Abstract
Reversible protein ubiquitination is an essential signaling mechanism within eukaryotes. Deubiquitinating enzymes are critical to this process, as they mediate removal of ubiquitin from substrate proteins. Ubiquitin-specific protease 7 (USP7) is a prominent deubiquitinating enzyme, with an extensive network of interacting partners and established roles in cell cycle activation, immune responses and DNA replication. Characterized USP7 substrates primarily interact with one of two major binding sites outside the catalytic domain. These are located on the USP7 N-terminal TRAF-like (TRAF) domain and the first and second UBL domains (UBL1-2) within the C-terminal tail. Here, we report that DNA polymerase iota (Pol ι) is a novel USP7 substrate that interacts with both TRAF and UBL1-2. Through the use of biophysical approaches and mutational analysis, we characterize both interfaces and demonstrate that bipartite binding to both USP7 domains is required for efficient Pol ι deubiquitination. Together, these data establish a new bipartite mode of USP7 substrate binding.
Keywords: Ubiquitin-specific protease, Herpesvirus-associated ubiquitin-specific protease, Deubiquitination, Ubiquitin-like domain, DNA polymerase iota
Introduction
The reversible ubiquitination of proteins is an important aspect of eukaryotic intracellular signaling. Deubiquitinating enzymes (DUBs) are central to this reversibility, as they can cleave the isopeptide bond formed between a ubiquitin moiety and a modified lysine residue of a substrate protein.1 Ubiquitin-specific proteases (USPs) represent the largest family of DUBs and are characterized by a conserved papain-like cysteine protease catalytic domain; many USPs also contain additional regulatory domains that modulate their substrate specificity, cellular localization and catalytic activity.2
USP7 is a prominent member of the USP family due to its ubiquitous high-level expression in tissues, as well as its extensive network of associating partners.3 USP7 is also frequently deregulated in cancer, while certain germline mutations in USP7 are associated with neurodevelopmental disorders.4 The functionality of USP7 is largely modulated by its N-terminal TRAF-like domain (TRAF), as well as by a C-terminal region containing five ubiquitin-like (UBL) domains. TRAF contains one of two major USP7 substrate-binding sites and binds to substrates that include p53 and MDM2,5–7 MDM4,8 MCM-BP9 and the Epstein-Barr viral protein, EBNA1.10 Each of these substrates encode P/A/ExxS motifs, which interact with USP7 residues D164 and W165, within a pocket on the TRAF domain surface.5 The C-terminal region of USP7 contains the second major substrate-binding site – located within UBL1-2 – while the terminal 19 amino acids interact with the catalytic domain and are required for activation of USP7 catalytic activity.11 Substrate binding via UBL1-2 is mediated by an acidic surface patch that interacts with highly basic (R/K)xKxxxK motifs within a separate subset of USP7 substrates.12 These substrates include GMP synthetase,13 DNMT1,14 UHRF1,15 RNF16916 and the Herpes simplex virus immediate-early protein, ICP0.12,17
Many known USP7 substrates function in DNA repair and replication.18 This includes PCNA,19 Rad18,20 Chk121 and DNA polymerase eta (Pol η)22 each of which contribute to the DNA damage tolerance pathway of translesion synthesis (TLS). Here, we demonstrate that the Pol η paralog, Pol ι, is also a substrate of USP7. Pol ι is a low fidelity DNA polymerase that is recruited to replication foci following DNA damage.23,24 Although the precise cellular function of Pol ι remains unclear, altered expression levels have been reported in many cancer types and correlated with decreased replication fidelity.25 Pol ι is also known to be monoubiquitinated at numerous lysine residues.26,27 In this work, we demonstrate that Pol ι monoubiquitination is regulated by USP7. We also find that, unlike other known USP7 substrates, Pol ι interacts with both USP7 TRAF and UBL1-2 substrate-binding sites and that this bipartite binding is essential for proper Pol ι deubiquitination.
Results
Pol ι is a substrate and interacting partner of USP7
The monoubiquitination of Pol ι is dependent on its ubiquitin-binding motifs (UBMs) and their capacity to bind and recruit ubiquitin-laden E2 enzymes.26,28 While these domains are major drivers of Pol ι monoubiquitination, we considered whether additional regulation of Pol ι ubiquitination might also be mediated by DUBs. To explore this possibility, we used mass spectrometry to identify proteins that co-immunoprecipitate (co-IP) with FLAG-tagged Pol ι, which we transiently expressed in HEK293T cells (Dataset 1). These analyses revealed a putative association between Pol ι and the deubiquitinating enzyme, USP7 (Figure 1(a)). Although we detected only a single peptide of USP7, we were able to confirm this association by immunoblotting similarly prepared FLAG co-IP eluents (Figure 1(b)). To ensure these findings were not an artefact of transient Pol ι overexpression, we also performed a co-IP of endogenous Pol ι and again immunoblotted for USP7 (Figure 1(c); these data supported the existence of a Pol ι-USP7 complex. To explore whether this association could be mediated by a direct interaction, we performed a pull-down assay using recombinant His-tagged Pol ι and GST-tagged USP7 (Figure 1(d)). The co-elution of His-Pol ι with GST-USP7, although not with GST alone, support that Pol ι and USP7 can interact directly.
Figure 1.
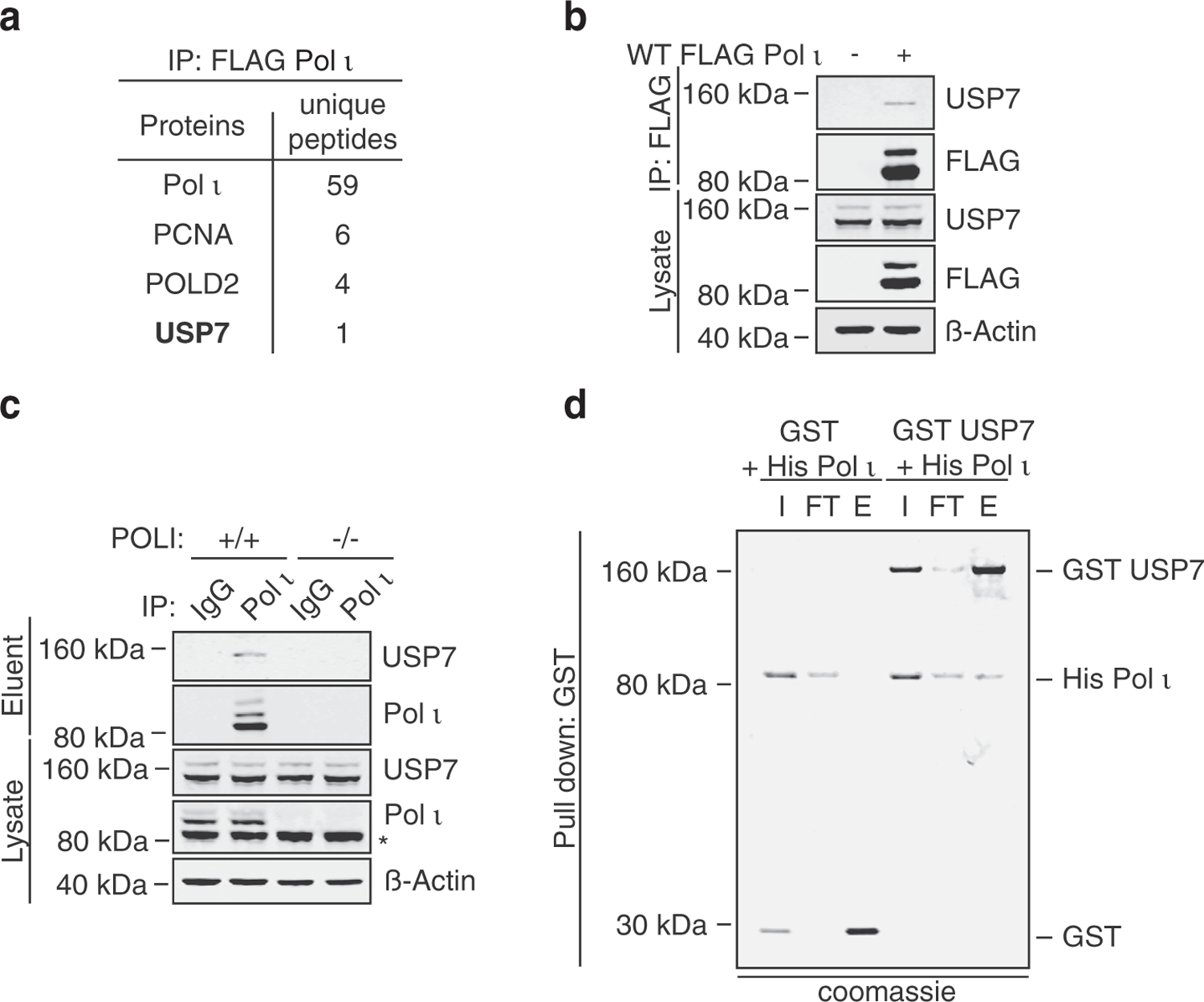
Pol ι interacts with the deubiquitinating enzyme, USP7. (a) Select proteins identified by GeLC-MS/MS analysis of proteins co-immunoprecipitated with FLAG Pol ι from 293T whole cell lysates (WCL). (b) Coimmunoprecipitated proteins and WCL prepared similarly to (a) were immunoblotted as indicated. (c) Endogenous Pol ι was immunoprecipitated from HEK293T whole cell lysate and the eluent and input immunoblotted as indicated. * = non-specific band (d) His Pol ι (1.5 µg) was incubated with equimolar quantities of GST (0.5 µg) or GST-tagged USP7 (2.3 µg). GST was captured on glutathione sepharose beads, washed, and eluted. Protein were separated by gel electrophoresis and proteins stained with colloidal Coomassie blue (shown in grayscale). I = 10% input, FT = 10% flow through, E = 50% eluent.
To assess whether Pol ι might be a substrate of USP7, we immunoprecipitated FLAG-Pol ι from 293T cells, incubated the protein with recombinant His-USP7 for 4 hours, then monitored Pol ι monoubiquitination by gel electrophoresis (Figure 2(a)). Pol ι was readily deubiquitinated by USP7 in this assay. We also tested whether USP7 might promote the deubiquitination of Pol ι in cells. To do so, we transiently expressed FLAG-Pol ι and HA-ubiquitin in cells co-expressing wild-type (WT) Myc-USP7, or a Myc-USP7 mutant where the catalytic cysteine residue was converted to serine (C223S) (Figure 2(b)). In the eluent samples, monoubiquitinated Pol ι was identifiable as a higher molecular weight Pol ι band, which was cross-reactive with FLAG- and HA-antibodies. Overexpression of WT USP7, although not C223S USP7, led to a clear reduction in Pol ι monoubiquitination.
Figure 2.
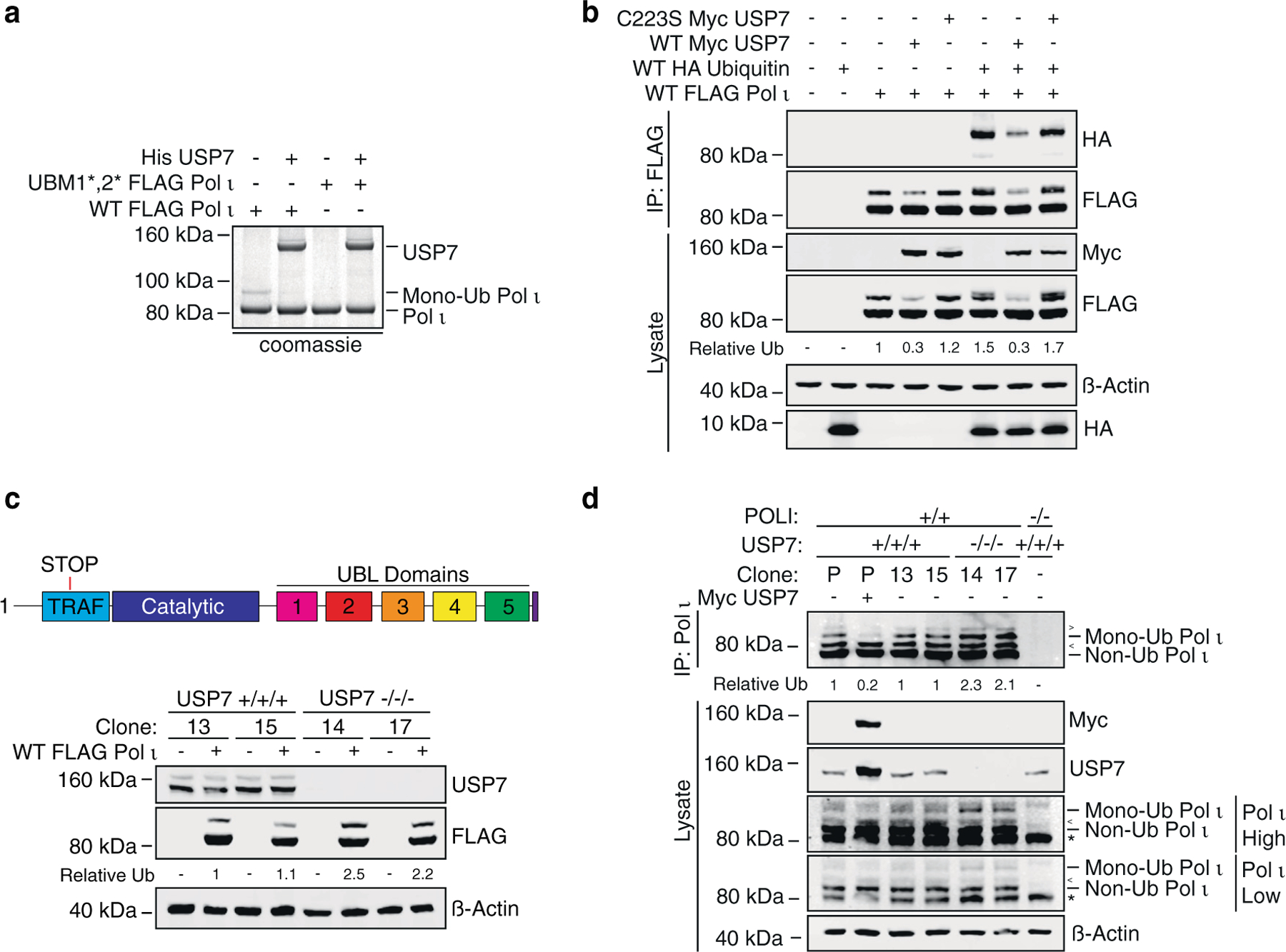
Monoubiquitinated Pol ι is a substrate of USP7. (a) WT or ubiquitin-binding mutant (UBM1*,2*) FLAG Pol ι was immunoprecipitated from HEK293T cells and incubated with recombinant human His6-USP7 for 4 hours at 37 C. Reactions were separated by gel electrophoresis and proteins stained with colloidal Coomassie blue (shown in grayscale). (b) WT FLAG Pol ι was co-expressed in 293T cells with WT HA ubiquitin and/or WT or catalytically inactive (C223S) Myc USP7. FLAG Pol ι was immunoprecipitated and eluents and WCL immunoblotted as indicated. (c) WT FLAG Pol ι was transiently expressed in two USP7 +/+/+ and two USP7 −/−/− clonal cell lines and WCL prepared and immunoblotted as indicated. (d) Endogenous Pol ι was immunoprecipitated from USP7 +/+/+ and −/−/− cell lines, POLI −/− cells or parental HEK293T cells transfected with Myc USP7. WCL and IP eluents were immunoblotted as indicated. * = non-specific band. The prominent, major isoform of Pol ι is indicated as ‘Non Ub Pol ι’. < = higher molecule weight Pol ι isoform and > = additional Pol ι of unclear identity.
Given the reduction in Pol ι monoubiquitination in cells overexpressing USP7, we questioned whether depletion of endogenous USP7 would have the opposing effect of increasing Pol ι monoubiquitination. We therefore used CRISPR-Cas9 to create 293T cell lines with frameshift mutations in exon 3 of USP7 (Supplemental Figure 1). We then transiently expressed FLAG-Pol ι in each cell line and compared the relative ratio of mono- to non-ubiquitinated Pol ι (Figure 2(c)). These data demonstrated increased Pol ι monoubiquitination in cells lacking USP7. We also examined the monoubiquitination of endogenous Pol ι. Although interpretation of these immunoblots was complicated by the presence of multiple Pol ι isoforms,29 as well as by non-specific antibody binding, monoubiquitinated Pol ι was readily detected as a band diminished by USP7 overexpression, as well as enhanced in USP7 knockout cells (Figure 2(d)). Together, these data support that Pol ι is a substrate and interacting partner of USP7.
Pol ι associates with the TRAF and UBL1-2 domains of USP7
Previous studies on USP7 protein–protein interactions have highlighted two major substrate-binding sites outside the catalytic domain, located on TRAF5,6 and within a negatively charged patch on the UBL1-2 interface.12,14,16,17,30 A secondary p53/MDM2 binding site (outside of TRAF), has also been predicted within UBLs 4–5.31 To gauge which of these domains might be involved in Pol ι-binding, we first created USP7 C-terminal truncation constructs to remove the UBL domains (ΔUBL), or to express just the TRAF domain (Figure 3(a)). We then performed immunoprecipitation studies between each of these constructs and WT FLAG Pol ι. In these experiments, Pol ι readily associated with each USP7 species, suggesting binding to the minimal TRAF domain (Figure 3(b)). By immunoblotting the whole cell lysate from these experiments, we also noted that unlike WT USP7, overexpression of the DUBL USP7 mutant did not cause a similar reduction in Pol ι monoubiquitination. This is most likely due to the previously described requirement for the far C-terminus of USP7 for activating catalysis.11 Indeed, a USP7 mutant lacking 19 amino acids from its C-terminus (Δ19 amino acid tail), was also unable to efficiently deubiquitinate Pol ι (Figure 3(c)). Given that the UBL domains are known to bind a subset of USP7 substrates, we nevertheless queried whether these domains might constitute an additional Pol ι-binding site. We therefore prepared a construct expressing UBL domains 1–5, as well as minimal sets of 2- to 3- UBL domains, with which we again performed co-immunoprecipitations with WT FLAG Pol ι. In these assays, Pol ι associated with the UBL1–5 fragment, as well as with UBL1-2 and UBL1–3 (Figure 3(d)). Although an association with UBL1–3 was more readily detected than with UBL1-2, this likely reflects the greater stability of the UBL1–3 fragment in cells. These data suggest that USP7 contains two Pol ι-binding sites, within the TRAF and UBL1-2 domains.
Figure 3.
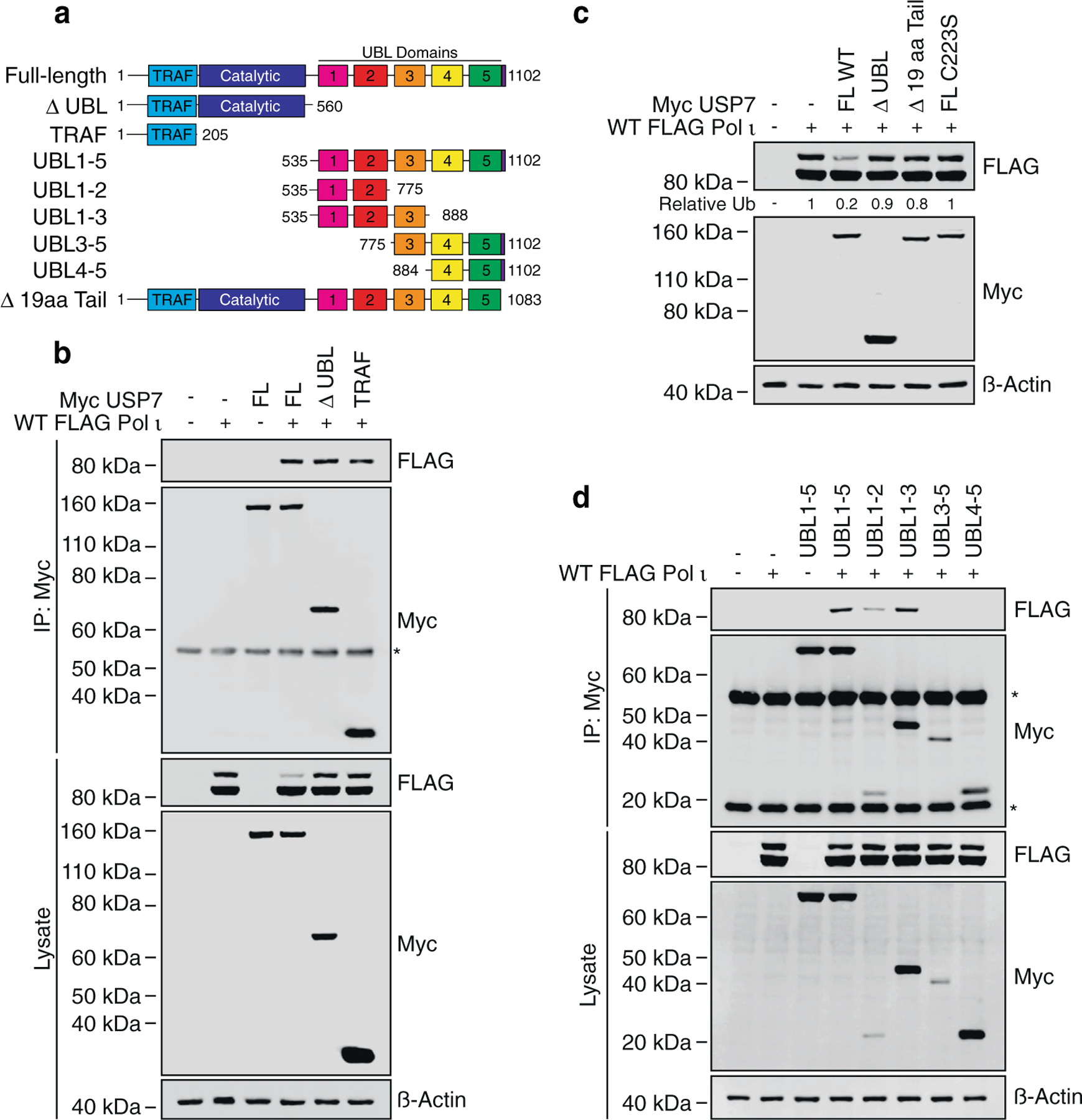
Pol ι associates with the TRAF and UBL1-2 domains of USP7. (a) Schematic illustrating USP7 truncation mutants employed in this figure. (b) Immunoprecipitation of the indicated USP7 truncations from HEK293T cells co-expressing WT FLAG Pol ι. Eluent and WCL (input) were immunoblotted as indicated. * = Myc antibody proteins. (c) WT FLAG Pol ι was co-expressed in 293T cells with WT Myc USP7, or a mutant lacking 19 aa from the C-terminal tail (Δ 19 aa Tail). WCL was then prepared and immunoblotted as indicated. (d) Immunoprecipitation of the indicated USP7 truncations from 293T cells co-expressing WT FLAG Pol ι. Eluent and WCL (input) were immunoblotted as indicated. * = Myc antibody proteins.
The USP7 TRAF domain interacts with the Pol ι C-terminus
Having determined that Pol ι associates with both the TRAF and UBL1-2 domains of USP7, we next set out to identify the regions of Pol ι that mediate these interactions. The USP7 TRAF domain is known to bind other substrates at P/A/ExxS motifs, via interactions centered around USP7 amino acids D164 and W165.6–8 We thus reasoned that Pol ι was likely to interact with TRAF similarly. As Pol ι contains 18 P/A/ExxS motifs, we first sought to narrow down the binding site, using co-immunoprecipitation experiments with C-terminal Pol ι truncation mutants. These studies revealed the USP7 TRAF domain is likely to interact with Pol ι between residues 421 and 590 (Figure 4(a)). As this region contains four putative TRAF-binding motifs, we next mutated the serine residue of each motif to alanine and performed further co-immunoprecipitations. In these assays, we also included a USP7 TRAF mutant, in which D164 and W165 were converted to alanine (DW164AA). Here, mutation of either the TRAF domain, or Pol ι residue S580, disrupted the association between these proteins (Figure 4(b)).
Figure 4.
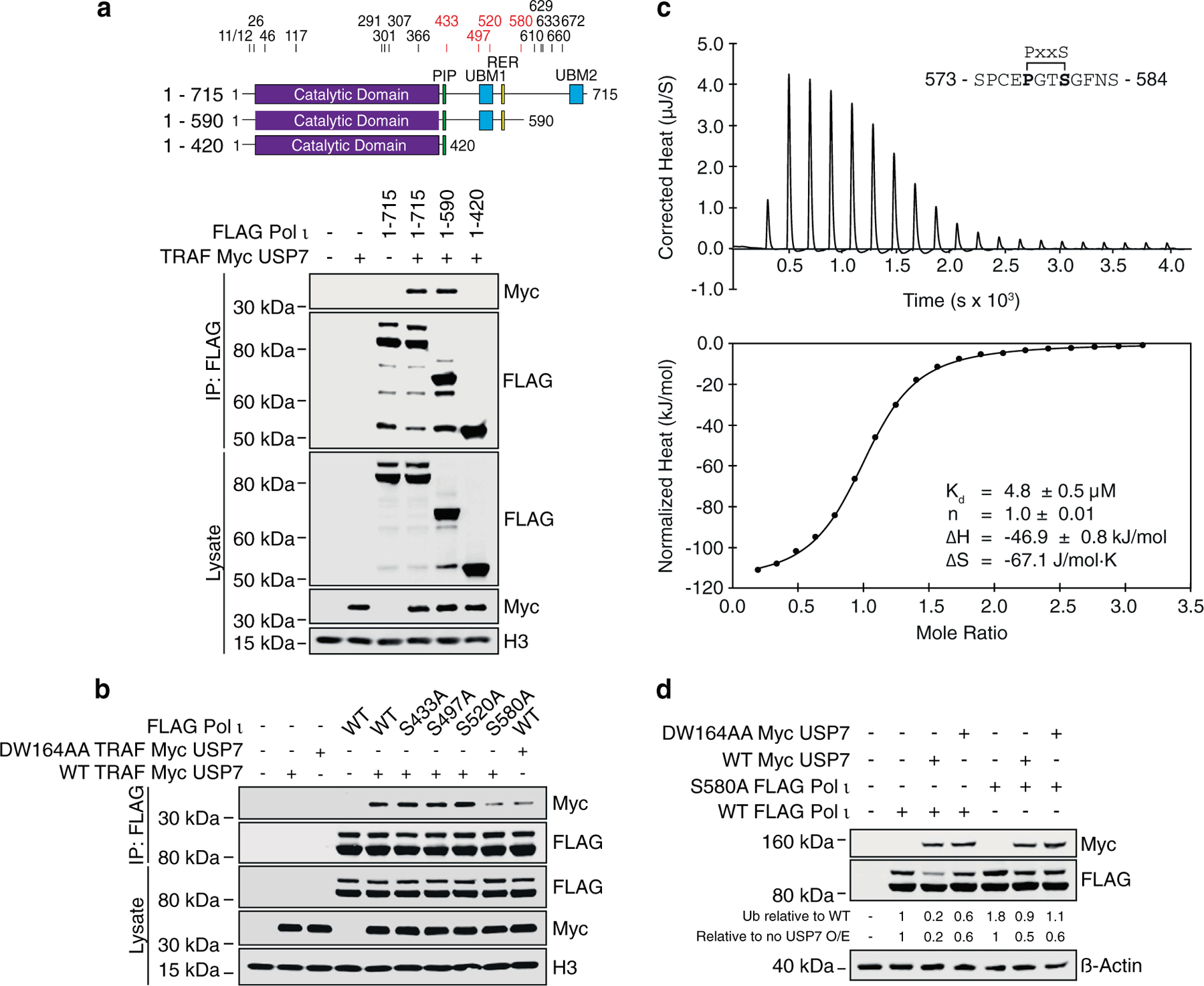
The USP7 TRAF domain interacts with the Pol ι C terminus. (a) Schematics of Pol ι truncation mutants used in this figure. The numbers above the schematics indicate the location of serine residues that are part of P/A/ExxS motifs. The numbers in red correspond to serine residues mutated in b. Pol ι truncation mutants were immunoprecipitated from cells co-expressing TRAF Myc USP7 and the eluent and WCL (input) immunoblotted as indicated. (b) WT or mutant Pol ι was immunoprecipitated from 293T cells co-expressing WT or DW164AA Myc TRAF USP7. Eluent and WCL (input) were immunoblotted as indicated. (c) ITC profile of binding between Pol ι peptide 573–584 and recombinant TRAF. (d) WT or S580A FLAG Pol ι was co-expressed in HEK293T cells with WT or DW164AA Myc USP7. Whole cell lysates were immunoblotted as indicated.
We also designed peptides representing each of the putative TRAF-binding motifs of Pol ι between residues 421 and 590 and used isothermal titration calorimetry (ITC) to measure their affinity to recombinant TRAF. Consistent with our mutational analysis, a Pol ι peptide representing residues 573–584 was readily bound by TRAF, with a Kd of 4.8 µM (Figure 4(c) and Supplemental Figure 2), a value similar to those reported for other TRAF-binding peptides (12,16–17) (Supplemental Figure 3). Together, these findings support localization of the Pol ι TRAF-binding site to residues proximal to S580.
Compared to WT Pol ι, we also noted an apparent higher ratio of monoubiquitination for the S580A mutant (Figure 4(d)). This is likely due to decreased association between this mutant and the TRAF domain of endogenous USP7. To confirm this, we co-transfected cells with WT or S580A Pol ι and WT or DW164AA USP7 and monitored Pol ι monoubiquitination by western blotting (Figure 4(d)). In these assays, mutation of either Pol ι or USP7 hindered Pol ι deubiquitination. These data suggest the interface between Pol ι and the USP7 TRAF domain is therefore important for proper Pol ι deubiquitination.
To further characterize the Pol ι-TRAF interface, we next used solution NMR spectroscopy to map the binding of the Pol ι 573–584 peptide on the surface of the TRAF domain. Here, 15N isotopically labeled TRAF was titrated with increasing amounts of unlabeled peptide and changes in 15N-HSQC spectrum monitored at each point of the titration. We then quantified and mapped the resulting chemical shift perturbations (CSPs) on the structure of USP7 TRAF, to reveal a continuous binding interface (Figure 5(a–c). Consistent with our mutational analysis, the peptide-binding region of TRAF is located around residues D164/W165, corresponding to a region recognized by other TRAF-binding peptides 12,16–17. Although NMR signals of D164 and W165 (located on β7 strand) are broadened beyond detection, other residues on β7 (R152, F167, N169 and I154) and residues on the adjacent β4 strand (F118, L119, Q120 and C121) demonstrated some of the largest CSPs caused by Pol ι.
Figure 5.
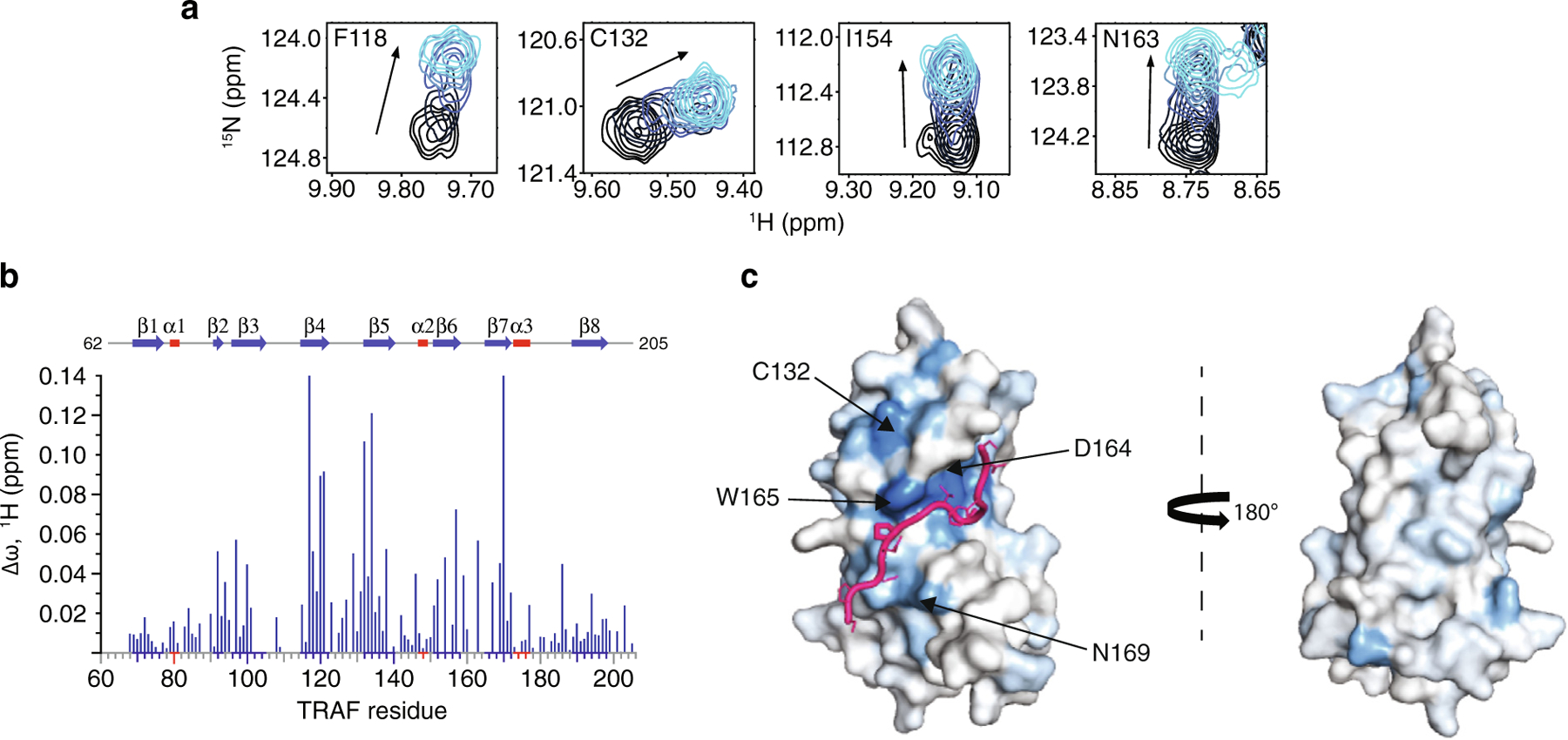
Pol ι interacts with the classic USP7 TRAF domain binding site. (a) Select residues of USP7 TRAF that demonstrate CSPs when titrated with increasing amounts of Pol ι peptide 573–584 (1:5 protein:peptide molar ratio). (b) Per-residue CSPs (Δω) are quantified and represented as bar plots. Residues that were only observed in “free” or “bound” states due to peak broadening were assigned to the highest observed CSP value. (c) HADDOCK model of the USP7 TRAF domain and Pol ι peptide 573–584 complex. CSPs from NMR experiments were used to map the binding interface, shown in blue.
We also used HADDOCK software32,33 to calculate a data-based structural model of the TRAF domain in complex with the Pol ι 573–584 peptide. This approach is based on experimental data including CSPs and allows docking of two molecules of known structure. For these calculations, we generated a structure of the disordered Pol ι peptide using MODELLER,34 which we docked onto the crystal structure of the USP7 TRAF domain (PDB ID 2fop) (Figure 5(c) and Movie 1). The residues with the largest NMR CSPs (>0.06 ppm) and those identified by mutational analysis of both TRAF and Pol ι, were used as experimental restraints to calculate the structure of the complex. These data further confirm that the USP7 TRAF domain utilizes its classic binding site to complex with Pol ι peptide 573–584.
UBL1-2 mediates a second USP7-Pol ι interaction
We next turned our attention to the interaction between Pol ι and USP7 UBL1-2. Previous studies have determined a substrate binding-site within a negatively charged surface patch on UBL1-2 involving residues D758, E759 and D764.14 In other substrates, these residues interact with a consensus binding motif, which can be oriented either N-terminal to C-terminal (K/RxKxxxK), or C-terminal to N-terminal (KxxxKxR/K). Within Pol ι, we identified two motifs that could potentially interact with UBL1-2; K440 – K446 and R524 – K530. To test these, we mutated the lysine and arginine residues of either motif (3KA and RKK/AAA, respectively) and performed co-immunoprecipitation assays with WT UBL1–3, or a UBL1–3 mutant in which we converted D758, E759 and D764 of UBL2 to alanine (DED/AAA). For these assays, we used UBL1–3, rather than UBL1-2, due to its greater stability in cells (Figure 3 (d)). Here, the Pol i-UBL1–3 association was disrupted by mutation of either the UBL2 surface patch, or the Pol ι motif from K440 – K446 (3KA Pol ι) (Figure 6(a)).
Figure 6.
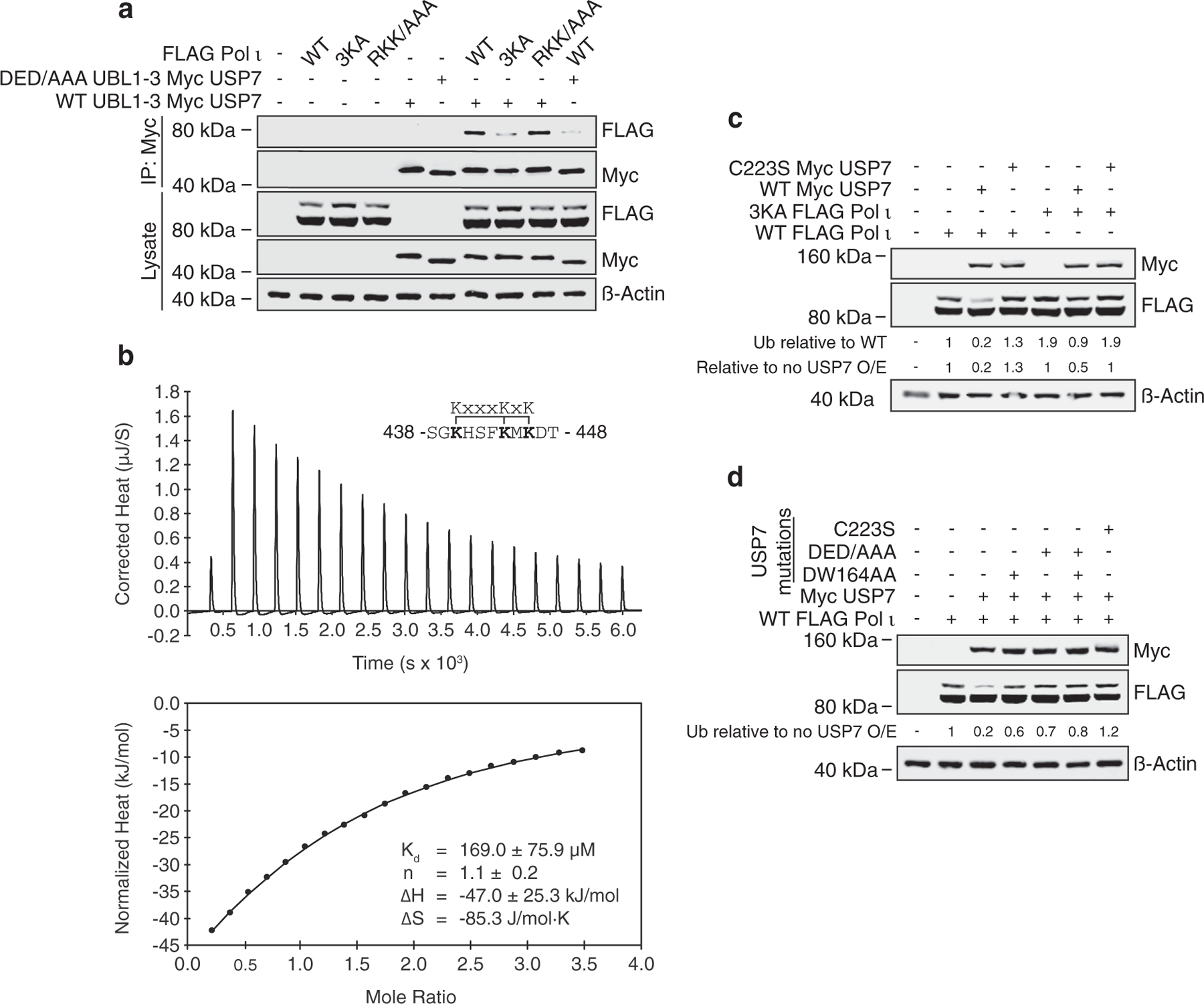
UBL1-2 mediates a second USP7-Pol ι interaction. (a) WT or mutant Pol ι was immunoprecipitated from 293T cells co-expressing WT or D758A_D759A_D764A (DED/AAA) Myc TRAF USP7. Eluent and WCL (input) were then immunoblotted as indicated. (b) ITC profile of binding between Pol ι peptide 438–448 and recombinant UBL1-2. (c) WT or 3KA FLAG-Pol ι was co-expressed in 293T cells with WT or C223S Myc USP7. Whole cell lysates were immunoblotted as indicated. (d) WT Pol ι was co-expressed in 293T cells with Myc USP7 containing C223S, DW164AA and/or DED/AAA mutations, as indicated. WCL was then prepared and immunoblotted as indicated.
To corroborate these findings, we used isothermal titration calorimetry (ITC) to assess binding of peptides containing either putative motif to recombinant UBL1-2. Consistent with our mutational analysis, these analyses revealed an interaction between UBL1-2 and Pol ι peptide 438–448 (Kd = 169 µM; Figure 6(b) and Supplemental Figure 4), confirming the location of the second Pol i-USP7 interface. Although this interaction is weaker than the TRAF Pol ι interaction (Kd = 4.8 µM), mutation of the lysine residues in this region caused an increase in Pol ι monoubiquitination similar to that observed by mutation of the TRAF-binding motif (S580A; Figure 6(c)). In addition, 3KA mutation of Pol ι disrupted deubiquitination by transiently expressed USP7. These findings suggest both Pol ι-USP7 interfaces are required for proper Pol ι deubiquitination. Indeed, mutation of either the TRAF or UBL2 USP7 domains, was sufficient to disrupt Pol ι deubiquitination (Figure 6(d)).
UBL1-2 binding of USP7 substrates can occur in two opposing orientations.35 To determine the orientation of Pol ι bound to USP7 UBL1-2, we titrated 15N isotopically labeled UBL1-2 with Pol ι peptide 438–448 and monitored changes in the 15N-HSQC spectrum at each point of the titration. We then quantified the resulting CSPs and mapped these on the USP7 UBL1-2 structure (Figure 7(a–c). In agreement with our mutational analysis, the largest CSPs were observed for residues I709, S710, D754, E759 and L760, all located on the acidic surface patch of UBL1-2.
Figure 7.
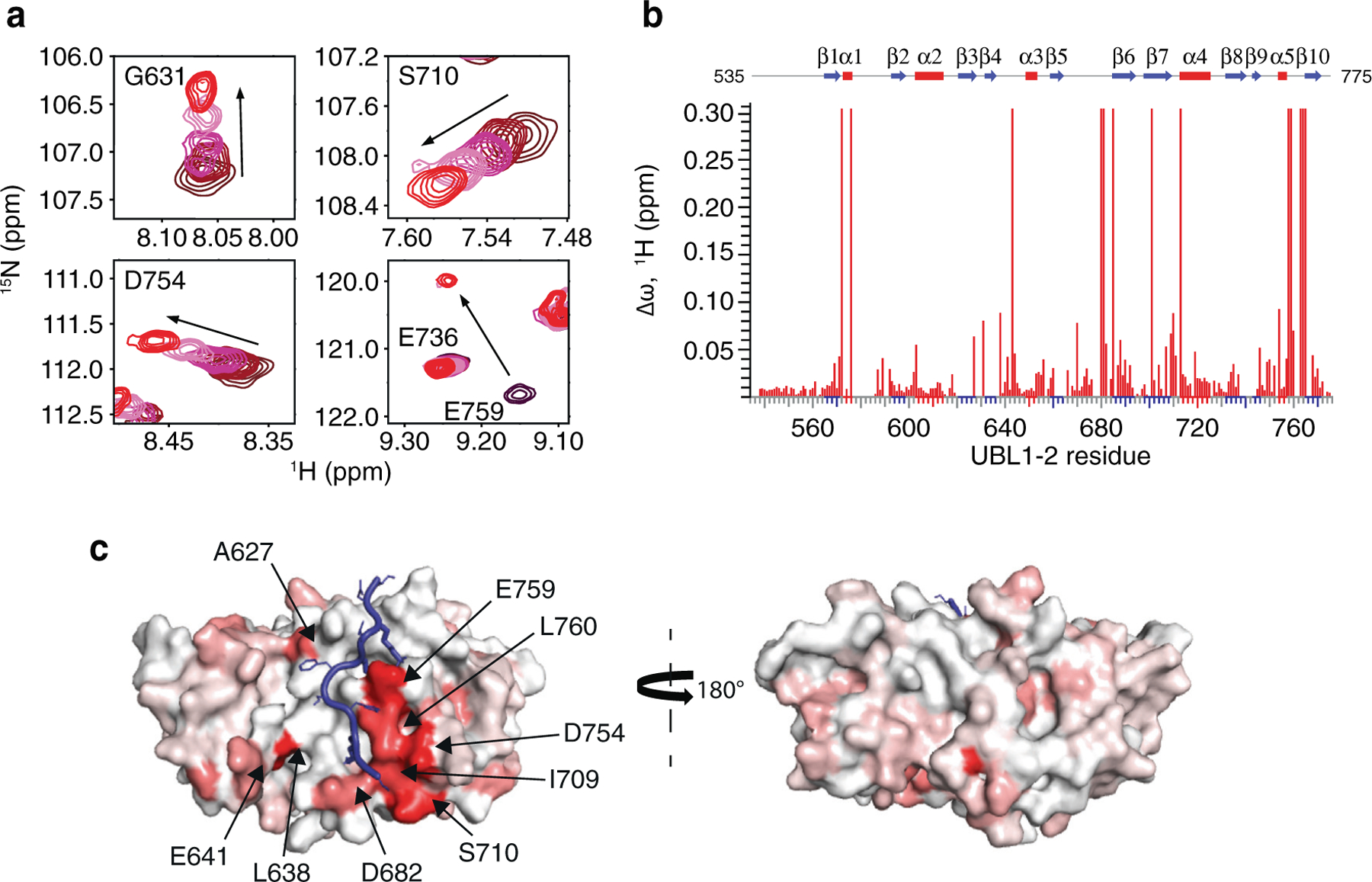
Pol ι binds to a crevice on the surface of USP7 UBL1-2. (a) Select residues of USP7 UBL1-2 domains that demonstrate CSPs when titrated with increasing amounts of Pol ι peptide 438–448 (1:5 protein:peptide molar ratio). (b) Per-residue CSPs (Δω) are quantified and represented as bar plots. Residues that were only observed in “free” or “bound” states due to peak broadening were assigned to the highest observed CSP value. (c) HADDOCK model of the USP7 UBL1-2 domains and Pol ι peptide 438–448 complex. CSPs from NMR experiments were used to map the binding interface, shown in red.
We then used HADDOCK32,33 to calculate a data-based structural model of USP7 UBL1-2 bound to Pol ι peptide 438–448 (Figure 7(c) and Movie 2). For docking, we again generated a structure of the Pol ι peptide using MODELLER. The residues of UBL1-2 with the largest CSPs (>0.08 ppm), and residues whose mutation disrupt the interaction (K440, K444 and K446 of Pol ι and D758, E759 and D764 of UBL1-2), were used as experimental restraints to drive the molecular docking. Both backbone and side chains of the peptide were treated as fully flexible during simulations. The lowest energy cluster of structures had the peptide docked to the acidic patch on UBL1-2, in an orientation similar to that found in USP7-RNF169 and USP7-ICP0 complexes (Supplemental Figure 5). In our model, the large positively charged side chains of K440, K444 and K446 of Pol ι peptide create a trident-like structure that interacts with UBL1-2. Here, K444 and K446 anchor Pol ι into a crevice on the surface of UBL1-2 formed by residues D758, E759, M761, D764 and R634, while K440 makes contacts with UBL1-2 residues D684 and D762. Overall, these data support that Pol ι interacts with both the TRAF and UBL1-2 domains of USP7.
Discussion
USP7 activity relies on a functional catalytic domain, as well as on substrate-binding sites that recognize and recruit substrate proteins. Two substrate-binding sites have previously been identified for USP7; one on the TRAF domain and one on UBL1-2.10 While previously characterized USP7 substrates have been found to interact with only one or the other, in this work, we identify Pol ι as a novel USP7 substrate that interacts with both sites. These interactions occur through distinct regions within the Pol ι C terminus, which although disordered, are well-conserved within mammalian species (Supplemental Figure 6). While we have been unable to assess whether both interactions occur simultaneously, our computational modelling of full-length USP7 bound to both Pol ι peptides, suggest such a bipartite interaction is sterically possible in the context of the full-length enzyme (Figure 8 and Movie 3). Interestingly, USP7 effectively deubiquitinates Pol ι, even though it can be monoubiquitinated at 27 dispersed lysine residues.27 This may reflect the flexibility of the Pol ι C-terminus, which allows for a range of ubiquitinated residues to be placed in proximity of the USP7 catalytic domain, once bound to the USP7 TRAF and UBL1-2 domains.
Figure 8.
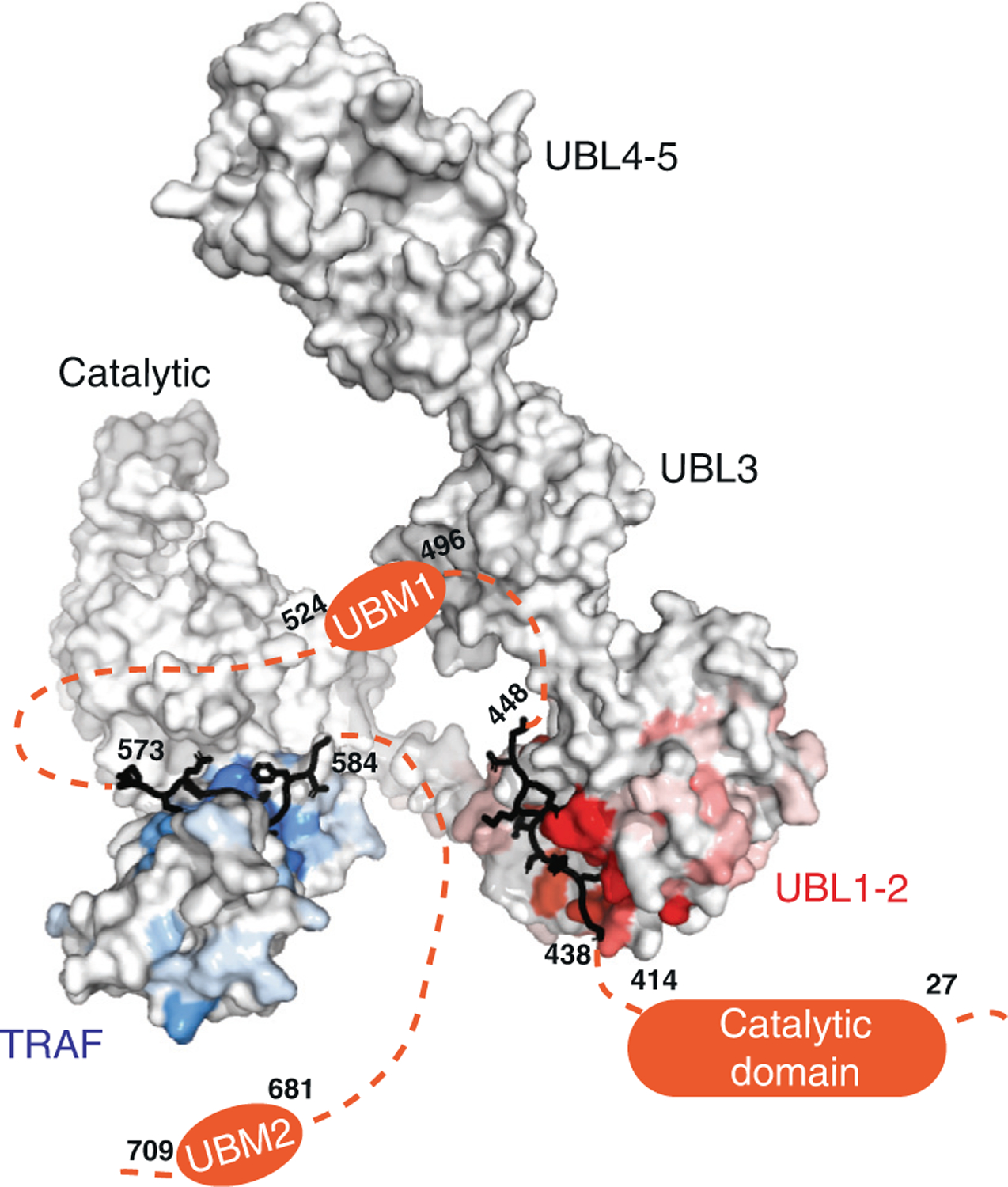
A model of full-length USP7 in complex with Pol ι peptides 573–584 (TRAF-binding) and 438–448 (UBL1-2-binding). The numbers indicate the residue location of Pol ι peptides and domains. The dashed lines indicate unstructured regions of Pol ι.
While Pol ι is the first characterized USP7 substrate that interacts with both substrate-binding sites, previous studies have alluded to dual interactions between USP7 and other substrates. For instance, while p53 and MDM2 have been considered as predominantly TRAF-binding,5–7 there is some evidence to suggest both proteins make secondary interactions with UBL4–5.31 UHRF1, which has largely been considered as a UBL1-2-binding substrate,15 may also bind the USP7 TRAF domain.36 In addition, the interaction between USP7 and the F-box protein FBXO38, was recently found to be impaired following mutation of the USP7 TRAF domain, as well as to a smaller extent, UBL2.37 Although additional work will be required to investigate bipartite binding between these proteins and USP7, these studies nevertheless suggest the possibility of a binding mechanism similar to that between USP7 and Pol ι. Our findings may therefore reflect a general mode of USP7 substrate binding, which will likely be a focus of future work.
Material and Methods
Mammalian expression plasmids
The wild-type FLAG-Pol ι (pCMV6-AN-DDK; pJRM46) mammalian expression vector has been described previously.38 The Cas9-expressing construct pX330-U6-Chimeric_BB-Cbh-hSpCas9 was a gift from Feng Zhang (Addgene plasmid # 42230)39 and was modified by cloning of the annealed primers 5′-caccgAGACCACAC CAAAAAAGCGT-3′ and 5′-aaacACGCTTTTTTGGTGTGGTCTc-3′, into the BbsI restriction sites upstream of the gRNA scaffold. The coding sequence of wild-type USP7 was synthesized by Genscript and subcloned into the KpnI and BamHI restriction sites of pcDNA3.1 (+)-N-Myc. The coding sequence of ubiquitin was similarly synthesized and subcloned into the AsiSI and MluI restriction sites of pCMV6-AN-HA (Origene). Pol ι and USP7 truncation and point mutants were constructed by synthesizing new gene fragments (Genscript) and subcloning into the wild-type plasmids using restriction sites flanking or contained within the coding sequences.
Mammalian cell culture, CRISPR and transfections
HEK293T cells were obtained from ATCC (cell line # CRL-3216). The POLI −/− HEK293T cell line was a gift from Kyungjae Myung and was prepared as a custom service by ToolGen, using Cas9 and guide RNA targeting POLI exon 5 (5′-T TGCACATCAGACTACTTGTTGG-3′). The USP7 −/−/− HEK293T cells were prepared using the Cas9-expressing construct pX330-U6-Chimeric_BB-Cbh-hSpCas9 and a guide RNA targeting USP7 exon 3 (5′-AGACCACAC CAAAAAAGCGT-3′). USP7 +/+/+ and −/−/− clones were identified by western blotting. Exon 3 of knock-out clones was subsequently amplified from genomic DNA by PCR, sequenced using sanger sequencing and indels identified using the Synthego ICE CRISPR Analysis Tool.40 Cell line stocks were ensured to be free of mycoplasma contamination by PCR analysis (ATCC Universal Mycoplasma Detection Kit). Cells were maintained in high glucose Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Thermo Fisher Scientific) containing 10 % fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific) and grown in a humidified incubator with 5% CO2 at 37 °C. Mammalian expression vectors were transfected into these cells with TurboFectin 8.0 (Origene) and typically expressed for 16–20 h prior to cell lysis.
Cell lysis, immunoblotting and antibodies
Whole cell lysates for western blotting were prepared by resuspending HEK293T cells in modified radioimmunoprecipitation buffer (RIPA buffer; 50 mM HEPES pH 8, 150 mM KCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100) supplemented with 1 × protease inhibitor cocktail (cOmplete, EDTA free; Roche, Sigma-Aldrich) and 50 µM PR-619 (Selleckchem), followed by sonication. 10 µg of whole cell lysate was typically separated by electrophoresis on a 15-well 1.5 mm 4–12% Bis-Tris NuPage precast gel (Thermo Fisher Scientific) and transferred to nitrocellulose, before incubation with primary antibodies. Antibodies against USP7/HAUSP (1:1000; clone D17C6, cat # 4833S), β-actin (1:5000; clone 13E5, cat # 4970S), Myc (1:2000; clone 9B11, cat # 2276S) HA (1:2000; clone C29F4, cat # 3724S) and H3 (1:4000; clone 1B1B2, cat # 14269S) were purchased from Cell Signaling Technology. The FLAG antibody was from Sigma-Aldrich (1:4000; clone M2, cat # F1804) and the Pol ι antibody was from Abnova (1:1000; clone M01, cat # H00011201-M01). Primary antibodies were detected using IRDye 680RD or 800CW-conjugated donkey anti-mouse or anti-rabbit fluorescent secondary antibodies (Li-Cor) and visualized using an Odyssey CLX infrared imaging system (Li-Cor). Where indicated, immunoblots were quantified using Image Studio Lite software (Li-Cor). Relative monoubiquitination of FLAG Pol ι was calculated as the ratio of monoubiquitinated to non-ubiquitinated Pol ι and was normalized to the ratio calculated for samples not overexpressing USP7. For endogenous Pol ι, the ratio of monoubiquitination was calculated against the sum of all endogenous Pol ι bands.
Immunoprecipitation
HEK293T cells were resuspended and sonicated in immunoprecipitation buffer (20 mM HEPES pH 7.5, 150 mM KCl, 5% glycerol, 10 mM MgCl2, 0.5% Triton X-100) supplemented with 1 × protease inhibitor cocktail, 50 µM PR-619 (Selleckchem) and Pierce Universal Nuclease for Cell Lysis (1:5000, Thermo Fisher Scientific). Where immunoprecipitated proteins were analyzed by mass spectrometry, 0.1% CHAPS was used in place of 0.5% Triton X-100. For immunoprecipitation of Pol ι or Myc-tagged proteins, protein G magnetic Dynabeads (Life Technologies) were prepared by incubation with the Pol ι or Myc antibodies, respectively. For Pol ι co-immunoprecipitation experiments, isotype control IgG-conjugated beads were also prepared by incubating Dynabeads with the Cell Signaling IgG XP isotype control rabbit mAb (clone DA1E, cat # 3900S). Magnetic anti-FLAG M2 beads (Sigma-Aldrich, cat # M8823) were used for the immunoprecipitation of FLAG-tagged proteins. In all cases, conjugated beads were washed in immunoprecipitation buffer, then incubated with whole cell lysates for 1 hour at 4 °C. Beads were then washed 4 × with immunoprecipitation buffer, 2 × with immunoprecipitation buffer modified to contain 250 mM KCl and then proteins eluted by incubating the beads in 100 mM pH 2.3 glycine for 10 min, followed by neutralization of the sample with 500 mM Tris pH 7.4.
Mass spectrometry
Wild-type FLAG-Pol ι, or an empty vector, was transiently expressed in 293T cells for 16 hours. Cells were then resuspended in CHAPS-containing immunoprecipitation buffer, lysed by sonication, and proteins immunoprecipitated from 3 mg of whole cell lysate, using 15 µL of magnetic anti-FLAG M2 beads. The immunoprecipitated samples were electrophoresed 1 cm into a 1 mm 4–12% Bis-Tris NuPage precast gel and then excised as a single gel slice. Gel slices were then washed twice for 2 minutes with 50% acetonitrile in water before being frozen at −20 °C. Samples were then transported on dry ice to the Harvard University Mass Spectrometry and Proteomics Resource Laboratory, where proteins were identified as a custom service. Here, gel pieces were reduced by incubation for 45 min at 37 °C in 20 mM TCEP (Tris-[2-catboxyethyl] phosphine) in 25 mM TEAB (triethylammonium bicarbonate). Samples were then cooled to room temperature, TCEP removed, and samples alkylated by incubation in 10 mM iodoacetamide for 45 min. To digest proteins, gel pieces were incubated in 0.06 µg/5 µL of trypsin (Promega cat # V5111) in 50 mM TEAB overnight at 37 °C. Peptides were extracted with 50 µL 5% formic acid in 50% acetonitrile and dried to a volume of 30 µL. Samples were then analyzed using a Thermo Scientific Orbitrap mass analyzer and a Thermo Scientific Velos Pro, using the following two databases to search on Thermo Electron’s Proteome Discoverer 2.1 software: Contaminants2Append.fasta and Uniprot_ Human2016_SPonly.fasta.
Constructs for bacterial expression
The USP7 UBL1-2 construct (residues 535–775)12 was a gift from Dr. Vivian Saridakis (York University, Toronto, Canada). This construct expresses USP7 UBL1-2 with an N-terminal 6-His tag, separated by a tobacco etch virus (TEV) protease cleavage site. The codon-optimized sequence of USP7 TRAF (residues 62–205) was chemically synthesized and subcloned into pET-28b + using NdeI and EcoRI cloning sites (Genscript).
USP7 expression and purification
USP7 TRAF and USP7 UBL1-2 were expressed and purified in a similar manner. Briefly, USP7 TRAF (62–205) and UBL1-2 (535–775) plasmids were transformed into Escherichia coli BL21(DE3) cells. Unlabeled proteins used for ITC experiments were expressed in 1 L of Luria broth (LB) media and 15N-labelled proteins used in NMR titration experiments were expressed in M9 minimal media containing 15NH4Cl as a sole source of nitrogen. Double labelled 15N/13C TRAF used in the backbone resonance assignment experiments was expressed in M9 media containing 15NH4Cl and 13C glucose as the sole nitrogen and carbon sources. Transformed cells were grown at 37 °C until A600 of 0.8–1.0 o. u. Protein expression was induced with either 500 µM (TRAF) or 1 mM (UBL1-2) isopropyl 1-thi σ-β-ᴅ-galactopyranoside (IPTG) overnight at 20 °C.
Cells were harvested by centrifugation, resuspended in a buffer containing 20 mM phosphate buffer pH 8, 250 mM NaCl, 10 mM imidazole, 1 mM PMSF, lysed by sonication, and centrifuged at 15,000 rpm for 1 hour. The supernatant was filtered and applied to a TALON HisPur cobalt resin (Thermo Scientific). Proteins were eluted in a buffer containing 20 mM phosphate buffer pH 8, 250 mM NaCl and 300 mM imidazole. TEV protease was then added to samples overnight at 4 °C to remove the 6-His tag. Proteins were then subjected to size-exclusion chromatography using a HiLoad Superdex 200 column (GE Healthcare) in a buffer containing 20 mM Tris pH 7.4, 100 mM NaCl and 10 mM dithiothreitol (DTT).
Commercial proteins
His6-USP7 was purchased from Boston Biochem (cat # E-519), while GST and GST-USP7 were purchased from UBP Bio (cat # I1100 and cat # H5710, respectively). His-Pol ι was purified as a custom service by Protein One.
Pull-down assay
To capture the USP7-Pol ι interaction, 1.5 lg of His Pol ι was incubated with equimolar quantities of GST (0.5 µg) or GST USP7 (2.3 µg) in 100 µL GST pull down buffer (50 mM HEPES, 150 mM KCl, 0.1 mM EDTA, 0.01% Triton X-100, pH 7.5) at 4 °C for 30 min, prior to addition of 5 µL glutathione sepharose 4 fast flow (GE) beads and resuspended in 100 µL of buffer. Samples were incubated at 4 °C for a further 2 h, washed 6x with GST pull down buffer and then proteins eluted by heating to 70 °C in 20 µL of loading buffer. The interaction was visualized by separating 10% of the input and flow through, as well as 50% of the eluent, on a NuPage gel followed by staining with colloidal Coomassie blue.
USP7 in vitro deubiquitination assay
Wild-type or UBM1*2* FLAG Pol ι was immunoprecipitated from 6000 µg of HEK293T cell lysate using 60 µL of M2 magnetic bead slurry and eluted with 40 µL glycine as above. Proteins were then diluted to 150 µL in DUB assay buffer (50 mM HEPES, pH 7.5, 50 mM NaCl 5 mM DTT, 1 mM EDTA, 0.01% Triton X-100) and aliquots snap frozen. For each reaction, 10% (15 µL) of the eluted FLAG Pol ι was incubated in the presence or absence of 1 µL recombinant human His6 USP7 for 4 hours at 37 °C. Reactions were stopped by heating in SDS loading dye, and reactions separated on a Bis-Tris NuPage precast gel. Proteins were then detected by staining with colloidal Coomassie blue and imaged using a LiCor Odyssey CLX.
Pol ι peptides
The following Pol ι peptides containing P/A/ExxS as well as forward and reverse KxxxKxK/R motifs were chemically synthesized (GenScript): Pol ι 421–440 (KGLIDYYLMPSLSTTSRSGK), Pol ι 491–501 (NEFPLCSLPEG), Pol ι 513–523 (DIQE EILSGKS), Pol ι 573–584 (SPCEPGTSGFN), Pol ι 438–448 (SGKHSFKMKDT) and Pol ι 523–532 (SREKFQGKGS). For binding studies, peptides were dissolved in the respective buffers listed under NMR and ITC experimental methods.
NMR assignment
The sample used for NMR backbone resonance assignment contained 0.9 mM 15N/13C TRAF in 20 mM phosphate buffer, pH 7.5, 250 mM NaCl, 10 mM DTT and 10% D2O (v/v). Standard two-dimensional 1H-15N HSQC and 1H-13C HSQC and three-dimensional HNCA, HNCACB, HNCO and 15N-NOESY NMR experiments41 were collected on 800 MHz (1H) Agilent VNMRS spectrometer at 30 °C. Data was processed using NMRPipe42 and analyzed using Sparky.43 All data processing and analysis was performed on the NMRbox platform44 (National Center for Biomolecular NMR Data Processing and Analysis, a Biomedical Technology Research Resource (BTRR) supported by NIH grant P41GM111135 (NIGMS)). 114 of the 136 non-proline residues were assigned, which constitutes 84% of backbone resonances. NMR resonance assignment was deposited to BMRB data bank (entry 50080). NMR backbone resonance assignment for USP7 UBL1-2 was transferred from BMRB data bank (entry 26782).
NMR titration experiments
All 15N USP7 TRAF NMR experiments were collected at 500 MHz (1H) Agilent VNMRS spectrometer at 30 °C. To test TRAF binding to Pol ι peptides containing P/A/ExxS motif, a 3 mM stock of unlabeled peptide was gradually added to 0.25 mM 15N TRAF to a molar ratio of 1:5 (protein: peptide). 1H-15N HSQC spectrum was collected at each of the six titration points. TRAF domain and the peptides were in a buffer containing 20 mM phosphate buffer pH 7.5, 250 mM NaCl, 10 mM DTT and 10% D2O (v/v).
All 15N USP7 UBL1-2 NMR experiments were collected at 800 MHz (1H) Agilent VNMRS spectrometer at 25 °C. To test UBL1-2 binding to Pol ι peptides containing forward and reverse KxxxKxK motifs, a 3 mM stock of unlabeled peptide was gradually added to 0.25 mM 15N UBL1-2 to a molar ratio of 1:5 (protein:peptide). 1H-15N HSQC spectrum was collected for each of the six titration points. UBL1-2 and the peptides were in a buffer containing 20 mM phosphate buffer pH 7, 100 mM NaCl, 10 mM DTT and 10% D2O (v/v).
Data was processed using NMRPipe42 and analyzed using Sparky.43 All data processing and analysis was performed on NMRbox44 platform. Observed frequency perturbations for each amino acid residue were calculated using the equation: where ΔωN and ΔωH are the frequency differences between free and peptide-bound samples measured in Hz for 15N and 1H, respectively. The frequency perturbations were converted to chemical shift perturbations by dividing Δωobs by 1H frequency of the spectrometer. The obtained Δωobs values in 1H ppm were mapped onto the HADDOCK model of the respective protein.
Isothermal titration calorimetry (ITC)
All measurements were collected using a Nano ITC instrument (TA Instruments). ITC data were analyzed with NanoAnalyze software (TA Instruments). Data fitting was performed using an “independent” model after correcting for the heat of peptide’s dilution.
USP7 TRAF was dialyzed into a buffer containing 20 mM phosphate buffer pH 8 and 250 mM NaCl. Pol ι peptides containing the P/A/ExxS motif were resuspended in this same buffer to 1 mM concentration and titrated into 100 mM USP7 TRAF. Experiments were performed at 4 °C with a total of 20 injections made with 200 second time intervals and 300 rpm mixing speed.
USP7 UBL1-2 was dialyzed into a buffer containing 20 mM phosphate buffer pH 7 and 100 mM NaCl. Pol ι peptides containing KxxxKxK motif were resuspended in the same buffer to 1 mM and titrated into 100 mM USP7 UBL1-2. A total of 20 injections were made with 300 second time intervals and 300 rpm mixing speed at 25 °C.
Homology modelling
The homology model of Pol ι peptide 573–584 was generated using MODELLER34 with TRAF/Mdm2 complex (PDB ID: 2fop) serving as a template. The homology model of peptide 438–448 was based on RNF169 peptide in complex with UBL1-2 (PDB ID: 5gg4). The model of the full-length USP7 was generated using MODELLER with overlapping structures of USP7 domains (PDB ID: 4yoc, 2flz and 2ylm) serving as a template.
Molecular docking
Pol ι peptides were docked onto USP7 TRAF and UBL1-2 domains using HADDOCK.32,33
TRAF: Active residues of TRAF were D164 W165 and passive residues were those with CSPs > 0.06 that were surface exposed (M100, F118, Q120, C121, N163, N169). Active residues for Pol ι peptide 573–584 were selected as the PGTS motif and all other residues of the peptide were selected as passive residues.
For the TRAF/Pol ι complex, HADDOCK clustered 107 structures in 3 clusters. The cluster with the best score contained 73/107 clusters. (HADDOCK score = −70.5 ± 1.8, Cluster size = 76, RMSD from the overall lowest-energy structure = 5.0 ± 0.1, = Van der Waals energy = −34.6 ± 3.6, Electrostatic energy = −84.7 ± 20.4, Desolvation energy = −19.0 ± 2.5, Restraints violation energy = 0.5 ± 0.26, Buried Surface Area = 893.7 ± 13.8, Z-Score = −1.3)
UBL1-2: Active residues: D758, E759 D764 were designated as “active” residues based on mutational analysis that demonstrates mutating these residues causes a loss of interaction between Pol ι and USP7 UBL1-2. Residues with CSPs > 0.08 (E572, C576, L638, D643, D680, K681, V685, Y701, S710, I713, D754, G763 and I765) were designated as “passive” residues in this interaction. Active residues for Pol ι peptide 438–448 include K440, K444, and K446 while the remaining residues of the peptide were designated as “passive” residues in this interaction. The entire peptide was designated as “flexible.”
For the UBL1-2/Pol ι complex, HADDOCK clustered 160 structures in 3 clusters. The cluster with the best score contained 95/160 clusters. (HADDOCK score −143.6 ± 5.6, cluster size = 95, RMSD from the overall lowest-energy structure = 0.5 ± 0.3, Van der Waals energy = −35.2 ± 1.5, Electrostatic energy = −564.1 ± 24.7, Desolvation energy = 4.4 ± 2.8, Restraints violation energy = 0. 0 ± 0.00, Buried Surface Area = 1432.7 ± 33.2, Z-S core = −1.3).
Phylogenetic alignment
The amino acid sequences of proteins homologous to human Poli were obtained from the NCBI Protein database. Accession numbers are: human (Homo sapiens) = NP_001338561.1; common chimpanzee (Pan troglodytes) = NP_001266739.1; Rhesus macaque (Macaca mulatta) = XP_014977486.1; house mouse (Mus musculus) = NP_036102.2; cattle (Bos taurus) = NP_001019863; domestic dog (Canis lupus familiaris) = XP_005615447.1; sperm whale (Physeter macrocephalus) = XP_023988058.1; southern white rhinoceros (Ceratotherium simum simum) = XP_014636911.1; black flying fox (Pteropus alecto) = XP_024894311.1; African elephant (Loxodonta africana) = XP_023399547.1; west Indian manatee (Trichechus manatus) = XP_023589726.1; koala (Phascolarctos cinereus) = XP_020849802.1; platypus (Ornithorhynchus anatinus) = XP_028916153.1. Sequences were initially aligned using Clustal Omega (ebi.ac.uk). Regions of interest were subsequently isolated and aligned using T-Coffee (tcoffee.crg.cat). Fasta_aln files of this analysis were then exported into and formatted using BoxShade (embnet.vital-it.ch/software/BOX_form.html).
Data availability
NMR assignment and chemical shift perturbations have been deposited to Biological Magnetic Resonance Data Bank (BMRB; entry 50080) and complex structures have been deposited to pdb-DEV. Mammalian expression vectors have been deposited in the Addgene public plasmid repository. Pol ι-associating proteins identified by GeLC-MS/MS in Figure 1 are presented in Dataset 1.
Supplementary Material
Acknowledgement
We thank Kyungjae Myung (Institute for Basic Research, UNIST, Ulsan, S. Korea) for providing us with the POLI −/− 293T cell line, Alexandra Pozhidaeva (UConn) for her help with NMR resonance assignments of USP7, Vivian Saridakis (York University, Canada) for the UBL1-2 and TRAF plasmids, Dmitry Korzhnev (UConn) for his help with NMR experiments and Justin Radolf (UConn) for critically reading the manuscript.
Funding
This work was supported by funds from the National Institute of Child Health and Human Development (NICHD)/National Institutes of Health (NIH) Intramural Research Program [to RW], as well by NIH grants [R35 GM128864, P41 GM111135 and R01 GM123249 to IB] and funding from the National Science Foundation [NSF 1616184 to IB]. Funding for open access charge: NICHD/NIH Intramural Research Program.
Footnotes
Conflict of interest
The authors declare they have no conflict of interest.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2020.166733.
References
- 1.Ronau JA, Beckmann JF, Hochstrasser M, (2016). Substrate specificity of the ubiquitin and Ubl proteases. Cell Res, 26, 441–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reyes-Turcu FE, Ventii KH, Wilkinson KD, (2009). Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem, 78, 363–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sowa ME, Bennett EJ, Gygi SP, Harper JW, (2009). Defining the human deubiquitinating enzyme interaction landscape. Cell, 138, 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fountain MD, Oleson DS, Rech ME, Segebrecht L, Hunter JV, McCarthy JM, Lupo PJ, Holtgrewe M, et al. , (2019). Pathogenic variants in USP7 cause a neurodevelopmental disorder with speech delays, altered behavior, and neurologic anomalies. Genet. Med, 21, 1797–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y, (2006). Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53–MDM2 pathway. PLoS Biol, 4, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, Frappier L, (2006). Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat. Struct. Mol. Biol, 13, 285–291. [DOI] [PubMed] [Google Scholar]
- 7.Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, Nguyen T, Zhang RG, Liao J, et al. , (2005). Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1. Mol. Cell, 18, 25–36. [DOI] [PubMed] [Google Scholar]
- 8.Sarkari F, La Delfa A, Arrowsmith CH, Frappier L, Sheng Y, Saridakis V, (2010). Further insight into substrate recognition by USP7: structural and biochemical analysis of the HdmX and Hdm2 interactions with USP7. J. Mol. Biol, 402, 825–837. [DOI] [PubMed] [Google Scholar]
- 9.Jagannathan M, Nguyen T, Gallo D, Luthra N, Brown GW, Saridakis V, Frappier L, (2014). A role for USP7 in DNA replication. Mol. Cell. Biol, 34, 132–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holowaty MN, Sheng Y, Nguyen T, Arrowsmith C, Frappier L, (2003). Protein interaction domains of the Ubiquitin-specific protease, USP7/HAUSP. J. Biol. Chem, 278, 47753–47761. [DOI] [PubMed] [Google Scholar]
- 11.Rougé L, Travis KM, Tong R, Paola I, Maurer T, James MJ, (2016). Molecular understanding of USP7 substrate recognition and C-terminal activation. Structure, 24, 1335–1345. [DOI] [PubMed] [Google Scholar]
- 12.Pfoh R, Lacdao IK, Georges AA, Capar A, Zheng H, Frappier L, Saridakis V, (2015). Crystal structure of USP7 ubiquitin-like domains with an ICP0 peptide reveals a novel mechanism used by viral and cellular proteins to target USP7. PLoS Path, 11, e1004950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faesen A, Dirac A, Shanmugham A, Ovaa H, Perrakis A, Sixma T, (2011). Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol. Cell, 44, 147–159. [DOI] [PubMed] [Google Scholar]
- 14.Cheng J, Yang H, Fang J, Ma L, Gong R, Wang P, Li Z, Xu Y, (2015). Molecular mechanism for USP7mediated DNMT1 stabilization by acetylation. Nat. Commun, 6, 7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma H, Chen H, Guo X, Wang Z, Sowa ME, Zheng L, Hu S, Zeng P, et al. , (2012). M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc. Natl. Acad. Sci. USA, 109, 4828–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An L, Jiang Y, Ng HHW, Man EPS, Chen J, Khoo U-S, Gong Q, Huen MSY, (2017). Dual-utility NLS drives RNF169-dependent DNA damage responses. Proc. Natl. Acad. Sci. USA, 114, E2872–E2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pozhidaeva AK, Mohni KN, Dhe-Paganon S, Arrowsmith CH, Weller SK, Korzhnev DM, Bezsonova I, (2015). Structural characterization of interaction between human ubiquitin-specific protease 7 and immediate-early protein ICP0 of herpes simplex virus1. J. Biol. Chem, 290, 22907–22918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pozhidaeva A, Bezsonova I, (2019). USP7: structure, substrate specificity, and inhibition. DNA Repair, 76, 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kashiwaba S-I, Kanao R, Masuda Y, KusumotoMatsuo R, Hanaoka F, Masutani C, (2015). USP7 is a suppressor of PCNA ubiquitination and oxidative-stressinduced mutagenesis in human cells. Cell Rep, 13, 2072–2080. [DOI] [PubMed] [Google Scholar]
- 20.Zlatanou A, Sabbioneda S, Miller ES, Greenwalt A, Aggathanggelou A, Maurice MM, Lehmann AR, Stankovic T, et al. , (2016). USP7 is essential for maintaining Rad18 stability and DNA damage tolerance. Oncogene, 35, 965–976. [DOI] [PubMed] [Google Scholar]
- 21.Alonso-De Vega I, Martín Y, Smits VA, (2014). USP7 controls Chk1 protein stability by direct deubiquitination. Cell Cycle, 13, 3921–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian J, Pentz K, Zhu Q, Wang Q, He J, Srivastava AK, Wani AA, (2015). USP7 modulates UV-induced PCNA monoubiquitination by regulating DNA polymerase eta stability. Oncogene, 34, 4791–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kannouche P, Fernández De Henestrosa AR, Coull B, Vidal AE, Gray C, Zicha D, Woodgate R, Lehmann AR, (2002). Localization of DNA polymerases η and ι to the replication machinery is tightly co-ordinated in human cells. EMBO J, 21, 6246–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petta TB, Nakajima S, Zlatanou A, Despras E, Couve-Privat S, Ishchenko A, Sarasin A, Yasui A, et al. , (2008). Human DNA polymerase iota protects cells against oxidative stress. EMBO J, 27, 2883–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J,Chen Z,Liu Y,Hickey RJ,Malkas LH,(2004). Altered DNA polymerase ι expression in breast cancer cells leads to a reduction in DNA replication fidelity and a higher rate of mutagenesis. Cancer Res, 64, 5597–5607. [DOI] [PubMed] [Google Scholar]
- 26.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, et al. , (2005). Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science, 310, 1821–1824. [DOI] [PubMed] [Google Scholar]
- 27.McIntyre J, McLenigan MP, Frank EG, Dai X, Yang W, Wang Y, Woodgate R, (2015). Posttranslational regulation of human DNA polymerase ι. J. Biol. Chem, 290, 27332–27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoeller D, Hecker C-M, Wagner S, Rogov V, Dötsch V, Dikic I, (2007). E3-independent monoubiquitination of ubiquitin-binding proteins. Mol. Cell, 26, 891–898. [DOI] [PubMed] [Google Scholar]
- 29.Frank EG, McLenigan MP, McDonald JP, Huston D, Mead S, Woodgate R, (2017). DNA polymerase ι: the long and the short of it!. DNA Repair, 58, 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z-M, Scott D, Cai Q, Joseph LL, Wang Y, Brian G, Song J, (2015). An allosteric interaction links USP7 to deubiquitination and chromatin targeting of UHRF1. Cell Rep, 12, 1400–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma J, Martin JD, Xue Y, Lor LA, Kennedy-Wilson KM, Sinnamon RH, Ho TF, Zhang G, et al. , (2010). C-terminal region of USP7/HAUSP is critical for deubiquitination activity and contains a second mdm2/p53 binding site. Arch. Biochem. Biophys, 503, 207–212. [DOI] [PubMed] [Google Scholar]
- 32.Dominguez C, Boelens R, Bonvin AMJJ, (2003). HADDOCK: a proteinprotein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc, 125, 1731–1737. [DOI] [PubMed] [Google Scholar]
- 33.Van Zundert GCP, Rodrigues JPGLM, Trellet M, Schmitz C, Kastritis PL, Karaca E, Melquiond ASJ, Van Dijk M, et al. , (2016). The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol, 428, 720–725. [DOI] [PubMed] [Google Scholar]
- 34.Šali A, Blundell TL, (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol, 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 35.Cheng J, Li Z, Gong R, Fang J, Yang Y, Sun C, Yang H, Xu Y, (2015). Molecular mechanism for the substrate recognition of USP7. Protein Cell, 6, 849–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felle M, Joppien S, Németh A, Diermeier S, Thalhammer V, Dobner T, Kremmer E, Kappler R, et al. , (2011). The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res, 39, 8355–8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Georges A,Coyaud E, Marcon E, Greenblatt J,Raught B, Frappier L, (2019). USP7 regulates cytokinesis through FBXO38 and KIF20B. Sci. Rep, 9, 2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McIntyre J, Vidal AE, McLenigan MP, Bomar MG, Curti E, McDonald JP, Plosky BS, Ohashi E, et al. , (2013). Ubiquitin mediates the physical and functional interaction between human DNA polymerases η and ι. Nucleic Acids Res, 41, 1649–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, et al. , (2013). Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsiau T, Conant D, Rossi N, Maures T, Waite K, Yang J, Joshi S, Kelso R, et al. , (2019). Inference of CRISPR edits from sanger trace data. bioRxiv, 251082 [DOI] [PubMed] [Google Scholar]
- 41.Sattler M, Schleucher J, Griesinger C, (1999). Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Mag. Res. Sp, 34, 93–158. [Google Scholar]
- 42.Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A, (1995). NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR, 6 [DOI] [PubMed] [Google Scholar]
- 43.Goddard TD, Kneller DG SPARKY 3. University of California, San Francisco: Available at: http://www.cgl.ucsf.edu/home/sparky/. [Google Scholar]
- 44.Maciejewski MW, Schuyler AD, Gryk MR, Moraru II, Romero PR, Ulrich EL, Eghbalnia HR, Livny M, et al. , (2017). NMRbox: a resource for biomolecular NMR computation. Biophys. J, 112, 1529–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NMR assignment and chemical shift perturbations have been deposited to Biological Magnetic Resonance Data Bank (BMRB; entry 50080) and complex structures have been deposited to pdb-DEV. Mammalian expression vectors have been deposited in the Addgene public plasmid repository. Pol ι-associating proteins identified by GeLC-MS/MS in Figure 1 are presented in Dataset 1.


