Abstract
DNA nanotechnology has progressed from proof-of-concept demonstrations of structural design towards application-oriented research. As a natural material with excellent self-assembling properties, DNA is an indomitable choice for various biological applications, including biosensing, cell modulation, bioimaging and drug delivery. However, a major impediment to the use of DNA nanostructures in biological applications is their susceptibility to attack by nucleases present in the physiological environment. Although several DNA nanostructures show enhanced resistance to nuclease attack compared with duplexes and plasmid DNA, this may be inadequate for practical application. Recently, several strategies have been developed to increase the nuclease resistance of DNA nanostructures while retaining their functions, and the stability of various DNA nanostructures has been studied in biological fluids, such as serum, urine and cell lysates. This Review discusses the approaches used to modulate nuclease resistance in DNA nanostructures and provides an overview of the techniques employed to evaluate resistance to degradation and quantify stability.
Subject terms: DNA, Nanobiotechnology, DNA nanostructures
DNA nanostructures are increasingly used in biological applications, in which nuclease resistance is a key parameter. This Review discusses the different strategies used to modulate and evaluate the nuclease resistance of DNA nanostructures.
Introduction
It has been almost 40 years since Ned Seeman published his thoughts on using DNA as a building block for the assembly of nanostructures, a field now called DNA nanotechnology1. Seeman’s original vision was to use DNA as a framework to crystallize other hard-to-crystallize macromolecules. Early work on DNA-based construction mostly involved the creation of DNA junctions, motifs and devices2–4, and, over the past four decades, multiple strategies have been developed to create diverse architectures using DNA5. The library of DNA nanostructures, ranging from the nanometre to the micrometre scale6–8, now includes geometric patterns9, polyhedral objects10, nanoscale bunnies11, a box with a lid12, a cargo-moving assembly line13 and even a miniature Mona Lisa14. More recently, the focus has moved from proof-of-concept construction using DNA towards the applications of these structures. Being a natural material, DNA nanostructures have found the most use in biological applications15–17. Indeed, nanocages built from DNA have been used in drug delivery18, reconfigurable DNA devices in biosensing19 and DNA structures conjugated to fluorophores and ligands for imaging20 and modulating cellular behaviour21. Although each of these applications has specific prerequisites, one common requirement is that the DNA nanostructures should remain stable under physiological conditions and be able to withstand degradation by nucleases.
For practical application, DNA nanostructures need to meet certain biostability thresholds. For example, in biosensing, DNA nanodevices must remain intact when mixed with samples (typically, biofluids such as blood, serum and urine), as unpredictable and sudden degradation can lead to false signals. In drug delivery, the biostability of DNA nanostructures is important to protect the drug from harsh physiological conditions until it reaches the destination site in the body. In cellular modulation and bioimaging, DNA nanostructures need to remain largely intact in the cellular or bodily environment to complete their functions (to deliver regulating proteins or accumulate at target tissues, respectively). A major reason for degradation of DNA nanostructures in these conditions is the presence of nucleases (enzymes that degrade nucleic acids). To address this issue, strategies have been developed to modulate the biostability of DNA nanostructures and to increase their suitability for biological applications.
In this Review, I discuss the inherent biostability of DNA nanostructures and the potential use of nanostructures with enhanced nuclease resistance in biological applications. In particular, I focus on the methods developed to tune the nuclease resistance of these structures, the techniques used to analyse their nuclease resistance and the comparative analysis of results from key studies. More general ideas of DNA nanotechnology are beyond the scope of this Review, and interested readers are directed to other reviews covering a range of topics, including biosensing22–24, drug delivery25–27, bioimaging28–30, cellular programming31,32 and scaffolding33,34.
Construction using DNA
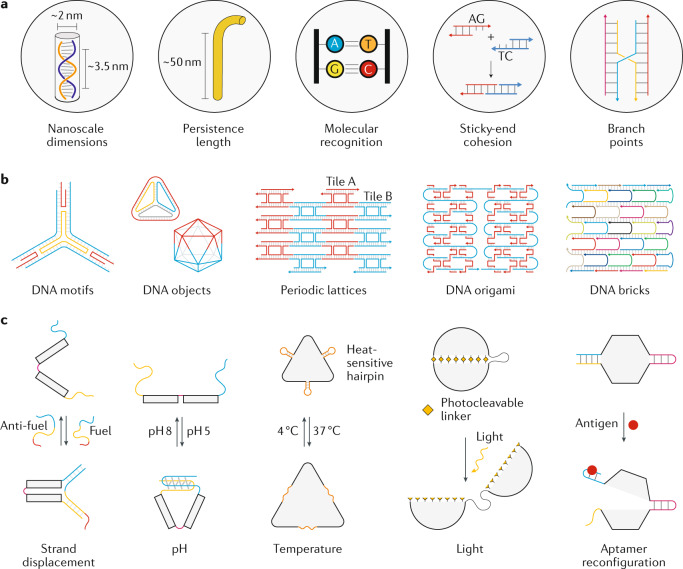
DNA is well known as the carrier of genetic information, but is also a natural nanoscale material. The characteristics of DNA that make it a suitable building block for nanoscale architectures include its nanoscale dimensions, persistence length of ~50 nm (that is, its ‘stiffness’ for robust construction) and predictable Watson–Crick base pairing, which allows for programmed assembly (Fig. 1a). Furthermore, multidimensional assembly is possible by creating branched DNA structures that can be connected by short single-stranded extensions (sticky ends) to create larger assemblies. Building using DNA has led to two areas of DNA nanotechnology: structural and dynamic. Structural DNA nanotechnology mainly involves the design and construction of DNA-based structures with different geometries, periodicity or spatial patterns (Fig. 1b). Researchers have created various DNA nanostructures using tile-based assembly35,36, DNA origami37 and DNA brick strategies38. In dynamic DNA nanotechnology, DNA structures are designed to reconfigure upon recognition of external moieties, such as nucleic acids39,40, antigens12,41 or ligands40,42, or in response to environmental changes, including changes in pH43,44, temperature45 or light46,47 (Fig. 1c). Such stimuli-responsive devices are useful in biosensing and as components for triggered release in drug-delivery carriers. Construction principles for DNA nanostructures, assembly strategies and the types of motifs and structures have been discussed elsewhere3,6,48,49.
Fig. 1. Concept, design and construction of DNA nanostructures.
a | Properties of DNA that make it suitable for the bottom-up construction of nanostructures. b | Structural DNA nanotechnology involves the construction of DNA-based structures with different geometries, periodicities or spatial patterns, including motifs36, objects such as polyhedra35,144, periodic 2D lattices145, DNA origami37 and DNA bricks38. c | Dynamic DNA nanotechnology involves the construction of DNA devices that operate in response to external stimuli, such as toehold-based strand displacement for nucleic acids107, pH changes43, temperature changes45, light46 (ultraviolet or near-infrared, for example) and antigen–aptamer interactions41.
DNA nanotechnology applications
Developments in DNA nanotechnology have not always adhered to the principle of ‘form follows function’: in most cases, the function of a DNA nanostructure has followed its form. Demonstrations of the applications of DNA in different fields have increased exponentially in the past decade50,51. In materials science, DNA can serve as a copolymer for creating integrated chips52,53, pooled DNA libraries are used in data storage and molecular computation54,55, and DNA-based moulds have been used for the assembly of metallized nanosheets and electrical nanowires56,57. In chemistry, DNA has been used in directing polymer construction58 and site-specific chemical reactions59, and as scaffolds for the structure determination of macromolecules60–62. However, most of the recent studies are in biological applications, some of which are briefly discussed below.
Biosensing
DNA-based biosensors are cost-effective and sensitive63,64, and are being developed for use as point-of-care diagnostic tools (Fig. 2a). DNA-based sensing strategies rely on the recognition event between the DNA nanostructure and a target analyte (such as in nucleic acid and protein sensing) or the conformational change of a DNA-based device (upon sensing, for example, pH changes or light)22. Advantages of DNA nanostructures for biosensing include the highly precise design based on base pairing, the large surface-to-volume ratios and the low-cost synthesis of functionalized DNA strands. So far, DNA nanostructures have been used to detect nucleic acids (including microRNAs19, viral RNAs65 and pathogenic DNA66), antigens67, antibodies68 and small molecules40, as well as in sensing in vivo pH changes43.
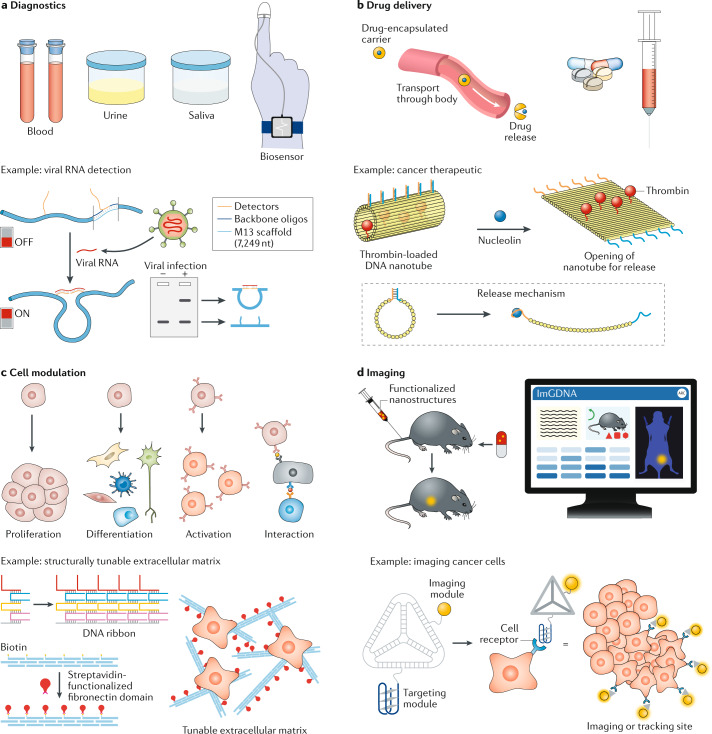
Fig. 2. Biological applications of DNA nanostructures.
a | DNA nanostructures can be used in diagnostics for disease detection and monitoring. The example shown is a DNA nanoswitch made from a long M13 scaffold strand and short complementary backbone oligonucleotides. When the single-stranded detectors of the nanoswitch bind to viral RNA, the nanoswitch reconfigures to a looped ‘on’ state that can be visualized on a gel65. b | DNA nanostructures are used as carriers to deliver drugs to specific sites in the body or to cellular compartments. For example, a DNA origami nanotube loaded with thrombin (an anticoagulant) can be targeted towards tumours using a nucleolin-targeting aptamer. On reaching the tumour, the nanotube is opened by interaction of the aptamer with the protein nucleolin18. c | DNA nanostructures functionalized with specific proteins or ligands can moderate cellular functions. As an example, DNA nanoribbons conjugated to fibronectin protein domains act as an extracellular matrix to enhance cell migration and proliferation79. d | DNA nanostructures functionalized with imaging modules can be used in bioimaging. For example, aptamer-conjugated DNA tetrahedra can be directed specifically to tumour sites in the body and then imaged using the attached fluorophores84. Part a (bottom) adapted with permission from ref.19, AAAS. Part b (bottom) adapted from ref.18, Springer Nature Limited. Part c (bottom) adapted with permission from ref.79, ACS.
Drug delivery
In biomedicine, DNA nanostructures have potential utility as drug-delivery carriers (Fig. 2b). DNA provides precise control over the size and geometry of the nanostructures, generating homogeneous populations of drug carriers — a unique advantage over other self-assembling materials and nanoparticles48. DNA strands can also be synthesized with functional moieties that are useful for imaging69, site-specific targeting70 and triggered delivery71. Furthermore, DNA nanostructures are biocompatible and exhibit quick renal clearance and minimal toxicity72. DNA nanostructures can be tailored to carry various drug cargos, including unmethylated cytosine–phosphate–guanine (CpG) sequences for immunotherapy73, monoclonal antibodies as immune checkpoint inhibitors74, small interfering RNA75 and antisense oligonucleotides76 for gene therapy, proteins for vaccine development77 and metallic nanoparticles for photodynamic therapy78. Functionalized DNA nanocarriers are, thus, useful in controlling the circulation time, drug-release rate and specificity to a particular target site, resulting in increased delivery efficiency and efficacy of the encapsulated drug25,26.
Cell modulation
DNA nanostructures have also been used in modulating cell behaviour and activity79 (Fig. 2c). For example, DNA tetrahedra promoted the proliferation and differentiation of stem cells80, and peptide-functionalized DNA nanotubes were used in the differentiation of neural stem cells into neurons81. In immunotherapy, DNA nanostructures can activate macrophages to provide anti-inflammatory and antioxidative responses82. In other studies, DNA nanostructures have been used to influence cell migration, a factor important in wound healing and tumour cell metastasis21. The activation of specific cellular pathways by DNA nanostructures and their effects are currently under investigation83, which may lead to the development of tools useful in regenerative medicine and gene therapy.
Bioimaging
DNA nanostructures tagged with different imaging modules, such as organic fluorophores, radioactive isotopes and quantum dots, are used for quantitative functional imaging in cells and live model organisms (Fig. 2d). The use of DNA polyhedra and many versions of DNA devices has been demonstrated in imaging tumour-related messenger RNAs84,85 and membrane proteins86, as well as in tracking cell-entry pathways of drug-carrying DNA nanostructures69. Functionalized DNA nanostructures are also used as imaging standards to calibrate super-resolution microscopes, such as in direct stochastic optical reconstruction microscopy (dSTORM) and stimulated emission depletion (STED)87. In addition, DNA-PAINT (DNA-based point accumulation for imaging in nanoscale topography) is being explored for imaging cellular architectures and the real-time trajectories of membrane proteins in live cells88.
DNA nanostructure stability
As a molecule, DNA is highly stable, with a half-life of ~500 years89. However, when assembled into a nanostructure, the stability of DNA can vary, depending on the environment. In each biological application described above, the stability of the assembled DNA nanostructure is a key parameter. To establish the viability of DNA nanostructures in different applications, the stability of these structures is often tested at elevated temperatures90, in high-salt and low-salt conditions91, and in the presence of nucleases92, chaotropic93 agents and crowding agents94. In most cases, biological operations are tested at the physiological temperature of 37 °C. Depending on the application, stability at 37 °C for multiple hours might suffice. In terms of the salt and cation concentration, there needs to be a compromise between the conditions required for assembly versus those in the body. In addition, the stability and functionality of DNA structures are also dependent on where they are used, and can be affected by the local density, molecular flexibility and interactions with other molecules present in the sample, such as biological nucleic acids and serum opsonin proteins (which can lead to opsonization)95.
A major challenge to the application of DNA-based nanostructures is their susceptibility to attack by a variety of nucleases present in body fluids such as blood, urine and saliva. Nucleases cleave various DNA and RNA substrates (Box 1), and are involved in biological processes such as DNA repair, replication and recombination96. Nuclease activity is also required for structural alterations of nucleic acids, such as in topoisomerization97, site-specific recombination98 and RNA splicing99. Furthermore, nucleases form an integral part of the host immune system in degrading foreign nucleic acids100 and have a role in programmed cell death101. Although nucleases have important biological functions, they also interfere with the function of DNA-based structures in biological applications, necessitating the study of DNA nanostructure biostability in vivo or in conditions that mimic body fluids. As substitutes for physiological conditions, the stability of DNA nanostructures is tested in vitro using cell and tissue culture media supplemented with mammalian serum (such as fetal bovine serum (FBS)), which contains various nucleases102.
Although not all applications require DNA nanostructures to be nuclease resistant, it would be an advantage in most. Exceptions include applications such as biomolecular analysis, in which the molecules under observation are usually isolated and purified before their interactions are tested. Thus, even if they are typically present in a biofluid or tissue, the experiments do not occur in these conditions and are rarely affected by nucleases in the system. There are also some applications in which the native stability of DNA nanostructures is an advantage, or even susceptibility to nuclease degradation might be beneficial (for quick or spontaneous drug release, for example). Nevertheless, in many cases, the stability of DNA nanostructures needs to be enhanced before they are used. In this regard, several strategies are being developed to create nuclease-resistant DNA nanostructures.
Box 1 Examples of nucleases and their activities.
Nucleases are enzymes that cleave phosphodiester bonds between the sugar and phosphate moieties of DNA. Depending on their activity, nucleases are categorized as exonucleases (cleavage reactions occur at the terminus) or endonucleases (cleavage occurs within the DNA strand). Exonucleases can be further classified as 5′-end processing (such as T7 exonuclease) or 3′-end processing (such as exonuclease III) according to their polarity of consecutive cleavage, with some enzymes preferring 5′-phosphorylated double-stranded DNA (for example, lambda exonuclease). Exonucleases can also be single-strand-specific (for example, exonuclease I) or double-strand-specific (such as exonuclease V). Endonucleases display similar specific activities, with some catalysing cleavage at mismatch sites within double-stranded DNA (for example, T7 endonuclease I), at the site of a ribonucleotide (such as RNase HII) or non-specifically at random sites (such as DNase I). The activity of nucleases can also be sequence-specific or structure-specific.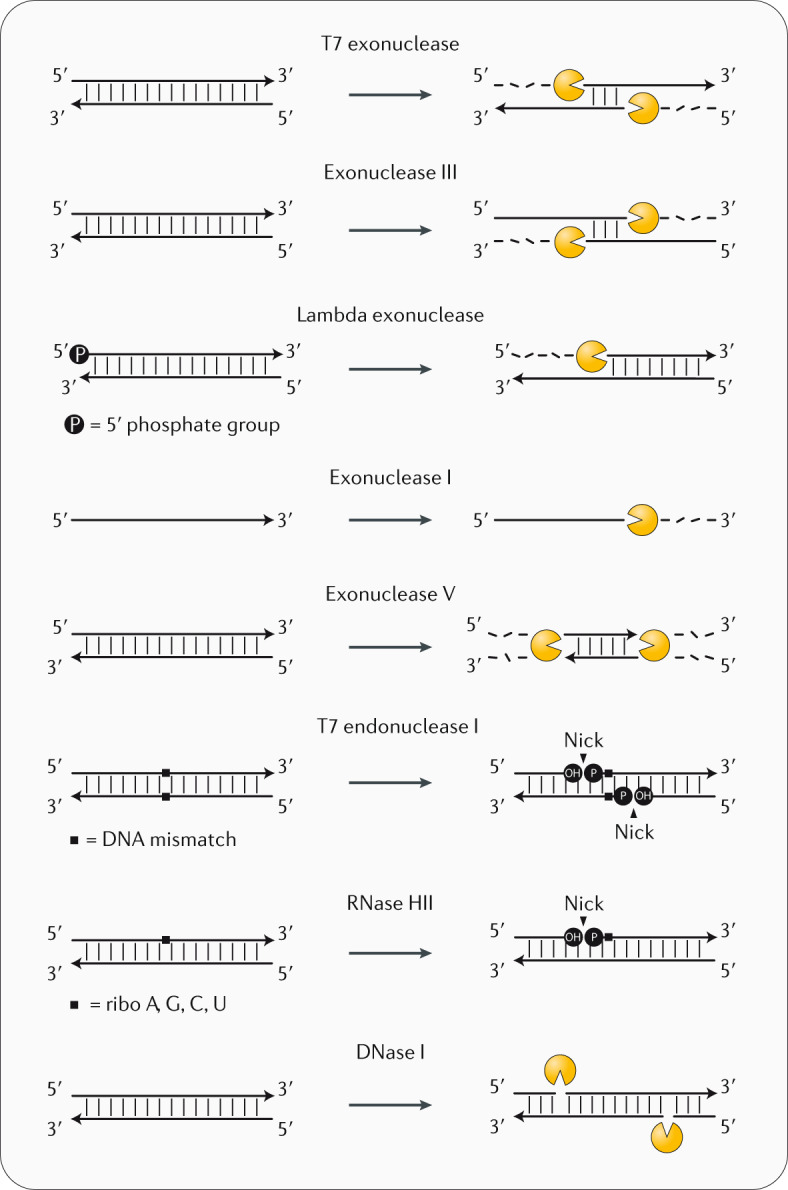
Strategies to modulate nuclease resistance
Strategies to modulate nuclease resistance can begin at the design stage, involve chemical modification or protective coating of the nanostructure, or occur at the point of use by solution treatment, each of which is discussed in more detail in the following sections. Unless otherwise mentioned, the studies discussed below were carried out at the physiological temperature of 37 °C. The structures tested for each strategy, the analysis technique and the resulting nuclease resistance are compiled in Table 1.
Table 1.
Reported nuclease resistance of various DNA nanostructures
| Strategy | Structure (size) | Test environment | Analysis technique | Stability metrics |
|---|---|---|---|---|
| Design | ||||
| Close-packed helices104 | 24-HB (100 nm) | 1 U DNase I | AGE |
Duplex plasmid DNA: degraded in 5 min Close-packed helices: degraded in 60 min |
| Topology106 | Tweezers (~14 nm) | 70% human serum | PAGE, FRET |
Closed state: degraded in ~37 h Open state: degraded in ~20 h |
| Restriction-site location105 | Ligated tetrahedron (7 nm) | DdeI restriction enzyme | PAGE |
Middle of edge: fully degraded Near vertex: fully protected |
| Increased crossovers92 | PX, DX and duplex DNA (~13 nm) | DNase I, exonuclease V, T5 and T7 exonucleases (for 1 h); 10% FBS, human serum and urine (for 24 h) | PAGE |
PX: ~100% intact DX: 0–30% intact Duplex: almost fully degraded |
| Solution treatment | ||||
| FBS heat treatment109 | Octahedron (50 nm) | Media + 10% FBS for 24 h | AGE |
Without heat treatment: 0% intact Heated FBS: almost 100% intact |
| Nuclease inhibitors (actin)109 | Octahedron (50 nm), nanotube (400 nm), nanorod (89 nm) | Media + 10% FBS for 24 h | AGE |
Without actin: 0% intact With actin: ~100% intact |
| Minor-groove binders (DAPI)110 | Pentagonal bipyramid (~40 nm) | 10% mouse serum | AGE |
Native structures: stable for 3 h With groove binders: stable for 24 h |
| Ethylenediamine buffer111 | Tetrahedron (14 nm) | 0.5 U DNase I | PAGE |
In TAE/Mg2+ buffer: 0% intact In ethylenediamine buffer: ~100% intact |
| Chemical modifications | ||||
| Ligation105,113 | Tetrahedron (7 nm), prism (7 nm) | 10% FBS | PAGE |
ssDNA: degraded in 0.8 h Ligated tetrahedron: degraded in 42 h |
| Crosslinking (click chemistry)114 | Nanotube (~30 nm) | Exonuclease I for 3 h | AGE |
Native: fully degraded Crosslinked: partially degraded |
| Crosslinking (UV-induced T–T dimers)115 | Brick-like DNA origami (~70 nm) | 0.4 U ml−1 DNase I | AGE |
Native: stable for 10 min Crosslinked: stable for 60 min |
| Hexanediol and hexaethylene glycol113,116 | Triangular prism (7 nm)113, tetrahedron (7 nm)116 | Media + 10% FBS | PAGE |
Lifetime of unmodified prism: 18 h Lifetime with hexanediol: 55 h |
| l-DNA117 | 4-Arm junction (~5 nm), nanotube (30–70-nm width, ~μm long) | 2 U μl−1 (exonuclease I) or 20 U μl−1 (exonuclease III) for 45 min | PAGE, AFM |
Native junction: completely degraded l-DNA junction: almost fully intact |
| Unnatural base pairs118 | 6-Arm junction (~14 nm) | T7 exonuclease for 12 h | PAGE |
Native structure: completely degraded Modified: partially degraded |
| Protective coatings | ||||
| HSA–DNA dendrite conjugates120 | Cube (~7 nm) | Media + 10% FBS for 48 h | PAGE |
ssDNA: 33-min half-life Protected cube: up to 22-h half-life |
| Dendritic oligonucleotides121 | DNA brick (~50 nm) | 100 U ml−1 DNase I for 1 h | AGE |
Native: fully degraded with 5 U ml−1 Coated: 50% degraded with 100 U ml−1 |
| PEGylated lipid bilayer122 | Octahedron (76 nm) | 20 U DNase I for 24 h | Fluorescence |
Without envelope: ~30% intact With envelope: ~85% intact |
| Cationic polysaccharides123 | Origami rod (350 nm), bottle (50 × 25 nm) | 10 U ml−1 DNase I | AGE, TEM |
Native structure: stable for 1 h With protection: stable for 24 h |
| PEG–polylysine block copolymers126 | Rectangle (~100 nm), 6-HB (~600 nm), truss (~20 × 200 nm) | 1 µl of 16 U ml−1 DNase I for 16 h | TEM, AFM |
Native: 0% intact Protected: 100% intact |
| 24-HB (~100 nm) | 1 µl of 2,000 U ml−1 DNase I for 16 h | |||
| Oligolysine–PEG copolymer124 | Origami barrel (~60 nm) | Media + 10% FBS | AGE, TEM |
Native structure: 5-min half-life With oligolysine: 50-min half-life |
| Crosslinking of oligolysine coating125 | Origami barrel (60–90 nm) | 1 U μl−1 DNase I | AGE, TEM |
Without crosslinking: 16-min half-life With crosslinking: ~66-h half-life |
| BSA–dendron conjugates127 | 60-HB (20 × 20 × 33 nm) | 10 U DNase I in 20 μl reaction for 1 h at RT | AGE |
Native: <20% intact With protection: ~100% intact |
| Peptoids128 | Octahedron (29 nm) | 15 μg ml−1 DNase I for 30 min | AGE, TEM |
Native structure: completely degraded With protection: almost fully intact |
| Silica coating130 | 24-HB (~100 nm) | 4 U ml−1 DNase I for 3 h | AGE, TEM |
Native structure: completely degraded Silicified structures: almost fully intact |
| 13-Helix ring (66 nm) | 0.5 U ml−1 DNase I for 3 h | |||
Unless mentioned otherwise, listed experiments were conducted at 37 °C. AFM, atomic force microscopy; AGE, agarose gel electrophoresis; BSA, bovine serum albumin; DAPI, 2-(4-amidinophenyl)-1H-indole-6-carboxamidine; DX, double crossover; EDTA, ethylenediaminetetraacetic acid; FBS, fetal bovine serum; FRET, Förster resonance energy transfer; HSA, human serum albumin; n-HB, n-helix bundle; PAGE, polyacrylamide gel electrophoresis; PEG, polyethylene glycol; PX, paranemic crossover; RT, room temperature; ssDNA, single-stranded DNA; TAE, tris-acetate-EDTA; TEM, transmission electron microscopy; UV, ultraviolet.
Design and inherent nuclease resistance
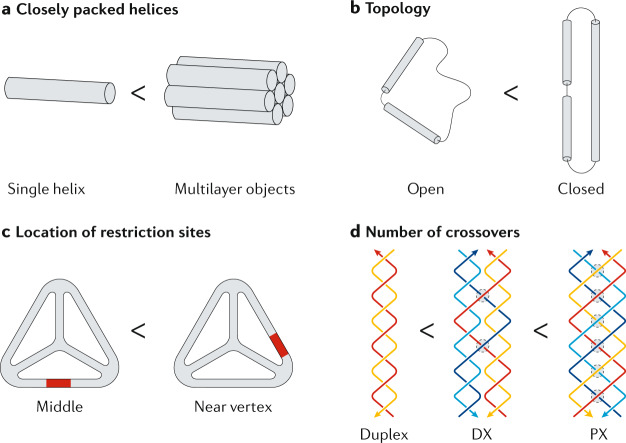
The structural design of DNA nanostructures influences the nuclease resistance. DNA tiles and motifs contain multiple strands hybridized together in a tightly woven fashion. Similarly, larger (and more complex) origami structures contain multiple helical domains packed into bundles (arranged in, for example, square or honeycomb lattices)37,103. This close-packed structure in DNA origami and other multi-helix structures is more nuclease resistant than linear duplexes or plasmid DNA104 (Fig. 3a). For example, 1 enzyme unit (U) of DNase I completely degraded 65 ng of duplex plasmid DNA (pET24b) in only 5 min, whereas it took ~60 min to degrade 2 ng of a 24-helix-bundle DNA origami structure104. This result indicates the potential of multilayer DNA origami objects as encapsulation agents for use with intercalating drugs, for example. However, DNA nanostructures are required to be hollow to encapsulate cargos such as nanoparticles and proteins. Hollow wireframe structures, such as DNA tetrahedra105, have also shown higher stability in 10% FBS (up to 42 h) than linear duplexes, possibly owing to their non-native geometries. The influence of molecular topology on nuclease resistance has also been noted106: a reconfigurable DNA device based on DNA tweezers107 was more nuclease resistant in the closed state than in the open state (~37 h versus 20 h in 70% human serum, respectively), possibly owing to the stacking of two helices side by side in the closed state106 (Fig. 3b).
Fig. 3. Nanostructure designs for enhanced nuclease resistance.
a | Close-packed helices, as used in DNA origami or DNA bundles104, are more nuclease resistant than linear duplexes or plasmid DNA. b | The biostability of nanostructures is also dependent on the topology, as seen in DNA tweezers106, for which the closed state is more nuclease resistant than the open state. c | Changing the location of enzyme-specific sequences such as restriction sites105 also influences the nuclease resistance. d | DNA motifs with a greater number of crossovers exhibit higher nuclease resistance92. For example, paranemic crossover (PX) DNA is more nuclease resistant than double crossover (DX) DNA.
Although, in the above examples, the structures are fortuitously nuclease resistant, this feature can also be intentionally designed into DNA nanostructures. In DNA tetrahedra, placement of restriction sites at specific locations on the duplex edges affected the activity of the enzyme105. Structures with the restriction site located near the vertex were protected against degradation by DdeI restriction enzyme, whereas those with the restriction site in the middle fully degraded (Fig. 3c). Another possible strategy to enhance the nuclease resistance is to increase the number of crossovers within the nanostructures. For example, a study92 on paranemic crossover (PX) DNA108 revealed the exceptional nuclease resistance of the motif, attributed to the continuous crossovers within the structure (Fig. 3d). Compared with duplexes, PX DNA was up to ~2,800-fold more resistant to four different nucleases (DNase I, exonuclease V, T5 exonuclease and T7 exonuclease), as well as to human serum and urine. Motifs with fewer crossovers showed reduced stability, suggesting that the increased number of crossovers in PX DNA confers additional biostability.
Solution treatment and nuclease inhibitors
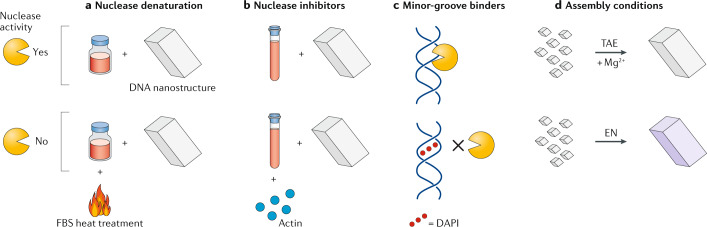
The effect of nucleases on the lifetime of DNA nanostructures can be minimized by using nuclease inhibitors or changing the environment in which the structures are assembled or used. One approach is to expose the samples to conditions that denature the nucleases. For example, DNA origami nanostructures (an octahedron, nanotube and nanorod) incubated in RPMI cell culture medium supplemented with 10% FBS degraded completely within 24 h (ref.109). By contrast, heating the FBS to 75 °C for 10 min before addition to the cell culture medium prolonged the nanostructure lifetime in the medium (with almost no degradation), owing to the inactivation of nucleases in the serum (Fig. 4a). However, heating the FBS might also have a more general effect on serum proteins, which could affect cell growth and phenotype.
Fig. 4. Solution treatment to prevent nuclease degradation.
a | Heating serum samples denatures the nucleases, thus rendering them inactive109. b | The addition of nuclease inhibitors, such as actin, to samples reduces nuclease activity, thus protecting DNA nanostructures from degradation109. c | The addition of minor-groove binders such as DAPI (2-(4-amidinophenyl)-1H-indole-6-carboxamidine) occludes nuclease binding to DNA nanostructures, thus minimizing degradation110. d | The assembly conditions can be varied to increase the biostability. For example, DNA nanostructures were minimally degraded when assembled in ethylenediamine (EN) buffer, but degraded fully when assembled in the commonly used buffer tris-acetate-EDTA (TAE) with Mg2+ (ref.111). EDTA, ethylenediaminetetraacetic acid; FBS, fetal bovine serum.
Nuclease inhibitors can also be used to reduce the effect of nucleases in solution. For example, the addition of a tenfold molar excess of actin, a known nuclease inhibitor, extended the lifetime of DNA nanostructures against DNase I (ref.109) (Fig. 4b). In contrast to heat treatment of FBS, which is incompatible with in vitro tissue culture, inclusion of actin (up to 200 nM) had no observable effect on cell growth and viability. In other cases, compounds that restrict access to the minor groove of double-stranded DNA can competitively inhibit DNase I activity (Fig. 4c). In a study reported in a recent preprint article, the efficiency of different classes of minor-groove binders on reducing the nuclease degradation of wireframe DNA origami structures (a two-helix pentagonal bipyramid) was tested110. Diamine 2-(4-amidinophenyl)-1H-indole-6-carboxamidine (DAPI) was the most potent stabilizer, increasing the protection of structures in 10% mouse serum to 24 h, compared with the 3-h stability of native structures.
The conditions in which the nanostructures are assembled also affect their biostability. DNA tetrahedra assembled in ethylenediamine buffer exhibited enhanced nuclease resistance compared with those assembled in one of the most commonly used buffers, tris-acetate-EDTA/Mg2+ (TAE/Mg2+; where EDTA is ethylenediaminetetraacetic acid)111) (Fig. 4d). In a 10-min reaction with DNase I, DNA tetrahedra assembled in TAE buffer completely degraded, whereas almost no digestion occurred in the ethylenediamine buffer. The increased resistance in ethylenediamine is possibly due to the absence of metal ions needed for enzymatic activity. Although this strategy increases the lifetime of DNA nanostructures in the presence of nucleases, further studies are needed to validate the compatibility of this buffer for the assembly of different DNA nanostructures and the toxicity of such organic compounds in vivo. For some structures, the inherent assembly units can be provided as fuel in the solution to heal the nanostructures as they are degraded by nucleases, thus reversing the degradation process. This approach was demonstrated using micrometre-scale DNA nanotubes assembled from double crossover (DX) tiles112. The DNA nanotubes degraded within 24 h in 10% FBS-supplemented medium. Upon introducing the monomer DX tiles into the solution, these units incorporated into the damaged regions of the DNA nanotubes and, thus, repaired the nanotubes, prolonging their lifetime to 96 h.
Chemical modifications
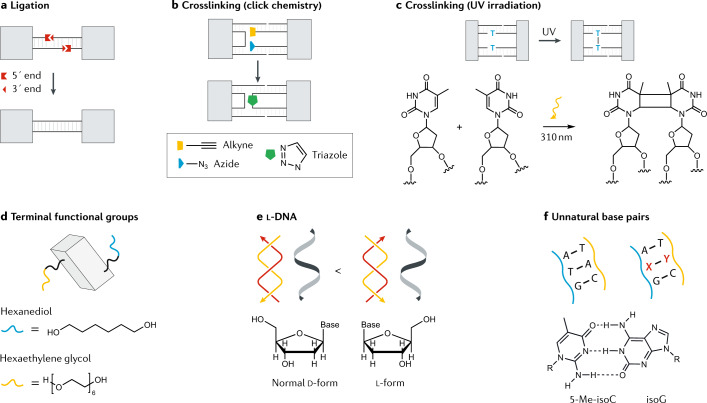
Numerous studies have reported chemical modifications to DNA nanostructures to enhance nuclease resistance. These modifications strengthen the weaknesses in the nanostructure design, making DNA nanostructures less susceptible to nuclease digestion. For example, the presence of single-stranded segments and internal nicks in DNA nanostructures make them vulnerable to nuclease attacks105. Once constructed, internal nicks can be eliminated by covalently linking the strand termini by ligation or crosslinking. Reducing the number of free termini by enzymatic ligation was effective in enhancing the nuclease resistance of DNA tetrahedra105 and prisms113, increasing the mean lifetime of these structures to ~42 h and ~200 h, respectively, in 10% FBS (Fig. 5a). Crosslinking the helices within nanostructures by azide–alkyne click reactions has also helped DNA nanostructures to withstand nuclease degradation114 (Fig. 5b). Crosslinked DNA nanotubes had greater protection against exonuclease I compared with native structures. In another example, thymidines were positioned in close proximity within DNA origami nanostructures and crosslinked using ultraviolet light, leading to the site-specific formation of covalent bonds and cyclobutane pyrimidine dimers115 (Fig. 5c). The additional covalent bonds increased the lifetime of the nanostructures from 10 min (for native structures) to 60 min in the presence of 0.4 U ml−1 DNase I.
Fig. 5. Chemical modifications to enhance nuclease resistance.
Various chemical modifications have been demonstrated to increase the nuclease resistance of DNA structures. These approaches include ligation of strand termini105 (part a); crosslinking of component strands by click chemistry114 (part b); crosslinking of strands through the formation of a thymidine dimer under ultraviolet (UV) irradiation115 (part c); the introduction of terminal functional groups, such as hexaethylene glycol and hexanediol113 (part d); the use of l-DNA117 (part e); and the use of unnatural base pairs (where 5-Me-isoC is 5-methyl-isocytidine; part f)118.
In addition to post-assembly modifications, the inherent stability of nanostructures can be increased by modifying the DNA backbone and nucleobases of component strands. In one example, DNA strand termini were functionalized with hexaethylene glycol and hexanediol groups, and then used in building DNA prisms113,116 (Fig. 5d). These structures had a lifetime of 55–62 h in FBS-supplemented culture medium, whereas the unmodified native DNA nanostructures degraded in less than 18 h. Another route to enhancing nuclease resistance is by using modified DNA as the component strands for assembly. Two-dimensional DNA arrays and a DNA tetrahedron built using l-DNA were more resistant to nucleases than their d-DNA (that is, regular right-handed DNA) counterparts117 (Fig. 5e). The l-DNA structures remained almost fully intact after treatment with exonuclease I and exonuclease III for 45 min, whereas the native structures fully degraded. In another study, six-arm junctions containing unnatural DNA base pairs (2-thiothymidine:A and 5-methyl-isocytidine:isoG) were only partially digested by T7 exonuclease after 12 h, whereas the unmodified junction completely degraded118 (Fig. 5f). In the context of chemical modifications, a less-explored option is the incorporation of 2´–5´-linked oligonucleotides into nucleic acid nanostructures to enhance resistance against ribonucleases119.
Protective coatings
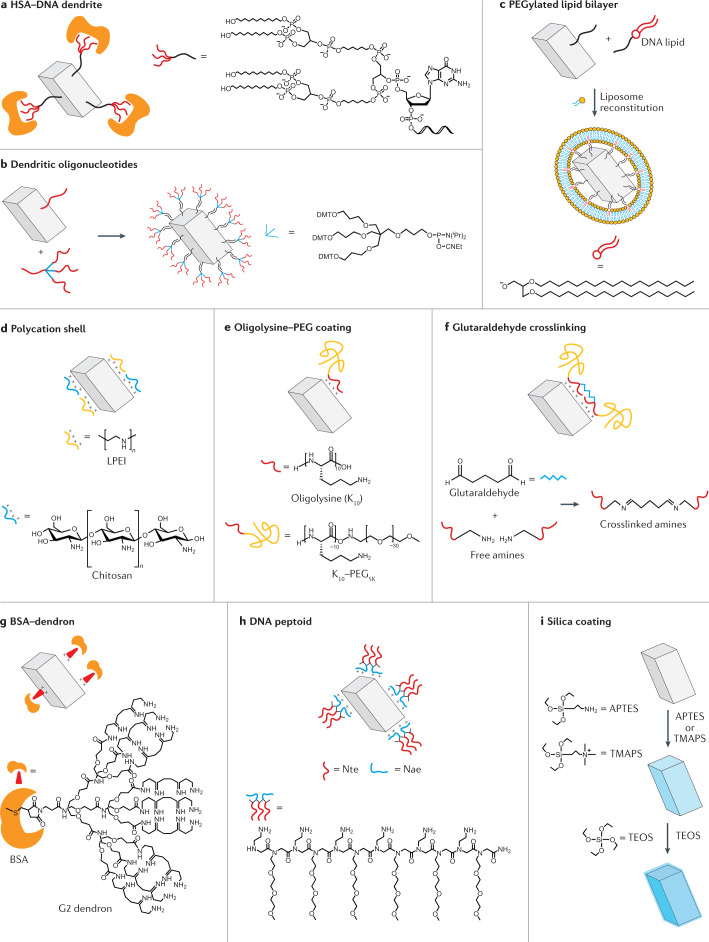
Modifying the component strands also allows the nanostructures to be coated with a protective layer that resists nuclease activity. In one such strategy, the component strands of DNA cubes were modified with dendritic alkyl chains to form amphiphiles that bind to human serum albumin (HSA)120 (Fig. 6a). In stability tests in Dulbecco’s modified Eagle medium supplemented with 10% FBS, single-stranded DNA modified with a dendritic alkyl chain and conjugated to HSA had a half-life of 33 min, increasing to 10 h for a DNA cube with four such modified strands conjugated to HSA and to 22 h for a DNA cube with eight HSA-conjugated strands. A protective coating formed from dendritic oligonucleotides that feature at one end a three-pronged group attached to three oligonucleotides has also been demonstrated121 (Fig. 6b). The stem of the dendritic oligonucleotide can hybridize to single-stranded handles on DNA brick structures, forming a thick DNA coating. DNA bricks with this coating were only 50% digested with 100 U ml−1 of DNase I in 1 h, whereas native structures were fully digested with only 5 U ml−1 of the nuclease. In another example, a DNA octahedron was enveloped by a PEGylated lipid bilayer, which increased the percentage of intact structures from 30% to 85% when incubated in 20 U DNase I for 24 h (ref.122) (Fig. 6c).
Fig. 6. Protective coatings to enhance nuclease resistance.
Various protective coatings have been developed to increase the nuclease resistance of DNA nanostructures. These approaches include a coating formed through the complexation of human serum albumin (HSA) and DNA dendrites120 (part a); coating nanostructures with dendritic DNA through hybridization to DNA handles121 (part b); a PEGylated (where PEG is polyethylene glycol) lipid bilayer protective envelope122 (part c); the charge-based accumulation of a polycationic shell123 (part d); an oligolysine–PEG coating124 (part e); a coating formed through glutaraldehyde crosslinking of oligolysines125 (part f); a bovine serum album (BSA)–dendron block copolymer coating127 (part g); a coating comprising DNA peptoids128 (part h); and silica-based coatings130 (part i). APTES, (3-aminopropyl)triethoxysilane; DMTO, dimethoxytrityloxy; LPEI, linear polyethyleneimine; Nae, N-(2-aminoethyl)glycine; Nte, N-2-(2-(2-methoxyethoxy)ethoxy)ethylglycine; TEOS, tetraethyl orthosilicate; TMAPS, N-trimethoxysilylpropyl-N,N,N-trimethylammonium chloride.
The high negative charge of DNA can also be exploited to create a polycationic coating through electrostatic interactions with DNA. For example, cationic polysaccharides such as chitosan and synthetic linear polyethyleneimine were used to coat DNA origami rods, yielding structures that were stable in 10 U ml−1 of DNase I for 24 h (ref.123) (Fig. 6d). Using a similar strategy, DNA origami barrels were coated with oligolysines to increase the lifetime of nanostructures in culture media to 50 min, compared with 5 min for native structures under the same conditions124. Furthermore, conjugating PEG to these oligolysines124 (Fig. 6e) or crosslinking the oligolysine–PEG coating using glutaraldehyde125 (Fig. 6f) further increased the nuclease resistance of these nanostructures to ~36 h in culture medium and ~66 h in 1 U µl−1 of DNase I, respectively. PEG-conjugated polylysines have also been used for coating DNA origami structures, forming DNA origami polyplex micelles126. The electrostatic interactions between PEG–polylysine and DNA enabled the origami structure to remain intact for 16 h in the presence of DNase I and in FBS-supplemented RPMI medium, whereas native structures completely degraded. This protection strategy was also compatible with functionalized DNA origami structures, causing no loss of functional moieties when applied to six-helix bundles decorated with gold nanoparticles or streptavidin-coated quantum dots.
Peptides and proteins can also be used to coat DNA nanostructures. In one example, a protein corona was formed around DNA origami nanostructures to make them nuclease resistant127. Protein–dendron conjugates were created by anchoring bovine serum albumin to a dendron through a cysteine–maleimide bond (Fig. 6g). The dendron part of the conjugate functions as a cationic binding domain that electrostatically attaches to the negatively charged DNA origami surface. Almost 100% of the protected DNA origami structures remained intact on exposure to 10 U of DNase I for 1 h at room temperature, whereas only 20% of the native structures remained intact. A peptoid-based coating has also been developed to protect DNA origami octahedral structures against nucleases128 (Fig. 6h). Compared with peptides, peptoids provide enhanced protection against proteolytic cleavage129. The peptoid sequences contained two types of monomer: positively charged N-(2-aminoethyl)glycine to facilitate electrostatic complexation with the DNA structure and neutral N-2-(2-(2-methoxyethoxy)ethoxy)ethylglycine for surface passivation. In the presence of DNase I, the bare octahedra completely degraded, whereas peptoid protection preserved the structures. To demonstrate the utility in biomedical applications, bovine serum albumin was encapsulated within the DNA octahedra, which protected the encapsulated protein from hydrolysis by trypsin. In another study, a solution-based method was used to coat 24-helix-bundle DNA origami nanostructures with a thin layer of silica130 (Fig. 6i). This coating enabled the DNA origami structures to withstand degradation by DNase I.
Analysis techniques
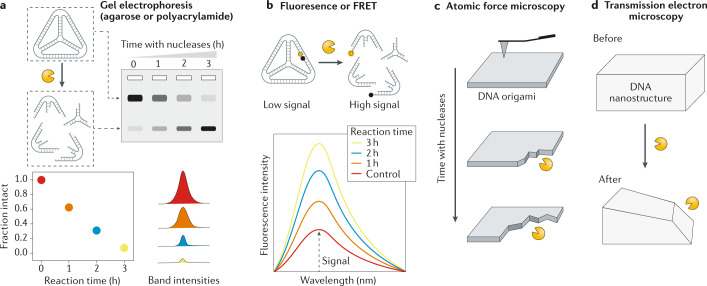
The predominant techniques used to analyse the nuclease digestion of DNA nanostructures are polyacrylamide or agarose gel electrophoresis (PAGE and AGE, respectively), fluorescence, transmission electron microscopy (TEM) and atomic force microscopy (AFM). All these techniques are commonly used to characterize the formation of DNA nanostructures, but gel electrophoresis is the most often-used technique to quantify the reduction in intact structures on incubation with nucleases or body fluids (Fig. 7a). Typically, nanostructures are incubated for different times with nucleases or serum (or with different concentrations of these), and the incubated samples run on PAGE or AGE. The band corresponding to the structure is then quantified to obtain the level of degradation131. Fluorescence and Förster resonance energy transfer (FRET) techniques are also often used to quantify degradation of nanostructures106,122. Upon degradation of DNA nanostructures conjugated to fluorescent dyes, the fluorophore and the quencher (or the FRET pair) separate, causing an increase in fluorescence levels (Fig. 7b). This increase can be monitored over time to study the degradation kinetics of DNA nanostructures.
Fig. 7. Techniques used to analyse nuclease degradation of DNA nanostructures.
The main techniques used to analyse DNA nanostructures are polyacrylamide or agarose gel electrophoresis (part a), fluorescence or Förster resonance energy transfer (FRET; part b), atomic force microscopy (AFM; part c) and transmission electron microscopy (TEM; part d).
Gel electrophoresis and fluorescence techniques provide information on the degradation levels of nanostructures but do not provide information on structural changes. To obtain such structural information, TEM and AFM are used to provide before and after snapshots of DNA nanostructures incubated with body fluids or nucleases111,132 (Fig. 7c,d). Real-time AFM was recently used to monitor the degradation of different 2D DNA origami shapes when incubated with nucleases133. This technique enables visualization of nanostructures as they degrade and is, thus, useful in determining the parameters for the design of improved DNA-based nanocarriers (for example, in determining whether nucleases affect vertices more than edges).
Overall, gel electrophoresis and fluorescence techniques are the easiest to adapt for most laboratories and provide timed degradation analysis. AFM and TEM require specific instrumentation and training, and are time-consuming, making them the least practical for monitoring the nuclease digestion of DNA nanostructures. The advances in real-time-AFM potentially address the issue of long experimental observation times but is still an expensive instrument not available in most laboratories. There are also challenges associated with the use of gel-based analysis techniques. For example, in the case of cationic block copolymers, polyplexed DNA origami need to be decomplexed (by dextran sulfate, for example) before they can be analysed on a gel (also applicable to TEM)126. Moreover, in studies involving FBS or human serum, the serum proteins could cause a background signal in both AGE and TEM, thus necessitating purification of treated samples before analysis.
Mechanism of nuclease resistance
There is still a lack of understanding of the mechanisms of reported nuclease resistance for different structures or protection strategies. Depending on the type of nuclease used, different parameters, such as DNA sequence, backbone geometry, groove width, curvature and flexibility, can all contribute to nuclease resistance134. For DNA nanostructures, the non-native geometries and sizes could have a role in inhibiting enzyme binding, thus decreasing the efficiency of specific or non-specific enzymatic cleavage105. Close-packed helices and frequent crossovers in DNA nanostructures can also occlude enzyme binding, thus negating the effect of nucleases92,133. Furthermore, nucleases such as DNase I cause distortion on binding to helices135, and, thus, the increased stiffness of shorter duplex regions or multi-helical edges within DNA nanostructures might prevent the binding of such enzymes. In the case of different modifications and coatings, strong multivalent interactions of the envelope with the DNA could hinder nuclease adsorption and, thus, reduce the degradation. In DNA origami, the scaffold strand already contains restriction sites. In a study on 2D structures, such as a DNA triangle and rectangle, the position of the restriction site post-folding and the introduction of defects (missing staple strands) affected the overall activity of restriction endonuclease on the structures136. In a follow-up computational study, global conformational fluctuations between metastable states of DNA origami structures were shown to affect the reactivity of restriction endonucleases on the structures137. Simulations of a 2D DNA origami triangle with four or eight staple strands omitted from the structure, each resulting in a metastable assembly, revealed that the accessibility of the enzyme to the structure is dependent on the interconversion between these metastable states. This analysis showed that the reactivity of restriction enzymes is dependent on the steric overlap between the enzyme and the adjacent helices, thus, providing a route to designing nanostructures with higher or lower nuclease resistance.
A checklist for biostability studies
Design principles
Researchers have designed a large library of DNA nanostructures, each requiring specific assembly conditions. Given this requirement, one has to choose a strategy for modulating the nuclease resistance of a DNA nanostructure on the basis of the design of the underlying DNA assembly. For example, DNA prisms in which the strand termini contained hexaethylene glycol and hexanediol groups showed enhanced nuclease resistance113,116, but the same strategy applied to a larger wireframe DNA origami pentagonal bipyramid did not further stabilize the structure in 10% FBS110. Although there is no general set of rules to make this choice, some studies have noted how the stability numbers reported could apply to similar structures. In DNA origami, most structures share a common scaffold strand (M13) and, thus, have similar GC content if completely double-stranded, eliminating sequence as a factor in stability variations. Therefore, the difference in stability arises from the design of the nanostructures, how closely the helical bundles are packed and the length of the staple strands104. Even if the stability reported for an origami structure is not representative for objects built with similar specifications, these results could be useful in designing new objects with an expected robustness. In designing other structures, adapting the edges to be multi-helical domains might increase the persistence length of the wireframe structures and, thus, provide additional stability.
Types of nucleases and concentrations
The nuclease of choice in most studies is DNase I. However, some studies have tested the degradation of DNA nanostructures in the presence of other nucleases, showing that the level of degradation is dependent on the exonuclease or endonuclease used92,104,118. In two studies, enzymes such as exonuclease VIII, lambda exonuclease, Mse I restriction endonuclease and T7 exonuclease did not have any effect on DNA origami objects, whereas DNase I, Escherichia coli exonuclease I, exonuclease T, T7 endonuclease and exonuclease III degraded them104,115. Similarly, PX DNA showed different levels of resistance to DNase I, exonuclease V, T7 exonuclease and T5 exonuclease; however, PX DNA displayed much higher nuclease resistance than DX or duplex structures in all cases92. Thus, although generic trends exist, the activity of different nucleases could differ. The concentration of nucleases tested is also an important factor in determining the extent of degradation. Through calibration of the nuclease levels in FBS by comparing DNA origami nanostructures incubated in 1–20% FBS and different concentrations of DNase I, it was estimated that typical tissue culture conditions may contain between 256 and 1,024 U l−1 equivalent of DNase I activity109. These studies indicate that it is important to know the specific levels of nucleases in different biofluids and to test relevant amounts of nucleases in such biostability studies.
Reagents
Another factor to consider when testing the stability of DNA nanostructures is the type of serum used and the freeze–thaw protocols. The level of nuclease activity in different FBS lots and frozen aliquots has been observed to vary109. In this report, the nuclease activity was highest after initial thawing of the FBS and was lost after a few weeks when FBS-supplemented medium was stored at 4 °C (ref.109). Thus, it is important to follow the specific reagent-handling protocol across multiple experiments to validate the nuclease-resistance values of different structures. Nuclease activity in body fluids also varies widely between species138,139, and studies on DNA nanostructures have also found the stability of nanostructures to vary in sera from different species110. Future studies could work with human-derived solutions instead of animal sera to make the results more relevant for human applications.
Choice of protection strategy
The type of application will determine the level of biostability needed and which strategy to use for modulating nuclease resistance. In drug delivery, partial digestion of DNA nanostructures could trigger release of the drug cargo or attached functional moieties (such as fluorophores for tracking). However, slow or delayed degradation of nanostructures could be useful in the spontaneous release of drugs. The addition of nuclease inhibitors or the heat treatment of samples are potential options for biosensing applications in which the sample can be preprocessed before addition of the DNA sensor. However, use of DNA nanostructures in vivo requires strategies that obviate the need to alter the environment, as the addition of external factors might influence other biomolecular processes. Furthermore, the choice of strategy will also depend on the type of biofluid the structures will be in (blood, urine, saliva), and prior knowledge of the types of nucleases present in these fluids and their levels will be useful in biostability studies. Once the stabilization strategy is chosen, it is imperative to test the functionality of these structures after the protection process; chemical modifications or coatings should not affect the binding of sensing elements or targeting moieties to DNA nanostructures. For example, the peptoid coating of DNA origami structures did not affect the encapsulation of cargos such as proteins and nanoparticles within the nanostructure, suggesting potential use in drug delivery128. In addition, polyplex micelles comprising cationic polysaccharides have been successfully used to stabilize plasmid DNA for gene therapy140, indicating the potential use of similarly stabilized DNA nanostructures.
In vivo stability and immune response
The integrity of DNA nanostructures in vivo affects the immune response in cells or animals. When used as drug carriers, the immune response elicited by intact and degraded nanostructures might differ in some cases, whereas in others, it might be dependent on the total mass of DNA and not the design or integrity of the nanostructures109. Thus, care has to be taken to test the intactness of DNA nanostructures in studies in which the specific immune response is important. For example, the oligolysine–PEG block copolymer coating of DNA nanostructures had negligible effect on cell viability or enzyme kinetics, indicating minimal immune response in the cells125,141. Studies to test in vivo stability and immune response might also require additional functionalities to track the nanostructure through the body or a cellular pathway, and can use newly developed techniques, such as a ‘hydroporator’, to deliver DNA nanostructures directly into cells for monitoring142.
Conclusion
As DNA nanotechnology moves towards real-life applications, enhancing the stability of nanostructures in biological environments is of increasing importance. The strategies discussed in this Review provide an overview of methods to modulate the nuclease resistance of DNA nanostructures, rendering them more useful in biological applications. There is already a large library of DNA nanostructures available, but future studies will need to focus on upgrading these structures to be application-ready.
The studies discussed herein have used different structures as examples in different environments (different culture media, with or without serum supplement, or in a combination of nucleases). From these studies, it is apparent that DNA nanostructures possess higher nuclease resistance than duplexes, and that all the strategies discussed can enhance this resistance. However, it is difficult to further compare results from these studies, as they report results using different metrics (lifetime, half-life, percentage intact) and use different nanostructures. To facilitate comparison, the DNA nanotechnology research community is encouraged to decide on a set of standardized reporting parameters for nuclease resistance and biostability. Such a system would benefit the generalization of design parameters and the choice of strategy for a specific structure and/or application. Future studies could also explore the effect of the shape and size of DNA nanostructures on nuclease resistance, reports on which are few.
DNA nanostructures have permeated many different fields and are closer to real-life applications than ever before. The ongoing development of stabilization strategies, construction methods, characterization tools and platforms that allow non-experts to design DNA nanostructures143 is contributing to the rapid progress towards achieving the potential that DNA nanotechnology promises.
Acknowledgements
The author thanks B.R. Madhanagopal and S. Selvam for critical reading and comments on the manuscript, as well as J. Mathivanan for help preparing the ChemDraw images of the chemical structures in the figures.
Glossary
- DNA origami
A technique that uses hundreds of short ‘staple’ strands to fold a long piece of single-stranded ‘scaffold’ DNA into desired shapes.
- Opsonization
Non-specific interactions of any foreign body, including DNA nanostructures, with plasma proteins, resulting in the formation of a protein corona.
- Enzyme unit
The amount of enzyme that will catalyse or produce a specific amount of a substrate or product, respectively, under the specified conditions of an assay.
- Restriction enzyme
An enzyme that recognizes a specific, short nucleotide sequence and cuts the DNA only at that specific site, which is known as a restriction site or target sequence.
- Paranemic crossover (PX) DNA
A four-stranded DNA motif that consists of two adjacent double-helical DNA domains connected by strand crossovers at every possible point between two side-by-side helices.
- l-DNA
Mirror-image form of naturally occurring DNA, forming a left-handed double helix, in contrast to the right-handed structure of ‘regular’ d-DNA.
- Dendron
Monodisperse, wedge-shaped branched molecule with multiple terminal groups and a single reactive function at the focal point.
- Peptoid
Small protein-like polymer in which the side chains are attached to backbone nitrogen atoms, rather than to the α-carbons, as in peptides.
Competing interests
The author declares no competing interests.
Footnotes
Peer review information
Nature Reviews Chemistry thanks K. Salaita, who co-reviewed with A. Bazrafshan, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Seeman NC. Nucleic acid junctions and lattices. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 2.Seeman NC, et al. New motifs in DNA nanotechnology. Nanotechnology. 1998;9:257–273. doi: 10.1088/0957-4484/9/3/018. [DOI] [Google Scholar]
- 3.Seeman NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 4.Chandrasekaran AR, Zhuo R. A ‘tile’ tale: hierarchical self-assembly of DNA lattices. Appl. Mater. Today. 2016;2:7–16. doi: 10.1016/j.apmt.2015.11.004. [DOI] [Google Scholar]
- 5.Seeman NC. DNA Nanotechnology at 40. Nano Lett. 2020;20:1477–1478. doi: 10.1021/acs.nanolett.0c00325. [DOI] [PubMed] [Google Scholar]
- 6.Seeman NC, Sleiman HF. DNA nanotechnology. Nat. Rev. Mater. 2018;3:17068. doi: 10.1038/natrevmats.2017.68. [DOI] [Google Scholar]
- 7.Xavier PL, Chandrasekaran AR. DNA-based construction at the nanoscale: emerging trends and applications. Nanotechnology. 2018;29:062001. doi: 10.1088/1361-6528/aaa120. [DOI] [PubMed] [Google Scholar]
- 8.Hong F, Zhang F, Liu Y, Yan H. DNA origami: scaffolds for creating higher order structures. Chem. Rev. 2017;117:12584–12640. doi: 10.1021/acs.chemrev.6b00825. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, Li Z, Li Y, Mao C. Rational design and self-assembly of two-dimensional, dodecagonal DNA quasicrystals. J. Am. Chem. Soc. 2019;141:4248–4251. doi: 10.1021/jacs.9b00843. [DOI] [PubMed] [Google Scholar]
- 10.He Y, et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 11.Benson E, et al. DNA rendering of polyhedral meshes at the nanoscale. Nature. 2015;523:441–444. doi: 10.1038/nature14586. [DOI] [PubMed] [Google Scholar]
- 12.Andersen ES, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 13.Gu H, Chao J, Xiao S-J, Seeman NC. A proximity-based programmable DNA nanoscale assembly line. Nature. 2010;465:202–205. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tikhomirov G, Petersen P, Qian L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature. 2017;552:67–71. doi: 10.1038/nature24655. [DOI] [PubMed] [Google Scholar]
- 15.Chandrasekaran AR, Anderson N, Kizer M, Halvorsen K, Wang X. Beyond the fold: emerging biological applications of DNA origami. ChemBioChem. 2016;17:1081–1089. doi: 10.1002/cbic.201600038. [DOI] [PubMed] [Google Scholar]
- 16.Chandrasekaran AR. DNA origami and biotechnology applications: a perspective. J. Chem. Technol. Biotechnol. 2016;91:843–846. doi: 10.1002/jctb.4826. [DOI] [Google Scholar]
- 17.Chandrasekaran AR, Levchenko O. DNA nanocages. Chem. Mater. 2016;28:5569–5581. doi: 10.1021/acs.chemmater.6b02546. [DOI] [Google Scholar]
- 18.Li S, et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018;36:258–264. doi: 10.1038/nbt.4071. [DOI] [PubMed] [Google Scholar]
- 19.Chandrasekaran AR, et al. Cellular microRNA detection with miRacles: microRNA-activated conditional looping of engineered switches. Sci. Adv. 2019;5:eaau9443. doi: 10.1126/sciadv.aau9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhatia D, Surana S, Chakraborty S, Koushika SP, Krishnan Y. A synthetic icosahedral DNA-based host–cargo complex for functional in vivo imaging. Nat. Commun. 2011;2:339. doi: 10.1038/ncomms1337. [DOI] [PubMed] [Google Scholar]
- 21.Shi S, et al. Modulation of chondrocyte motility by tetrahedral DNA nanostructures. Cell Prolif. 2017;50:e12368. doi: 10.1111/cpr.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao M, et al. Rationally engineered nucleic acid architectures for biosensing applications. Chem. Rev. 2019;119:11631–11717. doi: 10.1021/acs.chemrev.9b00121. [DOI] [PubMed] [Google Scholar]
- 23.Ye D, Zuo X, Fan C. DNA nanotechnology-enabled interfacial engineering for biosensor development. Annu. Rev. Anal. Chem. 2018;11:171–195. doi: 10.1146/annurev-anchem-061417-010007. [DOI] [PubMed] [Google Scholar]
- 24.Chandrasekaran AR, Wady H, Subramanian HKK. Nucleic acid nanostructures for chemical and biological sensing. Small. 2016;12:2689–2700. doi: 10.1002/smll.201503854. [DOI] [PubMed] [Google Scholar]
- 25.Hu Q, Li H, Wang L, Gu H, Fan C. DNA nanotechnology-enabled drug delivery systems. Chem. Rev. 2019;119:6459–6506. doi: 10.1021/acs.chemrev.7b00663. [DOI] [PubMed] [Google Scholar]
- 26.Madhanagopal BR, Zhang S, Demirel E, Wady H, Chandrasekaran AR. DNA nanocarriers: programmed to deliver. Trends Biochem. Sci. 2018;43:997–1013. doi: 10.1016/j.tibs.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Linko V, Ora A, Kostiainen MA. DNA nanostructures as smart drug-delivery vehicles and molecular devices. Trends Biotechnol. 2015;33:586–594. doi: 10.1016/j.tibtech.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Jiang D, England CG, Cai W. DNA nanomaterials for preclinical imaging and drug delivery. J. Control. Release. 2016;239:27–38. doi: 10.1016/j.jconrel.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathur D, Medintz IL. The growing development of DNA nanostructures for potential healthcare-related applications. Adv. Healthc. Mater. 2019;8:1801546. doi: 10.1002/adhm.201801546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakraborty K, Veetil AT, Jaffrey SR, Krishnan Y. Nucleic acid–based nanodevices in biological imaging. Annu. Rev. Biochem. 2016;85:349–373. doi: 10.1146/annurev-biochem-060815-014244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajwar A, Kharbanda S, Chandrasekaran AR, Gupta S, Bhatia D. Designer, programmable 3D DNA nanodevices to probe biological systems. ACS Appl. Bio Mater. 2020;3:7265–7277. doi: 10.1021/acsabm.0c00916. [DOI] [PubMed] [Google Scholar]
- 32.Zhao N, Chen Y, Chen G, Xiao Z. Artificial cells based on DNA nanotechnology. ACS Appl. Bio Mater. 2020;3:3928–3934. doi: 10.1021/acsabm.0c00149. [DOI] [PubMed] [Google Scholar]
- 33.Kuzuya A, Ohya Y. DNA nanostructures as scaffolds for metal nanoparticles. Polym. J. 2012;44:452–460. doi: 10.1038/pj.2012.38. [DOI] [Google Scholar]
- 34.Chandrasekaran AR. Programmable DNA scaffolds for spatially-ordered protein assembly. Nanoscale. 2016;8:4436–4446. doi: 10.1039/C5NR08685J. [DOI] [PubMed] [Google Scholar]
- 35.Bhatia D, et al. Icosahedral DNA nanocapsules by modular assembly. Angew. Chem. Int. Ed. 2009;48:4134–4137. doi: 10.1002/anie.200806000. [DOI] [PubMed] [Google Scholar]
- 36.He Y, Chen Y, Liu H, Ribbe AE, Mao C. Self-assembly of hexagonal DNA two-dimensional (2D) arrays. J. Am. Chem. Soc. 2005;127:12202–12203. doi: 10.1021/ja0541938. [DOI] [PubMed] [Google Scholar]
- 37.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 38.Ke Y, Ong LL, Shih WM, Yin P. Three-dimensional structures self-assembled from DNA bricks. Science. 2012;338:1177–1183. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandrasekaran AR, Halvorsen K. Controlled disassembly of a DNA tetrahedron using strand displacement. Nanoscale Adv. 2019;1:969–972. doi: 10.1039/C8NA00340H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pei H, et al. Reconfigurable three-dimensional DNA nanostructures for the construction of intracellular logic sensors. Angew. Chem. Int. Ed. 2012;51:9020–9024. doi: 10.1002/anie.201202356. [DOI] [PubMed] [Google Scholar]
- 41.Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 42.Dittmer WU, Reuter A, Simmel FC. A DNA-based machine that can cyclically bind and release thrombin. Angew. Chem. Int. Ed. 2004;43:3550–3553. doi: 10.1002/anie.200353537. [DOI] [PubMed] [Google Scholar]
- 43.Modi S, et al. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 2009;4:325–330. doi: 10.1038/nnano.2009.83. [DOI] [PubMed] [Google Scholar]
- 44.Idili A, Vallée-Bélisle A, Ricci F. Programmable pH-triggered DNA nanoswitches. J. Am. Chem. Soc. 2014;136:5836–5839. doi: 10.1021/ja500619w. [DOI] [PubMed] [Google Scholar]
- 45.Juul S, et al. Temperature-controlled encapsulation and release of an active enzyme in the cavity of a self-assembled DNA nanocage. ACS Nano. 2013;7:9724–9734. doi: 10.1021/nn4030543. [DOI] [PubMed] [Google Scholar]
- 46.Kohman RE, Han X. Light sensitization of DNA nanostructures via incorporation of photo-cleavable spacers. Chem. Commun. 2015;51:5747–5750. doi: 10.1039/C5CC00082C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandrasekaran AR, Punnoose JA, Valsangkar V, Sheng J, Halvorsen K. Integration of a photocleavable element into DNA nanoswitches. Chem. Commun. 2019;55:6587–6590. doi: 10.1039/C9CC03069G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinheiro AV, Han D, Shih WM, Yan H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones MR, Seeman NC, Mirkin CA. Programmable materials and the nature of the DNA bond. Science. 2015;347:1260901. doi: 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- 50.Dunn KE. The business of DNA nanotechnology: commercialization of origami and other technologies. Molecules. 2020;25:377. doi: 10.3390/molecules25020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Linko V, Dietz H. The enabled state of DNA nanotechnology. Curr. Opin. Biotechnol. 2013;24:555–561. doi: 10.1016/j.copbio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 52.Kershner RJ, et al. Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat. Nanotechnol. 2009;4:557–561. doi: 10.1038/nnano.2009.220. [DOI] [PubMed] [Google Scholar]
- 53.Takabayashi S, et al. Boron-implanted silicon substrates for physical adsorption of DNA origami. Int. J. Mol. Sci. 2018;19:2513. doi: 10.3390/ijms19092513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 55.Erlich Y, Zielinski D. DNA fountain enables a robust and efficient storage architecture. Science. 2017;355:950–954. doi: 10.1126/science.aaj2038. [DOI] [PubMed] [Google Scholar]
- 56.Sun W, et al. Casting inorganic structures with DNA molds. Science. 2014;346:1258361. doi: 10.1126/science.1258361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weichelt R, Ye J, Banin U, Eychmüller A, Seidel R. DNA-mediated self-assembly and metallization of semiconductor nanorods for the fabrication of nanoelectronic interfaces. Chem. Eur. J. 2019;25:9012–9016. doi: 10.1002/chem.201902148. [DOI] [PubMed] [Google Scholar]
- 58.Liao S, Seeman NC. Translation of DNA signals into polymer assembly instructions. Science. 2004;306:2072–2074. doi: 10.1126/science.1104299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y, Mao C. Reprogramming DNA-directed reactions on the basis of a DNA conformational change. J. Am. Chem. Soc. 2004;126:13240–13241. doi: 10.1021/ja045718j. [DOI] [PubMed] [Google Scholar]
- 60.Martin TG, et al. Design of a molecular support for cryo-EM structure determination. Proc. Natl Acad. Sci. USA. 2016;113:E7456–E7463. doi: 10.1073/pnas.1612720113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Douglas SM, Chou JJ, Shih WM. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl Acad. Sci. USA. 2007;104:6644–6648. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hernandez C, et al. Self-assembly of 3D DNA crystals containing a torsionally stressed component. Cell Chem. Biol. 2017;24:1401–1406.e2. doi: 10.1016/j.chembiol.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 63.Chandrasekaran AR, et al. DNA nanotechnology approaches for microRNA detection and diagnosis. Nucleic Acids Res. 2019;47:10489–10505. doi: 10.1093/nar/gkz580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coleridge EL, Dunn KE. Assessing the cost-effectiveness of DNA origami nanostructures for targeted delivery of anti-cancer drugs to tumours. Biomed. Phys. Eng. Express. 2020;6:065030. doi: 10.1088/2057-1976/abbe73. [DOI] [PubMed] [Google Scholar]
- 65.Zhou L, et al. Programmable low-cost DNA-based platform for viral RNA detection. Sci. Adv. 2020;6:eabc6246. doi: 10.1126/sciadv.abc6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y, Cu YTH, Luo D. Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes. Nat. Biotechnol. 2005;23:885–889. doi: 10.1038/nbt1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chandrasekaran AR, et al. DNA nanoswitch barcodes for multiplexed biomarker profiling. Nano Lett. 2021;21:469–475. doi: 10.1021/acs.nanolett.0c03929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Porchetta A, et al. Programmable nucleic acid nanoswitches for the rapid, single-step detection of antibodies in bodily fluids. J. Am. Chem. Soc. 2018;140:947–953. doi: 10.1021/jacs.7b09347. [DOI] [PubMed] [Google Scholar]
- 69.Liang L, et al. Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew. Chem. Int. Ed. 2014;53:7745–7750. doi: 10.1002/anie.201403236. [DOI] [PubMed] [Google Scholar]
- 70.Hu R, et al. DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery. Angew. Chem. Int. Ed. 2014;53:5821–5826. doi: 10.1002/anie.201400323. [DOI] [PubMed] [Google Scholar]
- 71.Valsangkar VA, et al. Click and photo-release dual-functional nucleic acid nanostructures. Chem. Commun. 2019;55:9709–9712. doi: 10.1039/C9CC03806J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Messaoudi S, Greschner AA, Gauthier MA. Progress toward absorption, distribution, metabolism, elimination, and toxicity of DNA nanostructures. Adv. Ther. 2019;2:1900144. doi: 10.1002/adtp.201900144. [DOI] [Google Scholar]
- 73.Schüller VJ, et al. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano. 2011;5:9696–9702. doi: 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- 74.Rosier BJHM, et al. Incorporation of native antibodies and Fc-fusion proteins on DNA nanostructures via a modular conjugation strategy. Chem. Commun. 2017;53:7393–7396. doi: 10.1039/C7CC04178K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stewart JM, et al. Programmable RNA microstructures for coordinated delivery of siRNAs. Nanoscale. 2016;8:17542–17550. doi: 10.1039/C6NR05085A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fakhoury JJ, McLaughlin CK, Edwardson TW, Conway JW, Sleiman HF. Development and characterization of gene silencing DNA cages. Biomacromolecules. 2014;15:276–282. doi: 10.1021/bm401532n. [DOI] [PubMed] [Google Scholar]
- 77.Liu X, et al. A DNA nanostructure platform for directed assembly of synthetic vaccines. Nano Lett. 2012;12:4254–4259. doi: 10.1021/nl301877k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song L, et al. DNA origami/gold nanorod hybrid nanostructures for the circumvention of drug resistance. Nanoscale. 2017;9:7750–7754. doi: 10.1039/C7NR02222K. [DOI] [PubMed] [Google Scholar]
- 79.Aldaye FA, Senapedis WT, Silver PA, Way JC. A structurally tunable DNA-based extracellular matrix. J. Am. Chem. Soc. 2010;132:14727–14729. doi: 10.1021/ja105431h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou M, et al. Effect of tetrahedral DNA nanostructures on proliferation and osteo/odontogenic differentiation of dental pulp stem cells via activation of the notch signaling pathway. Nanomed. Nanotechnol. Biol. Med. 2018;14:1227–1236. doi: 10.1016/j.nano.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 81.Stephanopoulos N, et al. Bioactive DNA-peptide nanotubes enhance the differentiation of neural stem cells into neurons. Nano Lett. 2015;15:603–609. doi: 10.1021/nl504079q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Q, et al. Anti-inflammatory and antioxidative effects of tetrahedral DNA nanostructures via the modulation of macrophage responses. ACS Appl. Mater. Interfaces. 2018;10:3421–3430. doi: 10.1021/acsami.7b17928. [DOI] [PubMed] [Google Scholar]
- 83.Peng Q, et al. Understanding the biomedical effects of the self-assembled tetrahedral DNA nanostructure on living cells. ACS Appl. Mater. Interfaces. 2016;8:12733–12739. doi: 10.1021/acsami.6b03786. [DOI] [PubMed] [Google Scholar]
- 84.Jia R, et al. Aptamer-functionalized activatable DNA tetrahedron nanoprobe for PIWI-interacting RNA imaging and regulating in cancer cells. Anal. Chem. 2019;91:15107–15113. doi: 10.1021/acs.analchem.9b03819. [DOI] [PubMed] [Google Scholar]
- 85.Wang S, Xia M, Liu J, Zhang S, Zhang X. Simultaneous imaging of three tumor-related mRNAs in living cells with a DNA tetrahedron-based multicolor nanoprobe. ACS Sens. 2017;2:735–739. doi: 10.1021/acssensors.7b00290. [DOI] [PubMed] [Google Scholar]
- 86.Liu X, Wu L, Wang L, Jiang W. A dual-targeting DNA tetrahedron nanocarrier for breast cancer cell imaging and drug delivery. Talanta. 2018;179:356–363. doi: 10.1016/j.talanta.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 87.Schmied JJ, et al. DNA origami–based standards for quantitative fluorescence microscopy. Nat. Protoc. 2014;9:1367–1391. doi: 10.1038/nprot.2014.079. [DOI] [PubMed] [Google Scholar]
- 88.Jungmann R, et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and exchange-PAINT. Nat. Methods. 2014;11:313–318. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Allentoft ME, et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proc. R. Soc. B Biol. Sci. 2012;279:4724–4733. doi: 10.1098/rspb.2012.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rajendran A, Endo M, Katsuda Y, Hidaka K, Sugiyama H. Photo-cross-linking-assisted thermal stability of DNA origami structures and its application for higher-temperature self-assembly. J. Am. Chem. Soc. 2011;133:14488–14491. doi: 10.1021/ja204546h. [DOI] [PubMed] [Google Scholar]
- 91.Kielar C, et al. On the stability of DNA origami nanostructures in low-magnesium buffers. Angew. Chem. Int. Ed. 2018;57:9470–9474. doi: 10.1002/anie.201802890. [DOI] [PubMed] [Google Scholar]
- 92.Chandrasekaran AR, et al. Exceptional nuclease resistance of paranemic crossover (PX) DNA and crossover-dependent biostability of DNA motifs. J. Am. Chem. Soc. 2020;142:6814–6821. doi: 10.1021/jacs.0c02211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ramakrishnan S, Krainer G, Grundmeier G, Schlierf M, Keller A. Structural stability of DNA origami nanostructures in the presence of chaotropic agents. Nanoscale. 2016;8:10398–10405. doi: 10.1039/C6NR00835F. [DOI] [PubMed] [Google Scholar]
- 94.Myhrvold C, Dai M, Silver PA, Yin P. Isothermal self-assembly of complex DNA structures under diverse and biocompatible conditions. Nano Lett. 2013;13:4242–4248. doi: 10.1021/nl4019512. [DOI] [PubMed] [Google Scholar]
- 95.Salvati A, et al. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013;8:137–143. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 96.Yang W. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 98.Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 99.Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 100.Barry ME, et al. Role of endogenous endonucleases and tissue site in transfection and CpG-mediated immune activation after naked DNA injection. Hum. Gene Ther. 1999;10:2461–2480. doi: 10.1089/10430349950016816. [DOI] [PubMed] [Google Scholar]
- 101.Samejima K, Earnshaw WC. Trashing the genome: the role of nucleases during apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- 102.Koizumi T. Deoxyribonuclease II (DNase II) activity in mouse tissues and body fluids. Exp. Anim. 1995;44:169–171. doi: 10.1538/expanim.44.169. [DOI] [PubMed] [Google Scholar]
- 103.Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Castro CE, et al. A primer to scaffolded DNA origami. Nat. Methods. 2011;8:221–229. doi: 10.1038/nmeth.1570. [DOI] [PubMed] [Google Scholar]
- 105.Keum, J.-W. & Bermudez, H. Enhanced resistance of DNA nanostructures to enzymatic digestion. Chem. Commun. 7036–7038 (2009). [DOI] [PubMed]
- 106.Goltry S, et al. DNA topology influences molecular machine lifetime in human serum. Nanoscale. 2015;7:10382–10390. doi: 10.1039/C5NR02283E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yurke B, Turberfield AJ, Mills AP, Jr, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 108.Wang X, et al. Paranemic crossover DNA: there and back again. Chem. Rev. 2019;119:6273–6289. doi: 10.1021/acs.chemrev.8b00207. [DOI] [PubMed] [Google Scholar]
- 109.Hahn J, Wickham SFJ, Shih WM, Perrault SD. Addressing the instability of DNA nanostructures in tissue culture. ACS Nano. 2014;8:8765–8775. doi: 10.1021/nn503513p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wamhoff, E.-C. et al. Controlling wireframe DNA origami nuclease degradation with minor groove binders. Preprint at bioRxivhttps://www.biorxiv.org/content/10.1101/2020.05.24.110783v1.full (2020). [DOI] [PMC free article] [PubMed]
- 111.Li Y, et al. Universal pH-responsive and metal-ion-free self-assembly of DNA nanostructures. Angew. Chem. Int. Ed. 2018;57:6892–6895. doi: 10.1002/anie.201804054. [DOI] [PubMed] [Google Scholar]
- 112.Li Y, Schulman R. DNA nanostructures that self-heal in serum. Nano Lett. 2019;19:3751–3760. doi: 10.1021/acs.nanolett.9b00888. [DOI] [PubMed] [Google Scholar]
- 113.Conway JW, McLaughlin CK, Castor KJ, Sleiman H. DNA nanostructure serum stability: greater than the sum of its parts. Chem. Commun. 2013;49:1172–1174. doi: 10.1039/c2cc37556g. [DOI] [PubMed] [Google Scholar]
- 114.Cassinelli V, et al. One-step formation of “chain-armor”-stabilized DNA nanostructures. Angew. Chem. Int. Ed. 2015;54:7795–7798. doi: 10.1002/anie.201500561. [DOI] [PubMed] [Google Scholar]
- 115.Gerling T, Kube M, Kick B, Dietz H. Sequence-programmable covalent bonding of designed DNA assemblies. Sci. Adv. 2018;4:eaau1157. doi: 10.1126/sciadv.aau1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lacroix A, Vengut-Climent E, de Rochambeau D, Sleiman HF. Uptake and fate of fluorescently labeled DNA nanostructures in cellular environments: a cautionary tale. ACS Cent. Sci. 2019;5:882–891. doi: 10.1021/acscentsci.9b00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lin C, et al. Mirror image DNA nanostructures for chiral supramolecular assemblies. Nano Lett. 2009;9:433–436. doi: 10.1021/nl803328v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu Q, et al. Enhanced stability of DNA nanostructures by incorporation of unnatural base pairs. ChemPhysChem. 2017;18:2977–2980. doi: 10.1002/cphc.201700809. [DOI] [PubMed] [Google Scholar]
- 119.Chandrasekaran AR, et al. Hybrid DNA/RNA nanostructures with 2′-5′ linkages. Nanoscale. 2020;12:21583–21590. doi: 10.1039/D0NR05846G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lacroix A, Edwardson TGW, Hancock MA, Dore MD, Sleiman HF. Development of DNA nanostructures for high-affinity binding to human serum albumin. J. Am. Chem. Soc. 2017;139:7355–7362. doi: 10.1021/jacs.7b02917. [DOI] [PubMed] [Google Scholar]
- 121.Kim Y, Yin P. Enhancing biocompatible stability of DNA nanostructures using dendritic oligonucleotides and brick motifs. Angew. Chem. Int. Ed. 2020;59:700–703. doi: 10.1002/anie.201911664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Perrault SD, Shih WM. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano. 2014;8:5132–5140. doi: 10.1021/nn5011914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ahmadi Y, Llano ED, Barišić I. (Poly)cation-induced protection of conventional and wireframe DNA origami nanostructures. Nanoscale. 2018;10:7494–7504. doi: 10.1039/C7NR09461B. [DOI] [PubMed] [Google Scholar]
- 124.Ponnuswamy N, et al. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 2017;8:15654. doi: 10.1038/ncomms15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Anastassacos FM, Zhao Z, Zeng Y, Shih WM. Glutaraldehyde cross-linking of oligolysines coating DNA origami greatly reduces susceptibility to nuclease degradation. J. Am. Chem. Soc. 2020;142:3311–3315. doi: 10.1021/jacs.9b11698. [DOI] [PubMed] [Google Scholar]
- 126.Agarwal NP, Matthies M, Gür FN, Osada K, Schmidt TL. Block copolymer micellization as a protection strategy for DNA origami. Angew. Chem. Int. Ed. 2017;56:5460–5464. doi: 10.1002/anie.201608873. [DOI] [PubMed] [Google Scholar]
- 127.Auvinen H, et al. Protein coating of DNA nanostructures for enhanced stability and immunocompatibility. Adv. Healthc. Mater. 2017;6:1700692. doi: 10.1002/adhm.201700692. [DOI] [PubMed] [Google Scholar]
- 128.Wang S-T, et al. DNA origami protection and molecular interfacing through engineered sequence-defined peptoids. Proc. Natl Acad. Sci. USA. 2020;117:6339–6348. doi: 10.1073/pnas.1919749117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Miller SM, et al. Proteolytic studies of homologous peptide and N-substituted glycine peptoid oligomers. Bioorg. Med. Chem. Lett. 1994;4:2657–2662. doi: 10.1016/S0960-894X(01)80691-0. [DOI] [Google Scholar]
- 130.Nguyen M-K, et al. Ultrathin silica coating of DNA origami nanostructures. Chem. Mater. 2020;32:6657–6665. doi: 10.1021/acs.chemmater.0c02111. [DOI] [Google Scholar]
- 131.Chandrasekaran AR, Halvorsen K. Nuclease degradation analysis of DNA nanostructures using gel electrophoresis. Curr. Protoc. Nucleic Acid. Chem. 2020;82:e115. doi: 10.1002/cpnc.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zagorovsky K, Chou LYT, Chan WCW. Controlling DNA–nanoparticle serum interactions. Proc. Natl. Acad. Sci. USA. 2016;113:13600–13605. doi: 10.1073/pnas.1610028113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ramakrishnan S, et al. Real-time observation of superstructure-dependent DNA origami digestion by DNase I using high-speed atomic force microscopy. ChemBioChem. 2019;20:2818–2823. doi: 10.1002/cbic.201900369. [DOI] [PubMed] [Google Scholar]
- 134.Suck D. DNA recognition by structure-selective nucleases. Biopolymers. 1997;44:405–421. doi: 10.1002/(SICI)1097-0282(1997)44:4<405::AID-BIP5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 135.Hogan ME, Roberson MW, Austin RH. DNA flexibility variation may dominate DNase I cleavage. Proc. Natl. Acad. Sci. USA. 1989;86:9273–9277. doi: 10.1073/pnas.86.23.9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stopar A, Coral L, Di Giacomo S, Adedeji AF, Castronovo M. Binary control of enzymatic cleavage of DNA origami by structural antideterminants. Nucleic Acids Res. 2018;46:995–1006. doi: 10.1093/nar/gkx1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Suma A, Stopar A, Nicholson AW, Castronovo M, Carnevale V. Global and local mechanical properties control endonuclease reactivity of a DNA origami nanostructure. Nucleic Acids Res. 2020;48:4672–4680. doi: 10.1093/nar/gkaa080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shaw J-P, Kent K, Bird J, Fishback J, Froehler B. Modified deoxyoligonucleotides stable to exonuclease degradation in serum. Nucleic Acids Res. 1991;19:747–750. doi: 10.1093/nar/19.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kurnick NB. Desoxyribonuclease activity of sera of man and some other species. Arch. Biochem. Biophys. 1953;43:97–107. doi: 10.1016/0003-9861(53)90088-5. [DOI] [PubMed] [Google Scholar]
- 140.Osada K. Development of functional polyplex micelles for systemic gene therapy. Polym. J. 2014;46:469–475. doi: 10.1038/pj.2014.49. [DOI] [Google Scholar]
- 141.Kiviaho JK, et al. Cationic polymers for DNA origami coating – examining their binding efficiency and tuning the enzymatic reaction rates. Nanoscale. 2016;8:11674–11680. doi: 10.1039/C5NR08355A. [DOI] [PubMed] [Google Scholar]
- 142.Kizer ME, et al. Hydroporator: a hydrodynamic cell membrane perforator for high-throughput vector-free nanomaterial intracellular delivery and DNA origami biostability evaluation. Lab Chip. 2019;19:1747–1754. doi: 10.1039/C9LC00041K. [DOI] [PubMed] [Google Scholar]
- 143.Jun H, et al. Autonomously designed free-form 2D DNA origami. Sci. Adv. 2019;5:eaav0655. doi: 10.1126/sciadv.aav0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Goodman RP, et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 145.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]