Abstract
Iron is a critical micronutrient for growth and development of plants and its deficiency limiting the crop productivity. MicroRNAs (miRNAs) play vital roles in adaptation of plants to various nutrient deficiencies. However, the role of miRNAs and their target genes related to Fe-deficiency is limited. In this study, we identified Fe-deficiency-responsive miRNAs from citrus. In Fe-deficiency conditions, about 50 and 31 miRNAs were up-regulated and down-regulated, respectively. The differently expressed miRNAs might play critical roles in contributing the Fe-deficiency tolerance in citrus plants. The miRNAs-mediated Fe-deficiency tolerance in citrus plants might related to the enhanced stress tolerance by decreased expression of miR172; regulation of S homeostasis by decreased expression of miR395; inhibition of plant growth by increased expression of miR319 and miR477; regulation of Cu homeostasis as well as activation of Cu/Zn superoxide dismutase activity due to decreased expression of miR398 and miR408 and regulation of lignin accumulation by decreased expression of miR397 and miR408. The identified miRNAs in present study laid a foundation to understand the Fe-deficiency adaptive mechanisms in citrus plants.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-02669-z.
Keywords: Citrus, Fe-deficiency, Illumina sequencing, miRNA
Introduction
Iron (Fe)-deficiency is a major nutritional disorder that limits the crop productivity (Zhang et al. 2019; López-Millán et al. 2013). The major symptoms associated with the Fe-deficiency are chlorosis and reduction in photosynthesis that severely effects the growth and yield of plants. To cope with Fe-deficiency conditions, plants have evolved an array of physiological and molecular adaptation mechanisms to facilitate the increased capacity of Fe uptake from soil. The strategy-I is a well-known mechanism established in dicotyledonous and non-graminaceous monocotyledonous species for efficient mobilization and acquisition of Fe mediated by plasma membrane localized H+-ATPases, ferric chelate reductase2 (FRO2) gene and iron-regulated transporter1 (IRT1) (Zhang et al. 2019; Robinson et al. 1999; Vert et al. 2002; Santi and Schmidt 2009). By contrast, the graminaceous plants employ strategy-II to improve Fe acquisition via chelation-based strategy with the help of yellow stripe-like (YSL) transporters (Curie et al. 2001; Mori 1999). Various studies demonstrated the altered expression of numerous genes under Fe-deficiency conditions for adoption and better survival of plants (Buckhout et al. 2009; Yang et al. 2010). Apart from these, microRNAs (miRNAs) also play a crucial role in the regulation of gene expression in plants under Fe-deficiency conditions (Kong and Yang et al. 2010). Identification and function prediction of iron-deficiency-responsive microRNAs may help to better understand the adaptation mechanisms of plant under low Fe stress.
miRNAs are endogenous, single stranded and small non-coding RNAs of approximately 21-nucleotide (nt) in length that regulate gene expression at the post-transcriptional level by cleavage or by translational repression of target mRNA (Millar 2020; Jones-Rhoades et al. 2006). Plant miRNAs are well known in regulating the growth, developmental processes and various environmental responses (Millar 2020). Moreover, recent studies also elucidated their functional role in regulating nutrient homeostasis in various plants (Hsieh et al. 2009; Liang et al. 2010; Zhao et al. 2011; Paul et al. 2015; Shahzad et al. 2018;). For instance, Fe- deficiency responsive miRNAs were identified in various plants and analyzed their expression under Fe-deficiency conditions (Agarwal et al. 2015; Kong and Yang 2010). However, little is known about Fe-deficiency-responsive miRNAs in citrus. Fe-deficiency is a common problem occurs in citrus plants resulted in yellowing of leaves and small fruit development (Tagliavini and Rombolà 2001). However, the most obvious symptom of iron deficiency in citrus is young leaf yellowing (Jin et al. 2017). Therefore, we have chosen citrus leaves to identify Fe-deficiency-responsive miRNAs. In this study, we identified Fe-deficiency miRNAs and predicted the role of their target genes in citrus plants.
Materials and methods
Plant material and Fe treatments
The fragrant citrus (Citrus. Junos) seedlings of 15 weeks old were grown in greenhouse conditions by irrigating with Hoagland nutrient solution in a greenhouse at Huazhong Agricultural University (30° 289′ N, 114° 219′ E), Wuhan, China. The Hoagland nutrient solution is composed of 2.5 mM KNO3, 2.5 mM Ca(NO3)2, 0.5 mM KH2PO4, 10 µM H3BO3, 2 µM MnCl2, 2 µM ZnSO4, 0.5 µM CuSO4, 0.065 µM (NH4)6Mo7O24, 1 mM MgSO4 and 0.1 µM (for Fe-deficiency) or 10 µM (for Fe-sufficiency) Fe-EDTA. The solution was ventilated for 20 min every 2 h and renewed twice a week. The pH of all the nutrient solutions was adjusted to 6.0. After 40 days of Fe-deficiency and -sufficiency treatment, leaves were harvested and immediately frozen in liquid N2, then stored at − 80 °C for further experiments.
Library construction and illumina sequencing
The frozen leaf samples (about 100 mg) of both Fe-deficient and Fe- sufficient plants were used to extract total RNA using TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. The total RNA quantity and purity were analyzed with Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, CA, USA). Initially, the RNA molecules of 18–30 nt length range were enriched by polyacrylamide gel electrophoresis. Then, the 36–44 nt length of RNAs were enriched by ligating the 3′ adapters and finally 5′ adapters were also ligated to the RNAs. The reverse transcription of ligation products was carried out by employing PCR, and the 140–160 bp size of PCR products were enriched to generate a cDNA library. The generated library was sequenced on Illumina HiSeq™ 2500 at Gene Denovo Biotechnology (Guangzhou, China).
Data cleaning of raw reads acquired from Illumina sequencing was performed by removing adaptors, low-quality tags, and various types of contaminate reads (Poly A, 5′ and 3′-adapter contaminants, and reads ≤ 18 nt length). Then, the miRNA prediction was done by aligning the obtained clean tags with orange (Citrus sinensis) genome (http://citrus.hzau.edu.cn/orange/) using SOAP (http://www.soap.genomics.org.cn). At the same time, the clean tags were also aligned with small RNAs in GeneBank database (Release 209.0 http://www.ncbi.nlm.nih.gov/genbank/) and Rfam database (11.0 ftp://sanger.ac.uk/pub/databases/Rfam/) for further identification and removal of rRNA, scRNA, snoRNA, snRNA, and tRNA. The remaining sequences were then searched against miRBase database (Release 21 http://www.mirbase.org/) to identify known miRNAs. The miRNAs of those perfectly match with citrus miRNAs were called as conserved miRNAs, and the miRNAs matched with other plant miRNAs were called as known miRNA. Unidentified sequences that did not match with any of the above databases were further analyzed to predict novel miRNAs using Mireap software (https://sourceforge.net/projects/mireap/).
Differential expression analysis of miRNAs
To determine expression patterns of miRNAs under Fe-deficient and Fe-sufficient conditions, the frequency of miRNA counts was normalized as transcripts per million (TPM). The fold change between Fe-deficient and Fe-sufficient conditions was calculated as: fold change = log2 (Fe-deficient/Fe-sufficient). The miRNAs expression with a fold change ≥ 2 and comparison p value < 0.05 were identified as significant differential expression of miRNAs.
Prediction of potential miRNA target genes and functional analysis
Further, target genes of differentially expressed miRNAs were predicted by employing the TargetFinder 1.6 (http://targetfinder.org/) software. Gene Ontology (GO) enrichment analysis was performed to probe the functions of target genes. For GO enrichment analysis, GO terms with a corrected p ≤ 0.05 were considered as significant enrichment.
Expression analysis of miRNAs by qRT-PCR
Total RNA was extracted from Fe-deficiency and Fe-sufficient treated citrus plants leaves as described above and cDNA was synthesized by reverse transcription, using the PrimeScript™ RT reagent Kit following the manufacturer’s instructions (TaKaRa, Dalian, China). cDNA products were used as templates to analyze the expression level of miRNAs and the reaction was performed in QuabtStudio 6 Flex (Life technologies). Expression of 16 randomly selected differently expressed miRNAs was analyzed using stem-loop qRT-PCR following the method of Chen et al (2005). Stem-loop primers for reverse transcription and primers for qRT-PCR were listed in supplementary file (Additional file 1). U6 snRNA was used as internal reference gene for normalizing the expression (Kou et al. 2012). Briefly, the primers for miRNAs and U6 were diluted in the SYBER GREEN PCR Master Mix (TaKaRa, Dalian, China) and 10 μl of the reaction mix was added to each well. Reactions were performed by an initial incubation at 50 °C for 2 min and at 95 °C for 1 min, and then cycled at 95 °C for 15 s and 60 °C for 1 min for 40 cycles.
Statistical analysis
The significant difference of qRT-PCR results between the control and the treatment were analyzed with t-test (LSD) using SAS software version 8 (SAS Institute, Cary, NC, USA). Differences were considered as significant at p < 0.05.
Results
Analysis of the small RNA libraries
Two miRNA libraries were constructed from the total RNAs extracted from leaves of Fe-sufficient and Fe-deficient treated citrus plants. After cleaning the data, we obtained 10,779,211 and 10,744,506 clean reads, from Fe-deficient and Fe-sufficient libraries respectively (Table 1). Approximately 8,163,243 (represents 405,497 unique tags) and 8,106,834 (represents 439,265 unique tags) clean tags were mapped to the Citrus sinensis genome sequences for miRNA prediction. The total mapping rate was 75.73% (unique tags 56.21%) and 75.45% (unique tags 55.23%), respectively. The rate of exon antisense, exon sense, intron antisense, and intron sense were ranged from 1 to 5%. The majority length distribution of the sRNAs was from 21 to 24 nt with 24 nt sRNAs as the major peak, followed by 21 nt sRNAs (Fig. 1). Compared with the Fe-sufficient library, a higher distribution in length with 21 and 24 nt was detected in the Fe-deficient library (Fig. 1). After annotation of the non-coding RNAs, 2,429,859 and 2,611,951 were found to be conserved miRNAs, 435,099 and 437,733 were known miRNA, 336,494 and 313,866 were novel miRNA from Fe-deficient and Fe-sufficient libraries, respectively.
Table 1.
Statistical analysis of sRNA sequencing data of citrus leaves. IS-S refers to Fe-sufficiency, ID-S refers to Fe-deficiency
| IS-S | ID-S | |||||||
|---|---|---|---|---|---|---|---|---|
| Unique | Rate | Total | Rate | Unique | Rate | Total | Rate | |
| Total | 721,360 | 100% | 10,779,211 | 100% | 795,307 | 100% | 10,744,506 | 100% |
| Mapping genome | 405,497 | 56.21% | 8,163,243 | 75.73% | 439,265 | 55.23% | 8,106,834 | 75.45% |
| exist_mirna | 565 | 0.08% | 2,429,859 | 22.54% | 582 | 0.07% | 2,611,951 | 24.31% |
| known_mirna | 2856 | 0.40% | 435,099 | 4.04% | 2880 | 0.36% | 437,733 | 4.07% |
| novel_mirna | 733 | 0.10% | 336,494 | 3.12% | 782 | 0.10% | 313,866 | 2.92% |
| exon_antisense | 34,608 | 4.80% | 537,952 | 4.99% | 37,633 | 4.73% | 593,857 | 5.53% |
| exon_sense | 42,734 | 5.92% | 567,896 | 5.27% | 45,528 | 5.72% | 585,095 | 5.45% |
| intron_antisense | 16,247 | 2.25% | 115,125 | 1.07% | 17,923 | 2.25% | 128,652 | 1.20% |
| intron_sense | 29,171 | 4.04% | 238,499 | 2.21% | 31,310 | 3.94% | 258,067 | 2.40% |
| rRNA | 67,538 | 9.36% | 2,739,782 | 25.42% | 54,886 | 6.90% | 2,075,381 | 19.32% |
| Repeat | 1632 | 0.23% | 27,902 | 0.26% | 1771 | 0.22% | 24,655 | 0.23% |
| snRNA | 551 | 0.08% | 4945 | 0.05% | 468 | 0.06% | 4095 | 0.04% |
| snoRNA | 395 | 0.05% | 2375 | 0.02% | 383 | 0.05% | 2312 | 0.02% |
| tRNA | 4814 | 0.67% | 141,626 | 1.31% | 3989 | 0.50% | 124,861 | 1.16% |
| Unann | 518,560 | 71.89% | 3,146,575 | 29.19% | 596,219 | 74.97% | 3,523,829 | 32.80% |
Fig. 1.
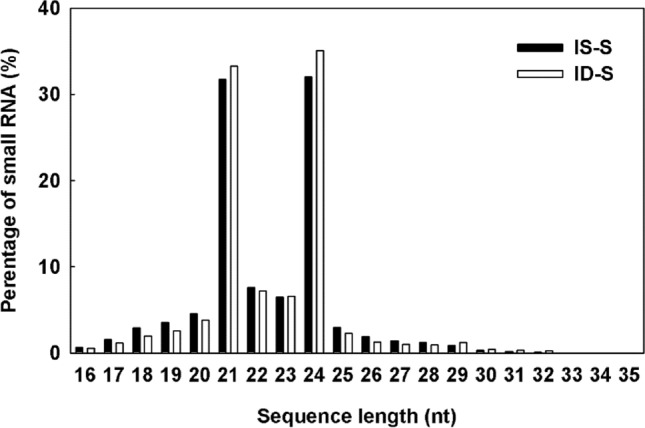
Length distribution of unique sequences of citrus leaves. IS-S refers to Fe-sufficiency, ID-S refers to Fe-deficiency
Identification of known and novel miRNAs
We found 147 known miRNAs belong to 74 annotated families from the two libraries based on their highly conserved sequences to the known plant miRNAs. Of the 147 known miRNAs, 50 miRNAs could be found in citrus and 97 miRNAs were found in other plants. The sequences, lengths and read counts of the known miRNAs are listed in Additional file 2. The comparison of miRNAs between the two libraries (IS-S and ID-S) and the abundance of each miRNA in two libraries was normalized to the transcripts per million (TPM). The results indicated that the known miRNAs exhibited extensive variation in their abundances between two libraries. For example, the TPM of miR166c was found to be 379,402.5 and 409,041.1 in the IS-S and ID-S libraries, respectively. Conversely, the TPM of miR399a was found to be 0.61 and 2.04 in the IS-S and ID-S libraries, respectively. This phenomenon also manifested between different miRNAs that belongs to the same family. There are 6 miRNAs in miR156 family, and the TPMs of them varied from 1.75 to 84,158.32.
About 427 secondary structures met the requirements to be considered as novel miRNAs. As some novel miRNAs showed a low abundance, the miRNAs with read count less than 4 in two miRNA libraries were removed (Additional file 3).
Differentially expressed miRNAs between IS_S and ID_S libraries
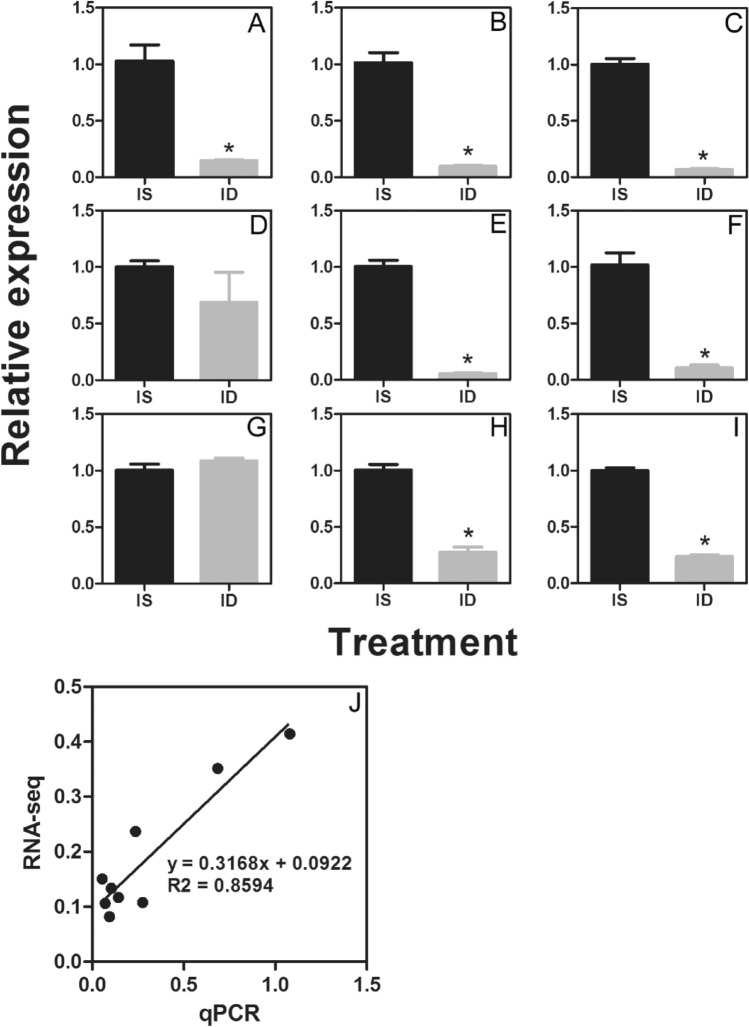
A total of 26 known miRNAs and 55 novel miRNAs were identified to be differentially expressed miRNAs between Fe-deficient and Fe-sufficient leaves (Fig. 2). Moreover, 10 known and 40 novel miRNAs were significantly up-regulated and 16 known and 15 novel miRNAs were down-regulated in Fe-deficient leaves compared to Fe-sufficient leaves (Additional file 3). To verify the expression patterns of the miRNAs obtained from the high-throughput sequencing, 16 miRNAs with different expression patterns from Illumina sequencing results were selected for stem-loop qRT-PCR analysis. As shown in Fig. 3a–i, qRT-PCR results coincide with the results obtained from high-throughput sequencing, and overall correlation coefficient was found to be 0.86 (Fig. 3j).
Fig. 2.
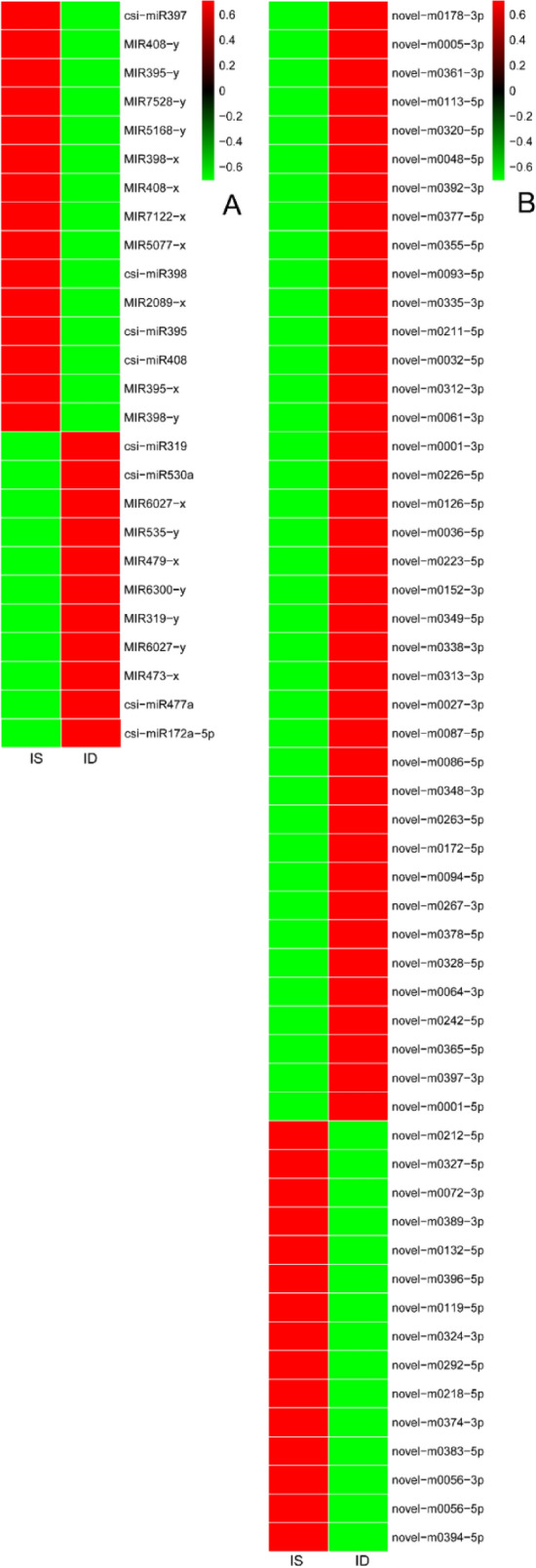
Heatmap of the differentially expressed miRNAs of citrus leaves. a Differentially expressed known miRNA, b differentially expressed novel miRNA. IS refers to Fe-sufficiency, ID refers to Fe-deficiency
Fig. 3.
qRT-PCR confirmation for differentially expressed genes from digital gene expression analysis. a–i refer to the transcript levels of 9 randomly selected different expressed miRNA, including 7 known and 2 novel miRNA. The bars represent SE (n = 4). J refers to the comparison between the log2 of gene expression ratios obtained from RNA-seq data and qRT-PCR. IS refers to Fe-sufficiency, ID refers to Fe-deficiency
Prediction of miRNA target genes
To better understand the biological functions of the known miRNAs in citrus, we employed the plant small RNA target prediction tool (Target Finder 1.6) to predict the putative target genes with the annotated transcripts of Citrus sinensis genome as the reference genome. Total, 3454 genes were predicted as the target genes of 462 miRNAs (Additional file 4).
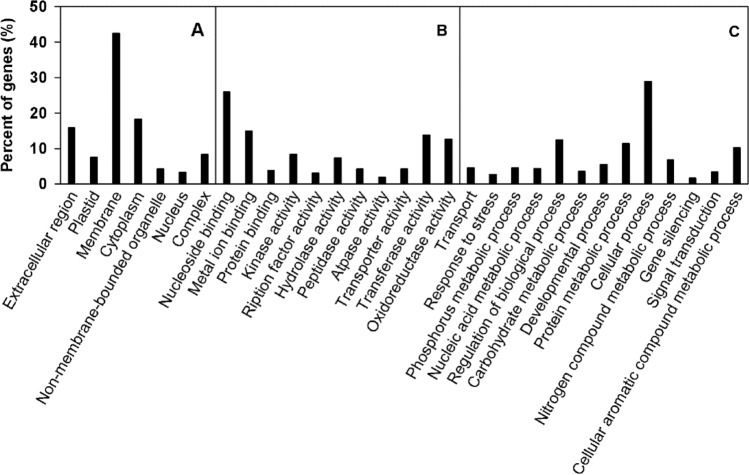
GO enrichment analysis assigned the predicted target genes of differently expressed miRNA to the cellular component, molecular function, and biological process. In the cellular component part, the target genes were grouped into 7 categories, including membrane, cytoplasm, extracellular region and so on (Fig. 4). In the molecular function part, the target genes were grouped into 11 categories including nucleoside binding, metal ion binding, and transferase activity and so on (Fig. 4). In the biological process part, the target genes were grouped into 13 categories, including cellular process, regulation of biological process, cellular aromatic compound metabolic process and so on (Fig. 4).
Fig. 4.
GO of the predicted target genes for differentially expressed miRNAs of citrus leaves. Categorization of miRNAs target genes was performed according to cellular component (a), molecular function (b) and biological (c)
To investigate the miRNAs regulation of target genes in response to Fe-deficiency, 227 target genes of 26 known and 8 novel miRNA (counts > 20 in any library) were selected from our previous RNA-seq data (Jin et al. 2017). Among them, 166 genes were detected in RNA-seq data. The expression pattern of 95 target genes was negatively correlated with miRNAs (Additional file 4). As miRNAs play a negative regulation on gene expression, the target genes with opposite expression patterns were analyzed further. The predicted target genes of miR172 were THO/TREX complex and Sodium-dependent phosphate transport protein. The predicted target genes of miR2089 were TMV resistance protein, DEAD-box ATP-dependent RNA helicase, tRNA pseudouridine synthase 1 and leucine-rich repeat-containing protein. The predicted target gene of miR319 was TCP2-like isoform X1. The predicted target genes of miR395 were pumilio, histidine kinase 2, calcium-dependent calmodulin-independent protein kinase isoform 1, pentatricopeptide repeat-containing protein, neutral ceramidase, calmodulin-binding transcription activator, ATP sulfurylase 1, LRR receptor-like serine/threonine-protein kinase and three uncharacterized proteins. The predicted target genes of miR397 were nudix hydrolase 2, copper/zinc superoxide dismutase (CSD), novel protein similar to vertebrate DDHD domain-containing protein 1, sacsin, LRR receptor-like serine/threonine-protein kinase GSO1 and nine laccases. The predicted target genes of miR398 were CSD, serine/threonine-protein kinase PBS1, calcium-transporting ATPase 11 and two uncharacterized proteins. The predicted target genes of miR408 were G-type lectin S-receptor-like serine/threonine-protein kinase, blue copper protein, 2-oxoglutarate and Fe(II)-dependent oxygenase-like protein, laccase-12 and RNA, and export factor-binding protein. The predicted target gene of miR477a was UDP-glycosyltransferase. The predicted target genes of miR479 were laccase-11 and an uncharacterized protein. The predicted target gene of miR5077 was ABC transporter F family member 3. The predicted target genes of miR5168-y were BTB/POZ domain-containing protein and two C2 and GRAM domain-containing proteins. The predicted target genes of miR530a was DNA double-strand break repair rad50 ATPase. The predicted target genes of miR6027 were hydrolases, protein CLEC16A, rhodanese-like family protein, histone-lysine N-methyltransferase, and disease resistance proteins. The predicted target genes of miR7122 were serine/threonine-protein kinase CTR1 and an uncharacterized protein. The predicted target genes of miR7528 were natural resistance associated macrophage protein, zinc finger CCCH domain-containing protein 44, trafficking protein particle complex subunit 8 and WRKY transcription factor 52.
Discussion
The length distribution of small RNAs in citrus varies from 16 to 35 nt. However the majority length of the small RNAs was 21–24 nt and sequences, which is in accordance with the results obtained in other plants (Qu et al. 2016). Moreover, Fe-deficiency increased the abundance of 21 nt and 24 nt sRNA, indicating that sRNA was affected by Fe-deficiency in citrus leaves. Evidences showed that miRNAs are involved in the adaptive regulation of nutrient-deficiency mechanisms in higher plants (Song et al. 2019; Paul et al. 2015). In our study, we identified 147 known miRNAs and 427 novel miRNAs from Fe-deficient and -sufficient leaves. Moreover, 26 known and 55 novel miRNAs were differentially expressed, suggesting that Fe-deficiency altered the expression profiles of miRNAs in leaves.
Our study also clearly demonstrated the occurrence of four miR172 family members, and only csi-miR172-5p was slightly down-regulated by Fe-deficiency. Previous studies have shown that miR172 was involved in regulation of flowering and growth phase conversion (Chen 2004; Wu et al. 2009). Moreover, miR172 was found to be increase in expression when infected with Tomato leaf curl New Delhi virus (Naqvi et al. 2010) in tomato plants. In Arabidopsis roots, expression of miR172c was decreased at 4-h Fe-deficient treatment and then gradually increased to normal levels (Waters et al. 2012). The target genes of csi-miR172-5p were THO/TREX complex and Sodium-dependent phosphate transport protein (SLC34) (Additional file 4). THO/TREX complex played an important role in transcription elongation and endogenous siRNA biosynthesis (Rondon et al. 2003; Yelina et al. 2010). The increase of THO/TREX complex gene promote the elongation of transcription and biosynthesis of siRNA that enhanced the regulation of gene expression to adapt Fe-deficiency. The decrease in miR172 expression may involve in the regulation of Fe-deficiency stress tolerance in citrus.
Three miR395 family members (csi-miR395, miR395-x and miR395-y) were identified in citrus leaves under Fe-deficiency conditions, and all of them were down-regulated under Fe-deficiency (Table 2). MiR395 is a general and critical component of the sulfate assimilation regulatory network in plants. Jones-Rhoades and Bartel (2004) found that the expression of Ath-miR395 was majorly governed by the concentration of sulfate. The expression of an Ath-miR395 target gene decreases with the reduced sulfate concentration. In this study, the target gene of miR395 was also predicted to be ATP sulfurylase (Additional file 4). However, the expression of ATP sulfurylase between Fe-deficient and -sufficient leaves was not significantly altered. These results indicated that there may be uncovering role of miR395 under Fe-deficiency conditions.
Table 2.
List of differentially expressed known miRNA of citrus leaves. TPM refers to transcripts per million, IS-S refers to Fe-sufficiency, ID-S refers to Fe-deficiency, FC refers to fold change which is the ratio of ID-S-TPM /IS-S-TPM
| miR-name | Length | Seq | IS-S-TPM | ID-S-TPM | FC | log2 (FC) | p value |
|---|---|---|---|---|---|---|---|
| csi-miR172a-5p | 21 | GCAGCGTCCTCAAGATTCACA | 37.77 | 16.65 | 0.44 | − 1.18 | 1.20E-07 |
| csi-miR319 | 21 | TTTGGACTGAAGGGAGCTCCT | 862.27 | 1740.51 | 2.02 | 1.01 | 9.61E-221 |
| csi-miR395 | 21 | CTGAAGTGTTTGGGGGAACTC | 37.77 | 0.58 | 0.02 | − 6.01 | 8.53E-36 |
| csi-miR397 | 21 | TCATTGAGTGCAGCGTTGATG | 70.01 | 8.18 | 0.12 | − 3.10 | 1.26E-42 |
| csi-miR398 | 21 | TGTGTTCTCAGGTCACCCCTT | 128.36 | 10.51 | 0.08 | − 3.61 | 4.41E-88 |
| csi-miR408 | 21 | ATGCACTGCCTCTTCCCTGGC | 3429.11 | 363.35 | 0.11 | − 3.24 | 0.00E + 00 |
| csi-miR477a | 21 | ACCTCCCTCGAAGGCTTCCAA | 7.37 | 15.77 | 2.14 | 1.10 | 1.40E-03 |
| csi-miR530a | 21 | TGCATTTGCACCTGCACCTTG | 1.23 | 3.80 | 3.09 | 1.63 | 4.00E-02 |
| MIR2089-x | 22 | TCTTACCTATGCCACCAATTCC | 9.52 | 0.58 | 0.06 | − 4.03 | 3.23E-08 |
| MIR319-y | 21 | ATCCAACGAAGCAGGAGCTGC | 339.32 | 761.46 | 2.24 | 1.17 | 3.67E-122 |
| MIR395-x | 21 | GTTCCTCCGAGCACTTCATTG | 1.84 | 0.01 | 0.01 | − 7.53 | 1.31E-02 |
| MIR395-y | 21 | TTGAAGTGTTTGGAGGAACTC | 54.05 | 18.99 | 0.35 | − 1.51 | 1.99E-14 |
| MIR398-x | 21 | GGGGCGACATGAGATCACATG | 41.46 | 5.84 | 0.14 | -2.83 | 1.11E-23 |
| MIR398-y | 21 | TGTGTTCTCAGGTCGCCCCTG | 17,198.65 | 2593.10 | 0.15 | − 2.73 | 0.00E + 00 |
| MIR408-x | 22 | ACGGGGAACAGGCAGAGCATGT | 5.22 | 0.01 | 0.00 | − 9.03 | 4.83E-06 |
| MIR408-y | 22 | TGCACTGCCTCTTCCCTGGCTT | 419.77 | 56.08 | 0.13 | − 2.90 | 9.19E-232 |
| MIR473-x | 22 | ACTCTCCCTCAAGGGCTTCGCT | 0.61 | 3.80 | 6.18 | 2.63 | 5.66E-03 |
| MIR479-x | 22 | TGTGATATTGTTTCGGCTCATC | 0.01 | 1.46 | 146.04 | 7.19 | 3.62E-02 |
| MIR5077-x | 21 | TTCTTCACGTCGGGTTCACCA | 54.97 | 22.78 | 0.41 | − 1.27 | 1.30E-11 |
| MIR5168-y | 21 | TCTCGGACCAGGCTTCAATTT | 4.30 | 0.58 | 0.14 | − 2.88 | 1.68E-03 |
| MIR535-y | 22 | CTGACAATGAGAGAGAGCACAC | 4.30 | 9.35 | 2.17 | 1.12 | 1.30E-02 |
| MIR6027-x | 22 | ATGGGTAGCACAAGGATTAATG | 0.01 | 2.34 | 233.67 | 7.87 | 4.88E-03 |
| MIR6027-y | 22 | TGAATCCTTCGGCTATCCATAA | 0.61 | 3.80 | 6.18 | 2.63 | 5.66E-03 |
| MIR6300-y | 18 | GTCGTTGTAGGATAGTGG | 4.61 | 45.27 | 9.83 | 3.30 | 3.05E-29 |
| MIR7122-x | 22 | TTATACAGAGAAATCACGGTCG | 5.22 | 1.17 | 0.22 | − 2.16 | 3.01E-03 |
| MIR7528-y | 22 | CCAAAATGCTAATCTGAGGCTT | 20.57 | 9.05 | 0.44 | − 1.18 | 9.47E-05 |
The transcript of miR319 (csi-miR319 and MIR319-y) was abundant in Fe-deficient leaves (Table 2). miR319 was reported as a positive regulator in response to abiotic stress and organ development. In sugarcane, miR319 was up-regulated subjected to 4 °C for 24 h; whereas, its target genes (PCF6 and GAMYB) were down-regulated (Thiebaut et al. 2012). In rice plants, overexpression of Osa-miR319b led to an enhanced tolerance to cold stress by increasing proline content (Wang et al. 2014). miR319 is also actively involved in regulation of compound leaf development via regulating lanceolate in tomato (Ori et al. 2007). In sugarcane, the predicted target genes of miR319 was proliferating cell factors (TCP/FCF) and GAMYB (Thiebaut et al. 2012). In this study, we predicted TCP2 as the target gene of csi-miR319 (Additional file 4). Members of the TCP gene family were reported to be involved in the regulation of growth and cell cycle, including axillary meristem activity (Cubas et al. 1999; Li et al. 2005). The transcript abundance of TCP2 in Fe-deficient leaves was lower compared to Fe-sufficient leaves (Additional file 4). To cope with Fe-deficient conditions in plants, plants have evolved many strategies including altered expressions of miRNAs. The up-regulation of miR319 expression and down-regulated expression of TCP2 gene may retard growth and enhanced the tolerance of citrus leaves under Fe-deficiency stress.
We found that three miR398 family members (csi-miR398, MIR398-x and MIR398-y) were down-regulated under Fe-deficiency conditions. (Table 2). Previous studies have shown that miR398 was down-regulated by both abiotic and biotic stresses (Sunkar et al. 2006; Jagadeeswaran et al. 2009). Moreover, miR398 was also involved in the regulation of nutritional-deficiency stress. The expression of miR398a is strongly reduced under nutrient-deficiency stress (P, N, and C deficiency) in Arabidopsis plants (Pant et al. 2009) and the target genes of miR398 were two Cu/Zn superoxide dismutase (CSD) genes in Arabidopsis (Sunkar et al. 2006). CSD is essential and play an important role in scavenging of reactive oxygen species that increase during nutrient limitations and other environmental stresses (Apel and Hirt 2004). CSD, Serine/threonine-protein kinase and an uncharacterized protein were predicted the target genes of miR398 family, and were up-regulated under Fe-deficiency (Additional file 4). Our DEG analysis showed that CSD gene (Cs3g12000) was 1.62-fold times increased in Fe-deficient leaves compared to Fe-sufficient leaves, indicating that CSD is the target gene of miR398. As Fe is essential to Chlorophyll biosynthesis, Fe-deficiency will block photosynthetic light energy and induces oxidative stress (Hänsch and Mendel 2009). In plant, Fe-containing superoxide dismutase (FSD) can scavenge reactive oxygen to protect the plants from oxidative stress damage (Pilon et al. 2011). And, some Fe- or Cu-containing enzymes can be functionally replaced, such as Cu-containing plastocyanin for cytochrome Fe-containing c6 (Ravet et al. 2009), and Cu/Zn SOD for FeSOD (Puig et al. 2007; Burkhead et al. 2009). The expression of FSD genes was down-regulated and the expression of CSD was up-regulated in Fe-deficiency leaves (Table3). These results suggested that the decrease of miR398 and increase of Cu/Zn SOD activity may play an important role in scavenging of reactive oxygen species and inhibition of oxidative damages under Fe-deficiency stress.
Table 3.
The target gene prediction and expression of differentially expressed miRNA. FC refers to the fold change which is the ratio of FPKM-ID/FPKM-IS
| miR-name | Accession | Target FC | Annotation |
|---|---|---|---|
| csi-miR172a-5p | Cs3g23470 | 1.19 | THO/TREX complex |
| Cs8g19810 | 1.05 | Sodium-dependent phosphate transport protein | |
| MIR2089-x | Cs2g30960 | 2.50 | TMV resistance protein |
| Cs2g30970 | 2.00 | TMV resistance protein | |
| Cs3g26290 | 1.16 | DEAD-box ATP-dependent RNA helicase | |
| Cs3g26810 | 1.12 | tRNA pseudouridine synthase 1 | |
| Cs5g17280 | 1.22 | Leucine-rich repeat-containing protein | |
| Cs5g19570 | 1.99 | Leucine-rich repeat-containing protein | |
| Cs5g19850 | 2.03 | Leucine-rich repeat-containing protein | |
| orange1.1t00706 | 1.32 | TMV resistance protein N | |
|
csi-miR319 MIR319-y |
Cs2g08080 | 0.86 | TCP2-like isoform X1 |
|
csi-miR395 MIR395-x MIR395-y |
Cs2g08400 | 1.18 | Pumilio |
| Cs2g19760 | 1.32 | Histidine kinase 2 | |
| Cs3g11190 | 1.32 | Uncharacterized protein | |
| Cs3g26650 | 1.44 | Calcium-dependent calmodulin-independent protein kinase isoform 1 | |
| Cs5g04130 | 1.39 | Uncharacterized protein | |
| Cs5g31350 | 1.01 | Pentatricopeptide repeat-containing protein | |
| Cs6g07540 | 1.24 | Neutral ceramidase | |
| Cs8g18130 | 1.19 | Uncharacterized protein | |
| Cs9g01510 | 1.22 | Calmodulin-binding transcription activator | |
| Cs9g07750 | 1.03 | ATP sulfurylase 1 | |
| Cs9g15900 | 1.14 | LRR receptor-like serine/threonine-protein kinase | |
| Cs9g18740 | 1.13 | Leucine-rich repeat receptor protein kinase | |
|
csi-miR397 MIR397-x |
Cs2g01700 | 1.05 | Nudix hydrolase 2 |
| Cs3g12000 | 1.62 | Copper/zinc superoxide dismutase | |
| Cs5g19700 | 1.95 | Novel protein similar to vertebrate DDHD domain-containing protein 1 | |
| Cs6g06880 | 1.62 | Laccase-17 | |
| Cs6g06890 | 7.22 | Laccase-17 | |
| Cs6g06920 | 1.32 | Laccase-17 | |
| Cs6g07800 | 1.51 | Laccase-4 | |
| Cs7g13980 | 1.07 | Sacsin | |
| Cs7g23490 | 1.83 | Laccase-17 | |
| Cs7g31620 | 1.49 | Laccase-22 | |
| Cs8g14840 | 1.32 | LRR receptor-like serine/threonine-protein kinase GSO1 | |
| Cs8g17630 | 1.02 | Laccase-17 | |
| Cs8g18800 | 1.06 | Laccase-17 | |
| Cs8g19850 | 1.55 | Laccase-4 | |
| orange1.1t01926 | 1.41 | Leucine-rich repeat-containing protein | |
|
csi-miR398 MIR398-x MIR398-y |
Cs2g29640 | 1.40 | Putative uncharacterized protein |
| Cs3g12000 | 1.62 | Cu/Zn superoxide dismutase | |
| Cs7g02980 | 1.17 | Serine/threonine-protein kinase PBS1 | |
| orange1.1t01967 | 2.03 | UN | |
| orange1.1t02346 | 2.66 | Calcium-transporting ATPase 11 | |
|
csi-miR408 MIR408-x MIR408-y |
Cs1g14910 | 1.23 | G-type lectin S-receptor-like serine/threonine-protein kinase |
| Cs3g20580 | 1.42 | Blue copper protein | |
| Cs5g13190 | 1.05 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase-like protein | |
| Cs7g30410 | 4.93 | Laccase-12 | |
| Cs8g18100 | 1.02 | RNA and export factor-binding protein | |
| csi-miR477a | Cs1g05220 | 0.43 | UDP-glycosyltransferase |
| MIR479-x | Cs1g24250 | 0.92 | Laccase-11 |
| Cs2g24350 | 0.89 | Uncharacterized protein | |
| MIR5077-x | Cs5g01810 | 1.11 | ABC transporter F family member 3 |
| MIR5168-y | Cs5g03420 | 1.26 | BTB/POZ domain-containing protein |
| Cs5g26880 | 1.35 | C2 and GRAM domain-containing protein | |
| Cs5g27350 | 2.14 | C2 and GRAM domain-containing protein | |
| csi-miR530a | Cs7g12670 | 0.96 | DNA double-strand break repair rad50 ATPase |
|
MIR6027-x MIR6027-y |
Cs1g07790 | 0.32 | Disease resistance RPP13-like protein 2 |
| Cs1g07860 | 0.69 | Disease resistance protein RDL6/RF9 | |
| Cs1g24300 | 0.67 | Hydrolases or acyltransferases | |
| Cs3g05560 | 0.88 | Disease resistance protein RDL6/RF9 | |
| Cs3g05870 | 0.57 | Disease resistance protein RDL6/RF9 | |
| Cs4g04250 | 0.95 | Protein CLEC16A | |
| Cs6g08440 | 0.66 | Rhodanese-like family protein-like protein | |
| orange1.1t04028 | 0.94 | Histone-lysine N-methyltransferase | |
| MIR7122-x | Cs1g09665 | 2.00 | Uncharacterized protein |
| Cs3g11580 | 1.30 | Serine/threonine-protein kinase CTR1 | |
| MIR7528-y | Cs1g22120 | 7.71 | Natural resistance associated macrophage protein |
| Cs3g03210 | 1.49 | Zinc finger CCCH domain-containing protein 44 | |
| Cs6g13600 | 1.10 | Trafficking protein particle complex subunit 8 | |
| orange1.1t03702 | 1.10 | WRKY transcription factor 52 | |
| m0001-5p | Cs1g13640 | 0.87 | ABC transporter G family member 40 |
| Cs9g03780 | 0.75 | Uncharacterized protein | |
|
miRN0056-5p miRN0056-3p |
Cs1g14690 | 11.22 | Spore coat protein A |
| Cs7g04730 | 1.08 | TMV resistance protein N | |
| orange1.1t04786 | 1.14 | Spore coat protein A | |
| Cs2g29640 | 1.40 | Uncharacterized protein | |
| miR0178-3p | Cs2g13630 | 0.55 | Cytochrome P450-dependent fatty acid hydroxylase |
| Cs6g08680 | 0.91 | Monothiol glutaredoxin-S16 | |
| Cs6g20630 | 0.64 | Hypersensitive-induced response protein 1 | |
| Cs8g17230 | 0.85 | Cytochrome P450-dependent fatty acid hydroxylase | |
| miR0212-5p | Cs2g06440 | 1.75 | Uncharacterized protein |
| Cs3g15360 | 1.08 | Thiosulfate sulfurtransferase/rhodanese-like domain-containing protein 2 | |
| Cs3g19210 | 1.32 | Pentatricopeptide repeat-containing protein | |
| Cs5g10910 | 1.07 | Galactoside 2-alpha-L-fucosyltransferase | |
| Cs6g02560 | 1.30 | GDT1-like protein 3 | |
| Cs9g07670 | 2.89 | Cold-inducible protein-like protein | |
| miRN0223-5p | Cs7g32240 | 0.82 | UDP-glycosyltransferase |
| miRN0324-3p | Cs1g15590 | 1.66 | GATA transcription factor 12 |
| Cs1g25380 | 1.45 | UN | |
| miRN0327-5p | Cs2g06440 | 1.76 | Uncharacterized protein |
| Cs3g15360 | 1.08 | Thiosulfate sulfurtransferase/rhodanese-like domain-containing protein 2 | |
| Cs3g19210 | 1.32 | Pentatricopeptide repeat-containing protein | |
| Cs5g10910 | 1.07 | Galactoside 2-alpha-L-fucosyltransferase | |
| Cs6g02560 | 1.30 | GDT1-like protein 3 | |
| Cs9g07670 | 2.89 | Cold-inducible protein-like protein |
miR397 is also known to involve in the various abiotic stress responses and in regulating various stages of plant growth and development. Dong and Pei (2014) reported that overexpression of miR397 in Arabidopsis improved the plant tolerance to cold stress. Recent studies have shown that the expression of miR397 was decreased under toxicity levels of boron in barley (Ozhuner et al. 2013) and citrus plants (Jin et al. 2016). Overexpression of OsmiR397 improves rice yield by increasing grain size and promoting panicle branching (Zhang et al. 2013). In this study, the predicted target genes of miR397 were laccase and CSD. The expression of two miR397 members was reduced in Fe-deficient leaves (Table 2); however, the abundance of target genes (laccase) transcript was increased. It is well known that laccase catalyzes the polymerization of lignin monomers in vitro (Sterjiades et al. 1992). Lignin plays multiple roles in stress responses (Moura et al. 2010). The reduced expression of miR397 may increase the lignin accumulation and enhance the Fe-deficiency tolerance in citrus plants.
The expression of three miR408 members (csi-miR408, MIR408-x, and MIR408-y) was down-regulated under Fe-deficiency (Table 2). miR408 has been reported to be involved in different abiotic stresses responses including cold, osmotic, drought and oxidative stress (Liu et al. 2008; Jovanovic et al. 2014; Ma et al. 2015). Under drought stress conditions, transcript level of miR408 was reduced in rice plants (Mutum et al. 2013). However, overexpression of miR408 increased the drought tolerance of chickpea (Hajyzadeh et al. 2015). miR408 was also involved in the nutritional homeostasis of plants. miR408 was down-regulated under nitrogen and boron deficiency conditions in Arabidopsis and citrus plants respectively (Buhtz et al. 2010; Yang et al. 2015). Moreover, miR408 was also induced under Cu deficiency (Abdel-Ghany and Pilon 2008). The target genes of miR408 were Cu/Zn SODs (CSDs), plantacyanin, peptide chain release factor and several laccases (Sunkar and Zhu 2004; Schwab et al. 2005). In this study, blue copper protein, RNA and export factor-binding protein, laccase-12, G-type lectin S-receptor-like serine/threonine-protein kinase and 2-oxoglutarate and Fe(II)-dependent oxygenase-like protein were predicted the target genes of miR408. In our previous study, we reported that the uptake of Cu content was increased under Fe-deficiency conditions in citrus plants (Jin et al. 2017). As the Fe-deficiency cause an oxidative stress, the reduced expression in miR408 might be advantageous for plant survival under Fe-deficiency.
In populus, miR477 plays an important role in the growth and formation of specialized woody tissue (Lu et al. 2008). In this study, UDP-glycosyltransferase (UGT) was also predicted as the target gene of miR477. The expression of UGT gene was decreased in Fe-deficient leaves and UGT was essential for growth and development of plants (Woo et al. 1999). The expression of miR477a was up-regulated in Fe-deficient citrus leaves, which is in coincidence with the suppression of growth. The predicted target genes of miR2089, miR6027 and novel-m0056-3p were disease resistance proteins, those are involved in disease resistance under Fe-deficiency. The target genes of miR473, miR535 and miR6300 were not dissected in this study and further investigations are needed to resolve their functions.
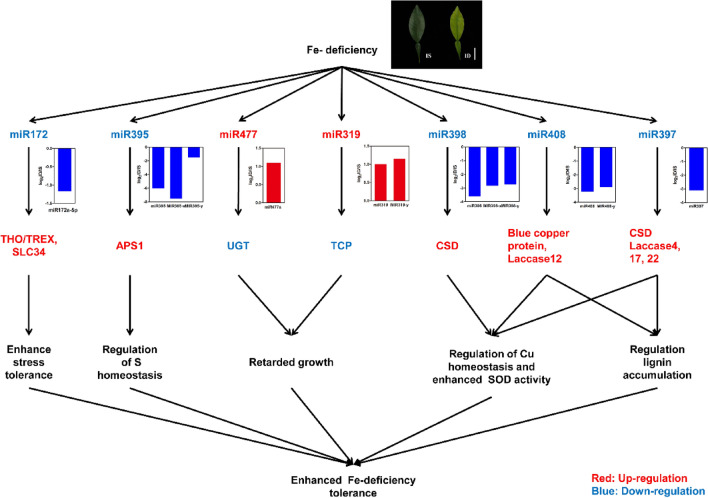
In conclusion, we identified 147 known and 427 novel miRNAs from Fe-deficient and Fe-sufficient citrus leaves. Among them, 50 miRNAs (10 known and 40 novel) were up-regulated and 31 miRNAs (16 known and 15 novel) were down-regulated by Fe-nutrient-deficiency conditions. As shown in Fig. 5, we proposed a model for the Fe-deficiency-responsive miRNAs in citrus plants. Our study clearly demonstrated that miR172, miR319, miR395, miR397, miR398, miR408, miR477 might play critical roles in regulating the Fe-deficiency adaptive mechanisms in citrus.
Fig. 5.
A proposed model for the possible roles of miRNAs in the tolerance to Fe-deficiency in citrus leaves. Red and blue colors represent the up-regulation and down-regulation of miRNAs or target genes
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgment
This work was supported by the earmarked fund for China Agriculture Research System (CARS-27).
Data availability
The raw data of the small RNA-Seq have been uploaded to the Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra/). The accession number is SRR5012112.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Abdel-Ghany SE, Pilon M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in arabidopsis. J Biol Chem. 2008;283(23):15932–15945. doi: 10.1074/jbc.M801406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S, Mangrauthia SK, Sarla N. Expression profiling of iron deficiency responsive microRNAs and gene targets in rice seedlings of Madhukar x Swarna recombinant inbred lines with contrasting levels of iron in seeds. Plant Soil. 2015;396:137–150. doi: 10.1007/s11104-015-2561-y. [DOI] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Buckhout TJ, Yang TJW, Schmidt W. Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genomics. 2009;10:147. doi: 10.1186/1471-2164-10-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhtz A, Janin P, Franziska S, et al. Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 2010;10(1):64. doi: 10.1186/1471-2229-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhead JL, Reynolds KA, Abdel-Ghany, et al. Copper homeostasis. New Phytol. 2009;182(4):799–816. doi: 10.1111/j.1469-8137.2009.02846.x. [DOI] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303(5665):2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 1999;18(2):215–222. doi: 10.1046/j.1365-313x.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, et al. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature. 2001;409:346. doi: 10.1038/35053080. [DOI] [PubMed] [Google Scholar]
- Dong CH, Pei HX. Over-expression of miR397 improves plant tolerance to cold stress in Arabidopsis thaliana. J Plant Biol. 2014;57(4):209–217. doi: 10.1007/s12374-013-0490-y. [DOI] [Google Scholar]
- Hajyzadeh M, Turktas M, Khawar KM, et al. miR408 overexpression causes increased drought tolerance in chickpea. Gene. 2015;555(2):186–193. doi: 10.1016/j.gene.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Hänsch R, Mendel RR. Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl) Curr Opin Plant Biol. 2009;12(3):259–266. doi: 10.1016/j.pbi.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Hsieh L-C, Lin S-I, Shih AC, et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151(4):2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeswaran G, Saini A, Sunkar R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta. 2009;229(4):1009–1014. doi: 10.1007/s00425-009-0889-3. [DOI] [PubMed] [Google Scholar]
- Jin LF, Liu YZ, Yin XX, et al. Transcript analysis of citrus miRNA397 and its target LAC7 reveals a possible role in response to boron toxicity. Acta Physiol Plant. 2016;38(1):18. doi: 10.1007/s11738-015-2035-0. [DOI] [Google Scholar]
- Jin LF, Liu YZ, Du W, et al. Physiological and transcriptional analysis reveals pathways involved in iron deficiency chlorosis in fragrant citrus. Tree Genet Genom. 2017;13(3):51. doi: 10.1007/s11295-017-1136-x. [DOI] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14(6):787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Jovanovic Z, Stanisavljevic N, Mikic A, et al. Water deficit down-regulates miR398 and miR408 in pea (Pisum sativum L.) Plant Physiol Biochem. 2014;83:26–31. doi: 10.1016/j.plaphy.2014.07.008. [DOI] [PubMed] [Google Scholar]
- Kong WW, Yang ZM. Identification of iron-deficiency responsive microRNA genes and cis-elements in Arabidopsis. Plant Physiol Biochem. 2010;48(2):153–159. doi: 10.1016/j.plaphy.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Kou S-J, Wu X-M, Liu Z, et al. Selection and validation of suitable reference genes for miRNA expression normalization by quantitative RT-PCR in citrus somatic embryogenic and adult tissues. Plant Cell Rep. 2012;31(12):2151–2163. doi: 10.1007/s00299-012-1325-x. [DOI] [PubMed] [Google Scholar]
- Li C, Potuschak T, Gutiérrez RA, et al. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA. 2005;102(36):12978–12983. doi: 10.1073/pnas.0504039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Yang F, Yu D. MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 2010;62(6):1046–1057. doi: 10.1111/j.1365-313X.2010.04216.x. [DOI] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, et al. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14(5):836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Millán A-F, Grusak MA, Abadía A, Abadía J. Iron deficiency in plants: an insight from proteomic approaches. Front Plant Sci. 2013;4:254. doi: 10.3389/fpls.2013.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Sun YH, Chiang VL. Stress-responsive microRNAs in populus. Plant J. 2008;55(1):131–151. doi: 10.1111/j.1365-313X.2008.03497.x. [DOI] [PubMed] [Google Scholar]
- Ma C, Burd S, Lers A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015;84(1):169–187. doi: 10.1111/tpj.12999. [DOI] [PubMed] [Google Scholar]
- Millar AA. The function of miRNAs in plants. Plants. 2020;9(2):198. doi: 10.1111/nph.14834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S. Iron acquisition by plants. Curr Opin Plant Biol. 1999;2(3):250–253. doi: 10.1016/S1369-5266(99)80043-0. [DOI] [PubMed] [Google Scholar]
- Moura JC, de BonineOliveira Fernandes Viana CAJ, et al. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol. 2010;52(4):360–376. doi: 10.1111/j.1744-7909.2010.00892.x. [DOI] [PubMed] [Google Scholar]
- Mutum RD, Balyan SC, Kansal S, et al. Evolution of variety-specific regulatory schema for expression of osa-miR408 in indica rice varieties under drought stress. FEBS J. 2013;280(7):1717–1730. doi: 10.1111/febs.12186. [DOI] [PubMed] [Google Scholar]
- Naqvi AR, Haq QMR, Mukherjee SK. MicroRNA profiling of tomato leaf curl new delhi virus (tolcndv) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virology Journal. 2010;7(1):1–16. doi: 10.1186/1743-422X-7-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Cohen AR, Etzioni A. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet. 2007;39(6):787–791. doi: 10.1038/ng2036. [DOI] [PubMed] [Google Scholar]
- Ozhuner E, Eldem V, Ipek A, et al. Boron stress responsive microRNAs and their targets in barley. PLoS ONE. 2013;8(3):e59543. doi: 10.1371/journal.pone.0059543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Musialak-Lange M, Nuc P, et al. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 2009;150(3):1541–1555. doi: 10.1104/pp.109.139139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Datta SK, Datta K. miRNA regulation of nutrient homeostasis in plants. Frontiers in Plant Science. 2015;6:232. doi: 10.3389/fpls.2015.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Ravet K, Tapken W, et al. The biogenesis and physiological function of chloroplast superoxide dismutases. Biochem Biophys Acta. 2011;1807(8):989–998. doi: 10.1016/j.bbabio.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Puig S, Andres-Colas N, Garcia-Molina A, et al. Copper and iron homeostasis in Arabidopsis: responses to metal deficiencies, interactions and biotechnological applications. Plant, Cell Environ. 2007;30(3):271–290. doi: 10.1111/j.1365-3040.2007.01642.x. [DOI] [PubMed] [Google Scholar]
- Qu D, Yan F, Meng R, et al. Identification of microRNAs and their targets associated with fruit-bagging and subsequent sunlight re-exposure in the “Granny Smith” apple exocarp using high-throughput sequencing. Front Plant Sci. 2016;7:27. doi: 10.3389/fpls.2016.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravet K, Touraine B, Boucherez J, et al. Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J. 2009;57(3):400–412. doi: 10.1111/j.1365-313X.2008.03698.x. [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, et al. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397(6721):694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- Rondon AG, Jimeno S, Garcia-Rubio M, et al. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J Biol Chem. 2003;278(40):39037–39043. doi: 10.1074/jbc.M305718200. [DOI] [PubMed] [Google Scholar]
- Santi S, Schmidt W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 2009;183(4):1072–1084. doi: 10.1111/j.1469-8137.2009.02908.x. [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, et al. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8(4):517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Shahzad R, W. Harlina P, Ayaad M, et al. Dynamic roles of microRNAs in nutrient acquisition and plant adaptation under nutrient stress: a review. Plant Omics. 2018;11(1):58–79. doi: 10.21475/poj.11.01.18.pne1014. [DOI] [Google Scholar]
- Song X, Li Y, Cao X, et al. MicroRNAs and their regulatory roles in plant-environment interactions. Annu Rev Plant Biol. 2019;70(1):489–525. doi: 10.1146/annurev-arplant-050718-100334. [DOI] [PubMed] [Google Scholar]
- Sterjiades R, Dean JFD, Eriksson KEL, et al. Laccase from sycamore maple (Acer pseudoplatanus) polymerizes monolignols. Plant Physiol. 1992;99:1162–1168. doi: 10.1104/pp.99.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16(8):2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18(8):2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliavini M, Rombolà AD. Iron deficiency and chlorosis in orchard and vineyard ecosystems. Eur J Agron. 2001;15(2):71–92. doi: 10.1016/S1161-0301(01)00125-3. [DOI] [Google Scholar]
- Thiebaut F, Rojas CA, Almeida KL, et al. Regulation of miR319 during cold stress in sugarcane. Plant, Cell Environ. 2012;35(3):502–512. doi: 10.1111/j.1365-3040.2011.02430.x. [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, et al. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14(6):1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ST, Sun XL, Hoshino Y, et al. MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.) PLoS ONE. 2014;9(3):e91357. doi: 10.1371/journal.pone.0091357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM, McInturf SA, Stein RJ. Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana. J Exp Bot. 2012;63(16):5903–5918. doi: 10.1093/jxb/ers239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H-H, Orbach MJ, Hirsch AM, et al. Meristem-localized inducible expression of a UDP-glycosyltransferase gene is essential for growth and development in pea and alfalfa. Plant Cell. 1999;11(12):2303–2315. doi: 10.1105/tpc.11.12.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138(4):750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TJW, Lin WD, Schmidt W. Transcriptional profiling of the Arabidopsis iron deficiency response reveals conserved transition metal homeostasis networks. Plant Physiol. 2010;152(4):2130–2141. doi: 10.1104/pp.109.152728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CQ, Liu T, Bai FX, et al. miRNAome analysis associated with anatomic and transcriptomic investigations reveal the polar exhibition of corky split vein in boron deficient Citrus sinensis. Mol Genet Genom. 2015;290(5):1639–1657. doi: 10.1007/s00438-015-1024-8. [DOI] [PubMed] [Google Scholar]
- Yelina N, Smith L, Jones A, et al. Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc Natl Acad Sci USA. 2010;107(31):13948–13953. doi: 10.1073/pnas.0911341107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YC, Yu Y, Wang CY, et al. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol. 2013;31(9):848–852. doi: 10.1038/nbt.2646. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang D, Sun W, et al. The adaptive mechanism of plants to iron deficiency via iron uptake, transport, and homeostasis. Int J Mol Sci. 2019;20(10):2424. doi: 10.3390/ijms20102424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Ding H, Zhu JK, et al. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011;190(4):906–915. doi: 10.1111/j.1469-8137.2011.03647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of the small RNA-Seq have been uploaded to the Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra/). The accession number is SRR5012112.