Abstract
The sense of smell enables insects to recognize olfactory signals crucial for survival and reproduction. In insects, odorant detection highly depends on the interplay of distinct proteins expressed by specialized olfactory sensory neurons (OSNs) and associated support cells which are housed together in chemosensory units, named sensilla, mainly located on the antenna. Besides odorant-binding proteins (OBPs) and olfactory receptors, so-called sensory neuron membrane proteins (SNMPs) are indicated to play a critical role in the detection of certain odorants. SNMPs are insect-specific membrane proteins initially identified in pheromone-sensitive OSNs of Lepidoptera and are indispensable for a proper detection of pheromones. In the last decades, genome and transcriptome analyses have revealed a wide distribution of SNMP-encoding genes in holometabolous and hemimetabolous insects, with a given species expressing multiple subtypes in distinct cells of the olfactory system. Besides SNMPs having a neuronal expression in subpopulations of OSNs, certain SNMP types were found expressed in OSN-associated support cells suggesting different decisive roles of SNMPs in the peripheral olfactory system. In this review, we will report the state of knowledge of neuronal and non-neuronal members of the SNMP family and discuss their possible functions in insect olfaction.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00441-020-03336-0.
Keywords: Olfactory sensilla, Olfactory sensory neuron, Pheromone detection, Sensory neuron membrane protein, CD36
Introduction
The recognition of olfactory signals is of crucial importance for survival, reproduction, and communication with conspecifics in almost all insects. As a consequence, many species of this animal group have evolved an extraordinarily powerful sense of smell enabling a highly sensitive and precise detection of informative odorants originating from odor sources as diverse as food, predators, and oviposition sites as well as conspecifics that release pheromones (Hansson and Stensmyr 2011, Andersson et al. 2015). On their body surface, insects comprise hundreds to up to several ten thousand chemosensory units, called sensilla (Steinbrecht 1996, Shanbhag et al. 1999), mainly concentrated on their antenna and found in lower densities on other body parts, such as the palps, the labellum, the legs, the wing margins, and the ovipositor. Each odor-detecting sensillum is equipped with a number of olfactory sensory neurons (OSNs) and associated support cells which jointly express the proteins that ensure a sensitive and specific peripheral recognition of relevant infochemicals.
In recent years, considerable progress has been made in enlightening the cellular and molecular basis of olfactory signal detection in insects (reviewed in Leal 2013, Kohl et al. 2015, Montagne et al. 2015, Zhang et al. 2015b, Fleischer and Krieger 2018). Support cells have been shown to express members of the odorant-binding protein (OBP) family and to secrete them into the sensillum lymph surrounding the dendritic (ciliary) extensions of OSNs. Soluble OBPs are supposed to bind odor molecules that enter the porous sensillum and mediate their protected transfer through the lymph towards ligand-matched receptors in the dendritic membrane of OSNs (Leal 2003, Pelosi et al. 2006, Brito et al. 2016). Diverse olfactory receptors are expressed across the population of OSNs of an insect and form the basis for the ability to accurately detect a variety of behavioral significant odorants, including pheromones. Each OSN generally expresses only one ligand-binding olfactory receptor type from one of the several chemosensory receptor gene families characterized in insects. Seven transmembrane domain odorant receptors (ORs) represent the vast majority of olfactory receptor types (Clyne et al. 1999, Vosshall et al. 1999), and ionotropic glutamate receptor–like ionotropic receptors (IRs) (Benton et al. 2009) are a second, larger group (Montagne et al. 2015, Wicher 2015, Fleischer et al. 2018). OR-expressing OSNs require the OR co-receptor Orco for proper function (Larsson et al. 2004, Benton et al. 2006) which heteromerizes with the ligand-binding OR forming a complex that functions as a ligand-activated cation channel required for chemoelectrical signal transduction of odorants and pheromones (Neuhaus et al. 2005, Benton et al. 2006, Sato et al. 2008, Wicher et al. 2008).
So-called sensory neuron membrane proteins (SNMPs) were discovered as abundant proteins in the dendritic membrane of pheromone-sensitive OSNs of moths (Rogers et al. 1997). Since then, they are considered to play a critical role in the pheromone detection process. The insect-specific SNMPs belong to a larger family of transmembrane receptors and transporters named according to the vertebrate protein CD36 (Rogers et al. 1997, Nichols and Vogt 2008). Several possible roles of SNMPs have been proposed based on their expression in pheromone-sensitive OSNs and their apparent evolutionary relationship to CD36 family proteins, including functions in the transmembrane transport of lipophilic compounds, in the docking of OBP/pheromone complexes to the membrane, and as co-receptors mediating the transfer of pheromones to odorant receptors (Vogt 2003).
Following the discovery of SNMPs in moths, orthologues have been identified in many insect species from various orders, utilizing available SNMP sequences in extensive homology-based searches of transcriptome and genome databases (Vogt et al. 2009, Zhang et al. 2020, Zhao et al. 2020). Within a given species, several SNMP-types have been revealed, which appeared to be differentially expressed in distinct subpopulations of the OR-expressing OSNs (Rogers et al. 2001a, Benton et al. 2007, Pregitzer et al. 2019) and (contrary to what the name SNMP implies) in the support cells of olfactory sensilla (Forstner et al. 2008, Gu et al. 2013, Blankenburg et al. 2019). While the role of SNMPs expressed by non-neuronal support cells is not yet understood, most recent results indicate that the neuronal-expressed SNMPs may act as co-receptors in the membrane of OSNs possibly involved in the transfer of lipophilic pheromones and distinct odorants to a nearby OR (Gomez-Diaz et al. 2016).
Altogether, the current data highlight the fundamental importance of SNMPs in the primary events of insect olfaction. In this review, we will give an overview on the diversity and expression of the SNMP gene family and the role of the proteins in the peripheral olfactory systems of insects. We will consider possible functions of SNMPs that are expressed in non-neuronal support cells, but direct our main focus on the relatively well-studied neuronal SNMPs. Regarding the latter, we will discuss results and current concepts on their functions in the dendritic membrane of pheromone-sensitive OSNs and their interplay with colocalized ORs and extracellular OBPs.
The discovery and diversity of the SNMP gene family
SNMPs were discovered in Lepidoptera. Analyses of purified dendritic membrane preparations of pheromone-specific sensilla of male Antheraea polyphemus moths revealed an abundant 69 kDa protein which was partially sequenced. This sequence information was used to screen a cDNA library of the antenna finally leading to a full-length clone encoding the respective protein (Rogers et al. 1997). Immunohistochemical studies using antibodies raised against the bacterially expressed protein demonstrated its abundancy in the dendritic membrane of OSNs housed in male moth pheromone-sensitive sensilla (Rogers et al. 1997, Rogers et al. 2001b). Fittingly, but somewhat general, the protein was named sensory neuron membrane protein 1 (ApolSNMP1). Noteworthy, in line with the presumption of ApolSNMP1 playing a central role in pheromone detection, earlier studies by the same group in search for pheromone receptors have disclosed a 69 kDa protein (SHMP69) specific to sensory hair dendritic membrane preparations that was photolabeled by a radioactive pheromone analog (Vogt et al. 1988).
The ApolSNMP1 sequence was the starting point for the identification of orthologues in other lepidopteran species such as the moths Bombyx mori, Manduca sexta, and Heliothis virescens by means of homology-based cloning and screening approaches (Rogers et al. 2001a). Besides SNMP1s, the screenings revealed genes encoding proteins of 25–30% sequence identity named SNMP2s (Rogers et al. 2001a, Forstner et al. 2008). More recently, a similarly related third SNMP type (SNMP3) was reported in the genome of several moth species (Liu et al. 2015).
The rapid progress in sequencing technologies and bioinformatics tools has significantly promoted the identification of new SNMPs within and beyond Lepidoptera (Table S1). Over the last decades, BLAST analyses of genomes and transcriptomes employing available SNMPs identified candidate SNMPs in various insects of holometabolous orders, namely, Diptera (flies and mosquitoes), Coleoptera (beetles), Hymenoptera (bees and wasps), and Neoptera (lacewings), as well as hemimetabolous insects, i.e., Hemiptera (bugs), Orthoptera (locusts), and Psocodea (lice) (overview in Nichols and Vogt 2008, Jiang et al. 2016, Zhang et al. 2020, Zhao et al. 2020).
Initial naming and classification of newly identified SNMPs were mainly based on their phylogenetic relationship to the moth SNMP types, indicating a subdivision of the insect SNMP clade into three main groups (SNMP1s to SNMP3s). However, the inclusion of more and more available SNMPs into their phylogenetic analyses revealed partly new relationships between SNMPs, regrouping SNMPs originally categorized as SNMP2s with SNMP3s and vice versa (Zhang et al. 2020).
Collectively, the data document a diverse SNMP gene family with variable numbers of SNMPs in species of the same and different insect orders ranging from two SNMPs in the vinegar fly Drosophila melanogaster (Nichols and Vogt 2008) to 16 in the dung beetle Onthophagus taurus (Zhao et al. 2020). Within a species, paralogues of the same SNMP type may exist; for example, the genome of the hessian fly Mayetiola destructor encodes six SNMP1 paralogues, five of which are transcribed in the antennae (Andersson et al. 2014). Similarly, the red flour beetle Tribolium castaneum comprises paralogous genes for four SNMP1s, three SNMP2s, and two SNMP3s (Zhao et al. 2020). Paralogous genes encoding SNMP1s, SNMP2s, and SNMP3s appear to be typical for beetles as impressively demonstrated by a recent study that annotated and characterized 128 SNMPs from the genomes and transcriptomes of 22 coleopteran species. In addition, lineage-specific expansions of SNMPs were found mainly in the family Scarabaeidae defining a novel group of SNMPs (named SNMP4s) (Zhao et al. 2020).
Why certain species, such as scarab beetles or the hessian fly, have adopted so many SNMP genes during insect evolution is unclear, yet the finding suggests an expanded functional role of SNMPs in these species.
Expression of SNMPs in the olfactory system and beyond
Analyzing the tissue-specific gene expression of the various SNMPs of an insect and characterizing the expressing cells in the antenna and other chemosensory appendages are a first step towards understanding their specific functions within in the olfactory system. In support of a central role in olfaction, the assessment of transcript levels has detected exclusive or abundant expression of the SNMP1 type in the antenna of numerous species from multiple orders, including Lepidoptera, Hymenoptera, Diptera, Coleoptera, Hemiptera, and Orthoptera (Table S1). In insects displaying paralogous SNMP1 genes, for example, the hessian fly Mayetiola destructor or the small brown planthopper Laodelphax striatellus, the transcript abundancy in the antenna differed significantly between the SNMP1 isoforms (Table S1). Beyond the antennae, pronounced levels of SNMP1 transcripts have been detected in other chemosensory appendages such as the palps or the proboscis of insects. Generally, only very low levels of SNMP1 transcripts were detected in other parts of the insect body with few exceptions in some coleopterans (high levels in the legs and abdomen) and Drosophila melanogaster (high levels in the legs, wings, and gut) (Table S1). Such broad expression patterns of SNMP1s might indicate that an insect uses the respective SNMP1 in an olfactory or another chemosensory context at different locations of the body.
Similar to SNMP1s, abundant transcripts of at least one SNMP2 type are found in the antenna. However, compared to SNMP1s, SNMP2s are generally more widely distributed across the insect body, as indicated by abundant transcripts in the chemosensory mouthparts, legs, wings, thorax, abdomen, and midgut (Table S1). These broader expression patterns suggest that SNMP2s, beyond being involved in olfactory processes, might serve non-olfactory functions in tissues of the insect body.
The tissue distribution of the novel coleopteran SNMP4 group (Zhao et al. 2020) still needs to be analyzed. Also, the tissue distribution of SNMP3 types has been assessed in only a few cases of moths so far (Table S1). In the beet army worm Spodoptera exigua, SNMP3 transcripts were found in various chemosensory and non-chemosensory tissues (Liu et al. 2015). In contrast, SNMP3 transcripts were detected only in the adult and larval gut of the silkmoth Bombyx mori and the cotton bollworm Helicoverpa armigera (Xu et al. 2020, Zhang et al. 2020); however, the specific role of the protein within the digestive system is unclear.
Expression of SNMPs in OSNs
More conclusive support for an olfactory role of distinct SNMP types has been gained by assessing their antennal expression topography on a cellular level using in situ hybridization (ISH) and immunohistochemical (IHC) approaches.
SNMP1s have originally been discovered as proteins specific to OSNs of pheromone-sensitive sensilla in the moth Antheraea polyphemus (Rogers et al. 1997). Accordingly, ISH and IHC investigations of SNMP1 orthologues in other moths (Rogers et al. 2001a, Forstner et al. 2008, Gu et al. 2013, Liu et al. 2014), as well as in flies (Benton et al. 2007), wasps (Shan et al. 2020), and locusts (Pregitzer et al. 2017, Pregitzer et al. 2019), revealed their neuronal expression in OSNs of the antenna.
Colocalization studies using SNMP1s and Orco (a marker for OR-expressing OSNs) revealed that SNMP1s are co-expressed only by a subset of the OR-expressing OSNs of the antenna (Benton et al. 2007, Pregitzer et al. 2017, Pregitzer et al. 2019). Also, in locusts, SgreSNMP1 is found in a subpopulation of the Orco-expressing neurons in the palps (Lemke et al. 2020). In Manduca sexta, ISH studies showed that the SNMP2 is also expressed in OSNs, but in different populations than SNMP1s (Rogers et al. 2001a). A similar expression pattern was visualized for Drosophila melanogaster SNMP1 and SNMP2 using the respective SNMP promotors and the Gal4/UAS system to drive GFP expression in SNMP-positive OSNs (Vogt et al. 2020).
In Drosophila melanogaster, DmelSNMP1 is expressed in Orco-positive OSNs of trichoid sensilla which do not express IRs (Benton et al. 2009). Moreover, two-color fluorescence ISH (FISH) performed on the antenna of the hymenopteran Microplitis mediator using MmedSNMP1 and IR specific probes revealed non-overlapping expression patterns (Shan et al. 2020). Together, these results indicate that neuronal SNMP1s are not associated with ionotropic receptors but are confined to a subpopulation of OSNs expressing ORs and Orco.
The number and distribution of SNMP1-expressing OSNs in the antenna can vary considerably across species and between sexes. In the Drosophila antenna, DmelSNMP1 is expressed by each OSN of all trichoid sensilla with no apparent differences between sexes, whereas OSNs of sensilla basiconica or sensilla coeliconica are devoid of the protein (Benton et al. 2007). In Heliothis virescens, HvirSNMP1 is present in OSNs innervating subsets of trichoid sensilla with sex-specific differences in the number of SNMP1-expressing OSNs per sensillum (Zielonka et al. 2018, Blankenburg et al. 2019). Remarkably, in females, all the 2–3 OSNs of the trichoid sensilla express HvirSNMP1, whereas in males, only a single cell of such a cluster possesses the protein (Zielonka et al. 2018). In males, the distribution of HvirSNMP1 cells matches the expression pattern of pheromone-sensitive ORs (Gohl and Krieger 2006, Zielonka et al. 2018) and is complementary with electrophysiological recordings of male Heliothis virescens trichoid sensilla, demonstrating that only one OSN of the sensillum is tuned to pheromone components (Almaas and Mustaparta 1991, Baker et al. 2004). HvirSNMP1-expressing neurons in females also express pheromone-sensitive ORs (Zielonka et al. 2018), yet it is unknown whether all the SNMP1 cells of a given trichoid sensillum are tuned to pheromone components. Ultimately, these sex-specific expression patterns indicate differences in the functional impact of HvirSNMP1 between males and females of Heliothis virescens.
The expression of SNMP1 is not restricted to OSNs of a morphologically distinct sensillum type. In extension of the initially described ApolSNMP1 expression in OSNs of long male-specific pheromone-sensitive sensilla trichodea, immunogold labelling experiments on antennal sections of male and female Antheraea polyphemus localized the protein in the dendrites of OSNs innervating intermediate sensilla and basiconic sensilla (Rogers et al. 2001b). Different labelling intensities were observed for ApolSNMP1 between sensilla, indicating differences in the abundancy and functional impact of the protein in the dendrites of OSNs of various sensilla. In Schistocerca gregaria, SgreSNMP1 is expressed in subsets of OSNs of basiconic and likely all OSNs of trichoid sensilla (Jiang et al. 2016). Among the 20–30 OSNs of a basiconic sensillum, a considerable number were SgreSNMP1 positive, in line with the relatively high number of ORs co-expressing SgreSNMP1 in the locust (Pregitzer et al. 2019). Finally, in the wasp Microplitis mediator, MmedSNMP1 was immunolocalized in OSNs of sensilla placodea (Shan et al. 2020). While SNMP1 expression is documented for OSNs in various sensilla types, it is absent from olfactory sensilla coeliconica of Drosophila and the desert locust Schistocerca gregaria (Benton et al. 2007, Jiang et al. 2016). This is consistent with the finding that the OSNs of this sensillum type generally express IRs (Benton et al. 2009, Guo et al. 2013) which are not colocalized with SNMP1s.
Expression of SNMPs in non-neuronal olfactory support cells
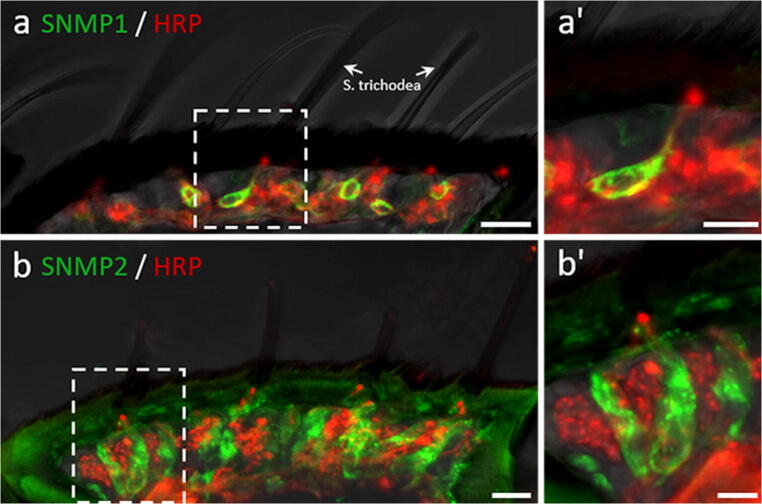
The SNMP1s of moths (Lepidoptera), the desert locust (Orthoptera), and the wasp Microplitis mediator (Hymenoptera) exhibit an OSN-specific expression. In contrast, in the antenna of these insects, the identified SNMP2 types were generally found selectively expressed in support cells surrounding OSNs (Forstner et al. 2008, Gu et al. 2013, Zhang et al. 2015a, Jiang et al. 2016, Blankenburg et al. 2019, Sun et al. 2019) revealing a differential expression of the SNMP1 and SNMP2 types and corroborating a novel function of SNMP2s in support cells. The differential expression of SNMP types became particularly obvious from studies in the moth Heliothis virescens (Forstner et al. 2008, Blankenburg et al. 2019), as exemplified by Fig. 1 showing an immunohistochemical analysis of sections from the antenna of a male analyzed with HvirSNMP1- and HvirSNMP2-specific antibodies. While in Heliothis virescens, HvirSNMP1 is expressed in a subset of antennal OSNs and within distinct sensilla trichodea OSNs (Fig. 1a, a´), HvirSNMP2 is expressed in support cells (Fig. 1b, b´), of most if not all trichoid (including pheromone-sensitive) and basiconic sensilla with no sex-specific differences (Blankenburg et al. 2019). This broad occurrence in various olfactory sensillum types, sensing a diversity of odorants as well as pheromones, indicates a more general function of the non-neuronal SNMP2 in the insect olfactory system.
Fig. 1.
Differential expression of SNMP1 in OSNs and SNMP2 in non-neuronal support cells in the antenna of male Heliothis virescens. Fluorescence immunohistochemical analyses of longitudinal sections through the male antenna using antibodies specific to SNMP1 (green) and SNMP2 (green) in combination with an anti-horseradish peroxidase (HRP) antibody (red) visualizing all neurons in the antenna. Experiments were performed as described in Blankenburg et al. 2019. The upper images (a, a′) demonstrate the expression of SNMP1 in a subset of antennal OSNs and within distinct sensilla trichodea. Only one of the 2–3 cells of a trichoid sensillum expresses SNMP1. The lower images visualize the broad expression of SNMP2 in multiple elongated support cells of the antenna surrounding clusters of neurons (b, b′). The areas boxed in a and b are shown at a higher magnification on the right (a′ and b′). Scale bars: left images = 10 μm, right images = 5 μm
The differential expression of SNMP1s and SNMP2s in OSNs and non-neuronal support cells of olfactory sensilla, respectively, suggests a functional specialization of SNMP types in insect olfaction which have been acquired and conserved in various holometabolous (moths, wasps) as well as hemimetabolous (locusts) insects during evolution. However, opposed to SNMP1s of other insects, in Drosophila melanogaster, expression of DmelSNMP1 has been reported in both OSNs and support cells of olfactory sensilla on the antenna (Benton et al. 2007). This finding indicates that one and the same SNMP plays a role in functionally different cell types of olfactory sensilla and may possibly serve in diverse processes. Noteworthy, reporter gene expression studies in Drosophila coupling the promoter of DmelSNMPs to the GAL4/UAS system to drive green fluorescent protein (GFP) expression in SNMP-positive cells revealed that DmelSNMP1 also largely associates with support cells but not neurons of taste or contact sensilla on the legs and wings whereas DmelSNMP2 was found expressed only by neurons of these sensilla (Vogt et al. 2020). These findings resemble the selective expression of SNMP types in OSNs and support cells of olfactory sensilla in other insects. In conclusion, the expression studies in Drosophila and other insects have drawn a complex picture with respect to the occurrence of SNMP types raising the question what different roles do SNMP1s and SNMP2s play in their respective neurons and support cells of olfactory and taste sensilla.
Function of SNMPs expressed by OSNs
Requirement of SNMP1s for pheromone and odorant detection
Expression studies have demonstrated the wide association of SNMP1s with pheromone-sensing OSNs (Benton et al. 2007, Forstner et al. 2008, Jin et al. 2008, Pregitzer et al. 2014). In line with a role of SNMP1 in pheromone detection, pheromone-sensitive ORs are co-expressed in OSNs with SNMP1s. For example, in the moth Heliothis virescens, FISH experiments have proven the colocalization of HvirSNMP1 with the pheromone receptors, HR13 and HR6, tuned to the major and minor female sex pheromone components, respectively (Krieger et al. 2002, Pregitzer et al. 2014, Zielonka et al. 2018). Similarly, in the vinegar fly, DmelSNMP1 is present in OR67d-expressing OSNs that detect the pheromone component cis-vaccenyl acetate (cVA) a compound released from the ejaculatory bulb of the males (Brieger and Butterworth 1970) and sensed by both males and females (Benton et al. 2007, Kurtovic et al. 2007).
In moths, SNMP1 expression in the antenna increases dramatically at the time point of adult emergence from pupae and continues well into later adult stages (Rogers et al. 1997, Gu et al. 2013, Liu et al. 2013, Sun et al. 2019). The onset of SNMP1 expression coincides with the time of expression of pheromone-detecting ORs in adult males’ OSNs (Gohl and Krieger 2006) and the onset of olfactory function (Schweitzer et al. 1976). Interestingly, analysis of the larval antenna of Heliothis virescens uncovered the presence of HvirSNMP1 in 6 out of 38 OSNs, two of which were co-expressed with the pheromone receptor HR6 (Zielonka et al. 2016). Since OSNs of the larval antenna respond to female sex pheromone components (Poivet et al. 2012, Zielonka et al. 2016), this finding indicates a critical role of SNMP1s in subsets of OSNs in earlier developmental stages and suggests a similar molecular machinery for pheromone detection in adult moths and larvae.
Yet, the function of SNMP1s appears not to be solely restricted to pheromone-sensing OSNs. In Drosophila melanogaster, SNMP1 is associated with OR83c, a receptor involved in the detection of the plant volatile farnesol (Ronderos et al. 2014). Furthermore, extensive FISH analyses conducted on the antenna of the desert locust Schistocerca gregaria revealed that out of the 83 tested ORs, 33 ORs were co-expressed with SgreSNMP1 (Pregitzer et al. 2019). While the functional aspects of these ORs have yet to be assessed and knowledge about pheromones in the desert locust is sparse, the large number of ORs co-expressed with SgreSNMP1 may suggest that not all of them are tuned to pheromones.
The essential requirement of SNMP1 for sensitive pheromone detection was first proven in 2007 using the powerful genetic model Drosophila melanogaster allowing for the generation of SNMP1-knockout animals. In single sensillum recordings (SSR), flies deficient for DmelSNMP1 displayed significantly diminished responses of OR67d-expressing OSNs to the pheromone cVA. These detection deficits in the SNMP1 mutants could be rescued by specific expression of DmelSNMP1 in OR67d neurons but not by the expression in support cells surrounding these OSNs, demonstrating a cell-autonomous and specific function of SNMP1 in the pheromone-sensing OSNs (Benton et al. 2007). More recently, pheromone detection deficits due to SNMP1 loss could be demonstrated for Bombyx mori. A deficiency of BmorSNMP1 evoked by RNA interference–based knockdown of the protein in males significantly reduced the ability of a male moth to locate the pheromone-releasing mating partner (Zhang et al. 2020).
While the requirement of SNMP1 for proper pheromone-evoked neural activity of OSNs and mating behavior have been demonstrated, the mechanisms of how SNMP1 acts in pheromone detection are largely unclear. SNMPs are glycosylated membrane proteins of about 510–560 amino acid residues (Rogers et al. 1997, Nichols and Vogt 2008, Gomez-Diaz et al. 2016). SNMPs comprise a subclade of insect genes related to a larger protein family of receptors and transporters characterized by the mammalian protein CD36 (cluster of differentiation 36) (Rogers et al. 1997, Nichols and Vogt 2008). Common for members of the CD36 gene family, SNMPs have two transmembrane domains, a large ectodomain, and rather short N- and C-terminal intracellular domains.
Insect and vertebrate members of the CD36 family exhibit diverse functions largely associated with the reception and transport of lipids and lipoproteins. For example, NINAD and Santa Maria have a role in the uptake of carotenoids in the gut and photoreceptor cells (Voolstra et al. 2006, Wang et al. 2007, Yang and O'Tousa 2007) and vertebrate CD36 family members function in lipoprotein scavenging, fatty acid transport, and lipid sensing (Silverstein and Febbraio 2009, Martin et al. 2011, Pepino et al. 2014, Oberland et al. 2015). Regarding chemosensory processes, CD36 acts as receptor for fatty acids in gustatory neurons of mammals and the zebrafish (Martin et al. 2011, Ozdener et al. 2014, Liu et al. 2017). Worthwhile emphasizing is the expression of CD36 in the cilia of a defined subset of OSNs in the nose of mice and a role indicated in the detection of oleic acid and oleic aldehyde (Lee et al. 2015, Oberland et al. 2015, Xavier et al. 2016). These findings for mouse CD36 are strikingly similar to the expression pattern of the neuronal SNMP1s in the insect antenna and their relevance for the detection of fatty acid–derived pheromones. In conclusion, they may point to analogous functions of neuronal-expressed SNMPs and CD36 in insects and vertebrates and possibly indicate an evolutionary conservation of olfactory mechanisms employed in the detection of lipophilic odorants.
Interplay of SNMP1s with OBPs and ORs
Based on the biological activities of CD36 family members in lipid sensing and transport, potential mechanisms have been envisaged for the action of SNMP1 in pheromone detection. Already early concepts suggested SNMP1 to bind pheromone molecules alone or in the complex with OBPs thereby just concentrating pheromones in the vicinity of cognate ORs. Alternatively, a more active role as co-receptor unloading pheromones from OBPs and passing the signal molecules directly to adjacent ORs was proposed (Rogers et al. 1997, Vogt 2003). Furthermore, SNMP1 was considered to interact with intracellular proteins and to have a role in the regulation of signal transduction cascades, which is a known feature of mammal CD36 (Stuart et al. 2005, Jaqaman et al. 2011); for example, taste bud cells exhibit CD36-mediated Ca2+-signaling in response to low concentrations of fatty acids (Ozdener et al. 2014).
The co-receptor model implies that SNMP1 is closely associated or even organized in a complex with ORs and Orco in the ciliary membrane of OSNs, thus ensuring rapid and sensitive pheromone detection. Accordingly, yeast two-hybrid system approaches indicate protein-protein interactions of Bombyx mori SNMP1 with both the pheromone receptor BmorOR1 and BmorOrco (Zhang et al. 2020), as well as a protein-protein interaction between Helicoverpa armigera SNMP1 and the sex pheromone receptor, HarmOR13 (Xu et al. 2020). In addition, in vivo fluorescent protein fragment complementation (PCA) experiments in Drosophila flies (Benton et al. 2007) and Förster resonance energy transfer (FRET) assays in insect cell cultures (German et al. 2013) identified heteromeric interactions between an OR and DmelSNMP1 indicating a close vicinity of the proteins in the membrane. These findings might be a first hint of an assembly of SNMP1, ORs, and Orco in a signaling complex, thus resembling other sensory detection systems, such as insect photoreceptor cells in which a supramolecular signal complex enables efficient and fast light reception (Huber 2001, Wang et al. 2007).
Besides membrane-bound proteins, pheromone signaling involves soluble OBPs which are supposed to encapsulate pheromone molecules entering the lymph and shuttle them to the receptive elements in the dendrites of OSNs (Vogt 2003, Leal 2013). Studies in OBP-deficient moths and Drosophila document the essential requirement of pheromone-specific OBPs for sensitive pheromone detection (Xu et al. 2005, Ye et al. 2017, Shiota et al. 2018, Dong et al. 2019, Zhu et al. 2019). Despite substantial efforts, the mechanisms of how pheromones are released from OBPs at the sensory neuron membrane are still enigmatic (Brito et al. 2016). Binding studies with pheromones and structural analyses of OBPs at various pH values suggest a conformational change in OBPs induced by local pH changes near the membrane (Wojtasek and Leal 1999, Horst et al. 2001, Damberger et al. 2007). Whether the large extracellular domain of SNMP1s via its ionization state may contribute to a local pH environment or whether the pheromone release process might possibly involve physical interactions of OBPs with SNMP1s and/or the OR/Orco assembly, thus providing the energy for conformational changes in OBPs, awaits further investigation.
Role of SNMP1s in OSN basal activity and odor response kinetics
While sensitive pheromone-evoked signaling clearly depends on SNMP1s and OBPs, pheromones can directly activate ORs expressed in the absence of these proteins in heterologous cell lines (Grosse-Wilde et al. 2007, Andersson et al. 2016, Yuvaraj et al. 2017) and Xenopus oocytes (Nakagawa et al. 2005, Wang et al. 2010, Jiang et al. 2014). Similarly, pheromones can induce OR-dependent responses in Drosophila mutants lacking SNMP1 (Li et al. 2014) or the OBP LUSH (Gomez-Diaz et al. 2013) at least when applied in high concentrations. These findings imply that pheromones must ultimately interact with ORs for OSN firing and disclose OBPs and SNMP1s as upstream elements critical for pheromone capture and delivery. Moreover, SNMP1 appears not to be required as an integral part of the molecular machinery necessary to generate the electrical response of a pheromone-sensitive neuron.
In conflict with this notion, loss of SNMP1 function due to gene defects or provoked by sensilla infusion of an SNMP1 antiserum was reported to increase the spontaneous activity of Drosophila OR67d-expressing neurons (Benton et al. 2007, Jin et al. 2008). Based on the elevated firing, SNMP1 was suggested as an inhibitory subunit of a receptor complex whose influence is suspended in the presence of pheromones resulting in the activation of the neurons (Jin et al. 2008, Ha and Smith 2009). However, an elevated firing of OR67d-expressing OSNs in SNMP1-deficient flies in the apparent absence of cVA was challenged by a later study suggesting that what has been interpreted as an increased “spontaneous activity” could rather represent a highly persistent ligand-induced activity initiated from exposure of grouped mutant flies to environmental cVA derived from Drosophila males. In support of this conclusion, SNMP1 mutant females raised in isolation from males did not display elevated spontaneous activity (Li et al. 2014). Also, considering a solely inhibitory function of SNMP1 would be rather incompatible with findings showing that the presence of SNMP1 enhances the sensitivity of responses of ORs to pheromones in heterologous cells (Benton et al. 2007, Pregitzer et al. 2014). For example, reconstitution of the OR/SNMP1 system of the moth Heliothis virescens in HEK293 cell lines showed that co-expression of HvirSNMP1 with the male-specific pheromone receptor HR13 increased the sensitivity of the cells to the female sex pheromone component (Z)-11-hexadecenal by about 1000-fold (Pregitzer et al. 2014).
Effective odorant sensing requires rapid responses of OSNs and then prompt inactivation after stimulus cessation. In this context, investigations of SNMP1-deficient Drosophila flies revealed some evidence for a decisive role of SNMP1 in controlling the signal response kinetics of pheromone-sensitive OSNs. At close-range stimulation of antennae with high cVA doses, pheromone-induced firing of OR67d neurons in SNMP1 mutant flies showed a slower activation and dramatic delay in the termination of the cVA-induced activity compared to wild-type flies (Li et al. 2014). Similarly, the OSN response kinetics differed between wild-type and SNMP1-deficient Drosophila when the silk moth pheromone receptor BmorOR1 was ectopically expressed in OR67d neurons and stimulated with its cognate ligand bombykol, albeit the delay in termination in the absence of SNMP1 was less pronounced. Thus, SNMP1, in addition to being required for sensitive neuronal responses to pheromones, appears to be important to achieve rapid “on/off” kinetics of receptors in response to ligands so that the receptor activation and inactivation are accelerated. Consistently, the bombykol-induced activation and deactivation of the BmorOR1/Orco receptor complex when expressed in Xenopus oocytes were accelerated when BmorSNMP1 was present (Li et al. 2014). How SNMP1 might facilitate the two opposing processes is unclear. In a model conception, SNMP1 has been suggested to act like an enzyme which can increase both the forward and reverse reaction rates by lowering the activation energy of a reversible reaction (Li et al. 2014).
Structure function dissection of SNMP1
Insights into functionally decisive molecular features of SNMP1 have been obtained by testing Drosophila flies expressing SNMP1 variants bearing distinct amino acid substitutions or short deletions in the protein sequence. Through analyzing the impact of the SNMP1 modifications on the cVA response of OR67d-expressing neurons and parallel testing of the ciliary localization of the proteins, it was found that a deletion of the cytosolic N- or C-terminal tails or their substitution by corresponding domains of the CD36 family member protein (NINAD) affected neither the ciliary localization nor the functionality of SNMP1 (Gomez-Diaz et al. 2016). Thus, at least in pheromone-sensitive OSNs of Drosophila, SNMP1 does not seem to couple with intracellular proteins and regulate downstream pathways as was suggested from findings on mammalian CD36 (Stuart et al. 2005, Jaqaman et al. 2011, Ozdener et al. 2014).
A more detailed structure-function dissection of the SNMP1 ectodomain indicated cysteines predicted to form structure-stabilizing disulfide bonds as essential for its functionality. Moreover, several amino acids representing possible N-glycosylation sites in the extracellular domain were found critical for SNMP1 ciliary targeting and thus proper pheromone responses. Similarly, all of the 17 short deletions introduced along the length of SNMP1 ectodomain (independent from their position) led to a loss of the pheromone response, which in most cases could be ascribed to a defect in ciliary targeting of the mutated proteins. Together, the analyses of the deletion mutants underlined the functional importance of the SNMP1 ectodomain in its entirety (Gomez-Diaz et al. 2016).
First insights into how the ectodomain might act in pheromone sensing were gained from the structure homology modeling of the DmelSNMP1 ectodomain using the available 3D structure of the mammalian CD36-related protein LIMP-2 as a template (Gomez-Diaz et al. 2016). According to the 3D modeling, the SNMP1 ectodomain forms a tunnel structure large enough to accommodate pheromone molecules and to form a putative passageway that might conduct pheromones in the direction of a cognate pheromone-detecting OR in the dendritic membrane.
Due to problems with expression of the DmelSNMP1 ectodomain in bacterial or insect cell systems, binding of pheromones to the SNMP1 ectodomain could not be demonstrated yet. However, measuring the interaction of ligands with the ectodomain of the SNMP1-related mammalian CD36 protein immobilized on a self-assembled monolayer by surface plasmon resonance (SPR) could demonstrate the ability of the CD36 ectodomain to bind farnesol and insect pheromones (cVA, (Z)-11-hexadecenal, bombykol) that activate SNMP1-expressing OSNs (Gomez-Diaz et al. 2016). Moreover, consistent with a tunneling concept, replacement of putative bottle neck amino acids in DmelSNMP1 by larger residues predicted to block the tunnel passageway led to either a strong or slight loss in the pheromone sensitivity of the OR67d-expressing OSNs in the mutant flies (Gomez-Diaz et al. 2016).
Model of SNMP1 function in pheromone sensing
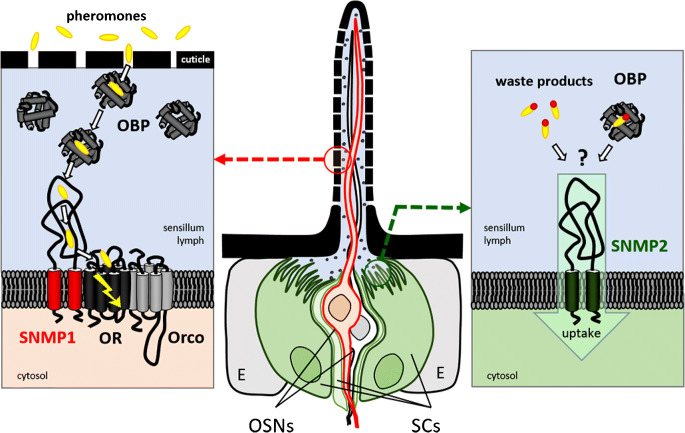
Altogether, the SNMP1 type turns out be a critical olfactory element expressed in a subset of OSNs for sensitive and rapid detection of pheromones. In addition, a role of SNMP1 is indicated in the detection of certain non-pheromonal odorants (Ronderos et al. 2014, Pregitzer et al. 2019). The current data, mainly obtained from studying its role in pheromone sensing, propose a model in which SNMP1 acts as a co-receptor that interplays with distinct OBPs and ORs in odor detection (Fig. 2). Accordingly, pheromone molecules enter an olfactory sensillum by pores in the cuticle. At the air/sensillum lymph interface, OBPs are thought to take up and solubilize the hydrophobic pheromones in the aqueous sensillum lymph (Vogt and Riddiford 1981, Vogt 2003, Leal 2013, Brito et al. 2016). The OBP/pheromone complex is supposed to diffuse towards the receptive membrane of the OR-expressing OSN where SNMP1s in the membrane may mediate the unloading of the OBP and pave the way to the OR (Rogers et al. 1997, Vogt 2003, Gomez-Diaz et al. 2016). In this process, the SNMP1 ectodomain might trigger the release of the pheromone from the OBP, either via its ionization state and creation of a local pH that induces conformational changes in the OBP or through a direct protein-protein interaction. Subsequently, the tunnel-like ectodomain of SNMP1 could take up the pheromone and pass it on to a heteromeric OR/Orco complex that is either directly associated with SNMP1 or lies in close vicinity to SNMP1 in the membrane (Benton et al. 2007, German et al. 2013, Zhang et al. 2020). In the first case (an association of SNMP1 and OR/Orco; the case shown in Fig. 2), the pheromone could be directly transferred via the SNMP1 ectodomain tunnel to the pheromone-detecting OR ligand–binding site, supposedly lying in the transmembrane domains of the OR (Hopf et al. 2015). Considering the importance of SNMP1 for rapid activation and termination of pheromone-evoked responses (Li et al. 2014), the SNMP1 ectodomain might pipe ligands to and from the binding site of an OR. In the second case (no association of SNMP1 and OR/Orco; not shown), the tunnel might release the hydrophobic pheromones into the lipid bilayer from where they reach the OR ligand–binding site by lateral diffusion in the membrane similar to the way by which free fatty acids are suggested to approach the mammalian receptor GPR40 (Srivastava et al. 2014).
Fig. 2.
Models for neuronal SNMP1 and non-neuronal SNMP2 activities in pheromone-sensing sensilla of insects based on findings in Drosophila and moths. For SNMP1s, the data suggest a role in pheromone detection by distinct subsets of OSN. After pheromone molecules have entered the porous sensillum, OBPs are supposed to take up the molecules into their hydrophobic binding pocket and shuttle the encapsulated pheromone towards SNMP1 and an OR/Orco complex in the ciliary membrane of an OSNs. At the membrane, SNMP1 is supposed to mediate the release of the ligand from the OBP and to forward it via its tunnel-like ectodomain to the ligand-binding site of a pheromone-detecting OR. Ultimately, the binding of the pheromone to the OR/Orco complex triggers signal transduction. For SNMP2s, the data suggest a role in the apical microvilli membranes of support cells, contacting the sensillum lymph. Based on its localization and the transport activities of other members of the CD36 family, we proposed a function of SNMP2 in clearance processes, i.e., the uptake of “waste products” from the sensillum lymph into the support cells. In this function, SNMP2 might directly accomplish the removal of soluble breakdown products of relevant pheromones and odorants or of odorants that accidentally entered a sensillum from the lymph. Alternatively, OBPs might bind “waste products” and deliver them to SNMP2 which mediates the transfer of the OBP ligand or the ligand-OBP complex into the support cell. SCs support cells, E epidermis cells
In the end, the question remains why only a subset of the OR-expressing OSNs of an insect requires SNMP1s for sensitive odorant detection, whereas SNMP1 is dispensable for robust odorant responses in the majority of the OR-expressing OSNs. Intuitively, following the current opinion that odor sensing in olfactory sensilla generally depends on OBPs and given that SNMP1 is in fact critical for ligand transfer from OBPs to ORs, one should expect that all OSNs express SNMP1. However, a recent study in Drosophila have challenged a general requirement of OBPs in odor detection, demonstrating robust responses of OR-expressing OSNs in six tested types of sensilla basiconica to odors of widely diverse chemical structure in the complete absence of OBPs (Xiao et al. 2019). This would suggest that in many olfactory sensilla of Drosophila in spite of the abundant expression of OBPs, odorants reach and activate the cognate OR-expressing neuron in an OBP-independent manner. In contrast, sensitive pheromone responses of OR67d-expressing OSNs in trichoid sensilla of Drosophila require the expression of the OBP LUSH (Xu et al. 2005). In the overall picture, this suggests the existence of OBP-independent and OBP-dependent odor detection processes in sensilla containing OR-expressing OSNs.
Thus, assuming that only an OBP-dependent odor detection process, as shown for pheromones (Xu et al. 2005, Ye et al. 2017, Shiota et al. 2018, Dong et al. 2019, Zhu et al. 2019), requires SNMP1 for ligand release and transfer to ORs, it is therefore possible that SNMP1 is only expressed in those subsets of OSNs which receive cognate ligands via indispensable OBPs that encapsulate the molecules to overcome the aqueous sensillum lymph. Consistent with this notion, in Drosophila, OSNs of sensilla basiconica that were shown to robustly respond in the absence of OBPs do not express SNMP1 (Benton et al. 2007, Xiao et al. 2019).
Possible role of SNMPs expressed by support cells of olfactory sensilla
But what might the non-neuronal SNMP2s do? SNMP2 types exhibit a broad expression in support cells of various sensilla types of the antenna (Gu et al. 2013, Blankenburg et al. 2019, Sun et al. 2019) indicating a more general function of the protein in olfactory sensilla. Detailed immunohistochemical analyses in moths revealed a subcellular localization of SNMP2s at the apical area of support cells where microvillar protrusions border the sensillum lumen (Gu et al. 2013, Blankenburg et al. 2019). These microvilli structures are considered sites of high transmembrane transport and exchange activities (Schweitzer et al. 1976; Steinbrecht and Gnatzy 1984; Keil 1989). Therefore, the support cells are thought to secrete the sensillum lymph and to control its composition (Thurm and Küppers 1980; Steinbrecht and Gnatzy 1984). In accordance with this view, support cells secrete OBPs into the sensillum lymph and are suggested to be involved in their turnover (Galindo and Smith 2001; Klein 1987; Steinbrecht et al. 1992). Somewhat surprisingly, two independent studies in the moths Agrotis ipsilon and Heliothis virescens, which used peptide-specific antibodies directed against short regions of the extracellular domain SNMP2, found antigenicity also in the sensillum lymph–filled lumen of different sensillum types (Gu et al. 2013, Blankenburg et al. 2019). Together, these findings suggest a function of SNMP2 in the microvillar membrane of support cells and in addition might hint on an unexpected role of the proteins in the extracellular sensillum lymph.
As mentioned before, vertebrate and insect members of the CD36 gene family have activities as membrane transporters and receptors in the selective uptake of lipids, including fatty acids, carotenoids, and cholesterol, as well as of lipid-protein complexes (Koonen et al. 2005, Levy et al. 2007, Silverstein and Febbraio 2009, Raheel et al. 2019). While these activities give no plausible clue about the function of extracellularly localized SNMP2s in the sensillum lymph, they do suggest possible functions for SNMP2 in the apical microvill imembranes of the support cells. Given its location in the membrane contacting the sensillum lymph, a role of the protein as lipid or lipid-protein transporter involved in the maintenance of the extracellular fluid is conceivable (Forstner et al. 2008, Blankenburg et al. 2019). Considering the broad repertoire of ligands of mammalian CD36 (Silverstein and Febbraio 2009, Cifarelli and Abumrad 2018), SNMP2s might contribute to the elimination of various lipophilic waste products from the sensillum lymph such as breakdown products of pheromones and odorants resulting from the activity of degrading enzymes in the lymph (Vogt 2003; Leal 2013) or of odorant molecules from the air that have entered the sensillum accidentally (Fig. 2). Since certain OBPs are suggested to have a function in odor deactivation by clearing specific odorants from the sensillum lymph (Scheuermann and Smith 2019), SNMP2 might alternatively take over these OBP ligands from OBPs, a scenario resembling the proposed interaction of pheromone/OBP complexes with SNMP1. Finally, based on the confirmed role of CD36 proteins in the internalization of the fatty acid carrier albumin into dermal endothelial cells (Raheel et al. 2019), SNMP2s might mediate the uptake of waste-loaded OBPs into the support cells for further decomposition. Altogether, we suggest that similar to the multifunctional vertebrate CD36 protein, SNMP2s in support cells act as transporters and receptors for lipids as well as lipid-protein complexes. In this way, they may operate as critical membrane proteins required for the clearance of the sensillum lymph thus ensuring sensillum homeostasis and the functionality of the olfactory unit.
Concluding remarks and perspective
Research of the last decades has emphasized the importance of SNMPs in insect olfaction. In particular, the proposed role of SNMP1s as co-receptors mediating the delivery of distinct odor molecules received from OBPs to ORs in the membrane of pheromone-sensitive OSNs has been corroborated. A different function for non-neuronal SNMP2 types, possibly in sensillum lymph clearance processes, is suggested based on their expression in support cells and a localization in the apical membrane facing the sensillum lymph. Despite this progress, a host of questions remains which await comprehensive investigations. Among the open issues are the protein-protein interactions between OBPs, SNMPs, and the OR/Orco complex and their dynamic interplay in odor detection, including proof of the proposed tunneling function of SNMP1. Also, the question to what extent does the detection of pheromonal and non-pheromonal odorants involve SNMP1s as well as the distinct roles of SNMP1 isoforms found in the olfactory system of various insect species needs examination. With respect to the proposed clearance function of SNMP2 types in olfactory sensilla, investigation towards their ligand binding and transport capabilities is urgently needed.
Experimental testing of the specific functions of the neuronal and non-neuronal SNMPs in heterologous expression systems may be technically demanding as it requires radioactively or fluorescently labeled ligands and/or OBPs as well as appropriate assay systems that permits monitoring the transient binding and dynamic transport capabilities of SNMPs. Moreover, the detailed analyses of SNMP1s need its functional reconstitution with other membrane proteins (ORs, Orco) and the involvement of soluble OBPs. While theses technical challenges have to be solved, future studies of neuronal SNMP1s and non-neuronal SNMP2s might not only illuminate their distinct roles in the peripheral olfactory processes, but might also uncover conserved and divergent mechanisms by which the paralogous proteins fulfill their specific tasks in cell types as functionally diverse as olfactory neurons and support cells of insect antenna.
Supplementary Information
(PDF 1.24 mb)
Acknowledgments
We apologize to all colleagues whose work we could not acknowledge enough or include due to space limitations. This work was supported by a grant to J.K. provided by the Deutsche Forschungsgemeinschaft (DFG) (KR1786/5-1).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
1/23/2021
This article was revised. In the original publication, the Supplementary Material was missing.
References
- Almaas TJ, Mustaparta H. Heliothis virescens: response characteristics of receptor neurons in sensilla trichodea type 1 and type 2. J Chem Ecol. 1991;17:953–972. doi: 10.1007/BF01395602. [DOI] [PubMed] [Google Scholar]
- Andersson MN, Corcoran JA, Zhang DD, Hillbur Y, Newcomb RD, Löfstedt C. A sex pheromone receptor in the hessian fly Mayetiola destructor (Diptera, Cecidomyiidae) Front Cell Neurosci. 2016;10:212. doi: 10.3389/fncel.2016.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MN, Löfstedt C, Newcomb RD. Insect olfaction and the evolution of receptor tuning. Front Ecol Evol. 2015;3:1–14. [Google Scholar]
- Andersson MN, Videvall E, Walden KKO, Harris MO, Robertson HM, Löfstedt C. Sex- and tissue-specific profiles of chemosensory gene expression in a herbivorous gall-inducing fly (Diptera: Cecidomyiidae) BMC Genomics. 2014;15:501. doi: 10.1186/1471-2164-15-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TC, Ochieng SA, Cosse AA, Lee SG, Todd JL, Quero C, Vickers NJ. A comparison of responses from olfactory receptor neurons of Heliothis subflexa and Heliothis virescens to components of their sex pheromone. J Comp Physiol A. 2004;190:155–165. doi: 10.1007/s00359-003-0483-2. [DOI] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450:289–293. doi: 10.1038/nature06328. [DOI] [PubMed] [Google Scholar]
- Blankenburg S, Cassau S, Krieger J. The expression patterns of SNMP1 and SNMP2 underline distinct functions of two CD36-related proteins in the olfactory system of the tobacco budworm Heliothis virescens. Cell Tissue Res. 2019;378:485–497. doi: 10.1007/s00441-019-03066-y. [DOI] [PubMed] [Google Scholar]
- Brieger G, Butterworth FM. Drosophila melanogaster: identity of male lipid in reproductive system. Science. 1970;167:1262. doi: 10.1126/science.167.3922.1262. [DOI] [PubMed] [Google Scholar]
- Brito NF, Moreira MF, Melo AC. A look inside odorant-binding proteins in insect chemoreception. J Insect Physiol. 2016;95:51–65. doi: 10.1016/j.jinsphys.2016.09.008. [DOI] [PubMed] [Google Scholar]
- Cifarelli V, Abumrad NA. Intestinal CD36 and other key proteins of lipid utilization: role in absorption and gut homeostasis. Compr Physiol. 2018;8:493–507. doi: 10.1002/cphy.c170026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Damberger FF, Ishida Y, Leal WS, Wuethrich K. Structural basis of ligand binding and release in insect pheromone-binding proteins: NMR structure of Antheraea polyphemus PBP1 at pH 4.5. J Mol Biol. 2007;373:811–819. doi: 10.1016/j.jmb.2007.07.078. [DOI] [PubMed] [Google Scholar]
- Dong XT, Liao H, Zhu GH, Khuhro SA, Ye ZF, Yan Q, Dong SL. CRISPR/Cas9-mediated PBP1 and PBP3 mutagenesis induced significant reduction in electrophysiological response to sex pheromones in male Chilo suppressalis. Insect Sci. 2019;26:388–399. doi: 10.1111/1744-7917.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer J, Krieger J. Insect pheromone receptors - key elements in sensing intraspecific chemical signals. Front Cell Neurosci. 2018;12:425. doi: 10.3389/fncel.2018.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer J, Pregitzer P, Breer H, Krieger J. Access to the odor world: olfactory receptors and their role for signal transduction in insects. Cell Mol Life Sci. 2018;75:485–508. doi: 10.1007/s00018-017-2627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstner M, Gohl T, Gondesen I, Raming K, Breer H, Krieger J. Differential expression of SNMP-1 and SNMP-2 proteins in pheromone-sensitive hairs of moths. Chem Senses. 2008;33:291–299. doi: 10.1093/chemse/bjm087. [DOI] [PubMed] [Google Scholar]
- German PF, van der Poel S, Carraher C, Kralicek AV, Newcomb RD. Insights into subunit interactions within the insect olfactory receptor complex using FRET. Insect Biochem Mol Biol. 2013;43:138–145. doi: 10.1016/j.ibmb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Gohl T, Krieger J. Immunolocalization of a candidate pheromone receptor in the antenna of the male moth, Heliothis virescens. Invertebr Neurosci. 2006;6:13–21. doi: 10.1007/s10158-005-0012-9. [DOI] [PubMed] [Google Scholar]
- Gomez-Diaz C, Bargeton B, Abuin L, Bukar N, Reina JH, Bartoi T, Graf M, Ong H, Ulbrich MH, Masson JF, Benton R. A CD36 ectodomain mediates insect pheromone detection via a putative tunnelling mechanism. Nat Commun. 2016;7:11866. doi: 10.1038/ncomms11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Diaz C, Reina JH, Cambillau C, Benton R. Ligands for pheromone-sensing neurons are not conformationally activated odorant binding proteins. PLoS Biol. 2013;11:e1001546. doi: 10.1371/journal.pbio.1001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse-Wilde E, Gohl T, Bouche E, Breer H, Krieger J. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur J Neurosci. 2007;25:2364–2373. doi: 10.1111/j.1460-9568.2007.05512.x. [DOI] [PubMed] [Google Scholar]
- Gu SH, Yang RN, Guo MB, Wang GR, Wu KM, Guo YY, Zhou JJ, Zhang YJ. Molecular identification and differential expression of sensory neuron membrane proteins in the antennae of the black cutworm moth Agrotis ipsilon. J Insect Physiol. 2013;59:430–443. doi: 10.1016/j.jinsphys.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Guo M, Krieger J, Grosse-Wilde E, Missbach C, Zhang L, Breer H. Variant ionotropic receptors are expressed in olfactory sensory neurons of coeloconic sensilla on the antenna of the desert locust (Schistocerca gregaria) Int J Biol Sci. 2013;10:1–14. doi: 10.7150/ijbs.7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha TS, Smith DP. Odorant and pheromone receptors in insects. Front Cell Neurosci. 2009;3:10. doi: 10.3389/neuro.03.010.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson BS, Stensmyr MC. Evolution of insect olfaction. Neuron. 2011;72:698–711. doi: 10.1016/j.neuron.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Hopf TA, Morinaga S, Ihara S, Touhara K, Marks DS, Benton R. Amino acid coevolution reveals three-dimensional structure and functional domains of insect odorant receptors. Nat Commun. 2015;6:6077. doi: 10.1038/ncomms7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst R, Damberger F, Luginbuhl P, Guntert P, Peng G, Nikonova L, Leal WS, Wuthrich K. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc Natl Acad Sci U S A. 2001;98:14374–14379. doi: 10.1073/pnas.251532998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K, Kuwata H, Touret N, Collins R, Trimble WS, Danuser G, Grinstein S. Cytoskeletal control of CD36 diffusion promotes its receptor and signaling function. Cell. 2011;146:593–606. doi: 10.1016/j.cell.2011.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Pregitzer P, Grosse-Wilde E, Breer H, Krieger J. Identification and characterization of two "sensory neuron membrane proteins" (SNMPs) of the desert locust, Schistocerca gregaria (Orthoptera: Acrididae) J Insect Sci. 2016;16:33. doi: 10.1093/jisesa/iew015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang XJ, Guo H, Di C, Yu S, Zhu L, Huang LQ, Wang CZ. Sequence similarity and functional comparisons of pheromone receptor orthologs in two closely related Helicoverpa species. Insect Biochem Mol Biol. 2014;48:63–74. doi: 10.1016/j.ibmb.2014.02.010. [DOI] [PubMed] [Google Scholar]
- Jin X, Ha TS, Smith DP. SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc Natl Acad Sci U S A. 2008;105:10996–11001. doi: 10.1073/pnas.0803309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl J, Huoviala P, Jefferis GS. Pheromone processing in Drosophila. Curr Opin Neurobiol. 2015;34:149–157. doi: 10.1016/j.conb.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonen DP, Glatz JF, Bonen A, Luiken JJ. Long-chain fatty acid uptake and FAT/CD36 translocation in heart and skeletal muscle. Biochim Biophys Acta. 2005;1736:163–180. doi: 10.1016/j.bbalip.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Krieger J, Raming K, Dewer YME, Bette S, Conzelmann S, Breer H. A divergent gene family encoding candidate olfactory receptors of the moth Heliothis virescens. Eur J Neurosci. 2002;16:619–628. doi: 10.1046/j.1460-9568.2002.02109.x. [DOI] [PubMed] [Google Scholar]
- Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446:542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Leal WS (2003) Proteins that make sense. In: Blomquist G, Vogt R (eds) Insect pheromone biochemistry and molecular biology. The biosynthesis and detection of pheromones and plant volatiles. Elsevier Academic Press, London, pp. 447–476
- Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- Lee S, Eguchi A, Tsuzuki S, Matsumura S, Inoue K, Iwanaga T, Masuda D, Yamashita S, Fushiki T. Expression of CD36 by olfactory receptor cells and its abundance on the epithelial surface in mice. PLoS One. 2015;10:e0133412. doi: 10.1371/journal.pone.0133412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke RS, Pregitzer P, Eichhorn AS, Breer H, Krieger J, Fleischer J. SNMP1 and odorant receptors are co-expressed in olfactory neurons of the labial and maxillary palps from the desert locust Schistocerca gregaria (Orthoptera: Acrididae) Cell Tissue Res. 2020;379:275–289. doi: 10.1007/s00441-019-03083-x. [DOI] [PubMed] [Google Scholar]
- Levy E, Spahis S, Sinnett D, Peretti N, Maupas-Schwalm F, Delvin E, Lambert M, Lavoie MA. Intestinal cholesterol transport proteins: an update and beyond. Curr Opin Lipidol. 2007;18:310–318. doi: 10.1097/MOL.0b013e32813fa2e2. [DOI] [PubMed] [Google Scholar]
- Li Z, Ni JD, Huang J, Montell C. Requirement for Drosophila SNMP1 for rapid activation and termination of pheromone-induced activity. PLoS Genet. 2014;10:e1004600. doi: 10.1371/journal.pgen.1004600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhang J, Liu Y, Wang G, Dong S. Expression of SNMP1 and SNMP2 genes in antennal sensilla of Spodoptera exigua (Hubner) Arch Insect Biochem Physiol. 2014;85:114–126. doi: 10.1002/arch.21150. [DOI] [PubMed] [Google Scholar]
- Liu HY, Xu YP, Wang Y, Zhong SJ, Wang M, Lin PY, Li HY, Liu ZH. Cd36 is a candidate lipid sensor involved in the sensory detection of fatty acid in zebrafish. Physiol & Behav. 2017;182:34–39. doi: 10.1016/j.physbeh.2017.09.015. [DOI] [PubMed] [Google Scholar]
- Liu NY, Zhang T, Ye ZF, Li F, Dong SL. Identification and characterization of candidate chemosensory gene families from Spodoptera exigua developmental transcriptomes. Int J Biol Sci. 2015;11:1036–1048. doi: 10.7150/ijbs.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Qiao F, Liang QM, Huang YJ, Zhou WW, Gong ZJ, Cheng J, Zhu ZR. Molecular characterization of two sensory neuron membrane proteins from Chilo supressalis (Lepidoptera: Pyralidae) Ann Entomol Soc Am. 2013;106:378–384. [Google Scholar]
- Martin C, Chevrot M, Poirier H, Passilly-Degrace P, Niot I, Besnard P. CD36 as a lipid sensor. Physiol Behav. 2011;105:33–42. doi: 10.1016/j.physbeh.2011.02.029. [DOI] [PubMed] [Google Scholar]
- Montagne N, de Fouchier A, Newcomb RD, Jacquin-Joly E. Advances in the identification and characterization of olfactory receptors in insects. Prog Mol Biol Transl Sci. 2015;130:55–80. doi: 10.1016/bs.pmbts.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307:1638–1642. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- Neuhaus EM, Gisselmann G, Zhang WY, Dooley R, Stortkuhl K, Hatt H. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat Neurosci. 2005;8:15–17. doi: 10.1038/nn1371. [DOI] [PubMed] [Google Scholar]
- Nichols Z, Vogt RG. The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem Mol Biol. 2008;38:398–415. doi: 10.1016/j.ibmb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Oberland S, Ackelst T, Gaab S, Pelz T, Spehr J, Spehr M, Neuhaus EM. CD36 is involved in oleic acid detection by the murine olfactory system. Front Cell Neurosci. 2015;9:366. doi: 10.3389/fncel.2015.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdener MH, Subramaniam S, Sundaresan S, Sery O, Hashimoto T, Asakawa Y, Besnard P, Abumrad NA, Khan NA. CD36- and GPR120-mediated Ca2+ signaling in human taste bud cells mediates differential responses to fatty acids and is altered in obese mice. Gastroenterology. 2014;146:995–1005. doi: 10.1053/j.gastro.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P, Zhou JJ, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cell Mol Life Sci. 2006;63:1658–1676. doi: 10.1007/s00018-005-5607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepino MY, Kuda O, Samovski D, Abumrad NA. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu Rev Nutr. 2014;34:281–303. doi: 10.1146/annurev-nutr-071812-161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poivet E, Rharrabe K, Monsempes C, Glaser N, Rochat D, Renou M, Marion-Poll F, Jacquin-Joly E. The use of the sex pheromone as an evolutionary solution to food source selection in caterpillars. Nat Commun. 2012;3:1047. doi: 10.1038/ncomms2050. [DOI] [PubMed] [Google Scholar]
- Pregitzer P, Greschista M, Breer H, Krieger J. The sensory neurone membrane protein SNMP1 contributes to the sensitivity of a pheromone detection system. Insect Mol Biol. 2014;23:733–742. doi: 10.1111/imb.12119. [DOI] [PubMed] [Google Scholar]
- Pregitzer P, Jiang X, Grosse-Wilde E, Breer H, Krieger J, Fleischer J. In search for pheromone receptors: certain members of the odorant receptor family in the desert locust Schistocerca gregaria (Orthoptera: Acrididae) are co-expressed with SNMP1. Int J Biol Sci. 2017;13:911–922. doi: 10.7150/ijbs.18402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregitzer P, Jiang X, Lemke RS, Krieger J, Fleischer J, Breer H. A subset of odorant receptors from the desert locust Schistocerca gregaria is co-expressed with the sensory neuron membrane protein 1. Insects. 2019;10:350. doi: 10.3390/insects10100350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raheel H, Ghaffari S, Khosraviani N, Mintsopoulos V, Auyeung D, Wang C, Kim YH, Mullen B, Sung HK, Ho M, Fairn G, Neculai D, Febbraio M, Heit B, Lee WL. CD36 mediates albumin transcytosis by dermal but not lung microvascular endothelial cells: role in fatty acid delivery. Am J Physiol Lung Cell Mol Physiol. 2019;316:L740–L750. doi: 10.1152/ajplung.00127.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers ME, Krieger J, Vogt RG. Antennal SNMPs (sensory neuron membrane proteins) of Lepidoptera define a unique family of invertebrate CD36-like proteins. J Neurobiol. 2001;49:47–61. doi: 10.1002/neu.1065. [DOI] [PubMed] [Google Scholar]
- Rogers ME, Steinbrecht RA, Vogt RG. Expression of SNMP-1 in olfactory neurons and sensilla of male and female antennae of the silkmoth Antheraea polyphemus. Cell Tissue Res. 2001;303:433–446. doi: 10.1007/s004410000305. [DOI] [PubMed] [Google Scholar]
- Rogers ME, Sun M, Lerner MR, Vogt RG. Snmp-1, a novel membrane protein of olfactory neurons of the silk moth Antheraea polyphemus with homology to the CD36 family of membrane proteins. J Biol Chem. 1997;272:14792–14799. doi: 10.1074/jbc.272.23.14792. [DOI] [PubMed] [Google Scholar]
- Ronderos DS, Lin CC, Potter CJ, Smith DP. Farnesol-detecting olfactory neurons in Drosophila. J Neurosci. 2014;34:3959–3968. doi: 10.1523/JNEUROSCI.4582-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Scheuermann EA, Smith DP. Odor-specific deactivation defects in a Drosophila odorant-binding protein mutant. Genetics. 2019;213:897–909. doi: 10.1534/genetics.119.302629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer ES, Sanes JR, Hildebrand JG. Ontogeny of electroantennogram responses in the moth, Manduca sexta. J Insect Physiol. 1976;22:955–960. doi: 10.1016/0022-1910(76)90078-0. [DOI] [PubMed] [Google Scholar]
- Shan S, Wang SN, Song X, Khashaveh A, Lu ZY, Dhiloo KH, Li RJ, Gao XW, Zhang YJ. Molecular characterization and expression of sensory neuron membrane proteins in the parasitoid Microplitis mediator (Hymenoptera: Braconidae) Insect Sci. 2020;27:425–439. doi: 10.1111/1744-7917.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag SR, Muller B, Steinbrecht RA. Atlas of olfactory organs of Drosophila melanogaster - 1. Types, external organization, innervation and distribution of olfactory sensilla. Int J Insect Morphol. 1999;28:377–397. [Google Scholar]
- Shiota Y, Sakurai T, Daimon T, Mitsuno H, Fujii T, Matsuyama S, Sezutsu H, Ishikawa Y, Kanzaki R. In vivo functional characterisation of pheromone binding protein-1 in the silkmoth, Bombyx mori. Sci Rep. 2018;8:13529. doi: 10.1038/s41598-018-31978-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal. 2009;2:1–8. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Yano J, Hirozane Y, Kefala G, Gruswitz F, Snell G, Lane W, Ivetac A, Aertgeerts K, Nguyen J, Jennings A, Okada K. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature. 2014;513:124–127. doi: 10.1038/nature13494. [DOI] [PubMed] [Google Scholar]
- Steinbrecht RA. Structure and function of insect olfactory sensilla. CIBA Found Symp. 1996;200:158–174. doi: 10.1002/9780470514948.ch13. [DOI] [PubMed] [Google Scholar]
- Stuart LM, Deng J, Silver JM, Takahashi K, Tseng AA, Hennessy EJ, Ezekowitz RA, Moore KJ. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J Cell Biol. 2005;170:477–485. doi: 10.1083/jcb.200501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wang Q, Zhang Y, Yan Y, Guo H, Xiao Q, Zhang Y. Expression patterns and colocalization of two sensory neurone membrane proteins in Ectropis obliqua Prout, a geometrid moth pest that uses type-II sex pheromones. Insect Mol Biol. 2019;28:342–354. doi: 10.1111/imb.12555. [DOI] [PubMed] [Google Scholar]
- Vogt RG (2003) Biochemical diversity of odor detection: OBPs, ODEs and SNMPs. In: Blomquist G, Vogt RG (eds) Insect pheromone biochemistry and molecular biology. The biosynthesis and detection of pheromones and plant volatiles. Elsevier Academic Press, London, pp. 391–445
- Vogt RG, Miller NE, Litvack R, Fandino RA, Sparks J, Staples J, Friedman R, Dickens JC. The insect SNMP gene family. Insect Biochem Mol Biol. 2009;39:448–456. doi: 10.1016/j.ibmb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Prestwich GD, Riddiford LM (1988) Sex pheromone receptor proteins. Visualization using a radiolabeled photoaffinity analog J Biol Chem 263:3952–3959 [PubMed]
- Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Sparks JT, Fandino RA, Ashourian KT. Reflections on antennal proteins: the evolution of pheromone binding proteins; diversity of pheromone degrading enzymes; and the distribution and behavioral roles of SNMPs. In: Blomquist G, Vogt RG, editors. Insect pheromone biochemistry and molecular biology. London: Elsevier Academic Press; 2020. [Google Scholar]
- Voolstra O, Kiefer C, Hoehne M, Welsch R, Vogt K, Von Lintig J. The Drosophila class B scavenger receptor NinaD-I is a cell surface receptor mediating carotenoid transport for visual chromophore synthesis. Biochemistry-Us. 2006;45:13429–13437. doi: 10.1021/bi060701u. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Wang G, Carey AF, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2010;107:4418–4423. doi: 10.1073/pnas.0913392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Jiao Y, Montell C. Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. J Cell Biol. 2007;177:305–316. doi: 10.1083/jcb.200610081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D. Olfactory signaling in insects. Prog Mol Biol Transl Sci. 2015;130:37–54. doi: 10.1016/bs.pmbts.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Wojtasek H, Leal WS. Conformational change in the pheromone-binding protein from Bombyx mori induced by pH and by interaction with membranes. J Biol Chem. 1999;274:30950–30956. doi: 10.1074/jbc.274.43.30950. [DOI] [PubMed] [Google Scholar]
- Xavier AM, Ludwig RG, Nagai MH, de Almeida TJ, Watanabe HM, Hirata MY, Rosenstock TR, Papes F, Malnic B, Glezer I. CD36 is expressed in a defined subpopulation of neurons in the olfactory epithelium. Sci Rep. 2016;6:25507. doi: 10.1038/srep25507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Sun JS, Carlson JR. Robust olfactory responses in the absence of odorant binding proteins. Elife. 2019;8:e51040. doi: 10.7554/eLife.51040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Atkinson R, Jones DNM, Smith DP. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Xu W, Zhang H, Liao Y, Papanicolaou A. Characterization of sensory neuron membrane proteins (SNMPs) in cotton bollworm Helicoverpa armigera (Lepidoptera: Noctuidae) Insect Sci. 2020;00:1–11. doi: 10.1111/1744-7917.12816. [DOI] [PubMed] [Google Scholar]
- Yang J, O’Tousa JE. Cellular sites of Drosophila NinaB and NinaD activity in vitamin a metabolism. Mol Cell Neurosci. 2007;35:49–56. doi: 10.1016/j.mcn.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Ye ZF, Liu XL, Han Q, Liao H, Dong XT, Zhu GH, Dong SL. Functional characterization of PBP1 gene in Helicoverpa armigera (Lepidoptera: Noctuidae) by using the CRISPR/Cas9 system. Sci Rep. 2017;7:8470. doi: 10.1038/s41598-017-08769-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaraj JK, Corcoran JA, Andersson MN, Newcomb RD, Anderbrant O, Löfstedt C. Characterization of odorant receptors from a non-ditrysian moth, Eriocrania semipurpurella sheds light on the origin of sex pheromone receptors in lepidoptera. Mol Biol Evol. 2017;34:2733–2746. doi: 10.1093/molbev/msx215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HJ, Xu W, Chen QM, Sun LN, Anderson A, Xia QY, Papanicolaou A. A phylogenomics approach to characterizing sensory neuron membrane proteins (SNMPs) in Lepidoptera. Insect Biochem Mol Biol. 2020;118:103313. doi: 10.1016/j.ibmb.2020.103313. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu Y, Walker WB, Dong SL, Wang GR. Identification and localization of two sensory neuron membrane proteins from Spodoptera litura (Lepidoptera: Noctuidae) Insect Sci. 2015;22:399–408. doi: 10.1111/1744-7917.12131. [DOI] [PubMed] [Google Scholar]
- Zhang J, Walker WB, Wang G. Pheromone reception in moths: from molecules to behaviors. Prog Mol Biol Transl Sci. 2015;130:109–128. doi: 10.1016/bs.pmbts.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Zhao YJ, Li GC, Zhu JY, Liu NY. Genome-based analysis reveals a novel SNMP group of the Coleoptera and chemosensory receptors in Rhaphuma horsfieldi. Genomics. 2020;112:2713–2728. doi: 10.1016/j.ygeno.2020.03.005. [DOI] [PubMed] [Google Scholar]
- Zhu GH, Zheng MY, Sun JB, Khuhro SA, Yan Q, Huang Y, Syed Z, Dong SL. CRISPR/Cas9 mediated gene knockout reveals a more important role of PBP1 than PBP2 in the perception of female sex pheromone components in Spodoptera litura. Insect Biochem Mol Biol. 2019;115:103244. doi: 10.1016/j.ibmb.2019.103244. [DOI] [PubMed] [Google Scholar]
- Zielonka M, Breer H, Krieger J. Molecular elements of pheromone detection in the female moth, Heliothis virescens. Insect Sci. 2018;25:389–400. doi: 10.1111/1744-7917.12434. [DOI] [PubMed] [Google Scholar]
- Zielonka M, Gehrke P, Badeke E, Sachse S, Breer H, Krieger J. Larval sensilla of the moth Heliothis virescens respond to sex pheromone components. Insect Mol Biol. 2016;25:666–678. doi: 10.1111/imb.12253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1.24 mb)