Abstract
Background & Aims
Zinc finger and BTB domain containing 20 (ZBTB20) has been implicated as a potential oncogene in liver cancer. However, knockout studies have shown it to be a transcriptional repressor of the alpha-foetoprotein (Afp) gene in adult liver, and reduced levels of ZBTB20 allow for upregulation of AFP with increased tumour severity in certain cases of hepatocellular carcinoma (HCC). As there are many discrepancies in the literature regarding its role in liver tumourigenesis, the aim of this study was to elucidate the role of ZBTB20 in HCC tumourigenesis.
Methods
A reverse genetic study using the Sleeping Beauty (SB) transposon system in mice was performed to elucidate the role of ZBTB20 in HCC tumourigenesis. In vitro ZBTB20 gain- and loss-of-function experiments were used to assess the relationship amongst ZBTB20, peroxisome proliferator activated receptor gamma (PPARG) and catenin beta 1 (CTNNB1).
Results
Transgenic overexpression of ZBTB20 in hepatocytes and in the context of transformation related protein (Trp53) inactivation induced hepatic hypertrophy, activation of WNT/CTNNB1 signalling, and development of liver tumours. In vitro overexpression and knockout experiments using CRISPR/Cas9 demonstrated the important role for ZBTB20 in downregulating PPARG, resulting in activation of the WNT/CTNNB1 signalling pathway and its downstream effectors in HCC tumourigenesis.
Conclusions
These findings demonstrate a novel interaction between ZBTB20 and PPARG, which leads to activation of the WNT/CTNNB1 signalling pathway in HCC tumourigenesis.
Lay summary
ZBTB20 has been implicated as a potential oncogene in liver cancer. Herein, we uncover its important role in liver cancer development. We show that it interacts with PPARG to upregulate the WNT/CTNNB1 signalling pathway, leading to tumourigenesis.
Keywords: Hepatocellular carcinoma, Sleeping Beauty, Reverse genetic screen, ZBTB20, PPARG, CTNNB1
Abbreviations: AFP, alpha-foetoprotein; BTB/POZ, broad complex; tramtrack, bric a brac/poxvirus and zinc finger; CTNNB1, catenin beta 1; Fah, fumarylacetoacetate hydrolase; GSK3B, glycogen synthase kinase 3 beta; HCC, hepatocellular carcinoma; HHL, immortalized human hepatic cell line; IF, immunofluorescence; NTBC, 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione; OFP, orange fluorescent protein; PHI, post-hydrodynamic injection; POK, POZ and Kruppel; PPARG, peroxisome proliferator activated receptor gamma; qPCR, quantitative RT-PCR; SB, Sleeping Beauty; ZBTB20, zinc finger and BTB domain containing 20
Graphical abstract
Highlights
-
•
Zbtb20 was identified as a candidate liver cancer gene by both forward and reverse genetics.
-
•
ZBTB20 promotes HCC tumorigenesis by suppressing PPARG expression
-
•
This results in activation of the WNT/CTNNB1 signalling pathway.
Introduction
Zinc finger and BTB domain containing 20 (ZBTB20) belongs to the POK (POZ and Kruppel) protein family, which functions primarily as transcriptional repressors via protein interactions mediated by DNA-binding C2H2 Kruppel-type zinc finger and broad complex, tramtrack, bric a brac/poxvirus and zinc finger (BTB/POZ) domains. ZBTB20 mRNA expression level is poorly detectable in the foetal liver, but gradually increases after birth.1 ZBTB20 is a nuclear protein and has two isoforms as a result of alternative translation initiation sites, both containing the N-terminal BTB/POZ and C-terminal zinc finger domains.2 Isoform 1 is predominantly found in the early postnatal development, whereas isoform 2 becomes dominantly expressed at the later postnatal stage.1
Zbtb20 knockout studies have demonstrated a severe phenotype characterised by postnatal retardation, metabolic dysfunction, and lethality.3 In addition, a liver phenotype was also observed in Zbtb20 mutant mice that included increased serum bilirubin and alanine aminotransferase levels, indicative of liver dysfunction.3 Zbtb20 has also been implicated as a positive regulator of hepatic replication for liver regeneration in mice.4 Liver-specific inactivation of Zbtb20 resulted in dramatic de-repression of the alpha-foetoprotein (Afp) gene in the liver throughout adult life.1 Although Afp is highly expressed in the foetal liver, Zbtb20 expression is developmentally activated in the liver after birth. It has been shown that ZBTB20 functions as a transcriptional repressor of Afp by specifically inhibiting Afp promoter-driven transcriptional activity.1
Zbtb20 has also been shown to be involved in liver intrinsic functions, possibly through regulating genes such as P450 family members, glucose metabolism, and somatotropic hormonal axis.3 It has been recently shown that ZBTB20 expression was increased in HCC and associated with poor prognosis in patients with high levels of ZBTB20.5 Interestingly, hepatocellular carcinoma (HCC) tissues with hepatitis B viral integration frequency were also found to have upregulation of ZBTB20 expression compared with normal liver tissues.6 We are particularly interested in the role of Zbtb20 in HCC since it was initially identified as a candidate liver cancer gene in a forward genetic screen using the Sleeping Beauty (SB) insertional mutagenesis system.7,8 Different studies involving similar SB insertional mutagenesis screen have also identified Zbtb20 as a candidate gene in the carcinogenesis of HCC.9,10 To elucidate the role of ZBTB20 in liver tumourigenesis, ZBTB20 was introduced into the livers of fumarylacetoacetate hydrolase (Fah)-deficient/SB transposase-expressing transgenic mice (Fah/SB11 mice) using the SB transposon system in a reverse genetic manner as previously described.7,8,11,12 Fah/SB11 mice co-injected with transposon vectors that overexpress ZBTB20 and a short-hairpin RNA directed against the Trp53 gene developed hepatic hypertrophy and developed tumours. In addition, we confirmed the enhanced hepatic hypertrophy leading to hepatomegaly was the result of WNT/catenin beta 1 (CTNNB1) pathway activation. Both in vivo and in vitro experiments suggest a novel interaction between ZBTB20 and peroxisome proliferator activated receptor gamma (PPARG) in regulating CTNNB1. Taken together, the current study confirms the novel role of ZBTB20 interaction and suppression of PPARG expression, resulting in the upregulation of CTNNB1 and contributing to HCC tumourigenesis.
Materials and methods
Hydrodynamic injection
A detailed description of the vectors and protocol used for hydrodynamic injections can be found in the Supplementary Materials and methods.
Liver tumour analyses
A detailed description of the protocol used for liver tumour and histopathological analyses can be found in the Supplementary Materials and methods.
Immunohistochemical analyses
A detailed description of the protocol used for immunohistochemical analyses can be found in the Supplementary Materials and methods.
RT-PCR
A detailed description of the protocol and primer sequences used for RT-PCR can be found in the Supplementary Materials and methods.
ZBTB20 overexpression in human liver cell lines
A detailed description of the vectors and protocol used for ZBTB20 overexpression transfection in human liver cell lines can be found in the Supplementary Materials and methods.
Targeted disruption of ZBTB20 in human liver cancer cell lines
A detailed description of the vectors and protocol used for targeted disruption of ZBTB20 in human liver cancer cell lines can be found in the Supplementary Materials and methods.
Analyses of the ZBTB20-disrupted cells
A detailed description of the protocol used for analysing targeted disruption of ZBTB20 in human liver cancer cell lines can be found in the Supplementary Materials and methods.
Quantitative RT-PCR for in vivo liver tissues
A detailed description of the protocol and primer sequences used for in vivo liver tissues quantitative RT-PCR (qPCR) can be found in the Supplementary Materials and methods.
qPCR for transfected cells
A detailed description of the protocol and primer sequences used for transfected cell qPCR can be found in the Supplementary Materials and methods.
Immunofluorescent staining
A detailed description of the protocol used for immunofluorescent (IF) staining can be found in the Supplementary Materials and methods.
Western blot analyses of transfected cells
A detailed description of the protocol used for Western blot analyses can be found in the Supplementary Materials and methods.
Promoter activity analyses
A detailed description of the vectors and protocol used for promoter activity analyses can be found in the Supplementary Materials and methods.
Statistical analyses
Values are given as mean ± SD. Statistical significance was assessed using the two-tailed unpaired Student t test with p values (Prism Software). Values of p <0.05 were considered statistically significant.
Results
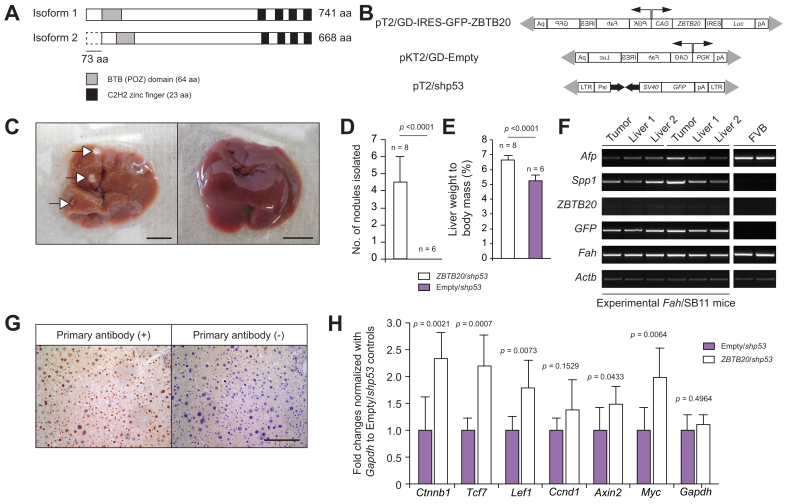
Identification of Zbtb20 as a candidate cancer gene associated with HCC
Current evidence in The Cancer Genome Atlas database (liver HCC, TCGA, provisional) suggest that ZBTB20 copy number and mRNA expression changes were altered in 22 (6%) of human HCCs (n = 360). In these altered sequenced patients (n = 22), 1 patient displayed gene amplification, 3 patients displayed deep deletions, 1 patient had a truncating mutation of unknown significance, 16 patients displayed mRNA upregulation and 1 patient displayed both gene amplification with mRNA upregulation. Taken together, these implicate the role of ZBTB20 as a potential liver oncogene. Schematic representation of the 2 known ZBTB20 isoforms are shown in Fig. 1A, with both isoforms containing the BTB/POZ and zinc finger domains. From a previous forward genetic screen for candidate liver cancer genes using the SB transposon insertional mutagenesis system, Zbtb20 was identified as a potential oncogene based on the insertion profiles of 65% mutagenic transposon vectors that were in the forward orientation before the exon with the start site of translation (Fig. S1A).7,8 Endogenous Zbtb20 was quantified using semiquantitative RT-PCR in liver tumours containing both orientations of mutagenic transposon insertions relative to the endogenous gene (Fig. S1B). These liver tumours all express Afp and/or secreted phosphoprotein 1 (Spp1), both known markers of liver cancer, and were positive for the SB transposase enzyme (SB11) indicating transposition event(s) were most likely responsible for liver tumourigenesis (Fig. S1B). Liver tumours with transposon insertions in a forward orientation relative to the endogenous Zbtb20 gene tended to show higher expression level compared with liver tumours with a reverse insertion profile (Figs. S1B and S1C).
Fig. 1.
In vivo validation of Zbtb20 as an oncogene involved in WNT/CTNNB1 pathway-associated HCC tumourigenesis. (A) Structural differences between the 2 major isoforms of ZBTB20. BTB, broad complex, tramtrack, bric a brac; POZ, poxvirus and zinc finger; C2H2, Kruppel-type zinc finger; aa, amino acid. (B) Gene delivery plasmids used for hydrodynamic tail vein injection. (C) Representative 120-day-old PHI livers taken from Fah/SB11 animals injected with ZBTB20/shp53 (left) and Empty/shp53 (right). Arrows, liver tumour nodules; scale bars, 0.5 cm. (D) Number of liver tumour nodules in ZBTB20/shp53 and Empty/shp53 cohorts. (E) Liver weight to body mass percentage of ZBTB20/shp53 and Empty/shp53 cohorts. (F) Representative RT-PCR for various genes in liver samples from Fah/SB11 mice co-injected with ZBTB20/shp53. FVB, 12-day-old wild-type FVB/N mouse liver; Tumour, tumour liver nodule; Liver, macroscopically normal liver. (G) Representative immunohistochemical staining for CTNNB1 in animals injected with ZBTB20/shp53 (left panel). Right panel, no primary antibody control. Scale bar, 250 μm. (H) Upregulation of Ctnnb1 and downstream target genes of the WNT/CTNNB1 pathway by qRT-PCR. Mean ± SD; p, unpaired t test; n, number of animals. HCC, hepatocellular carcinoma; PHI, post-hydrodynamic injection; (q)RT-PCR, (quantitative) reverse transcription PCR.
Validation of ZBTB20 as a novel oncogene involved in liver tumourigenesis
As Zbtb20 was identified as a candidate liver oncogene in the context of Trp53-deficient genetic background,7 SB transposon-based expression vectors for the ZBTB20 (pT2/GD-IRES-GFP-ZBTB20) and a short-hairpin RNA vector directed against the Trp53 gene (pT2/shp53) were co-administered to Fah/SB11 mice by high-volume rapid hydrodynamic tail vein injection (Fig. 1B). As a control, SB transposon-based empty vector (pKT2/GD-Empty) was co-administered with pT2/shp53 (Empty/shp53) (Fig. 1B). At 120-days post-hydrodynamic injection (PHI), experimental animals co-injected with pT2/GD-IRES-GFP-ZBTB20 and pT2/shp53 (ZBTB20/shp53) were sacrificed. Experimental Fah/SB11 animals injected with ZBTB20/shp53 (n = 8) developed significantly more liver tumour nodules compared with Fah/SB11 animals injected with Empty/shp53 (n = 6) (p <0.0001, Student t test) (Fig. 1C and D). The number of liver nodules isolated from injected cohorts are shown in Table S3.
Liver phenotype observed in Fah/SB11 cohorts injected with ZBTB20/shp53
Fah/SB11 animals injected with ZBTB20/shp53 had significantly enlarged livers relative to whole body weight (6.625% ± 0.306, w/w percentage ±SD) compared with control cohorts injected with Empty/shp53 (5.233% ± 0.367) (Fig. 1E). These results suggest that ZBTB20 overexpression, together with Trp53 inactivation, could induce hepatomegaly. Liver weight percentages of injected cohorts are shown in Table S3. Interestingly, Fah/SB11 mice injected with ZBTB20 overexpression vector only (n = 3) displayed significantly higher liver to body weight percentage than Empty only overexpression controls (p = 0.0098). The liver to body weight percentage was 5.27% ± 0.26 and 7.03% ± 1.45 for Empty only control and ZBTB20 only cohorts, respectively (Table S3). Although no tumours were observed in Empty control animals, only 1 animal displayed a single liver nodule for the ZBTB20 only cohort (Table S3).
Livers of 120-day-old PHI Fah/SB11 mice co-injected with ZBTB20 and short-hairpin directed against the Trp53 gene all express ZBTB20 and GFP (green fluorescent reporter gene found in the pT2/shp53 vector) (Fig. 1F). As expected, wild-type FVB controls do not express GFP or the ZBTB20 transgene (Fig. 1F). As the FVB liver controls were taken from young 12-day-old mice, Afp expression was positive but no detectable level of Spp1 was found, indicating normal liver physiology (Fig. 1F). A liver tumour from Fah/SB11 M1582 had a well-circumscribed nodule composed of sheets of pleomorphic hepatocytes with loss of organisation, some exhibiting large irregular bizarre nuclei. In addition, varying degrees of micro- and macrovesicular fat droplets (steatosis) were observed in most of the cells. Morphologically, this tumour is most compatible with moderate to poorly differentiated HCC (Fig. S2A). Liver tumours from Fah/SB11 M1584 and M1752 both had distinct nodules composed of hepatocytes that exhibited very mild atypia and no significant steatosis. Morphologically, these tumours would best be characterised as well-differentiated hepatic neoplasms (Fig. S2B and S2C).
Immunohistochemical analyses were performed to determine whether cell proliferation was increased in experimental (ZBTB20/shp53) and control (Empty/shp53) injected cohorts (Fig. S3A). Interestingly, Fah/SB11 animals (n = 8) injected with experimental plasmids (ZBTB20/shp53) had statistically significantly larger livers (6.625% ± 0.306, liver to whole body percentage) than Fah/SB11 animals (n = 6) injected with the control plasmids (Empty/shp53) (5.233% ± 0.364) (p <0.0001) (Fig. 1E). However, mitotic activity as determined by uptake of Ki67 staining, did not show any detectable increase in staining between the 2 different cohorts (Fig. S3A). Consistent with the histopathological results (Fig. S2), liver tumours induced by ZBTB20/shp53 in experimental animals displayed varying degrees of increased mitotic activity, indicating varying HCC tumour stage or grade (Fig. S3B).
As cellular proliferation was not deemed to be the cause of the hepatomegaly phenotype seen in ZBTB20/shp53 injected Fah/SB11 animals, arbitrary cell numbers in a fixed area were counted to determine whether cell area number density was decreased, indicating cellular hypertrophy. Cell numbers within a fixed field of view were counted for experimental and control cohorts. Highly significant differences were seen between control and experimental injected animals (p <0.0001) (Fig. S4A). These results suggest ZBTB20 induces hypertrophy as a liver phenotype. To confirm the hypertropic phenotype, cell measurements of experimental and control hepatocytes were taken using NIH ImageJ software (National Institutes of Health, USA). Highly significant differences were seen between control and experimental injected animals (p <0.0001) (Fig. S4B and S4C). These results suggest that the pronounced hepatic hypertrophy could explain the hepatomegaly phenotype seen in experimental injected animals.
Activated WNT/CTNNB1 signalling pathway in Fah/SB11 cohorts injected with ZBTB20/shp53
As activation of the WNT/CTNNB1 pathway has been previously associated with hepatomegaly,13 CTNNB1 immunohistochemical staining was performed on both experimental and control cohorts. Interestingly, activation of the WNT/CTNNB1 pathway was observed in all experimental animals injected with ZBTB20/shp53 according to the positive results of immunohistochemical staining for CTNNB1 (Table S4 and Fig. 1G). To confirm the activation of the WNT/CTNNB1 pathway in the animals injected with ZBTB20/shp53, WNT target genes were evaluated using qRT-PCR. Several downstream genes such as transcription factor 7, T cell specific (Tcf7), lymphoid enhancer binding factor 1 (Lef1), axin 2 (Axin2), and myelocytomatosis oncogene (Myc) were significantly upregulated compared with the control animals injected with Empty/shp53, confirming the activation of the WNT/CTNNB1 pathway (Fig. 1H).
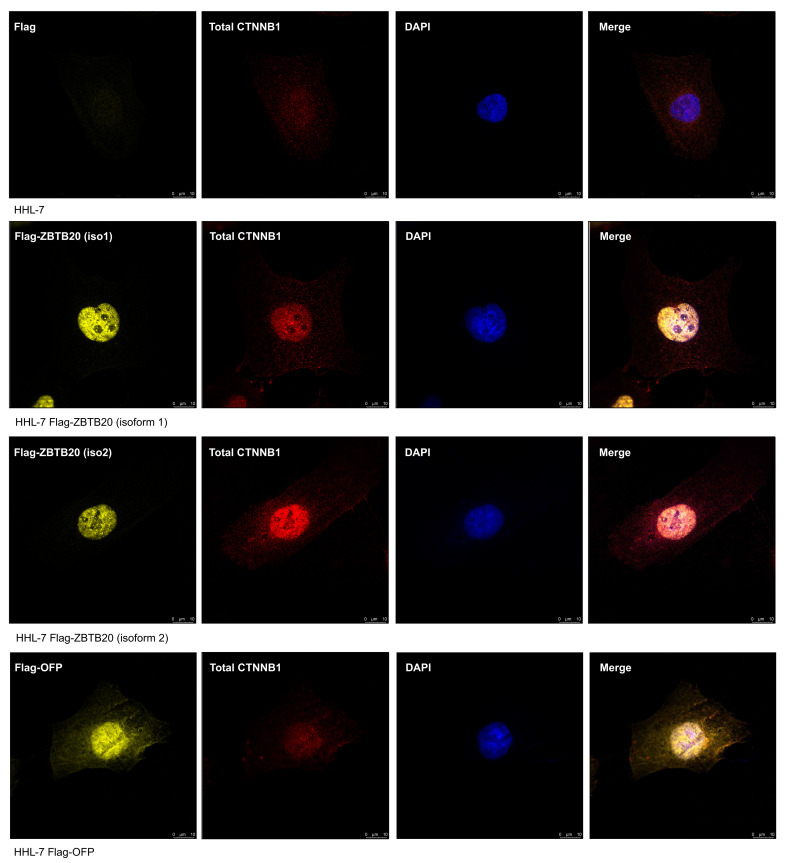
Colocalisation of ZBTB20 and CTNNB1
To confirm whether any interacting relationship between ZBTB20 and CTNNB1 exists, IF staining was performed on ZBTB20 overexpression transfected cells. The localisation of Flag-tagged ZBTB20 and CTNNB1 were determined using specific antibodies and both proteins were shown to be centralised in the cell nucleus, comparing with the orange fluorescent protein (OFP) control and non-transfected cells (Fig. 2). Interestingly, a higher fluorescent signal of CTNNB1 were detected in both isoform 1 and 2 of ZBTB20 overexpression transfected cells compared to the OFP control and non-transfected cells (Fig. 2). The IF results were consistent with the in vivo data that overexpression of ZBTB20 could induce the CTNNB1 signalling pathway. Nuclear staining for CTNNB1 also indicated that its active form was being detected in both isoforms of ZBTB20 overexpression transfected cells.
Fig. 2.
Immunofluorescent staining of ZBTB20 overexpression transfected cells. No Flag-tagged signal (yellow) could be detected in the wild-type HHL7 cell line, while strong Flag-tagged signals were detected in cells transfected with ZBTB20 isoforms 1, isoform 2 and OFP, under the same excitation parameters. Strong centralised CTNNB1 signals (red) were detected in both ZBTB20 isoforms 1 and 2 overexpression transfected cells, while only very weak CTNNB1 signals were detected in wild-type HHL7 and OFP control cell lines, under the same excitation parameters. Nuclei of cells were stained with DAPI (blue). Merged image shown in far-right panels. Scale bars, 10 μm. HHL, immortalized human hepatic cell line.
Validating the effects of ZBTB20 on CTNNB1 in human liver cell lines
To further study the effects of ZBTB20 on CTNNB1, both ZBTB20 isoforms were either overexpressed (Fig. 3) or disrupted (Fig. 4) in human liver cell lines.
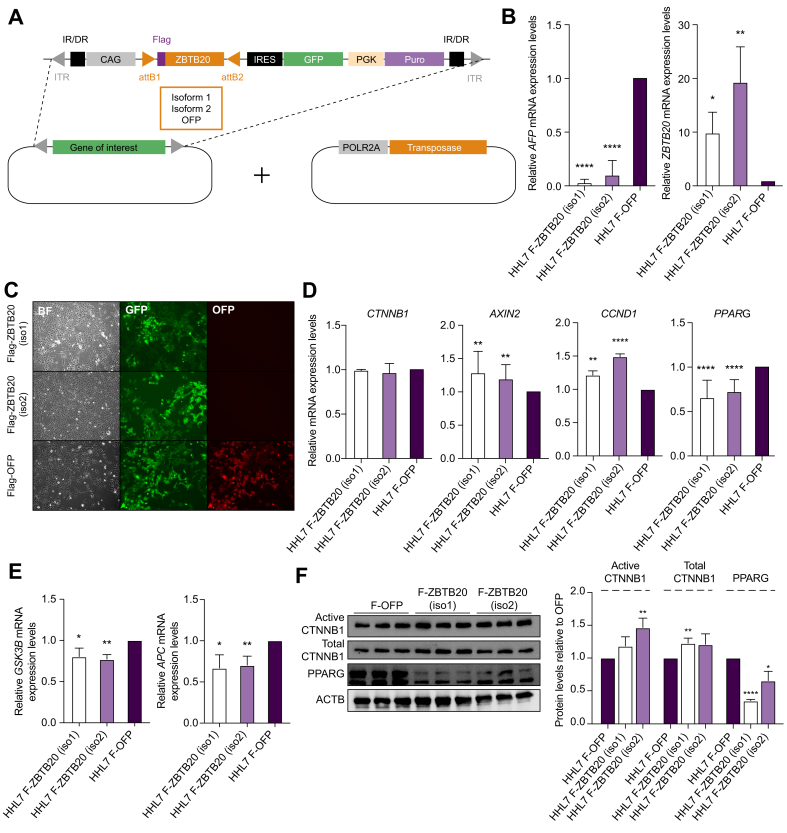
Fig. 3.
In vitro overexpression of ZBTB20 and its effect on the WNT/CTNNB1 pathway. (A) Schematic overexpression vectors for stable integration of ZBTB20 Flag-tagged isoforms 1 and 2 of into various human liver cell lines using the piggyBac transposon system. Flag-tagged OFP was used as a negative control. (B) Relative mRNA expression levels of AFP and ZBTB20 in transfected HHL cells. ∗∗∗∗p <0.00005; ∗∗p <0.005; ∗p <0.05. (C) Fluorescence detection of GFP and OFP in transfected HHL cells were observed under fluorescent microscopy. (D) Relative mRNA expression levels of CTNNB1, AXIN2, CCND1 and PPARG in HHL transfected cells. ∗∗∗∗p <0.00005; ∗∗p <0.005. (E) Relative mRNA expression levels of GSK3B and APC in transfected cells. ∗∗p <0.005; ∗p <0.05. (F) Representative Western blot and relative protein levels of active CTNNB1, total CTNNB1, PPARG, and ACTB in HHL transfected cells. ∗∗∗∗p <0.00005; ∗∗p <0.005; ∗p <0.05. p, unpaired t test; BF, bright-field microscopy.
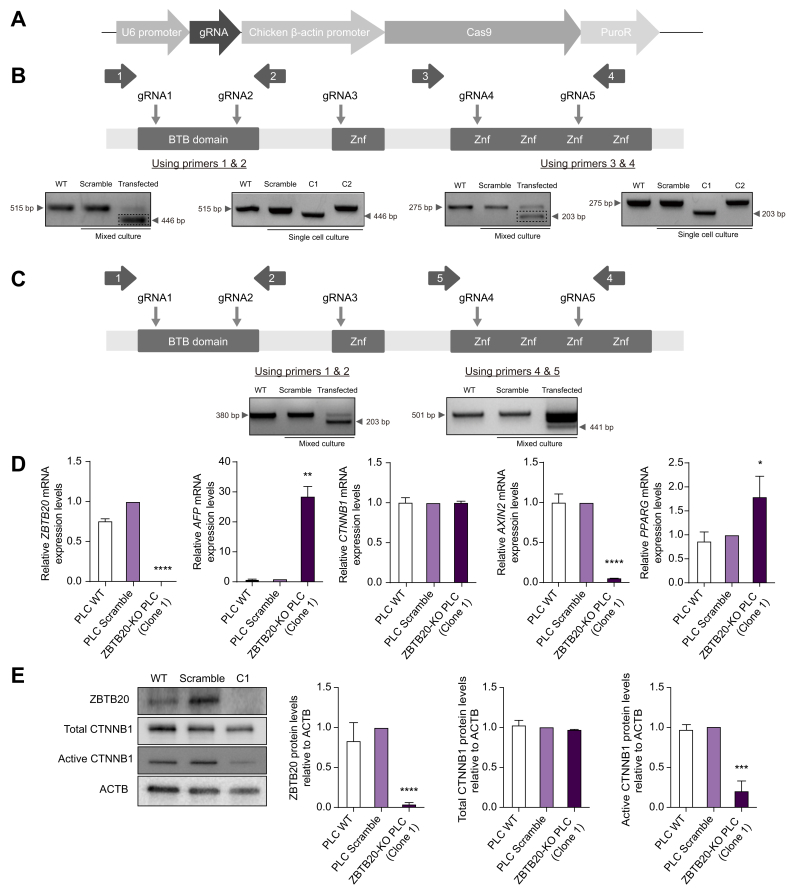
Fig. 4.
In vitro disruption of ZBTB20 and its effect on the WNT/CTNNB1 pathway. (A) Schematic knockout vector that carries the gRNA for targeting the BTB and zinc finger domains of ZBTB20. (B) Detection of knockout regions in genomic DNA using specific indicated primer pairs. PCR products from bands with deletion indels (dashed boxes) were extracted and sequenced. Representative PCR results from mixed and single cell cultures were shown. (C) Detection of knockout regions at the transcriptional level were also confirmed using specific primer pairs for cDNA. Representative PCR results from mixed and single cell cultures were shown. (D) Relative mRNA expression levels of AFP, CTNNB1, AXIN2, and PPARG in ZBTB20 disrupted cells. ∗∗∗∗p <0.00005; ∗∗p <0.005; ∗p <0.05. (E) Representative Western blot and protein levels of ZBTB20, total and active CTNNB1, relative to ACTB, in ZBTB20-disrupted cells. ∗∗∗∗p <0.00005; ∗∗∗p <0.0005. p, unpaired t test.
The piggyBac transposon system was used to stably transfect both isoforms individually into various human liver cell lines (Fig. 3A), with successful overexpression determined by qPCR (Fig. 3B) and detection of GFP signals in culture (Fig. 3C). To determine the function of ZBTB20 in human cells, the mRNA expression level of a negative target of ZBTB20, AFP, was evaluated by qPCR. Reduced expression of AFP was detected in both isoform 1 and isoform 2 ZBTB20 overexpression transfected immortalized human hepatic cell line (HHL) cells compared with OFP control vector (Fig. 3B). This confirmed the efficiency and function of overexpression of ZBTB20 in transfected HHL cells. Interestingly, there was no significant difference in mRNA expression level of CTNNB1 in ZBTB20 overexpression transfected cell lines but significantly higher expression of AXIN2 and cyclin D1 (CCND1) was detected in both isoform 1 and isoform 2 ZBTB20 overexpression transfected HHL cells (Fig. 3D). The components for the canonical WNT/CTNNB1 destruction complex, such as glycogen synthase kinase 3 beta (GSK3B) and APC regulator of WNT signalling pathway (APC), were also significantly reduced in ZBTB20 overexpressing HHL cells compared with OFP control vector (Fig. 3E). At the protein level, ZBTB20 overexpression transfected HHL cells resulted in significantly higher active CTNNB1 levels as determined by the results of Western blot (Fig. 3F), consistent with the IF staining (Fig. 2). Because PPARG has been suggested to be the intermediate protein that interacts with both ZBTB20 and CTNNB1, its expression level was also investigated.14 Interestingly, ZBTB20 overexpression transfected HHL cells do reflect this interaction as significantly lower PPARG were detected at both the mRNA (Fig. 3D) and protein levels (Fig. 3F), consistent with the CTNNB1 results. Similar results were also recapitulated in both PLC/PRF/5 and Hep3B cell lines stably transfected with both isoforms of ZBTB20. ZBTB20 overexpression transfected PLC/PRF/5 (Fig. S5A) and Hep3B (Fig. S5B) cell lines resulted in higher CTNNB1 levels and reduced PPARG levels as determined by Western blot analyses. In addition, TOP/FOP-Flash luciferase reporter assay was also conducted on Hep3B and PLC/PRF/5 cell lines to confirm the activation of the WNT/CTNNB1 signalling pathway. Cell lines transfected with both ZBTB20 isoforms displayed dose-dependent increase in TOP/FOP luciferase activity (Fig. S5C).
To investigate the role of ZBTB20 in HCC tumourigenesis, its function was disrupted by gene editing. Using the CRISPR/Cas9 system in PLC/PRF/5 cell line (Fig. 4A), successful knockout of ZBTB20 in mixed culture and single cell culture was determined by PCR using specific primer pairs located about 50 bp flanking the targeted regions by gRNAs. At the genomic level, gRNA1 and gRNA2 successfully generated a deletion indel in the BTB domain, whereas gRNA4 and gRNA5 generated a deletion indel in the zinc finger domain (Fig. 4B and Fig. S6). In addition, these indels could also be detected at the transcriptional level using cDNA (Fig. 4C). As expected, ZBTB20 disrupted cells demonstrated significantly increased levels of AFP compared with the scrambled control (Fig. 4D). However, consistent with the ZBTB20 overexpression results (Fig. 3D), AXIN2 levels were reduced in cells deficient for ZBTB20 (Fig. 4D). Inversely, significantly lower active CTNNB1 levels with unchanged total CTNNB1 levels were detected in ZBTB20 disrupted cells (Fig. 4E). Consistent with the ZBTB20 overexpression results (Fig. 3D), ZBTB20 disrupted cells demonstrated significantly increased levels of PPARG compared with the scrambled control (Fig. 4D). Similar gene disruption experiments were also performed on the human liver cancer cell line C3A (Fig. S7). Significant reduction in ZBTB20 mRNA levels was demonstrated in both C3A cell pools (Fig. S7A). Consistent with the PLC/PRF/5 results (Fig. 4D), ZBTB20 disrupted C3A cell pools demonstrated significantly increased PPARG levels compared with the scrambled control (Fig. S7A). While ZBTB20 disrupted C3A cell pools demonstrated no significantly changes in CTNNB1 levels compared with the scrambled control (Fig. S7A), downstream target genes of the WNT/CTNNB1 signalling pathway, such as CCND1 and MYC, were significantly reduced (Fig. S7B).
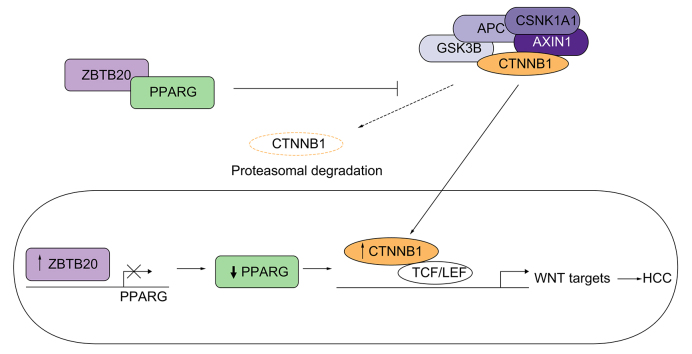
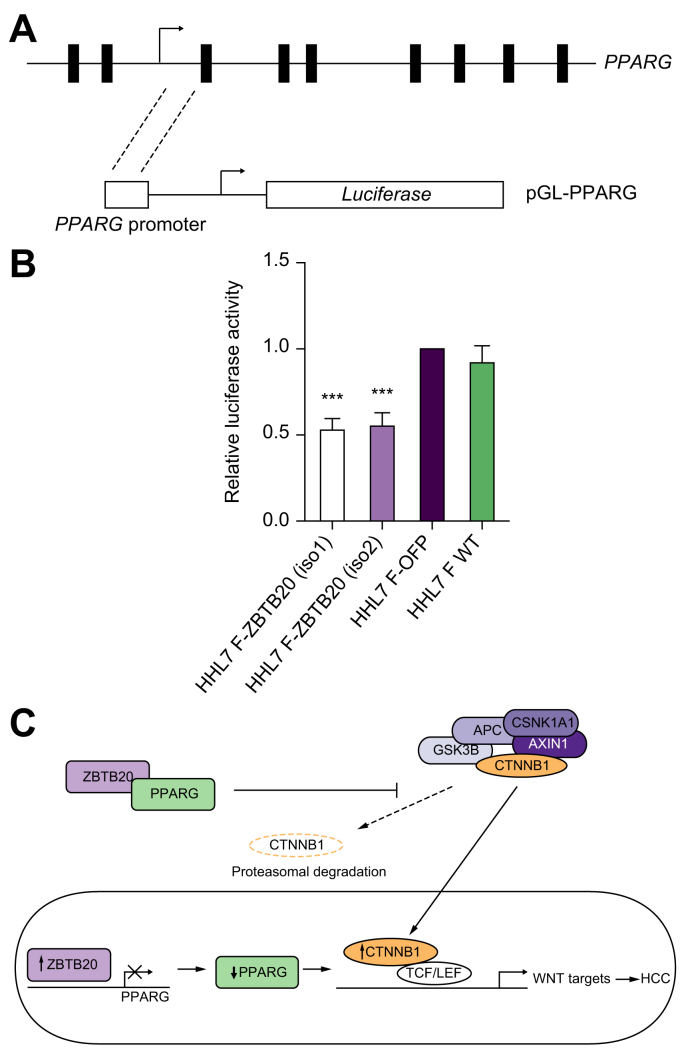
The pGL3-PPARG reporter plasmid containing the 1038 bp PPARG promoter sequence was used to determine if ZBTB20 acts as transcriptional repressor to PPARG expression (Fig. 5A). The 1038-bp PPARG promoter sequence is located on chromosome 3p25.2 12392051 to 12393088, whereas the putative binding motif for ZBTB20 is located from 12392366 to 12392374 (UCSC Genome Browser, GRCh37/hg19) (Fig. S8). Using PPARG promoter luciferase activity analyses containing the putative binding site for ZBTB20 (Fig. 5A), both ZBTB20 isoforms 1 and 2 overexpressing cells were able to decrease PPARG promoter activities by 47% and 45%, respectively, when compared with OFP control transfected or HHL7 wild-type cells (Fig. 5B). Taken together, ZBTB20 is suggested to repress PPARG expression to activate WNT/CTNNB1 signalling resulting in HCC tumourigenesis (Fig. 5C).
Fig. 5.
ZBTB20 acts as a transcriptional repressor for PPARG expression. (A) Schematic diagram for the PPARG gene locus and the promoter sequence used to construct pGL3-PPARG. (B) Luciferase promoter analyses showing both ZBTB20 isoforms 1 and 2 overexpressing cells were able to decrease PPARG promoter activities by 47% and 45%, respectively, compared with orange fluorescent protein control vector transfected cells. HHL7 wild-type cells (HHL7 WT) were also included for comparison. ∗∗∗, p <0.0005. (C) Summary diagram showing the role of ZBTB20 in suppressing PPARG expression and subsequently, allowing the activation of the canonical WNT/CTNNB1 signalling pathway and its downstream target genes.
Potential therapeutic implications
To investigate any potential therapeutic implications of our results, the online database Gene Expression Omnibus was used for comparison. The GSE6764 dataset contains gene expression profiles of 75 tissue samples comparing 4 neoplastic stages (very early HCC, early HCC, advanced HCC, and very advanced HCC) to individuals with normal livers (controls).23 Higher expression of ZBTB20 was detected at very early stages of HCC whereas no significant changes in AFP were detected (Fig. S9A). PPARG expression was also downregulated at very early stages of the disease although not statistically significant, whereas activation of WNT signalling pathway was evident at very early stages of the disease via the upregulation of CTNNB1 and its targets genes – such as LEF1, TCF7, and CCND1 (Fig. S9B).
Discussion
Zbtb20 is essential for liver development as mice deficient for this gene display a liver dysfunction phenotype.3 AFP is currently an accepted tumour marker for HCC as its level is generally increased in the diseased stage.15 Zbtb20 has been shown to be a transcriptional repressor of Afp using gene-targeting studies.1,3 However, it has also been shown that higher ZBTB20 expression in human HCC is associated with poor prognosis, suggesting its role as a potential oncogene.5 It has also been recently reported that another member of the POK transcription factor family, Zbtb7a, can act as an oncogene in some context but also has onco-suppressive activity in others.16 Zbtb20 was previously identified in forward genetic screens as a candidate driver gene for liver cancer using the SB transposon insertional mutagenesis system in a Trp53-predisposed genetic background.7,8 As tumor protein p53 (TP53) mutation is one of the most commonly observed molecular abnormalities in human HCC and because of the discrepancies in the role of ZBTB20 in liver tumourigenesis, ZBTB20 was co-introduced with a short-hairpin RNA directed against Trp53 (ZBTB20/shp53) into the livers of Fah/SB11 mice for functional validation. The results of this study implicate ZBTB20 as an oncogene, causing significantly more liver tumours than in control Fah/SB11 mice co-injected with an empty vector and shp53 (Empty/shp53) (Fig. 1D). These tumours have been classified as well-differentiated hepatic neoplasms to HCC (Fig. S2). Interestingly, ZBTB20/shp53 injected Fah/SB11 mouse livers also displayed hepatomegaly when compared with Empty/shp53 controls (Fig. 1E). Although ZBTB20 overexpression alone could induce hepatomegaly, Trp53 deficiency appears to cooperate with ZBTB20 overexpression in HCC tumourigenesis.
Therefore, this in vivo study strongly implements the oncogenic role of ZBTB20 in the development of HCC. Liver tumours that developed in ZBTB20/shp53 injected animals had varying levels of Ki67-intensity, indicating various stages of tumour development or grade (Fig. S2 and Fig. S3B). Although cell number density appears to be reduced in animals injected with ZBTB20/shp53 (Fig. S4A), hepatic hypertrophy could be a contributing factor to the hepatomegaly phenotype seen in experimental animals. ZBTB20/shp53 injected animals have significantly enlarged hepatocytes compared with animals injected with control vectors (Fig. S4B and S4C). Hepatocytes transgenic for ZBTB20, functioning as a transcriptional repressor, may have targeted downstream gene(s) involved in the feedback regulation of hypertrophy. These transgenic hepatocytes may remain constitutively hypertrophic even when proliferation had been initiated. Therefore, the combined enhanced hypertrophy and proliferation effect may have contributed to liver tumourigenesis. Interestingly, activation of the WNT/CTNNB1 pathway was observed in all experimental animals injected with ZBTB20/shp53 (Table S4). In vivo validation also demonstrated the activation of downstream genes of the WNT/CTNNB1 signalling pathway, such as Axin2, Lef1, Myc, and Tcf7 by ZBTB20/shp53 (Fig. 1H). Taken together, this suggests that WNT/CTNNB1 and Zbtb20 cooperate in the pathogenesis of liver cancer.
By overexpressing and disrupting ZBTB20 in human liver cell lines, the positive relationship between ZBTB20 and CTNNB1 could be confirmed. Novel results demonstrating the activity of CTNNB1 could be regulated by ZBTB20, further confirms the oncogenic role of ZBTB20 in HCC development acting on the WNT/CTNNB1 signalling pathway. Moreover, both the colocalisation and centralisation of ZBTB20 and CTNNB1 in the nucleus of ZBTB20 cells by IF staining further confirms this interaction (Fig. 2). Using PPARG as a bait protein expressed as a DNA-binding domain fusion and ZBTB20 as the prey expressed as a transcriptional activation domain fusion, it has been previously shown that unique physical interaction between these 2 proteins were measured by reporter gene activation in a 2-hybrid system study.14 Furthermore, expression and transcription of PPARG, the apparent intermediate protein for both ZBTB20 and CTNNB1, was downregulated in ZBTB20 overexpression transfected cells (Fig. 3D and F). Using PPARG promoter analyses, both ZBTB20 isoforms 1 and 2 overexpressing cells were able to decrease PPARG promoter activities (Fig. 5B). Consistent with other in vitro results, these results confirm the repressive role of ZBTB20 on PPARG expression. An inverse relationship of CTNNB1 activation and PPARG reduction has been previously reported in various diseases and cancers, such as neurodegenerative diseases, colorectal cancer, and breast cancer.[17], [18], [19] In these cancers, PPARG was downregulated whereas the WNT/CTNNB1 pathway was upregulated. Previous studies have demonstrated that PPARG can target CTNNB1 to proteasome through the interaction between the catenin-binding domain of PPARG and the TCF-binding domain of CTNNB1.20 Therefore, downregulation of PPAGR might increase accumulation of active CTNNB1 via inhibiting proteasomal degradation. Several studies had also reported that activation of PPARG can inhibit HCC growth and progression.21,22 Furthermore, the expression of PPARG is significantly reduced in HCC tumour tissues compared with adjacent non-tumour liver tissues.21 Taken together, our results demonstrate the overexpression of ZBTB20 could activate CTNNB1 via the suppression of PPARG expression and induce its translocation into the nucleus of the cells (Fig. 5C). Currently, the commonly used biomarker AFP is not effective in early diagnosis of HCC, particularly in AFP-negative HCC patients. In the GSE6764 dataset, higher expression of ZBTB20 was detected at very early stages of HCC whereas no significant changes in AFP were only detected at very advanced HCC stage (Fig. S9A).23 Therefore, ZBTB20 could be further evaluated as a potential marker for the early diagnosis of HCC. ZBTB20 also likely plays a role in the activation of the WNT signalling pathway at very early stages of the disease via the upregulation of CTNNB1 and its target genes (Fig. S9B).
In conclusion, the role of ZBTB20 in liver tumourigenesis appears to involve hepatic hypertrophy and hepatomegaly augmented by TP53 inactivation. Experimental animals co-injected with ZBTB20/shp53 have increased tumour multiplicity and hepatomegaly associated with enhanced hepatic hypertrophy associated with activation of the WNT/CTNNB1 pathway via the interaction and suppression of PPARG expression. Based on the several methods of reverse genetics for both in vivo and in vitro validation, the role of ZBTB20 as a novel oncogene and its mechanism involved with HCC tumourigenesis has been implicated.
Financial support
VWK is supported by the Early Career Scheme (ECS)/General Research Fund (GRF) (PolyU 25100815), Collaborative Research Fund Equipment Grant (PolyU C5012-15E), and Research Impact Fund (PolyU R5050-18) from the Research Grant Council, Hong Kong Government; NSFC/RGC Joint Research Scheme (N-PolyU 503/16). State Key Laboratory of Chemical Biology and Drug Discovery (1-BBX8), and Research Project Grants (1-ZVAG, G-UA94, and 1-ZVLC) funded by the Department of Applied and Chemical Technology, The Hong Kong Polytechnic University. DAL is supported by the grant R01 CA113636 from the National Cancer Institute and the American Cancer Society Research Professor Award.
Authors’ contributions
Experiments and procedure: JCT, APC, BRT, LHL, CHC, XXL, TPK, MAL, KA, WCC, JBB, BSM, VWK. Animal experiments: JCT, APC, BRT, LHL, CHC, XXL, TPK, JBB, VWK. Histopathology review and pathologic characterisation: MAL, KA. Concept and design: DAL, VWK. Financial support: DAL, VWK. Writing of manuscript: All authors.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its Supplementary Materials and methods. Any additional data are available from the corresponding author V.W.K. or D.A.L. upon reasonable request.
Conflict of interest
All authors declare no conflicts of interest.
Please refer to the accompanying ICMJE disclosure forms for further details.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2020.100223.
Contributor Information
David A. Largaespada, Email: larga002@umn.edu.
Vincent W. Keng, Email: vincent.keng@polyu.edu.hk.
Appendix A. Supplementary data
References
- 1.Xie Z., Zhang H., Tsai W., Zhang Y., Du Y., Zhong J. Zinc finger protein ZBTB20 is a key repressor of alpha-fetoprotein gene transcription in liver. Proc Natl Acad Sci USA. 2008;105:10859–10864. doi: 10.1073/pnas.0800647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchelmore C., Kjaerulff K.M., Pedersen H.C., Nielsen J.V., Rasmussen T.E., Fisker M.F. Characterization of two novel nuclear BTB/POZ domain zinc finger isoforms. Association with differentiation of hippocampal neurons, cerebellar granule cells, and macroglia. J Biol Chem. 2002;277:7598–7609. doi: 10.1074/jbc.M110023200. [DOI] [PubMed] [Google Scholar]
- 3.Sutherland A.P., Zhang H., Zhang Y., Michaud M., Xie Z., Patti M.E. Zinc finger protein Zbtb20 is essential for postnatal survival and glucose homeostasis. Mol Cel Biol. 2009;29:2804–2815. doi: 10.1128/MCB.01667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H., Shi J.H., Jiang H., Wang K., Lu J.Y., Jiang X. ZBTB20 regulates EGFR expression and hepatocyte proliferation in mouse liver regeneration. Cell Death Dis. 2018;9:462. doi: 10.1038/s41419-018-0514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q., Tan Y.X., Ren Y.B., Dong L.W., Xie Z.F., Tang L. Zinc finger protein ZBTB20 expression is increased in hepatocellular carcinoma and associated with poor prognosis. BMC Cancer. 2011;11:271. doi: 10.1186/1471-2407-11-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Z., Zhu J., Mo J., Zhao H., Chen Q. HBV DNA integrates into upregulated ZBTB20 in patients with hepatocellular carcinoma. Mol Med Rep. 2020;22:380–386. doi: 10.3892/mmr.2020.11074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keng V.W., Villanueva A., Chiang D.Y., Dupuy A.J., Ryan B.J., Matise I. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27:264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keng V.W., Sia D., Sarver A.L., Tschida B.R., Fan D., Alsinet C. Sex bias occurrence of hepatocellular carcinoma in Poly7 molecular subclass is associated with EGFR. Hepatology. 2013;57:120–130. doi: 10.1002/hep.26004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kodama T., Bard-Chapeau E.A., Newberg J.Y., Kodama M., Rangel R., Yoshihara K. Two-step forward genetic screen in mice identifies Ral GTPase-activating proteins as suppressors of hepatocellular carcinoma. Gastroenterology. 2016;151:324–337e312. doi: 10.1053/j.gastro.2016.04.040. [DOI] [PubMed] [Google Scholar]
- 10.Bard-Chapeau E.A., Nguyen A.T., Rust A.G., Sayadi A., Lee P., Chua B.Q. Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat Genet. 2014;46:24–32. doi: 10.1038/ng.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wangensteen K.J., Wilber A., Keng V.W., He Z., Matise I., Wangensteen L. A facile method for somatic, lifelong manipulation of multiple genes in the mouse liver. Hepatology. 2008;47:1714–1724. doi: 10.1002/hep.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keng V.W., Tschida B.R., Bell J.B., Largaespada D.A. Modeling hepatitis B virus X-induced hepatocellular carcinoma in mice with the Sleeping Beauty transposon system. Hepatology. 2011;53:781–790. doi: 10.1002/hep.24091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apte U., Zeng G., Muller P., Tan X., Micsenyi A., Cieply B. Activation of Wnt/beta-catenin pathway during hepatocyte growth factor-induced hepatomegaly in mice. Hepatology. 2006;44:992–1002. doi: 10.1002/hep.21317. [DOI] [PubMed] [Google Scholar]
- 14.Ravasi T., Suzuki H., Cannistraci C.V., Katayama S., Bajic V.B., Tan K. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Bisceglie A.M. Issues in screening and surveillance for hepatocellular carcinoma. Gastroenterology. 2004;127:S104–S107. doi: 10.1053/j.gastro.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 16.Wang G., Lunardi A., Zhang J., Chen Z., Ala U., Webster K.A. Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat Genet. 2013;45:739–746. doi: 10.1038/ng.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen Z.Y., Deng H.J., Fang Y., Zhu X.J., Ye G.T., Yan L. Identification of the interplay between SOX9 and S100P in the metastasis and invasion of colon carcinoma. Oncotarget. 2015;6:20672–20684. doi: 10.18632/oncotarget.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vallee A., Lecarpentier Y. Crosstalk between peroxisome proliferator-activated receptor gamma and the canonical WNT/beta-catenin pathway in chronic inflammation and oxidative stress during carcinogenesis. Front Immunol. 2018;9:745. doi: 10.3389/fimmu.2018.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vallee A., Lecarpentier Y., Guillevin R., Vallee J.N. Effects of cannabidiol interactions with Wnt/beta-catenin pathway and PPAR gamma on oxidative stress and neuroinflammation in Alzheimer’s disease. Acta Bioch Bioph Sin. 2017;49:853–866. doi: 10.1093/abbs/gmx073. [DOI] [PubMed] [Google Scholar]
- 20.Liu J., Wang H., Zuo Y., Farmer S.R. Functional interaction between peroxisome proliferator-activated receptor gamma and beta-catenin. Mol Cel Biol. 2006;26:5827–5837. doi: 10.1128/MCB.00441-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J., Qiao L., Zimmermann L., Ebert M.P., Zhang H., Lin W. Troglitazone inhibits tumor growth in hepatocellular carcinoma in vitro and in vivo. Hepatology. 2006;43:134–143. doi: 10.1002/hep.20994. [DOI] [PubMed] [Google Scholar]
- 22.Yu J., Shen B., Chu E.S., Teoh N., Cheung K.F., Wu C.W. Inhibitory role of peroxisome proliferator-activated receptor gamma in hepatocarcinogenesis in mice and in vitro. Hepatology. 2010;51:2008–2019. doi: 10.1002/hep.23550. [DOI] [PubMed] [Google Scholar]
- 23.Wurmbach E., Chen Y.B., Khitrov G., Zhang W., Roayaie S., Schwartz M. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45(4):938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its Supplementary Materials and methods. Any additional data are available from the corresponding author V.W.K. or D.A.L. upon reasonable request.