on the microarray, significant RFU values (> 300) were obtained for 5 fucosylated glycans using the highest protein concentration. Glycan ID 389 with α-Gal-LeA epitope displayed the highest RFU value (1072 ± 47) followed by two glycans, ID 249 and ID 526, containing sLeX epitopes. This recognition pattern therefore suggests that E1_10125 could recognize fucosylated substrates with diverse terminal modifications at the reducing end.
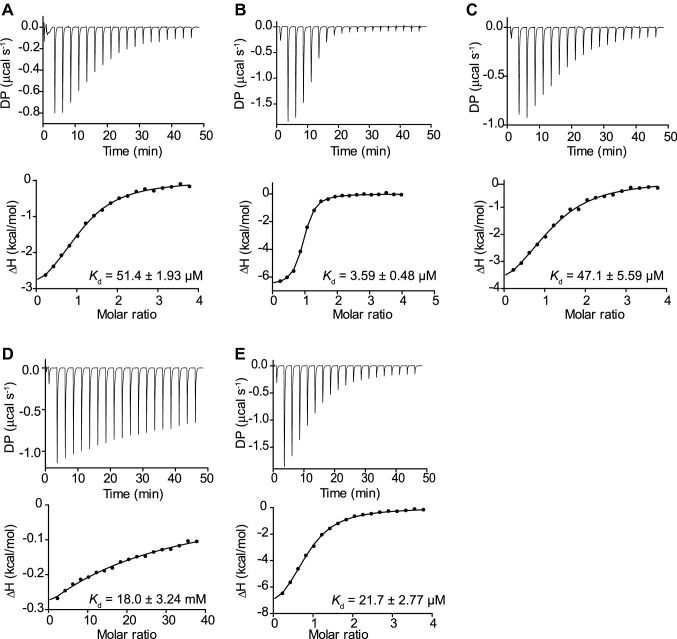
In order to further test this hypothesis, ITC was used to determine the binding parameters of E1_10125 D221A mutant towards these ligands (Fig. 5 and Table S5). The enzyme bound to LeX with a Kd of 51.43 ± 1.93 μM (Fig. 5a) and to sLeX with a Kd of 3.59 ± 0.48 μM (Fig. 5b). Further, a Kd of 47.13 ± 5.60 μM was obtained when αGal-LeX (Fig. 5c) was used as a ligand whereas a Kd of 17.98 mM and 21.7 μM were obtained with the monosaccharides Neu5Ac or Fuc used as a control (Fig. 5d, e). To compare the substrate specificity among LeX, sLeX and αGal-LeX, the kinetic parameters were determined against these substrates (Table 4). E1_10125 showed strongest affinity to sLeX with a Km of 163.1 µM and the presence of sialic acid or galactose on the non-reducing end of LeX significaly increased the catalytic efficienty up to 20-fold, consistent with the binding parameters (Table 4).
Fig. 5.
ITC isotherms of R. gnavus GH29 fucosidase E1_10125 binding to fucosylated ligands. a E1_10125 binding to LeX. b E1_10125 binding to sLeX. c E1_10125 binding to αGal-LeX. d E1_10125 binding to Neu5Ac. e E1_10125 binding to l-Fucose. DP differential power
Table 4.
Kinetic parameters of E1_10125 towards LeX, sLeX and aGal-LeX
| LeX | sLeX | αGal-LeX | |
|---|---|---|---|
| Catalytic efficiency (s−1 M−1) | 1416.67 ± 288.68 | 12,874.85 ± 1620.36 | 28,888.89 ± 2545.88 |
| Km (µM) | ND | 163.1 ± 16.67 | ND |
| kcat (s−1) | ND | 2.10 ± 0.07 | ND |
ND could not be determined under experimental conditions
To gain further structural insights into the unique ligand specificity of E1_10125, STD NMR studies [68] were conducted with E1_10125 D221A mutant in the presence of 2′FL, 3FL, LeA, LeX, sLeX and αGal-LeX (Fig. 6). Transfer of magnetization as saturation from the protein to the ligand was observed for all substrates tested, in agreement with the binding of E1_10125 to these substrates with medium-weak affinities (µM-mM). The enzyme intimately recognized the three sugar residues constituting 3FL and LeA (Fig. 6a, b) with no significant differences in their binding epitopes. 2′FL showed binding to E1_10125 D221A by STD NMR (Fig. 6c), but the main contacts were restricted to the fucose residue, whereas loose contacts were observed with the lactose disaccharidic moiety. The binding epitope of LeX also revealed a reduction in close contacts with the enzyme, particularly at the fucose residue. A comparison between 3FL (Fig. 6a) and LeX (Fig. 6d) supports an impact of the acetamido group (NHAc) at position 2 of GlcNAc on binding, which leads to changes in the contacts of the fucose ring with the protein in the bound state. This is highlighted by the reduction of the relative STD intensity of the methyl group at position 6 of this ring in the case of LeX. It is well established that the NHAc group in LeX limits the flexibility of the Fucα1–3GlcNAc linkage via steric hindrance with the adjacent fucose ring [69]. This leads to the observed changes in the contacts of the fucose ring in the bound state of LeX in comparison 3FL (Fig. 6d). These structural changes together with the advantageous reduction in entropy penalty upon binding expected for LeX due to the limited inter-glycosidic flexibility, are in good agreement with the observed differences in fucosidase activities (Table 2). The STD NMR results, in alignment with the activity assays and LC–MS/MS data, confirmed that the E1_10125 fucosidase shows a preference for α1-3/4 linkage (Fig. 3, Table 2), in which the fucose is linked at the reducing glucopyranose ring of the Gal β 1–3/4Glc(NAc) disaccharidic sequence. Interestingly, STD NMR revealed that the sialic acid moiety of sLeX makes contacts with the enzyme at C3 and C5 positions (Fig. 6e), suggesting that the sialic acid moiety is in part solvent exposed, and in part surrounded by residues at the protein surface, in agreement with the crystal structure showing a cavity that could accommodate such a sialic acid residue at the non-reducing end of the ligand. The molecular model (Fig. 4d) is also in excellent agreement with the experimental NMR data, as in this binding mode, protons at C3 and C5 are pointing towards the surface of the enzyme in the pocket. Likewise, in αGal-LeX, weak contacts were observed for the non-reducing αGal and βGal rings in the bound state, whereas fucose was the main ligand recognition moiety, followed by GlcNAc (Fig. 6f). Together, these data support the X-ray crystal structure that the binding pocket of E1_10125 could accomodate terminal residues although with a clear preference for sialic acid.
Fig. 6.
Binding epitope mapping from STD NMR spectroscopy depicting interactions of R. gnavus GH29 fucosidase E1-10125 with fucosylated oligosaccharides. Normalized saturation transfer intensities (0–100%) from STD NMR experiments mapped onto the chemical structures of a 2′FL, b 3FL, c LeA, d LeX, e sLeX and f αGal-LeX. Stronger normalized STD intensities correlate with closer ligand contacts with the surface of the protein in the bound state. Legend indicates normalized STD intensities: blue, 0–24%; yellow, 25–50%, red 51–100%. The enzyme intimately recognizes 3FL and LeA, whereas looser contacts are observed for 2′FL, LeX, sLeX and αGal-LeX. For the latter, a much higher degree of proton chemical shift overlapping implied lower binding epitope resolution and a normalized STD intensity value was assigned for each ring, as an average of the STD intensities of its isolated protons
Discussion
Fucose decorating glycan chains in HMOs or mucins contributes to shaping the composition of the gut microbiota in adults and infants. Previous studies in mice showed that the loss of the α-1,2-fucosyltransferase FUT2, and therefore fucosylated host glycans, leads to a decreased diversity and differences in intestinal microbial community [70–73], whereas an association between the composition of the intestinal microbiota and the ABO blood group or FUT2 secretor status was reported in humans [72, 74–77]. Human fetal mucins along the GI tract harbor a repertoire of O-glycans similar to HMOs [78, 79] which may also contribute to the differences in gut microbiota composition as compared to adults [80]. The ability to utilize fucosyllactose is a trait of early inhabitants of the human GI tract, such as R. gnavus [27] or various bifidobacteria species [81] as well as probiotic strains, such as Lactobacillus casei [82]. To access this nutrient source, gut bacteria have evolved to express a wide range of fucosidases with distinct ligand specificity, contributing to their fitness across nutritional niches [5, 6]. Furthermore, in its free form, fucose released by bacterial fucosidases may affect gut homeostasis. For example, B. thetaiotaomicron produces multiple fucosidases that cleave fucose from host glycans, resulting in high fucose availability in the gut lumen [83] which can then act as a signal to modulate the pathogenicity and metabolism of the pathogen enterohaemorrhagic E. coli (EHEC) [84].
Complexity in HMOs or mucins lies in the diversity of glycosidic bonds in these molecules, rendering a large number of potential combinations. Since fucosylation varies across and along the intestine and that fucosidase activity is dependent on the type of linkages present in the glycans or glycoconjugates, it is critical to understand the ligand specificity of the fucosidases encoded by major gut symbionts. Recently, the substrate specificities of two structure-solved GH29 fucosidases from B. thetaiotaomicron VPI-5482 were determined showing that the protein with locus tag BT 2970 belongs to GH29-A while BT 2192 belongs to GH29-B [15]. R. gnavus is a human gut symbiont of the infant and adult microbiota [23–25]. Here, we showed that R. gnavus strains encodes a range of fucosidases belonging to GH95 and GH29-A and GH29-B families with varied specificities, highlighting the versatility of fucosylated substrates they can access, such as those present in HMOs or intestinal mucins. The enzymatic characterization of R. gnavus fucosidases focused on previously uncharacterized bacterial fucosidases revealed that fucosidase ATCC_03833 belongs to GH29-A while E1_10125 belongs to GH29-B subfamily. GH29-B E1_10125 and E1_10180 showed strict substrate specificity towards α1,3/4 fucosylated linkages while fucosidases GH95 ATCC_00842 and GH29-A ATCC_03833 showed preference for α1,2 linkages, as reported for B. bifidum AfcA fucosidase [85]. O-glycan analyses of human fetal mucins showed that fucose is present in a large variety of terminal linkages, including blood group H as well as LeA (Galβ(1–3[Fucα1-4]GlcNAc), LeB (Fucα1-2Galβ1-3[Fucα1-4]GlcNAc), LeX (Galβ(1–4 [Fucα1-3]GlcNAc) and LeY (Fucα1-2Galβ1-4[Fucα1-3] GlcNAc) determinants [79]. The diversity of fucosidases may confer R. gnavus strains with an advantage in colonizing the infant gut [25].
In addition, we showed that E1_10125 could act on LeA and LeX even when the galactose moiety was linked to a sialic acid residue or other decorations. In particular, E1_10125 showed highest affinity towards sLeX. The ability to accommodate the sialic acid moiety, as also confirmed by STD NMR, appears to be enabled by an open region adjacent to the + 2 site (Gal residue in Le antigens), which comprises residues Met220, Phe259, Gly260, Ala261 and Thr265 with additional stabilizing interactions likely to be provided by Trp269 and Glu262. This structural arrangement lacks the incoming loops present in the S. pneumoniae and B. longum GH29 enzymes [22, 67], supporting the unique specificity of R. gnavus E1_10125 fucosidase. Glycan array analyses suggested that E1_10125 could recognize fucosylated glycans with diverse terminal modifications as also supported by ITC showing binding of E1_10125 to both sialic acid and αGal linked to Gal of LeX. Extensive differences in the glycosylation profile of mucins occur along the GI tract, characterized by the presence of decreasing gradients of fucose and ABH blood group and increasing gradients of sialic acid from ileum to rectum [7, 86]. In human colonic mucin, more than 100 complex O-linked oligosaccharides were identified, mostly based on the core 3 structure with sialic acid at the 6-position of the GalNAc [9]. The most abundant components were -Gal-(Fuc)GlcNAc-3(NeuAc-6)GalNAcol, GalNAc-(NeuAc-)Gal-4/3GlcNAc-3(NeuAc-6)GalNAcol, GalNAc-3(NeuAc-6) GalNAcol and GlcNAc-3(NeuAc-6)GalNAcol [9]. The unusual specificity of E1_10125 may, therefore, contribute to the fitness and spatial adaptation of R. gnavus strains into the adult human GI tract [23, 24].
The specificities of R. gnavus fucosidases could be exploited for diagnostic assays. For example, changes in the abundance of antennary fucosylation in plasma N-glycans have been associated with diabetes [87, 88] and with colorectal cancer [89–91]. The quantitation of these low abundant antennary fucosylated glycans in the plasma N-glycome is complex because the structural diversity of its component glycans [92, 93] that rely on chromatographic platforms requiring extensive measurement time [91–95]. However, the recent technological advances integrating the use of fucosidases or other glycosidases and analysis on a MALDI-MS platform enabled identification and quantification of glycans of specific fucose isomers [96, 97]. The antennary fucosidase specificity reported in this work could therefore be used as a discriminatory tool to identify N-glycan biomarkers of diseases and as a valuable tool for the purpose of glycoprofiling biopharmaceutical glycoproteins.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors gratefully acknowledge the support of the Biotechnology and Biological Sciences Research Council (BBSRC); this research was mostly funded by the Innovate UK Biocatalyst grant Glycoenzymes for Bioindustries (BB/M029042/) with contribution from the Royal Society and the BBSRC Institute Strategic Programmes BB/J004529/1 ‘The Gut Health and Food Safety’ and BB/R012490/1 ‘Gut Microbes and Health’ and its consituent project BBS/E/F/000PR10353 (Theme 1, Determinants of microbe-host responses in the gut across life). We would like to thank Diamond Light Source beamlines VMXi, I03 and I04 for beamtime and assistance, as well as the crystallization facility at Harwell for access and support. We wish to acknowledge the Consortium for Functional Glycomics grant number GM62116 and GM098791 for glycan screening. Osmond Rebello received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement GlySign No 722095. Serena Monaco and Jesús Angulo
acknowledge financial support from BBSRC (BB/P010660/1). Jesús Angulo also acknowledges
financial support from the Universidad de Sevilla (Acciones Especiales del VI Plan Propio de
Investigación y transferencia). Samuel Walpole and Thomas Hicks acknowledge BBSRC DTP
studentships.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474(11):1823–1836. doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22(9):1147–1162. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabrera-Rubio R, Kunz C, Rudloff S, Garcia-Mantrana I, Crehua-Gaudiza E, Martinez-Costa C, Collado MC. Association of maternal secretor status and human milk oligosaccharides with milk microbiota: an observational pilot study. J Pediatr Gastr Nutr. 2019;68(2):256–263. doi: 10.1097/Mpg.0000000000002216. [DOI] [PubMed] [Google Scholar]
- 4.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10(5):323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015;6:ARTN81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ndeh D, Gilbert HJ. Biochemistry of complex glycan depolymerisation by the human gut microbiota. FEMS Microbiol Rev. 2018;42(2):146–164. doi: 10.1093/femsre/fuy002. [DOI] [PubMed] [Google Scholar]
- 7.Robbe C, Capon C, Maes E, Rousset M, Zweibaum A, Zanetta JP, Michalski JC. Evidence of regio-specific glycosylation in human intestinal mucins: presence of an acidic gradient along the intestinal tract. J Biol Chem. 2003;278(47):46337–46348. doi: 10.1074/jbc.M302529200. [DOI] [PubMed] [Google Scholar]
- 8.Jensen PH, Kolarich D, Packer NH. Mucin-type O-glycosylation—putting the pieces together. Febs J. 2010;277(1):81–94. doi: 10.1111/j.1742-4658.2009.07429.x. [DOI] [PubMed] [Google Scholar]
- 9.Larsson JM, Karlsson H, Sjovall H, Hansson GC. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology. 2009;19(7):756–766. doi: 10.1093/glycob/cwp048. [DOI] [PubMed] [Google Scholar]
- 10.Mollicone R, Bara J, Le Pendu J, Oriol R. Immunohistologic pattern of type 1 (Lea, Leb) and type 2 (X, Y, H) blood group-related antigens in the human pyloric and duodenal mucosae. Lab Invest. 1985;53(2):219–227. [PubMed] [Google Scholar]
- 11.Kelly RJ, Rouquier S, Giorgi D, Lennon GG, Lowe JB. Sequence and expression of a candidate for the human Secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J Biol Chem. 1995;270(9):4640–4649. doi: 10.1074/jbc.270.9.4640. [DOI] [PubMed] [Google Scholar]
- 12.Henry SM, Jovall PA, Ghardashkhani S, Gustavsson ML, Samuelsson BE. Structural and immunochemical identification of Leb glycolipids in the plasma of a group O Le(a-b-) secretor. Glycoconj J. 1995;12(3):309–317. doi: 10.1007/bf00731334. [DOI] [PubMed] [Google Scholar]
- 13.Larsson JMH, Thomsson KA, Rodriguez-Pineiro AM, Karlsson H, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. III. Gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveal a regiospecific distribution. Am J Physiol-Gastr L. 2013;305(5):G357–G363. doi: 10.1152/ajpgi.00048.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sano M, Hayakawa K, Kato I. Purification and characterization of alpha-L-fucosidase from Streptomyces species. J Biol Chem. 1992;267(3):1522–1527. doi: 10.1016/S0021-9258(18)45977-7. [DOI] [PubMed] [Google Scholar]
- 15.Sakurama H, Tsutsumi E, Ashida H, Katayama T, Yamamoto K, Kumagai H. Differences in the substrate specificities and active-site structures of two alpha-L-fucosidases (glycoside hydrolase family 29) from Bacteroides thetaiotaomicron. Biosci Biotechnol Biochem. 2012;76(5):1022–1024. doi: 10.1271/bbb.111004. [DOI] [PubMed] [Google Scholar]
- 16.Sulzenbacher G, Bignon C, Nishimura T, Tarling CA, Withers SG, Henrissat B, Bourne Y. Crystal structure of Thermotoga maritima alpha-L-fucosidase. Insights into the catalytic mechanism and the molecular basis for fucosidosis. J Biol Chem. 2004;279(13):13119–13128. doi: 10.1074/jbc.M313783200. [DOI] [PubMed] [Google Scholar]
- 17.Lezyk M, Jers C, Kjaerulff L, Gotfredsen CH, Mikkelsen MD, Mikkelsen JD. Novel alpha-L-Fucosidases from a soil metagenome for production of fucosylated human milk oligosaccharides. PLoS ONE. 2016;11(1):e0147438. doi: 10.1371/journal.pone.0147438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wongmadden ST, Landry D. Purification and characterization of novel glycosidases from the bacterial genus xanthomonas. Glycobiology. 1995;5(1):19–28. doi: 10.1093/glycob/5.1.19. [DOI] [PubMed] [Google Scholar]
- 19.Megson ZA, Koerdt A, Schuster H, Ludwig R, Janesch B, Frey A, Naylor K, Wilson IB, Stafford GP, Messner P, Schaffer C. Characterization of an alpha-l-fucosidase from the periodontal pathogen Tannerella forsythia. Virulence. 2015;6(3):282–292. doi: 10.1080/21505594.2015.1010982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, Katayama T, Yamamoto K. Two distinct alpha-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19(9):1010–1017. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- 21.Fan S, Zhang H, Chen X, Lu L, Xu L, Xiao M. Cloning, characterization, and production of three alpha-L-fucosidases from Clostridium perfringens ATCC 13124. J Basic Microbiol. 2016;56(4):347–357. doi: 10.1002/jobm.201500582. [DOI] [PubMed] [Google Scholar]
- 22.Hobbs JK, Pluvinage B, Robb M, Smith SP, Boraston AB. Two complementary alpha-fucosidases from Streptococcus pneumoniae promote complete degradation of host-derived carbohydrate antigens. J Biol Chem. 2019;294(34):12670–12682. doi: 10.1074/jbc.RA119.009368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Meta HITC, Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraal L, Abubucker S, Kota K, Fischbach MA, Mitreva M. The prevalence of species and strains in the human microbiome: a resource for experimental efforts. PLoS ONE. 2014;9(ARTNe97279):7279. doi: 10.1371/journal.pone.0097279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sagheddu V, Patrone V, Miragoli F, Puglisi E, Morelli L. infant early gut colonization by lachnospiraceae: high frequency of Ruminococcus gnavus. Front Pediatr. 2016;4:57. doi: 10.3389/fped.2016.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall AB, Yassour M, Sauk J, Garner A, Jiang XF, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, Bertha M, Cohen M, Garber J, Khalili H, Gevers D, Ananthakrishnan AN, Kugathasan S, Lander ES, Blainey P, Vlamakis H, Xavier RJ, Huttenhower C. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9:ARTN103. doi: 10.1186/s13073-017-0490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS ONE. 2013;8(10):ARTNe76341. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crost EH, Tailford LE, Monestier M, Swarbreck D, Henrissat B, Crossman LC, Juge N. The mucin-degradation strategy of Ruminococcus gnavus: The importance of intramolecular trans-sialidases. Gut Microbes. 2016;7(4):302–312. doi: 10.1080/19490976.2016.1186334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tailford LE, Owen CD, Walshaw J, Crost EH, Hardy-Goddard J, Le Gall G, de Vos WM, Taylor GL, Juge N. Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nat Commun. 2015;6:7624. doi: 10.1038/ncomms8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell A, Brunt J, Crost E, Vaux L, Nepravishta R, Owen CD, Latousakis D, Xiao A, Li W, Chen X, Walsh MA, Claesen J, Angulo J, Thomas GH, Juge N. Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut. Nat Microbiol. 2019;4(12):2393–2404. doi: 10.1038/s41564-019-0590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berrow NS, Alderton D, Sainsbury S, Nettleship J, Assenberg R, Rahman N, Stuart DI, Owens RJ. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35(6):e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez-Weatherby J, Sandy J, Mikolajek H, Lobley CMC, Mazzorana M, Kelly J, Preece G, Littlewood R, Sorensen TLM. VMXi: a fully automated, fully remote, high-flux in situ macromolecular crystallography beamline. J Synchrotron Radiat. 2019;26(Pt 1):291–301. doi: 10.1107/S1600577518015114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 34.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr Sect D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter G, Lobley CMC, Prince SM. Decision making in xia2. Acta Crystallogr Sect D Biol Crystallogr. 2013;69:1260–1273. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winter G, Waterman DG, Parkhurst JM, Brewster AS, Gildea RJ, Gerstel M, Fuentes-Montero L, Vollmar M, Michels-Clark T, Young ID, Sauter NK, Evans G. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr D. 2018;74:85–97. doi: 10.1107/S2059798317017235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Method Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P. Tools for ligand validation in Coot. Acta Crystallogr D. 2017;73:203–210. doi: 10.1107/S2059798317003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Beusekom B, Joosten K, Hekkelman ML, Joosten RP, Perrakis A. Homology-based loop modeling yields more complete crystallographic protein structures. IUCrJ. 2018;5(Pt 5):585–594. doi: 10.1107/S2052252518010552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, Jain S, Lewis SM, Arendall WB, Snoeyink J, Adams PD, Lovell SC, Richardson JS, Richardson DC. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27(1):293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schrödinger Release 2020–1: Maestro S, LLC, New York, NY, 2020.
- 43.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 44.Olsson MH, Sondergaard CR, Rostkowski M, Jensen JH. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput. 2011;7(2):525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 45.Case DAIYB-S, Brozell SR, Cerutti DS, Cheatham TE, III, Cruzeiro VWD, Darden TA, Duke RE, Ghoreishi D, Gilson MK, Gohlke H, Goetz AW, Greene D, Harris R, Homeyer N, Izadi S, Kovalenko A, Kurtzman T, Lee TS, LeGrand S, Li P, Lin C, Liu J, Luchko T, Luo R, Mermelstein DJ, Merz KM, Miao Y, Monard G, Nguyen C, Nguyen H, Omelyan I, Onufriev A, Pan F, Qi R, Roe DR, Roitberg A, Sagui C, Schott-Verdugo S, Shen J, Simmerling CL, Smith J, Salomon-Ferrer R, Swails J, Walker RC, Wang J, Wei H, Wolf RM, Wu X, Xiao L, York DM, Kollman PA. AMBER 2018. San Francisco: University of California; 2018. [Google Scholar]
- 46.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23(3):327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 47.Roe DR, Cheatham TE., 3rd PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9(7):3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 48.Schrödinger Release 2020–1: LigPrep S, LLC, New York, NY, 2020.
- 49.Schrödinger Release 2020–1: MacroModel S, LLC, New York, NY, 2020.
- 50.Schrödinger Release 2020–1: Glide S, LLC, New York, NY, 2020.
- 51.Davis MI, Shen M, Simeonov A, Hall MD. Diaphorase Coupling Protocols for Red-Shifting Dehydrogenase Assays. Assay Drug Dev Techn. 2016;14(3):207–212. doi: 10.1089/adt.2016.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsui I, Ishikawa K, Matsui E, Miyairi S, Fukui S, Honda K. Subsite structure of Saccharomycopsis α-amylase secreted from Saccharomyces cerevisiae. J Biochem. 1991;109:566–569. doi: 10.1093/oxfordjournals.jbchem.a123420. [DOI] [PubMed] [Google Scholar]
- 53.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26(5):680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR. Whalen KL (2015) Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim Biophys Acta. 1854;8:1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RDAB (2005) Protein identification and analysis tools on the ExPASy server. In: The proteomics protocols handbook. Humana Press, pp 571–607. 10.1385/1-59259-890-0:571
- 57.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47(W1):W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Armenteros JJA, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37(4):420. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 59.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 60.Fimereli DK, Tsirigos KD, Litou ZI, Liakopoulos TD, Bagos SJHPG. CW-PRED: A HMM-based method for the classification of cell wall-anchored proteins of Gram-positive bacteria. Springer's Lect Notes Comput Sci. 2012;7297(2012):285–290. doi: 10.1007/978-3-642-30448-4_36. [DOI] [Google Scholar]
- 61.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26(13):1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom HJ, Joachimiak A, Lebrilla CB, Mills DA. Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol. 2012;78(3):795–803. doi: 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeuner B, Muschiol J, Holck J, Lezyk M, Gedde MR, Jers C, Mikkelsen JD, Meyer AS. Substrate specificity and transfucosylation activity of GH29 alpha-l-fucosidases for enzymatic production of human milk oligosaccharides. N Biotechnol. 2018;41:34–45. doi: 10.1016/j.nbt.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 64.Ono A, Suzuki T, Gotoh S, Kono H, Matsui M, Aoki D, Matsuda M, Kawagishi H, Ogata M. Structural investigation of alpha-l-fucosidase from the pancreas of Patiria pectinifera, based on molecular cloning. Carbohydr Res. 2019;475:27–33. doi: 10.1016/j.carres.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 65.Shaikh FA, Lammerts van Bueren A, Davies GJ, Withers SG. Identifying the catalytic acid/base in GH29 alpha-L-fucosidase subfamilies. Biochemistry. 2013;52(34):5857–5864. doi: 10.1021/bi400183q. [DOI] [PubMed] [Google Scholar]
- 66.Zeleny R, Leonard R, Dorfner G, Dalik T, Kolarich D, Altmann F. Molecular cloning and characterization of a plant alpha1,3/4-fucosidase based on sequence tags from almond fucosidase I. Phytochemistry. 2006;67(7):641–648. doi: 10.1016/j.phytochem.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 67.Sakurama H, Fushinobu S, Hidaka M, Yoshida E, Honda Y, Ashida H, Kitaoka M, Kumagai H, Yamamoto K, Katayama T. 1,3–1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 Chains. J Biol Chem. 2012;287(20):16709–16719. doi: 10.1074/jbc.M111.333781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mayer M, Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew Chem Int. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 69.Imberty A, Perez S. Structure, conformation, and dynamics of bioactive oligosaccharides: theoretical approaches and experimental validations. Chem Rev. 2000;100(12):4567–4588. doi: 10.1021/cr990343j. [DOI] [PubMed] [Google Scholar]
- 70.Meng D, Newburg DS, Young C, Baker A, Tonkonogy SL, Sartor RB, Walker WA, Nanthakumar NN. Bacterial symbionts induce a FUT2-dependent fucosylated niche on colonic epithelium via ERK and JNK signaling. Am J Physiol Gastrointest Liver Physiol. 2007;293(4):G780–787. doi: 10.1152/ajpgi.00010.2007. [DOI] [PubMed] [Google Scholar]
- 71.Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, Knight R, Gordon JI, Sonnenburg JL. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci USA. 2013;110(42):17059–17064. doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tong M, McHardy I, Ruegger P, Goudarzi M, Kashyap PC, Haritunians T, Li X, Graeber TG, Schwager E, Huttenhower C, Fornace AJ, Sonnenburg JL, McGovern DPB, Borneman J, Braun J. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn's disease risk polymorphism. Isme J. 2014;8(11):2193–2206. doi: 10.1038/ismej.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rausch P, Kunzel S, Suwandi A, Grassl GA, Rosenstiel P, Baines JF. Multigenerational influences of the fut2 gene on the dynamics of the gut microbiota in mice. Front Microbiol. 2017;8:ARTN991. doi: 10.3389/fmicb.2017.00991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rausch P, Rehman A, Kunzel S, Hasler R, Ott SJ, Schreiber S, Rosenstiel P, Franke A, Baines JF. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci USA. 2011;108(47):19030–19035. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wacklin P, Tuimala J, Nikkila J, Tims S, Makivuokko H, Alakulppi N, Laine P, Rajilic-Stojanovic M, Paulin L, de Vos WM, Matto J. Faecal microbiota composition in adults is associated with the FUT2 gene determining the secretor status. PLoS ONE. 2014;9(4):ARTNe94863. doi: 10.1371/journal.pone.0094863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davenport ER, Goodrich JK, Bell JT, Spector TD, Ley RE, Clark AG. ABO antigen and secretor statuses are not associated with gut microbiota composition in 1,500 twins. Bmc Genom. 2016;17:ARTN 941. doi: 10.1186/s12864-016-3290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turpin W, Bedrani L, Espin-Garcia O, Xu W, Silverberg MS, Smith MI, Guttman DS, Griffiths A, Moayyedi P, Panaccione R, Huynh H, Steinhart H, Aumais G, Shestopaloff K, Dieleman LA, Turner D, Paterson AD, Croitoru K. FUT2 genotype and secretory status are not associated with fecal microbial composition and inferred function in healthy subjects. Gut Microbes. 2018;9(4):357–368. doi: 10.1080/19490976.2018.1445956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hounsell EF. Novel core, backbone and peripheral region sequences of the oligosaccharides of foetal gastrointestinal mucins present in human meconium. Symp Soc Exp Biol. 1989;43:149–154. [PubMed] [Google Scholar]
- 79.Robbe-Masselot C, Maes E, Rousset M, Michalski JC, Capon C. Glycosylation of human fetal mucins: a similar repertoire of O-glycans along the intestinal tract. Glycoconj J. 2009;26(4):397–413. doi: 10.1007/s10719-008-9186-9. [DOI] [PubMed] [Google Scholar]
- 80.Rodriguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, Avershina E, Rudi K, Narbad A, Jenmalm MC, Marchesi JR, Collado MC. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis. 2015;26:26050. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bunesova V, Lacroix C, Schwab C. Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. Bmc Microbiol. 2016;16:ARTN 248. doi: 10.1186/s12866-016-0867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodriguez-Diaz J, Monedero V, Yebra MJ. Utilization of natural fucosylated oligosaccharides by three novel alpha-L-fucosidases from a probiotic Lactobacillus casei strain. Appl Environ Microbiol. 2011;77(2):703–705. doi: 10.1128/AEM.01906-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299(5615):2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 84.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492(7427):113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, Sakata K, Yamanoi T, Kumagai H, Yamamoto K. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-alpha-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95) J Bacteriol. 2004;186(15):4885–4893. doi: 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robbe C, Capon C, Coddeville B, Michalski JC. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J. 2004;384(Pt 2):307–316. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Juszczak A, Pavic T, Vuckovic F, Bennett AJ, Shah N, Pape Medvidovic E, Groves CJ, Sekerija M, Chandler K, Burrows C, Rojnic Putarek N, Vucic Lovrencic M, Cuca Knezevic J, James TJ, Gloyn AL, Lauc G, McCarthy MI, Owen KR, Gornik O. Plasma fucosylated glycans and c-reactive protein as biomarkers of HNF1A-MODY in young adult-onset nonautoimmune diabetes. Diabetes Care. 2019;42(1):17–26. doi: 10.2337/dc18-0422. [DOI] [PubMed] [Google Scholar]
- 88.Thanabalasingham G, Huffman JE, Kattla JJ, Novokmet M, Rudan I, Gloyn AL, Hayward C, Adamczyk B, Reynolds RM, Muzinic A, Hassanali N, Pucic M, Bennett AJ, Essafi A, Polasek O, Mughal SA, Redzic I, Primorac D, Zgaga L, Kokic I, Hansen T, Gasperikova D, Tjora E, Strachan MWJ, Nielsen T, Stanik J, Klimes I, Pedersen OB, Njolstad PR, Wild SH, Gyllensten U, Gornik O, Wilson JF, Hastie ND, Campbell H, McCarthy MI, Rudd PM, Owen KR, Lauc G, Wright AF. Mutations in HNF1A result in marked alterations of plasma glycan profile. Diabetes. 2013;62(4):1329–1337. doi: 10.2337/db12-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Holst S, Deuss AJ, van Pelt GW, van Vliet SJ, Garcia-Vallejo JJ, Koeleman CA, Deelder AM, Mesker WE, Tollenaar RA, Rombouts Y, Wuhrer M. N-glycosylation profiling of colorectal cancer cell lines reveals association of fucosylation with differentiation and caudal type homebox 1 (CDX1)/Villin mRNA expression. Mol Cell Proteom. 2016;15(1):124–140. doi: 10.1074/mcp.M115.051235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Vroome SW, Holst S, Girondo MR, van der Burgt YEM, Mesker WE, Tollenaar R, Wuhrer M. Serum N-glycome alterations in colorectal cancer associate with survival. Oncotarget. 2018;9(55):30610–30623. doi: 10.18632/oncotarget.25753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Doherty M, Theodoratou E, Walsh I, Adamczyk B, Stockmann H, Agakov F, Timofeeva M, Trbojevic-Akmacic I, Vuckovic F, Duffy F, McManus CA, Farrington SM, Dunlop MG, Perola M, Lauc G, Campbell H, Rudd PM. Plasma N-glycans in colorectal cancer risk. Sci Rep. 2018;8(1):8655. doi: 10.1038/s41598-018-26805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Royle L, Campbell MP, Radcliffe CM, White DM, Harvey DJ, Abrahams JL, Kim YG, Henry GW, Shadick NA, Weinblatt ME, Lee DM, Rudd PM, Dwek RA. HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal Biochem. 2008;376(1):1–12. doi: 10.1016/j.ab.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 93.Stumpo KA, Reinhold VN. The N-glycome of human plasma. J Proteome Res. 2010;9(9):4823–4830. doi: 10.1021/pr100528k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Knezevic A, Bones J, Kracun SK, Gornik O, Rudd PM, Lauc G. High throughput plasma N-glycome profiling using multiplexed labelling and UPLC with fluorescence detection. Analyst. 2011;136(22):4670–4673. doi: 10.1039/c1an15684e. [DOI] [PubMed] [Google Scholar]
- 95.Pivac N, Knezevic A, Gornik O, Pucic M, Igl W, Peeters H, Crepel A, Steyaert J, Novokmet M, Redzic I, Nikolac M, Hercigonja VN, Curkovic KD, Curkovic M, Nedic G, Muck-Seler D, Borovecki F, Rudan I, Lauc G. Human plasma glycome in attention-deficit hyperactivity disorder and autism spectrum disorders. Mol Cell Proteom. 2011;10(1):M110004200. doi: 10.1074/mcp.M110.004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Benicky J, Sanda M, Pompach P, Wu J, Goldman R. Quantification of fucosylated hemopexin and complement factor H in plasma of patients with liver disease. Anal Chem. 2014;86(21):10716–10723. doi: 10.1021/ac502727s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rebello OD, Nicolardi S, Lageveen-Kammeijer GSM, Nouta J, Gardner RA, Mesker WE, Tollenaar RAEM, Spencer DIR, Wuhrer M, Falck D. A matrix-assisted laser desorption/ionization—mass spectrometry assay for the relative quantitation of antennary fucosylated n-glycans in human plasma. Front Chem. 2020;8:138–10.3389/fchem.2020.00138. doi: 10.3389/fchem.2020.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T, Prestegard JJ, Schnaar RL, Freeze HH, Marth JD, Bertozzi CR, Etzler ME, Frank M, Vliegenthart JF, Lutteke T, Perez S, Bolton E, Rudd P, Paulson J, Kanehisa M, Toukach P, Aoki-Kinoshita KF, Dell A, Narimatsu H, York W, Taniguchi N, Kornfeld S. Symbol nomenclature for graphical representations of glycans. Glycobiology. 2015;25(12):1323–1324. doi: 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.