Abstract
Plant–microbiome interactions are significant determinant for plant growth, fitness and productivity. Depending upon the specific habitat, plants' microbial communities are classified as the rhizo-, phyllo-, and endospheric regions. Understanding the plant microbiome interactions could provide an opportunity to develop strategies for sustainable agricultural practices. There is a necessity to decipher the complex structural and functional diversity within plant microbiomes to reveal its immense potential in agriculture. The plant microbiota harbors enormous microbial communities that defy analytical methodologies to study dynamics underlying plant microbiome interactions. Findings based on conventional approaches have ignored many beneficial microbial strains, which creates a serious gap in understanding the microbial communications along with the genetic adaptations, which favors their association with host plant. The new era of next generation sequencing techniques and modern cost-effective high-throughput molecular approaches can decipher microbial community composition and function. In this review, we have presented the overview of the various compartments of plants, approaches to allow the access to microbiome and factors that influence microbial community composition and function. Next, we summarize how plant microbiome interactions modulate host beneficial properties particularly nutrient acquisition and defense, along with future agricultural applications.
Supplementary Information
The online version contains supplementary material available at. 10.1007/s12298-021-00927-1.
Keywords: Agriculture, Endosphere, Microbiome, Phyllosphere, Rhizosphere
Introduction
Last decade experienced substantial advancements in plant microbiome research owing to its influence on the overall plant productivity and development (Compant et al. 2019; Bhatt et al. 2020). Plant microbiome study aims to decipher the structural and functional diversity of microbial communities associated with a specific host plant in a particular habitat. Plant associated microbial communities are generally related to a specific set of gene pool interconnected with the host irrespective of taxonomy and functionality of the microbial diversity. Further, there is great variability in microbial diversity for different plant regions/organs for example the phyllosphere, rhizosphere, and endosphere (Dutta and Bora 2019; Liu et al. 2019, 2020). In general, plants employ diverse strategies such as presence of specialized structures like trichomes and hairs or secretion of secondary metabolites to favor and support microbial colonization. However, there is a need to investigate a wider range of plant species associated microflora to get an insight into the potential of microbiome in enhancing the crop productivity. Attempts have been made to uncover plant-microbe interactions by studying the plant organs enriched with specific microbial species/genotypes using modern technologies (Schlaeppi and Bulgarelli 2015; Müller et al. 2016).
A holistic approach for understanding the microbial community and its association with plant is an emerging area of research and has several dimensions well taken by several researchers globally (Bulgarelli et al. 2013; Levy et al. 2018). It has been reported that diversity in plant microbial community is dependent not only on host species and selection pressure but also on developmental stage and environmental conditions. Various researchers have targeted the unexplored niche to elucidate the potential role of microbial community in enhancing plant resistance towards stress (reviewed in Teixeira et al. 2019). Agler et al. (2016) have observed that microbiome composition and its interactions with other microbial community are dependent on host genetic constitution, abiotic factors, and the function of specific microbial species or family. It has been reported that plant genetic makeup acts on keystone microbial populations, which communicates these effects to the entire microbial population by modifying microbe–microbe interactions and thus changing the overall plant performance (Agler et al. 2016). This finding clearly indicates that there is a need to have more detailed information of microbial compositions of plants' to unravel the underlying mechanism of microbiome assembly (Fitzpatrick et al. 2020; Trivedi et al. 2020). There have been substantial enhancements in technologies to study the microbial composition of a particular plant microbiome in totality. The conventional cultivation-based methods have limitations which provides only a partial outlook of microbiomes. Also, they are not sensitive enough to notice changes in the microbial community. New sequencing technologies have immense potential to understand the functional plant microbiome.
Sequencing techniques such as sanger's method, with various modifications: 454 pyrosequencing, Illumina (reverse dye terminator), PacBio (phospholinked fluorescent nucleotides), and Ion Torrent (proton detection) have been used to detect and analyse microbial population, belonging to even extremely rare taxa, extracting information beyond the results provided by specific microbes (Fadiji and Babalola 2020). Although each sequencing method varies in upstream and down-stream inspection, they all share common prospective to discover uncommon microorganisms in specific niche. By performing phylogenetic analysis of gene sequences present in a particular population, one can gain insight about type and diversity of the microbial communities. However, they do not distinguish between live and dead cells, and possibly carry sequence errors that often lead to overestimation of microbial diversity. Modern technical advancements utilize specific tagging of DNA molecule to manage PCR errors and biases (Jongman et al. 2020).
There are several key questions regarding plant microbiomes like how host plant gain microflora. Whether definite microbial communities are transferred generation to generation or they are engaged from the environment? In addition, the potential makeup of the least known microbial communities needs to be deciphered in light of technological advances. Despite the clear functions of microorganisms in plant fitness, we still need more detailed findings regarding the connection of microbiome with plant health. Such an effort in this field provides the opportunity for a better understanding of the microbial community regarding shifts in environmental conditions and adopting appropriate agricultural practices in coherence with the proper functioning of plant microbiomes. In this review the strategies to sample, process and analyze the plant microbiome by using novel molecular approaches, understanding the significance of the functional microbiome that are beneficial to plant health along with future prospects are discussed.
Plant associated microbial communities
Plants are associated with a plethora of microbial communities showing diversity in terms of number and types. The interactions of microbes with their host are very crucial not only for the plant development but also for existence of plant variety in natural habitats (Schlaeppi and Bulgarelli 2015). Plant associated microbes assist in seed germination and vigor, cellular development, stress tolerance, nutrient uptake, improved productivity and synthesis of therapeutically important compounds (Compant et al. 2019; Yadav et al. 2020). Owing to this close association, the host plants completely rely on their microbiome for definite task or trait. Plant microbiome also includes an array of biological datasets including genetic information, transcripts, proteins and metabolites which represent the host associated microbial communities (Busby et al. 2017; Singh et al. 2020). Still, in depth studies and competent techniques to decipher the microbes associated with plants in totality is needed.
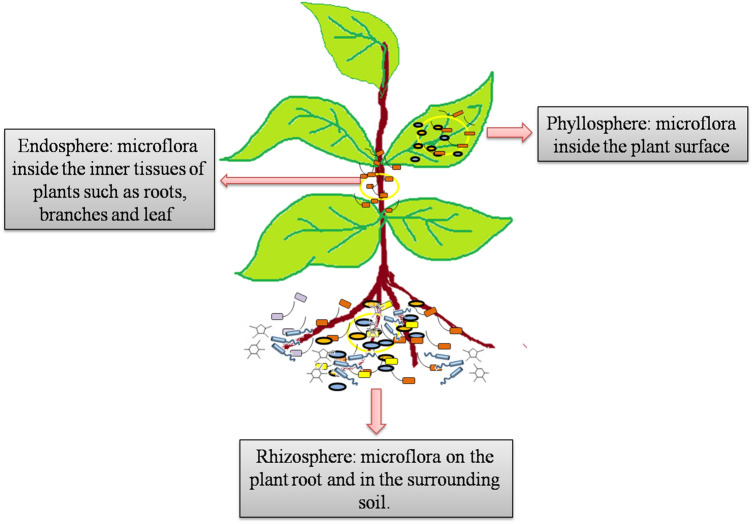
Microbial communities are dispersed on each and every sphere (air, water and soil) on earth and can easily interact with plant and different compartment of plant parts which possessed their own microbial communities. It is a new consensus that the microbial dynamics is highly variable and totally relies on the plant species, stage of development and genetic variation. The conditions experienced by host plants in their life span can also intervene the modulation of microbial diversity by the activation of host innate immune system. Moreover, the compromised immunity of host plants driven by several biotic and abiotic stresses can also affect the diversity of its microbiome. Therefore, in order to explore the efficacy of these factors on the microbial matrix at functional as well as phylogenetic level, it is important to analyze the heterogeneity of plant microbiome based on different niche individually (Fig. 1).
Fig. 1.
Schematic illustration of the major sources for various microflora that form the plant-associated communities: the phyllosphere, endosphere, and rhizosphere
Rhizosphere
The soil ecosystem is the primary niche of microbes exhibiting about 108 to 109 taxa g−1 of soil (Huang et al. 2014). The rhizosphere is the area of soil around the plant root which is regularly affected by the rhizo-depositions i.e. root exudates, mucilages and sloughed cells (Paterson et al. 2007). The complex of microbial population inhabiting the rhizosphere is quite different than the bulk soil microbial communities as evidenced by two different patterns of recruitment: primary is the impact of the root secretomes, enhancing the availability of nutrients and secondary the effluence of enhanced microbiome which alters rhizospheric environment. In consequence of root secretomes, some molecules are common for the initiation of metabolic activity of soil inhabiting microbes whereas, few are subjected to attract the specific microbial communities (Paterson et al. 2007).
Efforts are being made to understand the taxonomy based selection criteria of rhizospheric microbiome along with the role of the microbial community in rhizosphere. It has been reported that several plant species possess different type of communities in the same soil (Berg and Smalla 2009; Leff et al. 2018). Even different genotypes of same species can also have different rhizospheric microbial communities. These studies reveal that plants are able to design their own microbial dynamics in the rhizosphere. However, higher variations in microbial dynamics observed at different rhizospheric environment reveal high unstablilty at genetic level. The recent higher availability of genetic data of rhizospheric microorganisms lead to explore the unique metabolic pathways along with higher genetic drift even in the similar species. It reveals that the genetic variation in microflora is the consequence of life style, multifarious interactions between other communities, co-evolution with other organisms existing in same environment and impact of environmental conditions (Andreote et al. 2014). The interaction between microbial communities is the main driving force for the horizontal gene transfer (HGT). However, a possibility exist that the mutual interaction between plant and microbes can lead to the re-modulation of genomes. In that case, the host plant selects highly susceptible microbial community for reframing the genetic material by imposing the local condition and finally acquires novel metabolic features. Rhizospheric region is known to be highly susceptible region for genetic level communication. It could possibly be due to the presence of higher complexity of microbial communities. Moreover, the higher abundance of viruses, plasmids and other highly transmitted genetic materials at plant borders can also provide several novel insights for microbiome.
Endosphere
The existence of endophytes in the inner plant tissues is known to be necessary for their existence (Singh and Gaur 2017; Adeleke and Babalola 2020). The cross talk between plant and endophytes to establish symbiotic relationship is still debatable. The endosphere differ extensively from the rhizosphere, which maintains the perception of coevolution among plant and microbial symbionts, in which the host is involved in recognition and selection of microbial community that endorse a homeostatic association with the plant (Compant et al. 2010). Recently, researchers are beginning to identify appropriate molecular techniques that can be applied to study the ability of microbial diversity of the plant microbiome colonizing the plant tissues. The microbial colonization process that occurs within host involves exudate-signaling communications through plant–microbe interactions (Hardoim et al. 2015). To support above stated fact, chemotaxis mechanism has been recognized in a number of microbes, and expression of motility genes is considered to be a prerequisite step towards adherence and colonization of the microbes (Santoyo et al. 2016). Here again, one critical question that arises at the nub of the signaling pathways of the plant microbiome is how the host identifies microbe showing mutualism against those acting as pathogens. This comprises complex communications between plants and microbes that trigger plant immune responses (Liu et al. 2019).
One perplexing influence that endopsheric microbes can express on the plant genome is articulated through horizontal and vertical transmission of the symbionts. Horizontal transmission allows host-to-host transfer of endosymbionts without involvement of plant sexual reproduction whereas vertical transmission relies upon host fitness and is well characterized in fungi as those indicating an asymptomatic lifestyle in the host (Verma et al. 2017). Majority of plant endophytes are known to be horizontally transmitted and present a possible clash of health due to antagonistic coevolution of functional trait expression between the plant–microbe symbionts. Vertically transmitted endophytes provide better host benefits than horizontally transmitted as correlated with plant density (Sneck et al. 2019). The importance of endosphere microbiome components indicating noticeable plant-growth promoting genes in addition to genes involved in establishment, persistence, and thriving of plant-beneficial microbes are of considerable attention to biotechnology and agriculture.
Phyllosphere
It represents an aerial part of the plant and considered to be third most prominent compartment enriched with microbial community (Parasuraman et al. 2019). Phyllosphere microflora are known to be associated with plant growth in terms of N2 fixation, biosynthesis of various phytohormones and protecting plant against pathogenic intruders (Cappelletti et al. 2016). The microbial communities found in phyllosphere are vulnerable in stringent conditions like restricted availability of organic substances; variable pH, O2 concentration, temperature, UV, humidity etc. (Thapa and Prasanna 2018). Bacterial communities are predominant in this region though significant alterations in microbial dynamics are observed due to the great versatility in the nutritional depositions. It has been reported that narrow and wax containing leaf plants generally have less microbial load as compared to other regions (Remus‐Emsermann and Schlechter 2018). Further, it has been observed that different microbial communities are associated with plants at specific sites presumably because of difference in light or UV intensity, air flow rate, humidity etc. For instance, pigment producing bacterial strains are mostly found at the epiphytic region whereas, mineral and humic acid utilizing bacterial communities are found at rhizosphere. Another important factor for microbial colonization at phyllosphere is mainly the origin of microbes. It is evident that a particular microbial community might be observed at a distant place from the plant habitat but their spores might migrate through the flow of wind and colonize the aerial part of the plant by means of vectors which were horizontally transformed in the microbial cells during co-evolution (Bulgarelli et al. 2013). Plant genotypic variation is also a significant driver of microbial diversity found at the phyllosphere. Several plant species that are found in the same habitat and environmental conditions have specific microbial communities owing to the genetic and metabolic diversity (Laforest-Lapointe et al. 2016). Geographical parameters also drive a constitutive role in the designing of the microbial matrix which influences the quality of the end products formed by host plant (Copeland et al. 2015). These fluctuations arise due to the variations in carbon substrates viz., amino acids and carbohydrates as well as nutrients. Biofilm formation is a general trait of phyllospheric bacterial communities which is well known phenomenon to induce the heterogeneity of phyllospheric microbial communities by protecting the microbial cells under adverse circumstances (Bridier et al. 2017). Despite the above stated facts, the most common phyllosphere colonizing bacterial communities belongs to Proteobacteria, Actinobacteria, Bacteroidetes and Firmicutes (Darlison et al. 2019). The predominance of these bacterial communities in the phyllosphere needs to be investigated by considering diverse plant species under variable environmental conditions. Further, there is a need to develop appropriate technology for assessing the microbial diversity for rhizosphere, endosphere and phyllosphere compartment of host plants in totality. Identifying the key microbial communities associated with different regions might be beneficial for the developing strategies for enhancing crop productivity and promoting sustainable agriculture.
Summary of plant microflora associated to different compartments of host plants are listed in Supplementary Table S1.
Conventional techniques for assessing the plant associated microbiota
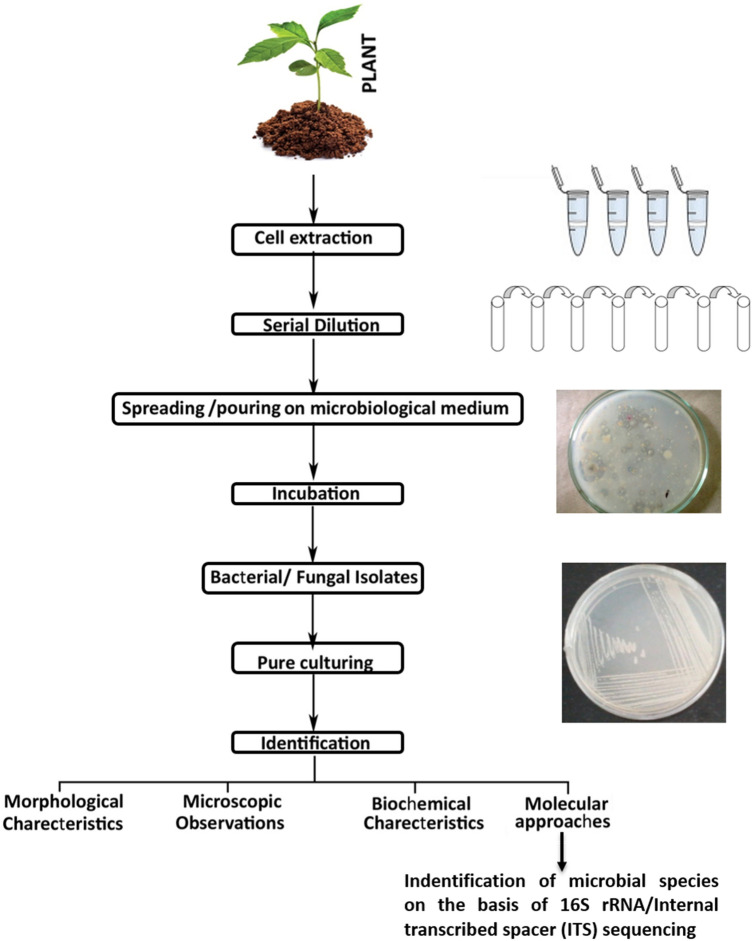
Although only a minor portion of microbial communities are known to be culturable, isolation, characterization and biochemical identifications of these are very feasible and convenient and is applicable to diverse microbial communities from different environmental samples (Fig. 2). The major limitation of this technique is that it does not represent the total microbial communities and only those which are culturable are represented. Apart from the limitations, culture-dependent techniques have several benefits. It allows culturing the microbes in laboratory conditions and purification of single strains from a group of microbes present in environmental samples. Over 150 years, researchers have been exploring the microbial diversity present at natural habitat by applying several culture dependent techniques. With the use of traditional plating techniques, several microbiologists have performed advanced selective enrichment techniques for the isolation and enumeration of microbes in selective or enriched medium under laboratory conditions (Hahn et al. 2019). This has led to the recovery of wide range of pure cultures from the rhizospheric, endophytic and phyllospheric regions of plants grown at different natural habitats. Finally attempts have been made to reveal the diversity by adopting traditional taxonomic parameters to the level of species.
Fig. 2.
Outline of culture-dependent techniques for evaluating the microbial communities associated with different parts of plants
However, investigators revealed that above mentioned traditional approaches for the analysis of microbial diversity have uncertainty due to several physiological and morphological variations. Hence there is a need to develop molecular tools which could be more precise and can overcome the limitations of the traditional methods used to decipher the microbial diversity. Hereafter, research on microbial diversity by several molecular approaches took place in past two decades which helped to combat these difficulties. In this direction, methods based on sequencing seem to be promising as it has the potential to reveal the phylogenetic relationship. This technique also facilitates secure taxonomic structures along with putative identification that are based on evolutionary variations at molecular level.
Modern approaches for assessing functional plant microbiome
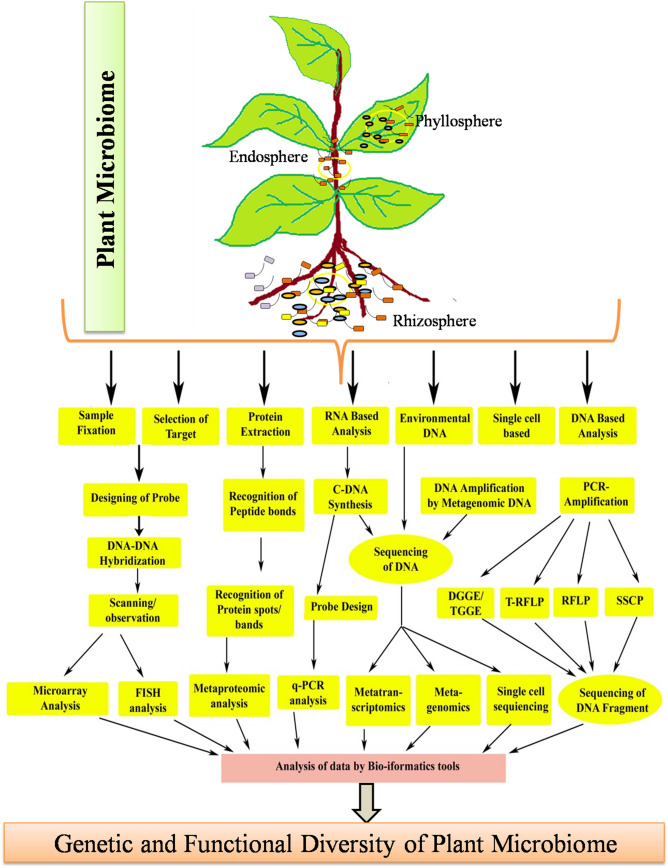
The huge diversity of uncultivable microorganisms in microbial communities associated with plant microbiome is a major limitation in selecting the appropriate methodology for identification (Lareen et al. 2016). However, a number of culture-independent approaches developed in the recent years provides valuable data, and are presently being useful either to interpret the unseen microbial diversity in plant environments, or to discover the molecular techniques revealing functional plant microbiome. In the current scenario of scientific revolution these molecular technique based methodologies are necessary to assess the effects of perturbations triggered by various stresses on diversity of microflora associated with plant. A great advancement in culture independent techniques for analyzing plant microflora relies on direct environmental DNA cloning (del Carmen Orozco-Mosqueda et al. 2018). Several culture independent molecular biology based techniques like terminal restriction /restriction fragment length polymorphism (T-RFLP/RFLP), single strand conformation polymorphism (SSCP), denaturing/temperature gradient gel electrophoresis (DGGE/TGGE), fatty acid methyl esters (FAME) and next generation sequencing (NGS) technology have been reported to be used for plant-associated microbial community studies (Gupta et al. 2018; Bodor et al., 2020). Application of other omics techniques, like metatranscriptomics, metaproteomics, metabolomics, or even single-cell genomics can support the development of new enzymes/proteins, supported by recombinant DNA technology as well as metabolic engineering. The advancement of these modern culture-independent techniques have substantially been applied in diverse microbial diversity studies for genetic data based microbial distribution (metagenomics) without disrupting the texture of ecosystem. Furthermore, the new era of advanced techniques also facilitate the analysis of intricate cases like, functional drift of microbial matrix in response to various stresses (Risely 2020). The genomic studies of environmental samples help us to unwind the large and complicated structure of ecology through the diversity of associated microbial communities.
The initial step of the culture-independent method involves processes for isolating DNA/RNA from different parts of plant samples. The total DNA isolated from different microbial communities associated with a plant constitutes an entity designated as a metagenome. The omics based approaches for assessment of this metagenome is referred as metagenomics and this could provide an insight into the phylogeny of microflora in specific ecological communities (Fadiji and Babalola 2020). The schematic representation of diverse approaches and methodology for culture-independent techniques for assessing microbial communities associated with a targeted host plant is shown in Fig. 3. To obtain consistent metagenomics data, preparation of a metagenomics library is important. Metagenomics library can then be selected for secondary metabolites and enzymes profiling. The quantitative PCR (qPCR) using microbial DNA extracted from plant samples as template has a great potential to assess diversity in microflora populations. The gene based identification of 16s/18s rRNA (ribosomal RNA) are target molecular markers for assessment of diversity, richness, evenness, and structure of microbial communities. Further, there have been several technological innovations like molecular fingerprint methods involving cloning and sequencing of PCR amplicons to access the structure of a target microbial community. These approaches are now being facilitated by the advancements in NGS resulting in the generation of massive quantity of nucleotide sequences (Agrahari et al. 2020). Advancements in sequencing technologies along with bioinformatics intervention by developing appropriate softwares for analysis has expedited the structural and functional assessment of miocrobiome associated with a plant. The diverse sequencing technologies, bioinformatics approaches used in metagenomics and for the study of community genomics is reviewed recently (Fadiji and Babalola 2020).
Fig. 3.
Molecular approaches applied to decipher the microbial diversity associated with plants
The functional microbial diversity can be studied by targeting particular genes and markers linked to the desired metabolic processes. For functional level studies, some insights on microbial processes are required, hence transcript abundance based method viz., qRT-PCR is very useful. To access the transcript expression profile from different genes, functional gene arrays have been employed to evaluate specific functional microbial activities. In this regard, DNA microarrays containing ecological cDNA based shotgun sequencing has been developed to analyze the several responses of plant microbiome at transcriptional level under various conditions (Bharati et al. 2020). However, this method can show only the species not the functions of microflora such as genomes and protein levels, which require assessment by another techniques. Metagenomics can also be combined with stable-isotope probing which can generate useful data on the metabolic properties of specific species in the microbial community (Achouak and Zahar Haichar 2019). In plant system, isotope labelling can help to recognize microflora that actively use plant secondary metabolites. Recently, sequencing-based metatranscriptomics techniques (NanoString technology based on hybridization) have been developed for the enrichment and identification of microbial transcripts (Goytain and Ng 2020).
Different methodologies are currently applied to understand the molecular basis of interactions among plant microbiome. A basic concept is that the various functions of microbial communities affecting plant functioning can be explored by high throughput plant phenotyping, however, the impact of plant, microbial and environmental factors on the functioning and diversity of microbial compositions can be analyzed by various molecular ecology techniques. The culture-independents techniques have immense potential for studying microbial communities associated with a plant but suffers from few limitations and hence cannot fully substitute conventional techniques. The less availability of assembled microbial species genomes, absence of functional and community dynamics data, hitches with data analysis owing to huge frequency of natural community inconsistencies and discerning between inter- and intra-species dissimilarities (Wolfe 2018).
Dynamics of plant–microbiome interactions
Plant microbiome interaction is considered highly complex and dynamic process. Plants and their interactions with microbes have usually been considered with respect to individual or small collections of microorganisms. Also, the research has been inclined towards the impact of microorganisms on plant fitness and response rather than plant factors that influence the selection of microbes. However, the plant microbial community is affected by the reciprocal nature of interaction between the host plant and its microbiome. It is important to define how specific microbes and their host plant may impact the microbial community. It has been suggested that apart from plant and its microbes, various environmemtal and abiotic factors such as climate, soil type and structure, human practices etc. are also important in selection of plant microbiome.
The type of plant species has pronounced impact on the identity of its microbiome associated with rhizosphere, endosphere and phyllosphere. Different plant species growing close to one another can have different microbiomes. Different members of family Poaceae, namely, wheat, sorghum and maize possess distinct rhizobacterial community (Bouffaud et al. 2014). It was found that, more the phylogenetic distance between the host plants more distinct is the associated rhizospheric bacterial community. When rhizospheric community of grape vine and weed species growing close to one another was analyzed, it has been observed that these plants harboured different microbiomes in the rhizosphere (Samad et al. 2017). Microbiome associated to weeds showed more plant growth-promoting characteristics compared to grapevine microbes. Not only rhizosphere but phyllosphere and endosphere microbial community is also influenced by the plant species. It has been observed that bacterial community present on the tree leaves of neotropical forest and fungal endophytic community of three Nicotiana species was highly influenced by host plant evolutionary relatedness (Dastogeer et al. 2018; Kembel et al. 2014).
Plant genotype significantly determines the associated microbial community. Findings suggest a difference in microbiome composition between genotypes of a specific plant species. A. thaliana genotype is an important factor in recruitment of root microbiome (Schlaeppi et al. 2014). Plant genotype dependent microbial community shaping has been defined for sweet potato, wheat, pea, soybean and oat (Turner et al. 2013; Zhong et al. 2019). Variations in plant genotype affects phenotyping properties for example root and leaf growth, their structure, and composition of exudate that in turn influence overall microbiome assembly. Even a minor genetic variation can impact plant microbiome. For example, introduction of the pm3b gene responsible for resistance to mildew had effect on pseudomonads and mycorrhizal colonization in the wheat rhizosphere (Meyer et al. 2013). Evidence also suggest that genetic variations that alter the host immune signaling pathway have an impact on microbiome assembly. Plants deficient in jasmonate mediated defense exhibited higher epiphytic diversity while stimulation of salicylic acid mediated defense response, reduced the endophytic diversity (Kniskern et al. 2007).A.thaliana mutants deficient of systemic acquired resistance (SAR) show differences in the bacterial communities near the root zone (Hein et al. 2008). Evidences suggest that the effects of various defenses signaling on the phyllosphere microbiome of A. thaliana are flexible. For example plants lacking in jasmonate mediated defense showed higher epiphytic diversity, while stimulation of salicylic acid mediated defense reduced the diversity of endophytes (Kniskern et al. 2007). In another study on Arabidopsis plants with disrupted jasmonate pathway, archeal and bacterial community distinct from wild type was reported. Mutant plants had abundance of Streptomyces, Bacillus, Lysinibacillus and Enterobacteriaceae and less abundance of Clostridiales population in the rhizosphere (Carvalhais et al. 2015). Some reports suggests alterations in the defense-related signaling, as revealed during pathogen attack on plant tissues, which can potentially affect the secretions of root exudates and hence the plant microbiome. The alterations in salicylic acid and jasmonic acid defense pathway significantly affected the natural microbiome of Arabidopsis (Kniskern et al. 2007; Reinhold-Hurek et al. 2015; Lebeis et al. 2015). A study also shows that exogenous production of glucosinolate significantly shifted the microbial community of the transgenic Arabidopsis root region (Bressan et al. 2009).
Plant roots exudates which include amino acids, phenolic compounds, organic acids, sugars, and other small secondary metabolites are another important factor in shaping microbiome especially rhizosphere. Root exudates differ substantially among various plant species and genotypes. These variations in exudates can either be substrate, chemotactic or signaling molecules and they can influence the structure and dynamics of microbial communities (Lareen et al. 2016). Diverse group of natural compounds released from the roots interact synergistically in recruiting the microbial communities (Hu et al. 2018; Jacoby et al. 2020). It has been reported that the selection applied in the root region can be driven not only by the plant species but, more precisely, by an integrated evolutionary mechanism directed by the host and associated microbiome interactions (Huang et al. 2014; Sasse et al. 2017). Therefore, the development of specific and sensitive profiling approaches is essential to identify such dynamic modifications in the plant microbiome.
Other than the factors discussed above, environmental factors, such as soils and climatic conditions could also influence plant associated microbial community composition (da Silva et al. 2018). Environmental changes results in alteration of plant physiology which ultimately results in distinct microbiome profile. Soil composition like, soil carbon, water content and carbon–nitrogen ratio may influence the microbiome composition (Peiffer et al. 2013). Soil characteristics not only influence microbial inoculum but also affect plant growth and morphology by influencing nutrient availability to plants. Rhizospheric bacterial community of A. thaliana is strongly influenced by soil characteristics (Schlaeppi et al. 2014). Microbial community composition of grapevine leaf and root has been shown as influenced by soil carbon, pH and carbon–nitrogen ratio (Zarraonaindia et al. 2015). Climatic variables like precipitation is also reported to influence bacterial and fungal community composition (Naylor et al. 2020). It has been observed that increasing mean annual precipitation increases bacterial and fungal load of the microbiome (De Vries et al. 2012).
Role of plant–microbiome interactions in agriculture
Nutrient acquisition
Microbiomes associated with their host plants have remarkable capacity to improve plant nutrition. There are large number of studies on plant symbiosis with arbuscular mycorrhizal fungi (AMF), and Rhizobium that benefit plant by nutrient acquisition (Dellagi et al. 2020). Besides, non-symbiotic beneficial, rhizospheric and endophytic fungi and bacteria can develop the root system architecture and increase the bioavailability of insoluble minerals to plants, thus enhancing the exploratory capacity of the host root for minerals and water (Tao et al. 2019). A recent finding revealed that differences in the nitrogen uptake of rice varieties Oryza indica and O. japonica is due to the higher abundance of nitrogen-solubilizing bacteria, leading to better efficient nitrogen transformation processes in the root of indica variety over to japonica (Zhang et al. 2019). Interestingly, using indica-enriched SynCom (16-member) substantially provided larger shoot and root growth of an indica variety than a japonica-enriched SynCom (3-member) in the presence of organic nitrogen. These results suggested that plant–microbiome cooperation effectively contribute to enhanced transformation of organic nitrogen in the host rhizosphere for mutual benefit. Lu et al. (2018) suggested that rhizospheric microorganisms that enhanced nitrogen availability via nitrification, delay time of flowering and induce plant growth via conversion of tryptophan to indole acetic acid that represses genes responsible for flowering. A recent study revealed that different types of mycorrhizal fungi, arbuscular vs. ectomycorrhizal colonization of plants, signify adaptations to high vs. low nutrient acquisition (Averill et al. 2019). Hestrin et al. (2019) observed that multifaceted microbial synergies between mycorrhizal fungi, Rhizophagus irregularis and soil microbial communities, resulted tenfold rise in the nitrogen acquisition in Brachypodium distachyon compared to those without soil microbial communities.
Plant associated microbiomes can transport, solubilized and mineralized nutrients such as inorganic phosphate and iron that are not freely available to host. Similar to mutualistic association, under low inorganic phosphate conditions, host depend on microbial cooperation partners viz., AMF and their symbiotic endophytes, to fulfill needs for availability of phosphate (Bodenhausen et al. 2019). Castrillo et al. (2017) used reconstituted microbial communities of different complexity which enhanced the uptake of inorganic phosphate by host plants via inducing transcription of host genes responsible for phosphate starvation responses. Recently it has been observed that fine-tuned relationships between plant immunity and nutritional homeostasis of host regulate the beneficial traits mediated by the microbiome (Herlihy et al. 2020). For example, coumarins (plant secondary metabolites) structured root bacterial community of A. thaliana particularly by facilitating iron mobilization and inhibiting the growth of Pseudomonas strain (that competes with host plant for nutrient) via producing reactive oxygen species (Voges et al. 2019). Interestingly, Stringlis et al. (2018) showed the volatile compounds from induced systemic resistance (ISR) activating rhizobacteria and Trichoderma trigger the expression of MYB72 (root-specific transcription factor) in order to stimulate iron uptake by plant roots. The findings of the work discussed above shed light on the genetic and molecular cues dictating microbiome assembly on nutrient availability and open up various prospects to extrapolating these results to identify the genetic and metabolic pathways involved in nutrient acquisition.
Plant immunity
The impact of resident microbiome based plant immunity on plant can occur via ISR or enhanced activation, whereby natural microbiota activate the plant immune system in order to induce resistance to phyto-pathogens (Fitzpatrick et al. 2020). Direct inhibition of fungal phyto-pathogens by the plant microbiome can also improve plant resistance, independent of the plant's inherent immune system (Santhanam et al. 2019). There are considerable amount of findings of diseases resistance against pathogen via the production of antimicrobial compounds such as coumarin (Stringlis et al. 2018), benzoxazinoids (Hu et al. 2018) and lytic enzymes as well as pathogen-inhibiting volatile compounds (Carrión et al. 2018; Vannier et al. 2019). Antimicrobial production is widespread among phyllosphere bacterial communities and protect the plants from pathogens. Berg and Koskella (2018) stated that phyllosphere microbiota mediated protection depend on competitive dynamics along with antimicrobial compounds. Recently, the role of rhizospheric bacteriophages was observed in plant health and protection (Pratama et al. 2020). The classical research emphasis is disease suppressive soils (Schlatter et al. 2017), where the resident microorganisms inhibit the pathogen growth (Trivedi et al. 2017). Additionally, management practices viz., rotation of crop, soil transplantations, residue retention and additions of compost can also stimulate disease suppression by altering microbiome composition (Wei et al. 2019). Disease-resistant variety of a tomato have been develop by transplantation of rhizospheric microbiota that can suppress the wilt pathogen, Ralstonia solanacearum from resistant plants to susceptible plants (Kwak et al. 2018). The mechanisms responsible for microbiome mediated stress tolerance against abiotic stress remain unknown. Plants generally select a stress tolerant microbiome under stress conditions. The plant hormone abscisic acid inhibits the plant immune response under drought conditions, thus allowing large endophytic community shifts in the root (Fitzpatrick et al., 2018). Thus, the alterations in the plant via plant hormone production and host biochemical activity alleviate water stress. Plants respond to various stresses by activating the ethylene signaling pathway that causes growth rate and stress resistance trade-offs (Ravanbakhsh et al. 2018). These highlight the exciting opportunity of reconstituting microbiome construct based on knowledge of the cultivated microorganisms in disease-suppressive soils to manage diseases in natural condition. In summary, findings based on SynComs under controlled conditions demonstrates that plant defense system can be influenced by the recruited microorganisms in response to various stresses that offer established plant immunity over manifold generations. Manipulation of microbiome associated with plants to enhanced plant immunity will continue to entail in a depth knowledge of the plant–microbiome–environment interactions among wide range of crops.
Plant microbiome in sustainable agriculture
Implication of the plant microbiome in agriculture will require a comprehensive understanding of the interactions between host, microbes and environment at functional and mechanistic levels. The elucidation of specific role of plant–microbiome interactions in agriculture by influencing plant health and productivity needs appropriate experimental approaches and advanced characterization techniques as reviewed in recent years (Singh et al. 2020; Jongman et al. 2020). Large number of microbes still remains unidentified at physiological and molecular levels. New high-throughput approaches that enable empirical and theoretical basis of microbial community structure and function across environmental variation are necessary for filling these information gaps. Understanding the plants microbiome interactions is particularly important for elucidating its diverse application in agriculture such as nutrient acquisition, enhancing disease resistance and stress tolerance (Trivedi et al. 2020). Large-scale experimental systems are necessary to reveal the dynamic microbial functions that drive microbial colonization in various plant parts such as rhizosphere, rhizoplane, and endophytic compartments along with alterations in plant–microbiome interactions. Understanding these intricate complex interactions to identify community structure and function could lead to deciphering of potential mechanisms for enhancing resistance in synthetic communities and thereby providing prognostic tools for farmers and agricultural agencies (Arif et al. 2020).
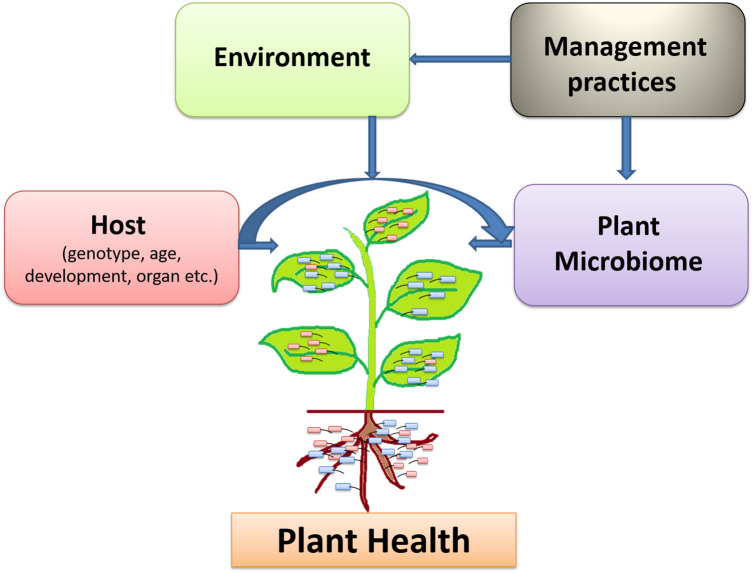
Defining the goal of core microbiome depends upon host’s genetic make-up and environmental challenges of the host plant. Microbiome with beneficial properties may enhance overall plant productivity and host immunity to manage variety of challenges. The environmental challenges faced by a plant can be controlled by judicial use of local beneficial microbiomes, soil additives, tillage, and cropping systems. Therefore, the cross talk among the microbes, environment, and management practices seems to be prerequisite for engineering microbiome structure to enhance overall plant health and production. Host genotype-by-environment interactions in the microbiome may be influenced by different environments (including abiotic and biotic) and diverse plant tissues. Microbiome engineering in terms of microbial consortium resilience also depends on various factors of management practices (Dastogeer et al. 2020). Thus, an important challenge is to design microbiome treatments that can persist in broader environmental variability.
Well-designed experiments based on host genotype/organ/age/plant derived compounds, environmental variations (climatic variables), microbiome and management tactics are necessary to unravel the interactions among them. To understand which interactions among microbiome and environment offers better productivity, experiments with synthetic microbiome (combination of beneficial microorganisms into agricultural and breeding management practices) need to be performed. The application of SynComs in breeding management practices need to be applied in order to develop beneficial microorganism-optimized plants. Selection of microbiome assembly depending upon the host genotype and various environmental (abiotic and biotic) factors could provide a guideline for elucidating application of beneficial microbial communities towards different stress tolerance. The results based on the microbiome engineering needs to be validated at field level, across a large scale of cultivation practices, soil, climates variables and management practices prior to its implementation for sustainable agriculture. The application of various experiments based on new molecular and analytical approaches will expedite this upcoming area of research of plant microbiome and will pave way for transformation of basic research to application-oriented research directly benefitting the farmers.
Future perspectives and information gaps
Despite the enormous potential of plant microbiome interactions in agriculture, the associated microbial communities and their interactions are poorly investigated. Substantial experimental work carried out by different researchers globally notion the significance of the microflora associated with plant in enhancing plant productivity and also the possibility of plants manipulating specific microbiome. This might be particularly interesting that plants occupying extreme natural ecosystems have the potential to control their microbiome in light of evolutionary selection. A plethora of studies associated with plant microbiome have been carried out but still fundamental and practical methods to explain the processes leading to community and function in and on plants are required. Metagenomic analysis of the plant microbiome have until now focused on phylogenetic and functional insights, leading to partial information on the presence of specific operational taxonomic units. Future research will bring many innovative insights into the selective factors that shape the plant microbiome based on the advancement of next-generation sequencing techniques helping us to understand the interplay between the plant-microflora. Improved knowledge of the plant microbiome interactions could fuel advances in sustainable agriculture such as the development of microbial inoculants as biocontrol and biofertilizer agents. Technological innovations in metagenomics research will deliver insights into the activities of functional plant microbiome. The ultimate aim of researchers in this field is to unravel the mechanisms through which host plants manage their microbiome and vice-versa as this could provide immense opportunities for improvement in agriculture mainly focusing on improved crop yield and developing biotic and abiotic stress tolerant crops. A deep knowledge of the whole plant microbiome community communications and dynamics has the potential to allow more efficient utilization of this largely unexploited resource. It is quite evident that various approaches are now being utilized to address various queries associated with plant microbiome research and its potential for promoting sustainable agriculture. This review has focused on some important concerns in microbiome research for various plants preventing the development of model host–microbiome systems. Explanation of plant microbiomes in these standard systems has the potential to reveal the guidelines of artificially and, functionally define microbiome assembly, to delineate functional plant microbiome interactions and illustrate interactions between host (genotype/species/age)-environment (climatic variables)-cultivation practices-microbiome (Fig. 4). In conclusion, the holistic approach in the area of plant microbiome especially upcoming functional plant microbiome with the aid of state-of-the art technologies could be applied for enhancing crop productivity in sustainable manner.
Fig. 4.
Interactions of microbiome with host, environment, and management practices to overall strengthen plant health
Supplementary Information
Below is the link to the electronic supplementary material.
Compliance with ethical standards
Conflict of interest
All authors declare that there exists no conflict of interest among them.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rupali Gupta and Gautam Anand have contributed equally to this work.
References
- Achouak W, Haichar FZ. Stable isotope probing of microbiota structure and function in the plant rhizosphere. In: Dumont MG, García MH, editors. Stable isotope probing. New York: Humana; 2019. pp. 233–243. [DOI] [PubMed] [Google Scholar]
- Adeleke BS, Babalola OO (2020) The endosphere microbial communities, a great promise in agriculture. Int Microbiol. 10.1007/s10123-020-00140-2 [DOI] [PubMed]
- Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, Weigel D, Kemen EM. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016;14:e1002352. doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrahari RK, Singh P, Koyama H, Panda SK. Plant-microbe interactions for sustainable agriculture in the postgenomic era. Curr Genom. 2020;21:168–178. doi: 10.2174/1389202921999200505082116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreote FD, Gumiere T, Durrer A. Exploring interactions of plant microbiomes. Sci Agríc. 2014;71:528–539. [Google Scholar]
- Arif I, Batool M, Schenk PM. Plant microbiome engineering: Expected benefits for improved crop growth and resilience. Trends Biotechnol. 2020 doi: 10.1016/j.tibtech.2020.04.015. [DOI] [PubMed] [Google Scholar]
- Averill C, Bhatnagar JM, Dietze MC, Pearse WD, Kivlin SN. Global imprint of mycorrhizal fungi on whole-plant nutrient economics. Proc Natl Acad Sci. 2019;116:23163–23168. doi: 10.1073/pnas.1906655116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS microbiology ecology. 2009;68(1):1–13. doi: 10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- Berg M, Koskella B. Nutrient-and dose-dependent microbiome-mediated protection against a plant pathogen. Curr Biol. 2018;28:2487–2492. doi: 10.1016/j.cub.2018.05.085. [DOI] [PubMed] [Google Scholar]
- Bharati AP, Kumar A, Kumar S, Maurya DK, Kumari S, Agarwal DK, Kumar SJ. Role of biotechnology in the exploration of soil and plant microbiomes. In: Solanki MK, Kashyap PL, Kumari B, editors. Phytobiomes: current insights and future vistas. Singapore: Springer; 2020. pp. 335–355. [Google Scholar]
- Bhatt P, Verma A, Verma S, Anwar M, Prasher P, Mudila H, Chen S. Understanding phytomicrobiome: a potential reservoir for better crop management. Sustainability. 2020;12:5446. [Google Scholar]
- Bodenhausen N, Somerville V, Desiro A, Walser JC, Borghi L, van der Heijden MG, Schlaeppi K. Petunia-and Arabidopsis-specific root microbiota responses to phosphate supplementation. Phytobiomes J. 2019;3:112–124. [Google Scholar]
- Bodor A, Bounedjoum N, Vincze GE, Kis ÁE, Laczi K, Bende G, Szilágyi Á, Kovács T, Perei K, Rákhely G. Challenges of unculturable bacteria: environmental perspectives. Rev Environ Sci Biol Technol. 2020;19:l1–22. [Google Scholar]
- Bouffaud ML, Poirier MA, Muller D, Moënne-Loccoz Y. Root microbiome relates to plant host evolution in maize and other Poaceae. Environ Microbial. 2014;16:2804–2814. doi: 10.1111/1462-2920.12442. [DOI] [PubMed] [Google Scholar]
- Bressan M, Roncato MA, Bellvert F, Comte G, el ZaharHaichar F, Achouak W, Berge O. Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J. 2009;3:1243. doi: 10.1038/ismej.2009.68. [DOI] [PubMed] [Google Scholar]
- Bridier A, Piard JC, Pandin C, Labarthe S, Dubois-Brissonnet F, Briandet R. Spatial organization plasticity as an adaptive driver of surface microbial communities. Front Microbial. 2017;8:1364. doi: 10.3389/fmicb.2017.01364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Ann Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A, Morsy M, Eisen JA, Leach JE, Dangl JL. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017;15:e2001793. doi: 10.1371/journal.pbio.2001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelletti M, Perazzolli M, Antonielli L, Nesler A, Torboli E, Bianchedi PL, Pindo M, Puopolo G, Pertot I. Leaf treatments with a protein-based resistance inducer partially modify phyllosphere microbial communities of grapevine. Front Plant Sci. 2016;7:1053. doi: 10.3389/fpls.2016.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrión VJ, Cordovez V, Tyc O, Etalo DW, de Bruijn I, de Jager VC, Medema MH, Eberl L, Raaijmakers JM. Involvement of Burkholderiaceae and sulfurous volatiles in disease-suppressive soils. ISME J. 2018;12:2307–2321. doi: 10.1038/s41396-018-0186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalhais LC, Dennis PG, Badri DV, Kidd BN, Vivanco JM, Schenk PM. Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol Plant Microbe Int. 2015;28:1049–1058. doi: 10.1094/MPMI-01-15-0016-R. [DOI] [PubMed] [Google Scholar]
- Castrillo G, Teixeira PJPL, Paredes SH, Law TF, de Lorenzo L, Feltcher ME, Finkel OM, Breakfield NW, Mieczkowski P, Jones CD, Paz-Ares J. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543:513–518. doi: 10.1038/nature21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S, Clément C, Sessitsch A. Plant growth-promoting bacteria in the rhizo-and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem. 2010;4:669–678. [Google Scholar]
- Compant S, Samad A, Faist H, Sessitsch A. A review on the plant microbiome: ecology, functions and emerging trends in microbial application. J Adv Res. 2019;19:29–37. doi: 10.1016/j.jare.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland JK, Yuan L, Layeghifard M, Wang PW, Guttman DS. Seasonal community succession of the phyllosphere microbiome. Mol Plant Microbe Interact. 2015;28:274–285. doi: 10.1094/MPMI-10-14-0331-FI. [DOI] [PubMed] [Google Scholar]
- da Silva FE, Peixoto RS, Rosado AS, de Carvalho BF, Tiedje JM, da Costa Rachid CTC. The microbiome of Eucalyptus roots under different management conditions and its potential for biological nitrogen fixation. Microbial Ecol. 2018;75:183–191. doi: 10.1007/s00248-017-1014-y. [DOI] [PubMed] [Google Scholar]
- Darlison J, Mogren L, Rosberg AK, Grudén M, Minet A, Liné C, Mieli M, Bengtsson T, Håkansson Å, Uhlig E, Becher PG. Leaf mineral content govern microbial community structure in the phyllosphere of spinach (Spinacia oleracea) and rocket (Diplotaxis tenuifolia) Sci Total Environ. 2019;675:501–512. doi: 10.1016/j.scitotenv.2019.04.254. [DOI] [PubMed] [Google Scholar]
- Dastogeer KM, Li H, Sivasithamparam K, Jones MG, Wylie SJ. Host specificity of endophytic mycobiota of wild Nicotiana plants from arid regions of Northern Australia. Microbial Ecol. 2018;75:74–87. doi: 10.1007/s00248-017-1020-0. [DOI] [PubMed] [Google Scholar]
- Dastogeer KM, Tumpa FH, Sultana A, Akter MA, Chakraborty A. Plant microbiome—an account of the factors that shape community composition and diversity. Curr Plant Biol. 2020;23:100161. [Google Scholar]
- De Vries FT, Manning P, Tallowin JR, Mortimer SR, Pilgrim ES, Harrison KA, Hobbs PJ, Quirk H, Shipley B, Cornelissen JH, Kattge J. Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecol Lett. 2012;15:1230–1239. doi: 10.1111/j.1461-0248.2012.01844.x. [DOI] [PubMed] [Google Scholar]
- del Carmen O-M, del Carmen R-G, Glick BR, Santoyo G. Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol Res. 2018;208:25–31. doi: 10.1016/j.micres.2018.01.005. [DOI] [PubMed] [Google Scholar]
- Dellagi A, Quillere I, Hirel B. Beneficial soil-borne bacteria and fungi: a promising way to improve plant nitrogen acquisition. J Exp Bot. 2020;71:4469–4479. doi: 10.1093/jxb/eraa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta J, Bora U. Rhizosphere microbiome and plant probiotics. In: Gupta VK, editor. New and future developments in microbial biotechnology and bioengineering. New York: Elsevier; 2019. pp. 273–281. [Google Scholar]
- Fadiji AE, Babalola OO. Metagenomics methods for the study of plant-associated microbial communities: a review. J Microbiol Methods. 2020;170:105860. doi: 10.1016/j.mimet.2020.105860. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick CR, Copeland J, Wang PW, Guttman DS, Kotanen PM, Johnson MT. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc Natl Acad Sci. 2018;115:E1157–E1165. doi: 10.1073/pnas.1717617115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick CR, Salas-González I, Conway JM, Finkel OM, Gilbert S, Russ D, Teixeira PJPL, Dangl JL. The plant microbiome: from ecology to reductionism and beyond. Ann Rev Microbiol. 2020;74:81–100. doi: 10.1146/annurev-micro-022620-014327. [DOI] [PubMed] [Google Scholar]
- Goytain A, Ng T. NanoString ncounter technology: high-throughput RNA validation. In: Li H, Elfman J, editors. Chimeric RNA. New York: Humana; 2020. pp. 125–139. [DOI] [PubMed] [Google Scholar]
- Gupta R, Singh A, Srivastava M, Shanker K, Pandey R. Plant-microbe interactions endorse growth by uplifting microbial community structure of Bacopa monnieri rhizosphere under nematode stress. Microbiol Res. 2018;218:87–96. doi: 10.1016/j.micres.2018.10.006. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Koll U, Schmidt J. Isolation and cultivation of bacteria. In: Hurst CJ, editor. The structure and function of aquatic microbial communities. Cham: Springer; 2019. pp. 313–351. [Google Scholar]
- Hardoim PR, Van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015;79:293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein JW, Wolfe GV, Blee KA. Comparison of rhizosphere bacterial communities in Arabidopsis thaliana mutants for systemic acquired resistance. Microbial Ecol. 2008;55:333–343. doi: 10.1007/s00248-007-9279-1. [DOI] [PubMed] [Google Scholar]
- Herlihy JH, Long TA, McDowell JM. Iron homeostasis and plant immune responses: recent insights and translational implications. J Biol Chem. 2020;120:13444–13457. doi: 10.1074/jbc.REV120.010856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hestrin R, Hammer EC, Mueller CW, Lehmann J. Synergies between mycorrhizal fungi and soil microbial communities increase plant nitrogen acquisition. Comm Biol. 2019;2:1–9. doi: 10.1038/s42003-019-0481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Robert CA, Cadot S, Zhang X, Ye M, Li B, Manzo D, Chervet N, Steinger T, Van Der Heijden MG, Schlaeppi K. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun. 2018;9:1–13. doi: 10.1038/s41467-018-05122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XF, Chaparro JM, Reardon KF, Zhang R, Shen Q, Vivanco JM. Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany. 2014;92(4):267–275. [Google Scholar]
- Jacoby RP, Chen L, Schwier M, Koprivova A, Kopriva S. Recent advances in the role of plant metabolites in shaping the root microbiome. F1000Res. 2020;9:151. doi: 10.12688/f1000research.21796.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongman M, Carmichael PC, Bill M. Technological advances in phytopathogen detection and metagenome profiling techniques. Curr Microbiol. 2020;77:1–7. doi: 10.1007/s00284-020-01881-z. [DOI] [PubMed] [Google Scholar]
- Kembel SW, O’Connor TK, Arnold HK, Hubbell SP, Wright SJ, Green JL. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc Natl Acad Sci. 2014;111:13715–13720. doi: 10.1073/pnas.1216057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniskern JM, Traw MB, Bergelson J. Salicylic acid and jasmonic acid signaling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana. Mol Plant Microbe Int. 2007;20:1512–1522. doi: 10.1094/MPMI-20-12-1512. [DOI] [PubMed] [Google Scholar]
- Kwak MJ, Kong HG, Choi K, Kwon SK, Song JY, Lee J, Lee PA, Choi SY, Seo M, Lee HJ, Jung EJ. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol. 2018;36:1100–1109. doi: 10.1038/nbt.4232. [DOI] [PubMed] [Google Scholar]
- Laforest-Lapointe I, Messier C, Kembel SW. Host species identity, site and time drive temperate tree phyllosphere bacterial community structure. Microbiome. 2016;4:27. doi: 10.1186/s40168-016-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareen A, Burton F, Schäfer P. Plant root-microbe communication in shaping root microbiomes. Plant Mol Biol. 2016;90:575–587. doi: 10.1007/s11103-015-0417-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, Del Rio TG, Jones CD, Tringe SG, Dangl JL. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 2015;349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- Leff JW, Bardgett RD, Wilkinson A, Jackson BG, Pritchard WJ, Long JR, Oakley S, Mason KE, Ostle NJ, Johnson D, Baggs EM. Predicting the structure of soil communities from plant community taxonomy, phylogeny, and traits. ISME J. 2018;12:1794–1805. doi: 10.1038/s41396-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Conway JM, Dangl JL, Woyke T. Elucidating bacterial gene functions in the plant microbiome. Cell Host Microbe. 2018;24:475–485. doi: 10.1016/j.chom.2018.09.005. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhu A, Tan H, Cao L, Zhang R. Engineering banana endosphere microbiome to improve Fusarium wilt resistance in banana. Microbiome. 2019;7:74. doi: 10.1186/s40168-019-0690-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Brettell LE, Singh B. Linking the phyllosphere microbiome to plant health. Trends Plant Sci. 2020;25:841–844. doi: 10.1016/j.tplants.2020.06.003. [DOI] [PubMed] [Google Scholar]
- Lu T, Ke M, Lavoie M, Jin Y, Fan X, Zhang Z, Fu Z, Sun L, Gillings M, Peñuelas J, Qian H. Rhizosphere microorganisms can influence the timing of plant flowering. Microbiome. 2018;6:1–12. doi: 10.1186/s40168-018-0615-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JB, Song-Wilson Y, Foetzki A, Luginbühl C, Winzeler M, Kneubühler Y, Matasci C, Mascher-Frutschi F, Kalinina O, Boller T, Keel C. Does wheat genetically modified for disease resistance affect root-colonizing pseudomonads and arbuscular mycorrhizal fungi? PLoS ONE. 2013;8:e53825. doi: 10.1371/journal.pone.0053825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller DB, Vogel C, Bai Y, Vorholt JA. The plant microbiota: systems-level insights and perspectives. Ann Rev Gen. 2016;50:211–234. doi: 10.1146/annurev-genet-120215-034952. [DOI] [PubMed] [Google Scholar]
- Naylor D, Sadler N, Bhattacharjee A, Graham EB, Anderton CR, McClure R, Lipton M, Hofmockel KS, Jansson JK. Soil microbiomes under climate change and implications for carbon cycling. Ann Rev Environ Res. 2020;45:29–59. [Google Scholar]
- Parasuraman P, Pattnaik S, Busi S. Phyllosphere microbiome: functional importance in sustainable agriculture. In: Gupta VK, editor. New and future developments in microbial biotechnology and bioengineering. London: Elsevier; 2019. pp. 135–148. [Google Scholar]
- Paterson E, Gebbing T, Abel C, Sim A, Telfer G. Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 2007;173:600–610. doi: 10.1111/j.1469-8137.2006.01931.x. [DOI] [PubMed] [Google Scholar]
- Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Edward SB, Ley RE. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci. 2013;110:6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratama AA, Terpstra J, de Oliveria ALM, Salles JF. The role of rhizosphere bacteriophages in plant health. Trends Microbiol. 2020;28:709–718. doi: 10.1016/j.tim.2020.04.005. [DOI] [PubMed] [Google Scholar]
- Ravanbakhsh M, Sasidharan R, Voesenek LA, Kowalchuk GA, Jousset A. Microbial modulation of plant ethylene signaling: ecological and evolutionary consequences. Microbiome. 2018;6:52. doi: 10.1186/s40168-018-0436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Bünger W, Burbano CS, Sabale M, Hurek T. Roots shaping their microbiome: global hotspots for microbial activity. Ann Rev Phytopathol. 2015;53:403–424. doi: 10.1146/annurev-phyto-082712-102342. [DOI] [PubMed] [Google Scholar]
- Remus-Emsermann MN, Schlechter RO. Phyllosphere microbiology: at the interface between microbial individuals and the plant host. New Phytol. 2018;218:1327–1333. doi: 10.1111/nph.15054. [DOI] [PubMed] [Google Scholar]
- Risely A. Applying the core microbiome to understand host–microbe systems. J Anim Ecol. 2020;89:1549–1558. doi: 10.1111/1365-2656.13229. [DOI] [PubMed] [Google Scholar]
- Samad A, Trognitz F, Compant S, Antonielli L, Sessitsch A. Shared and host-specific microbiome diversity and functioning of grapevine and accompanying weed plants. Environ Microbiol. 2017;19:1407–1424. doi: 10.1111/1462-2920.13618. [DOI] [PubMed] [Google Scholar]
- Santhanam R, Menezes RC, Grabe V, LiD BIT, Groten K. A suite of complementary biocontrol traits allows a native consortium of root-associated bacteria to protect their host plant from a fungal sudden-wilt disease. Mol Ecol. 2019;28:1154–1169. doi: 10.1111/mec.15012. [DOI] [PubMed] [Google Scholar]
- Santoyo G, Moreno-Hagelsieb G, del Carmen O-M, Glick BR. Plant growth-promoting bacterial endophytes. Microbiol Res. 2016;183:92–99. doi: 10.1016/j.micres.2015.11.008. [DOI] [PubMed] [Google Scholar]
- Sasse J, Martinoia E, Northen T. Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 2017;23:25–41. doi: 10.1016/j.tplants.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Schlaeppi K, Bulgarelli D. The plant microbiome at work. Mol Plant Microbe Interact. 2015;28:212–217. doi: 10.1094/MPMI-10-14-0334-FI. [DOI] [PubMed] [Google Scholar]
- Schlaeppi K, Dombrowski N, Oter RG, Loren V, van Themaat E, Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci. 2014;111:585–592. doi: 10.1073/pnas.1321597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlatter D, Kinkel L, Thomashow L, Weller D, Paulitz T. Disease suppressive soils: new insights from the soil microbiome. Phytopathology. 2017;107:1284–1297. doi: 10.1094/PHYTO-03-17-0111-RVW. [DOI] [PubMed] [Google Scholar]
- Singh SP, Gaur R. Endophytic Streptomyces spp. underscore induction of defense regulatory genes and confers resistance against Sclerotium rolfsii in chickpea. Biol Control. 2017;104:44–56. [Google Scholar]
- Singh A, Kumar M, Verma S, Choudhary P, Chakdar H. Plant microbiome: trends and prospects for sustainable agriculture. In: Varma A, Tripathi S, Prasad R, editors. Plant microbe symbiosis. Cham: Springer; 2020. pp. 129–151. [Google Scholar]
- Sneck ME, Rudgers JA, Young CA, Miller TE. Does host outcrossing disrupt compatibility with heritable symbionts? Oikos. 2019;128:892–903. [Google Scholar]
- Stringlis IA, Yu K, Feussner K, de Jonge R, Van Bentum S, Van Verk MC, Berendsen RL, Bakker PA, Feussner I, Pieterse CM. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc Natl Acad Sci. 2018;115:5213–5222. doi: 10.1073/pnas.1722335115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K, Kelly S, Radutoiu S. Microbial associations enabling nitrogen acquisition in plants. Curr Opin Microbiol. 2019;49:83–89. doi: 10.1016/j.mib.2019.10.005. [DOI] [PubMed] [Google Scholar]
- Teixeira PJP, Colaianni NR, Fitzpatrick CR, Dangl JL. Beyond pathogens: microbiota interactions with the plant immune system. Curr Opin Microbiol. 2019;49:7–17. doi: 10.1016/j.mib.2019.08.003. [DOI] [PubMed] [Google Scholar]
- Thapa S, Prasanna R. Prospecting the characteristics and significance of the phyllosphere microbiome. Ann Microbial. 2018;68:229–245. [Google Scholar]
- Trivedi P, Delgado-Baquerizo M, Trivedi C, Hamonts K, Anderson IC, Singh BK. Keystone microbial taxa regulate the invasion of a fungal pathogen in agro-ecosystems. Soil Biol Biochem. 2017;111:10–14. [Google Scholar]
- Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. Plant–microbiome interactions: from community assembly to plant health. Natl Rev Microbiol. 2020;18:1–15. doi: 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- Turner TR, James EK, Poole PS. The plant microbiome. Gen Biol. 2013;14:209. doi: 10.1186/gb-2013-14-6-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier N, Agler M, Hacquard S. Microbiota-mediated disease resistance in plants. PLoS Pathog. 2019;15:e1007740. doi: 10.1371/journal.ppat.1007740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma SK, Gond SK, Mishra A, Sharma VK, Kumar J, Singh DK, Kumar A, Kharwar RN. Fungal endophytes representing diverse habitats and their role in plant protection. In: Satyanarayana T, Deshmukh SK, Johri BN, editors. Developments in fungal biology and applied mycology. Singapore: Springer; 2017. pp. 135–157. [Google Scholar]
- Voges MJ, Bai Y, Schulze-Lefert P, Sattely ES. Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc Natl Acad Sci. 2019;116:12558–12565. doi: 10.1073/pnas.1820691116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Gu Y, Friman VP, Kowalchuk GA, Xu Y, Shen Q, Jousset A. Initial soil microbiome composition and functioning predetermine future plant health. Sci Adv. 2019;5:eaaw0759. doi: 10.1126/sciadv.aaw0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe BE. Using cultivated microbial communities to dissect microbiome assembly: challenges, limitations, and the path ahead. Msystems. 2018;3:e00161–e217. doi: 10.1128/mSystems.00161-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav AN, Singh J, Rastegari AA, Yadav N, editors. Plant microbiomes for sustainable agriculture. Berlin: Springer; 2020. [Google Scholar]
- Zarraonaindia I, Owens SM, Weisenhorn P, West K, Hampton-Marcell J, Lax S, Bokulich NA, Mills DA, Martin G, Taghavi S, van der Lelie D. The soil microbiome influences grapevine-associated microbiota. MBio. 2015;6:e02527–e2614. doi: 10.1128/mBio.02527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu YX, Zhang N, Hu B, Jin T, Xu H, Qin Y, Yan P, Zhang X, Guo X, Hui J. NRT1. 1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat Biotechnol. 2019;37:676–684. doi: 10.1038/s41587-019-0104-4. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Yang Y, Liu P, Xu R, Rensing C, Fu X, Liao H. Genotype and rhizobium inoculation modulate the assembly of soybean rhizobacterial communities. Plant Cell Environ. 2019;42:2028–2044. doi: 10.1111/pce.13519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.