Abstract
Cancer therapy is a strategic measure in inhibiting breast cancer stem cell (BCSC) pathways. Naringenin, a citrus flavonoid, was found to increase breast cancer cells’ sensitivity to chemotherapeutic agents. Bioinformatics study and 3D tumorsphere in vitro modeling in breast cancer (mammosphere) were used in this study, which aims to explore the potential therapeutic targets of naringenin (PTTNs) in inhibiting BCSCs. Bioinformatic analyses identified direct target proteins (DTPs), indirect target proteins (ITPs), naringenin-mediated proteins (NMPs), BCSC regulatory genes, and PTTNs. The PTTNs were further analyzed for gene ontology, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, protein–protein interaction (PPI) networks, and hub protein selection. Mammospheres were cultured in serum-free media. The effects of naringenin were measured by MTT-based cytotoxicity, mammosphere forming potential (MFP), colony formation, scratch wound-healing assay, and flow cytometry-based cell cycle analyses and apoptosis assays. Gene expression analysis was performed using real-time quantitative polymerase chain reaction (q-RT PCR). Bioinformatics analysis revealed p53 and estrogen receptor alpha (ERα) as PTTNs, and KEGG pathway enrichment analysis revealed that TGF-ß and Wnt/ß-catenin pathways are regulated by PTTNs. Naringenin demonstrated cytotoxicity and inhibited mammosphere and colony formation, migration, and epithelial to mesenchymal transition in the mammosphere. The mRNA of tumor suppressors P53 and ERα were downregulated in the mammosphere, but were significantly upregulated upon naringenin treatment. By modulating the P53 and ERα mRNA, naringenin has the potential of inhibiting BCSCs. Further studies on the molecular mechanism and formulation of naringenin in BCSCs would be beneficial for its development as a BCSC-targeting drug.
Keywords: Naringenin, Breast cancer stem cells, Targeted therapy, Bioinformatics, P53, ERα
Abbreviations: BCSCs, Breast cancer stem cells; CSC, Cancer stem cell; DAVID, Database for Annotation, Visualization, and Integrated Discovery; DTPs, Direct target proteins; DXR, Doxorubicin; EGF, Epidermal growth factor; EMT, Epithelial to mesenchymal transition; FITC, fluorescein isothiocyanate; GO, Gene ontology; ITPs, Indirect target proteins; KEGG, Kyoto Encyclopedia of Genes and Genomes; MET, Metformin; MFP, Mammosphere forming potential; NAR, Naringenin; NMPs, Naringenin-mediated proteins; PE, phycoerythrin; PPI, Protein-protein interaction; PTTN, Potential target of naringenin in inhibition of BCSCs; q-RT PCR, Quantitative real-time polymerase chain reaction; ROS, Reactive oxygen species
1. Introduction
Cancer is a deadly disease triggered by impaired cell proliferation and apoptosis (Hanahan and Weinberg, 2011). According to the World Health Organization, breast cancer is the second leading cause of death in women worldwide, and its incidence is predicted to increase (WHO, 2015). Along with surgery and radiotherapy, chemotherapy is widely used as a neoadjuvant in breast cancer therapy (Alvarez et al., 2010). The emergence of acquired resistance and the progression of cancer cells and metastasis resulted in treatment failure, where only 50% of the patients have shown good response to chemotherapy (Liu, 2009).
The existence of resistance to the use of chemotherapy agents is partly due to the presence of a population of cancer stem cells (CSCs) that can differentiate, causing relapse and metastasis and death in patients (Peitzsch et al., 2017). Breast cancer stem cell (BCSC) emergence is characterized by the cell’s ability to inactivate drugs, alter target cells, and suppress the accumulation of drugs entering the cells through increased efflux pumping (Badve and Nakshatri, 2012, Moitra, 2015). Therefore, breast cancer therapy, with inhibition of BCSC pathways, is a strategic and targeted approach.
Naringenin is a citrus flavonoid compound potentially developed as a BCSC-targeted drug. Previously, naringenin (Fig. 1A), a citrus flavonoid, has been shown to have cytotoxic and antiproliferative effects on several cancer cell types, one of which is the MCF-7 cells (Kanno et al., 2005). Naringenin inhibits proliferation, migration, and invasion, triggers apoptosis of SGC7901 gastric cancer cells through downregulation (Bao et al., 2016). Naringenin also induces apoptosis, increases intracellular reactive oxygen species (ROS), and upregulates ASK-1, p53, p38, and JNK in SNU-213 pancreatic cancer cells (Park et al., 2017).
Fig. 1.
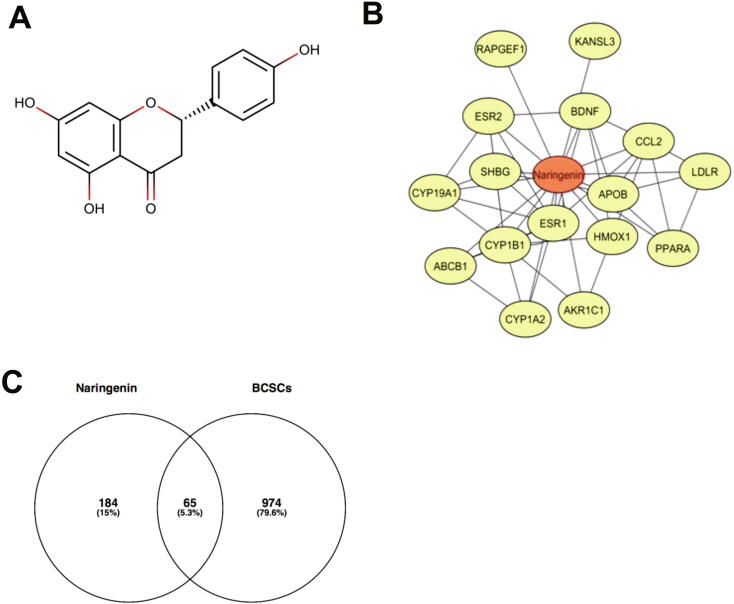
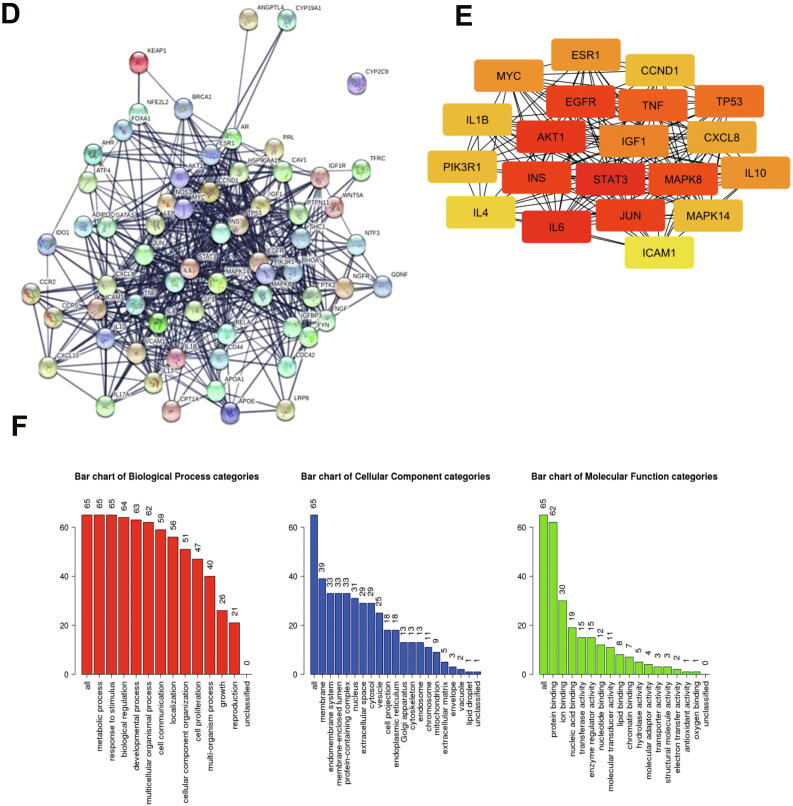
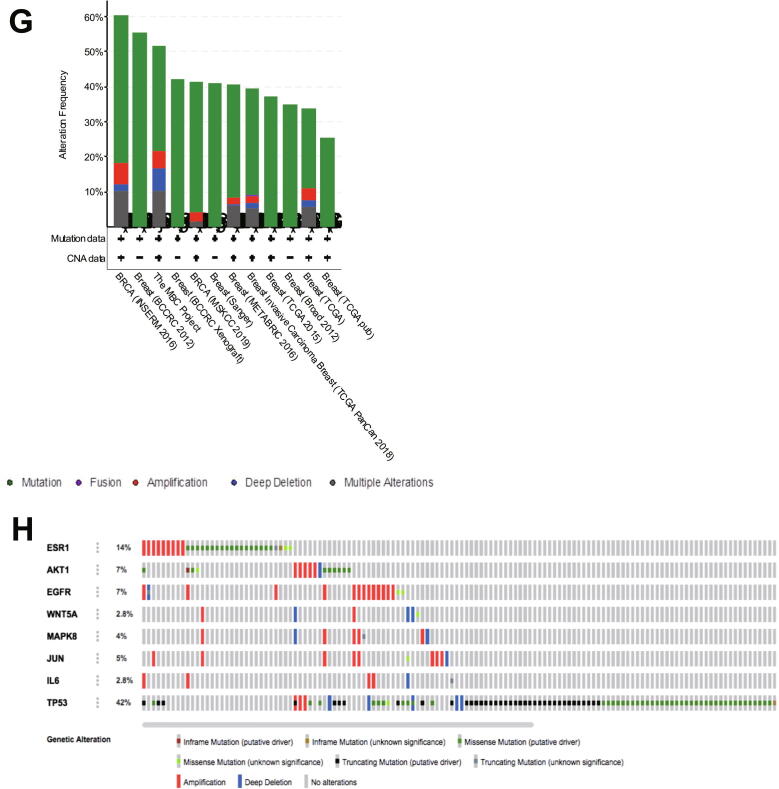
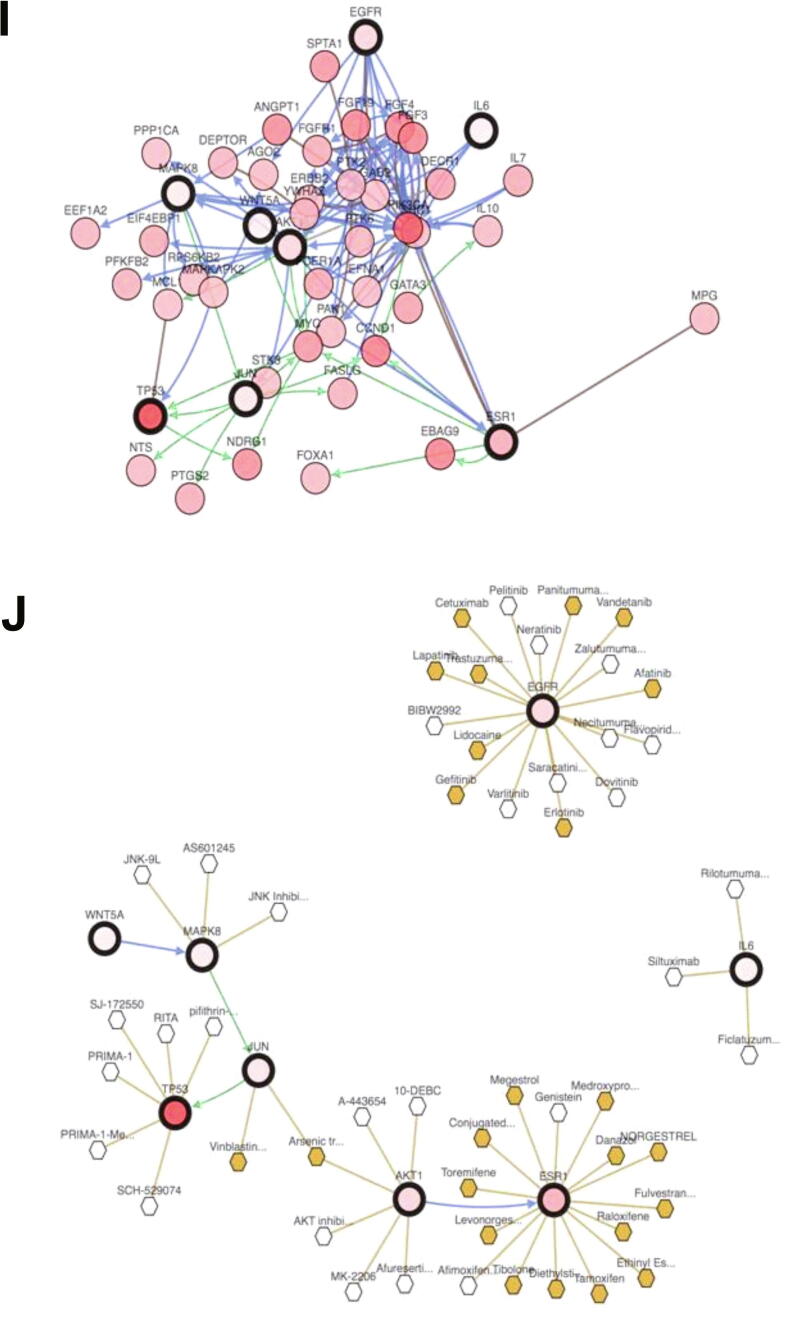
(A). The structure of naringenin. (B). Interactions of naringenin and its DTPs. (C). A Venn diagram of PTT related to Naringenin and BCSCs. (C). PPI network of naringenin protein targets, analyzed using STRING. (D). Top 10 hub proteins based on degree score, analyzed by CytoScape. (E). GO enrichment of PTTNs, analyzed by WebGestalt. (F). Overview of changes in PRKCA, EGFR, ERBB4, AREG, ESR1, and STAT1 in genomics dataset from 16 studies of breast cancer. (G). Summary alterations of ESR1, AKT1, EGFR, WNT5A, MAPK8, JUN, IL6, and TP53 across breast cancer samples (based on a study by Levebvre et al., 2016). (H). Gene network and (I). Drug-gene network connected to ESR1, AKT1, EGFR, WNT5A, MAPK8, JUN, IL6, and TP53 in breast cancer samples (based on a study by Lefebvre et al., 2016).
ROS play a critical role in cancer progression, in which it induces angiogenesis, metastasis, and chemoresistance in low concentrations and promotes cell death in high concentrations (Aggarwal et al., 2019). On the one hand, naringenin exhibits anti-inflammatory activity by decreasing ROS and antioxidant content in CD45+ hematopoietic cells (Bussmann et al., 2019) and RAW 264.7 cell line (Teng et al., 2018); and on the other hand, it inhibits diethylnitrosamine/acetylaminofluorene-induced hepatocarcinogenesis by inducing apoptosis and modulating oxidative stress (Ahmed et al., 2019).
To overcome chemotherapy resistance, studies on naringenin as a combination agent with daunomycin, mitoxantrone, doxorubicin, and vincristine in MCF-7/ADR breast cancer cells, NCI-H460 lung cancer cells, and A549 lung cancer cells were carried out (Mitsunaga et al., 2000, Chung et al., 2005). The combination of naringenin with doxorubicin shows an increase in the effectiveness of doxorubicin treatment in monolayer MCF-7 cells (Fitriasari et al., 2010). Moreover, combining naringenin and TNF-related apoptosis-inducing ligand (TRAIL) is an appropriate and safe strategy in treating apoptosis-resistant non-small cell lung cancer (Jin et al., 2011). The combination of naringenin and ABT-737 (Bcl-2 and Bcl-xL dual inhibitor) increases the effect of ABT-737 on gastric cancer cells (Zhang et al., 2016). Although naringenin promotes apoptosis in cancer cells, it can prevent apoptosis in normal cells stimulated by radiation by means of p53-, Bax-, and Bcl-2-mediated expansion modulation (Kumar and Tiku, 2016). However, research on the effects of naringenin as well as the molecular mechanisms of its action in BCSCs has not been conducted.
This study aimed to examine the potential of naringenin in BCSC inhibition using a 3D tumorsphere from MCF-7 breast cancer cells. Moreover, an integrated bioinformatics analysis was also used in predicting the potential therapeutic target of naringenin (PTTN) in BCSCs, therefore highlighting the association between naringenin and BCSC inhibition. The results of this study are available as reference for furture studies in the development of BCSC-targeted drugs capable of overcoming chemoresistance.
2. Methods
2.1. Bioinformatics analysis
In cells, a compound can target or interact with proteins. Proteins that interact or become targets of compounds are called direct target proteins (DTPs). Naringenin DTPs were searched from STITCH (http://stitch.embl.de) (Kuhn et al., 2007) and DrugBank (https://www.drugbank.ca) (Wishart et al., 2018), and the results were used for the following analysis. Proteins that interact with DTPs are defined as indirect target proteins (ITPs). The ITPs of each DTP were obtained from the STRING database (https://string-db.org) (Szklarczyk et al., 2015), with a minimum interaction score of 0.4 and a maximum number of interactors of 20. After removing repetitive proteins, the ITPs of all DTPs were generated. All DTPs and ITPs are naringenin-mediated proteins (NMPs). Breast cancer stem cell regulatory genes were retrieved from the PubMed gene database with the keywords “breast cancer stem cells.“ The PTTN against BCSCs are proteins that potentially target BCSCs and poses as NMPs. Analysis of protein–protein interaction (PPI) network of the PTTN was constructed with STRING-DB v11.0 (Szklarczyk et al., 2015), with confidence scores of > 0.7 and visualized by Cytoscape software (Shannon et al., 2003). As analyzed by CytoHubba plugin, proteins with a degree score of > 10 were selected as hub proteins (Chin et al., 2014). Analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of the PTTN was conducted using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (Huang da et al., 2009), with p < 0.05 selected as the cutoff value.). The genetic alterations of selected genes were analyzed using cBioPortal (http://www.cbioportal.org) (Cerami et al., 2012, Gao et al., 2013). In the present study, protein genes such as EGFR, AKT1, WNT5A, MAPK8, JUN, TP53, IL6, and ESR1 were screened for genetic alterations in all breast cancer studies available in the cBioportal database, and the study with the highest number of genetic alterations was chosen for further connectivity analysis.
2.2. Cell culture and mammosphere generation
MCF-7 cells (2D cell culture) were cultured in Dulbecco's Modified Eagle's medium (DMEM) (Gibco) with a 10% fetal bovine serum (FBS) (Sigma-Aldrich), 1.5% penicillin–streptomycin (Sigma-Aldrich), and 0.5% fungizone (Sigma-Aldrich) supplementation. To generate the mammosphere (3D cell culture), MCF-7 cells were cultured in 5% polyHEMA (Sigma-Aldrich)-coated plate using DMEM (Gibco) with the addition of 10 ng/mL of epidermal growth factor (EGF) (Sinobiological), 10 ng/mL of B27 supplement (Gibco), 5 µg/mL of insulin (Gibco), 1.5% of penicillin–streptomycin (Sigma-Aldrich), and 0.5% of fungizone (Thermo Fischer Scientific), as previousy described (Grimshaw et al., 2008, Pickl and Ries, 2009, Oak et al., 2012). Cells were incubated at 37 °C with 5% CO2 and were routinely tested and confirmed as mycoplasma free.
2.3. Drugs
Naringenin or NAR (N5893, purity ≥ 95%), metformin (MET, D150959, purity 97%), and doxorubicin (DXR, 44583, purity 98.0%–102.0%) were purchased from Sigma-Aldrich. In previous studies, MET and DXR were used as a positive control for the mammosphere forming experiment and for the clonogenic assay, respectively (Lou et al., 2006, Hirsch et al., 2009).
2.4. CD44+/CD24− phenotype enrichment assay
The CD44+/CD24− population phenotype enrichment assay was performed according to the previous study with modifications (Seo et al., 2015). The mammosphere was separated into a single cell by trypsinization. The cell suspension was incubated with phycoerythrin (PE)-conjugated anti-human CD44 antibody and fluorescein isothiocyanate (FITC)-conjugated anti-human CD24 mouse antibody (Molecular Probes) on ice and dark conditions for 20 min. Cells were washed after incubation, and were measured by flow cytometer (BD FACS Calibur) at a wavelength of 561 and 488 nm, respectively.
2.5. Cytotoxic assay
Cytotoxicity testing was performed based on a previous study (Mosmann, 1983). Cells were cultured on 96-well plates for the 2D model and polyHEMA-coated 96-well plate for the 3D model. These cells were treated with samples in a series concentration for 72 h, followed by MTT reagent addition, and were incubated for 2–4 h. In each well, SDS was added after incubation to stop the reaction. Using a microplate reader set at a 570-nm wavelength, the absorbance was measured, and the cell viability percentage was calculated.
2.6. Mammosphere forming potential
With reference to Oak et al., mammosphere forming potential (MFP) was calculated (Oak et al., 2012). Briefly, cells were treated with samples for 72 h then cultured on polyHEMA-coated 96-well plates for 96 h. The number of mammospheres formed was calculated manually.
2.7. Clonogenic survival assay
Cells were cultured on 6-well plates and treated with samples for 72 h. The media was replaced with fresh media. Cells were incubated for the following 14 days, and were fixed with methanol and stained with gentian violet solution after incubation. The Colony Area plugin of ImageJ was used to analyze the remaining colony formation, as previously described (Guzman et al., 2014).
2.8. Cell cycle modulation assay
Cells were cultured on 6-well plates (as both 2D and 3D models) and were treated with samples for 72 h. These cells were harvested and separated into cell suspensions, which were stained using propidium iodide (PI) reagent, transferred into a cytometric tube, and analyzed using a flow cytometer (BD FACS Calibur, BD Bioscience).
2.9. Apoptosis assay
As both 2D and 3D models, cells were cultured on 6-well plates and were treated with samples for 96 h. These cells were harvested, separated into a cell suspension, and stained using an FTIC Annexin V (Roche) staining kit according to the manufacturer’s instructions and were analyzed using a flow cytometer (BD FACS Calibur, BD Bioscience).
2.10. Scratch wound-healing assay
Mammosphere-derived MCF-7 cells (70,000 cells/well) were seeded in a 24-well plate and incubated for 24 h and were starved with the serum-free medium for another 24 h. The cells were scratched after starvation using a sterile pipette tip and were treated with naringenin. Images of the cells were captured at 0, 18, 24, 42, and 48 h after treatment by a digital camera (Nikon, Japan). ImageJ software was used to analyze the results, presented as percentage closure.
2.11. Quantitative real-time polymerase chain reaction (q-RT PCR)
Total mRNA was isolated using GeneJet RNA Purification Kit (Thermoscientific). A RevertAid First Strand cDNA Synthesis Kit (Thermoscientific) was utilized to transcribe RNA to cDNA. To quantify real-time polymerase chain reaction (RT PCR), SsoFast Evagreen Supermix (Bio-Rad) was used. Stem cell markers SOX2, CD133, and ALDH1 were examined, whereas GAPDH was used as a housekeeping gene. The results were analyzed using a comparative threshold cycle (ΔΔCT). GAPDH was used as an internal control. RT PCR was performed using the selected primers (Table S1).
2.12. Statistical analysis
From at least three independent experiments, data were presented as mean ± SD and were analyzed using GraphPad Prism 5.0 software. To analyze the CD44+/CD24− population enrichment assay and cytotoxic effect of NAR, two-way ANOVA with post-hoc Bonferroni’s multiple comparisons test was used, whereas one-way ANOVA with post-hoc Bonferroni’s multiple comparisons test was used to analyze the effect of NAR on mammosphere and colony formation. Student’s t-test was used to analyze the anti-migratory effect of NAR and quantitative RT PCR (q-RT PCR) results.
3. Results
3.1. Bioinformatics analysis
To obtain DTPs, we searched the DTPs of naringenin in STITCH and DrugBank databases and obtained 17 DTPs such as ESR1, AKR1C1, and CYP1B1 (Table 1). Fig. 1B shows the analysis of the interactions between naringenin and DTPs. A total of 249 NMPs containing 17 DTPs and 232 ITPs were obtained (Table S2), whereas 1041 BCSC regulatory genes from PubMed were retrieved (Table S3). A Venn diagram showed 65 PTTNs in BCSCs (Fig. 1C; Table S4).
Table 1.
Direct target protein of naringenin, from DrugBank and STITCH.
| No | Protein symbol | Protein name | Database |
|---|---|---|---|
| 1 | ESR1 | Estrogen receptor alpha | DrugBank |
| 2 | AKR1C1 | Aldo-keto reductase family 1 member C1 | DrugBank |
| 3 | CYP1B1 | Cytochrome P450 1B1 | DrugBank |
| 4 | KANSL3 | KAT8 regulatory NSL complex subunit 3 | DrugBank |
| 5 | SHBG | Sex hormone-binding globulin | DrugBank |
| 6 | CYP19A1 | Cytochrome P450 19A1 | DrugBank |
| 7 | ESR2 | Estrogen receptor beta | DrugBank |
| 8 | RAPGEF1 | Rap guanine nucleotide exchange factor 1 | STITCH |
| 9 | ABCB1 | ATP binding cassette subfamily B1 | STITCH |
| 10 | CYP1A2 | Cytochrome P450 1A2 | STITCH |
| 11 | CYP1B1 | Cytochrome P450 1B1 | STITCH |
| 12 | LDLR | Low-density lipoprotein receptor | STITCH |
| 13 | APOB | Apolipoprotein B-100 | STITCH |
| 14 | PPARA | Peroxisome proliferator-activated receptor alpha | STITCH |
| 15 | CCL2 | C-C motif chemokine 2 | STITCH |
| 16 | HMOX1 | Heme oxygenase 1 | STITCH |
| 17 | BDNF | Brain-derived neurotrophic factor | STITCH |
A PPI network of PTTNs was generated using a STRING database consisting 65 nodes, 553 edges, average node degree of 17, high confidence interaction of 0.7, average local clustering coefficient of 0.576, and PPI enrichment value < 1.10e-16 (Fig. 1D). Furthermore, hub proteins were selected from the PPI network based on a degree score (Fig. 1E). ESR1 was the only DTP among the hub proteins with a degree score of 26, indicating strong correlation with the biological effect of naringenin (Table 2).
Table 2.
The top 20 hub protein by degree score.
| No | Protein symbol | Degree score |
|---|---|---|
| 1 | STAT3 | 41 |
| 2 | IL6 | 40 |
| 3 | AKT1 | 36 |
| 4 | JUN | 34 |
| 5 | EGFR | 34 |
| 6 | INS | 34 |
| 7 | MAPK8 | 32 |
| 8 | TNF | 31 |
| 9 | TP53 | 28 |
| 10 | IGF1 | 27 |
| 11 | MYC | 26 |
| 12 | ESR1 | 26 |
| 13 | IL10 | 26 |
| 14 | CXCL8 | 25 |
| 15 | PIK3R1 | 24 |
| 16 | CCND1 | 24 |
| 17 | IL1B | 24 |
| 18 | MAPK14 | 24 |
| 19 | IL4 | 23 |
| 20 | ICAM1 | 22 |
GO analysis of PTTNs was classified into three groups: biological process, cellular component, and molecular function. Among the PTTNs (1F) were ones taking part in the biological process of apoptosis and intracellular signaling cascades. The PTTNs were located in the extracellular space and cytosol. Moreover, the PTTNs played a molecular function in growth factor and transcription factor activity. Pathway enrichment analysis using KEGG of the NMPs (Table S5) showed the regulation of approximately 53 pathways. Among these are pathways in cancer, Wnt/ß-catenin signaling, and TGF-beta signaling.
In exploring genomic alterations in breast cancer, a total of 8 PTTNs were analyzed using cBioportal. Regulators of cancer and the Wnt pathway are PTTNs EGFR, AKT1, WNT5A, MAPK8, JUN, TP53, and IL6, whereas ESR1 is the only hub protein from DTP based on highest degree score. Due to high alterations (60%), Lefebvre et al. (2016) was selected for further analysis among 12 breast cancer studies (Fig. 1G; (Lefebvre et al., 2016). Moreover, OncoPrint analysis showed that genetic alterations of the 8 PTTNs occurred in 2.8% to 42% of patients (Fig. 1H), and mutation is the most common gene alteration. From a study by INSERM 2016, additional mutual exclusivity analysis showed that gene pairs MAPK8-JUN, ESR1-TP53, EGFR-IL6, WNT5A-MAPK8, EGFR-MAPK8, WNT5A-JUN, and EGFR-JUN exhibited significant (p < 0.05) co-occurrence in breast cancer samples (Table 3). Further analysis of the relationship between the 7 PTTN and the neighboring genes can be found in the INSERM 2016 study (Fig. 1I). Moreover, TP53, ESR1, and EGFR were the main target of most cancer drugs, indicating the potential of TP53, ESR1, and EGFR as naringenin targets in BCSCs (Fig. 1J)
Table 3.
Mutual exclusivity analysis of selected genes in metastatic breast cancer study.
| A | B | Log2 Odds Ratio | p-Value | Tendency |
|---|---|---|---|---|
| MAPK8 | JUN | >3 | <0.001 | Co-occurrence |
| ESR1 | TP53 | −2.583 | <0.001 | Mutual exclusivity |
| EGFR | IL6 | >3 | <0.001 | Co-occurrence |
| WNT5A | MAPK8 | >3 | <0.001 | Co-occurrence |
| EGFR | MAPK8 | >3 | 0.001 | Co-occurrence |
| WNT5A | JUN | >3 | 0.001 | Co-occurrence |
| EGFR | JUN | 2.67 | 0.029 | Co-occurrence |
| IL6 | TP53 | 2.85 | 0.049 | Co-occurrence |
| AKT1 | IL6 | 2.807 | 0.065 | Co-occurrence |
| AKT1 | TP53 | 1.292 | 0.074 | Co-occurrence |
| AKT1 | EGFR | 1.731 | 0.103 | Co-occurrence |
| AKT1 | MAPK8 | 2.208 | 0.111 | Co-occurrence |
| WNT5A | IL6 | >3 | 0.157 | Co-occurrence |
| ESR1 | AKT1 | 1.095 | 0.181 | Co-occurrence |
| ESR1 | EGFR | 1.095 | 0.181 | Co-occurrence |
| ESR1 | IL6 | 1.642 | 0.207 | Co-occurrence |
| JUN | IL6 | 2.159 | 0.25 | Co-occurrence |
| MAPK8 | TP53 | −1.166 | 0.268 | Mutual exclusivity |
| JUN | TP53 | −0.799 | 0.326 | Mutual exclusivity |
| EGFR | TP53 | 0.495 | 0.342 | Co-occurrence |
| AKT1 | WNT5A | 1.379 | 0.373 | Co-occurrence |
| EGFR | WNT5A | 1.379 | 0.373 | Co-occurrence |
| ESR1 | JUN | 0.61 | 0.434 | Co-occurrence |
| WNT5A | TP53 | 0.471 | 0.5 | Co-occurrence |
| AKT1 | JUN | 0.501 | 0.545 | Co-occurrence |
| ESR1 | WNT5A | 0.263 | 0.61 | Co-occurrence |
| ESR1 | MAPK8 | −0.239 | 0.678 | Mutual exclusivity |
| MAPK8 | IL6 | <−3 | 0.795 | Mutual exclusivity |
3.2. Enrichment of BCSCs
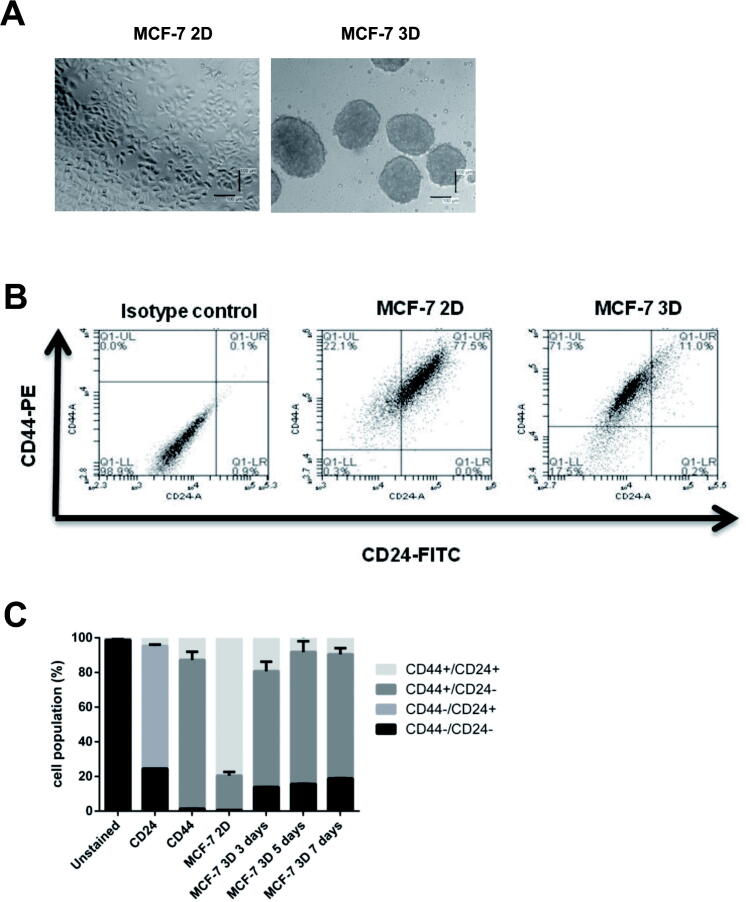
As described in Section 2, BCSCs enrichment was conducted by generating mammospheres using different methods from previous studies with modification. Cell preparation was initiated by harvesting previously cultured MCF-7 cells in DMEM (Fig. 2A). Cells (40,000 cells/well) were seeded in 50 mg/mL polyHEMA-coated 24-well plate, in SFM supplemented with EGF, insulin, and B27. Mammosphere was stable until day 7 after seeding.
Fig. 2.
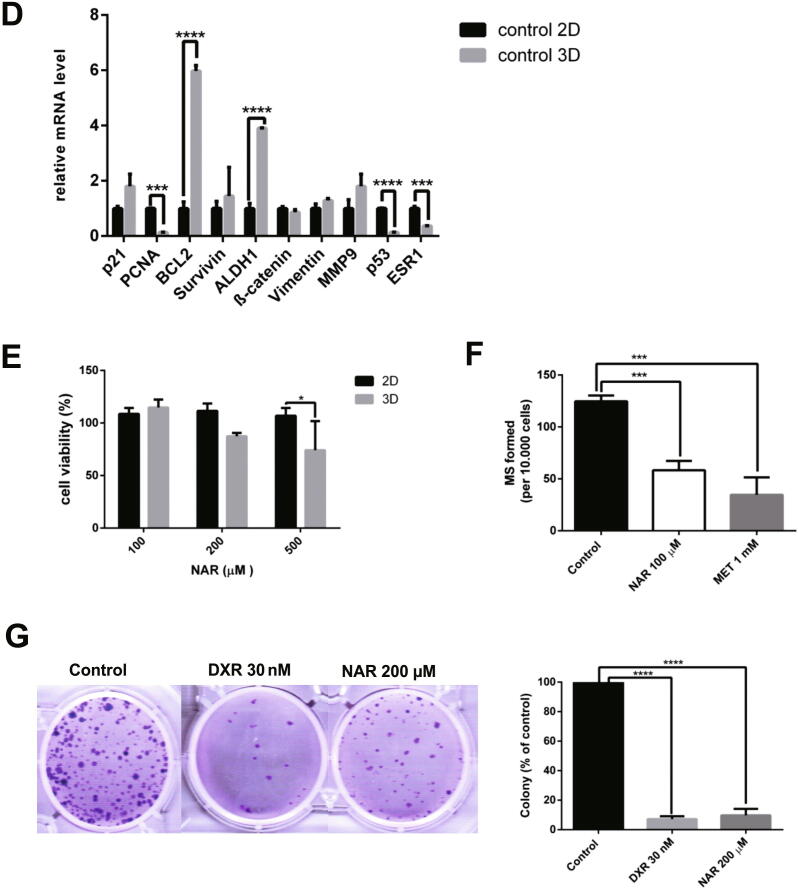
(A). Generation of mammospheres from MCF-7 cells. Formation of mammospheres from MCF-7 cells in serum-free media, as described in the methods section. Cells were seeded in a poly HEMA-coated plate (B). Identification of CD44+/CD24− population in MCF-7 cells using flow cytometry. (C). Quantification of CD44+/CD24− population in MCF-7 cells using flow cytometry. Cells in Q1-UL corresponded to CD44+/CD24− cells. An isotype control of MCF-7 cells (2D) was shown. (D). Gene expression of apoptosis, cell cycle, EMT, and stemness markers in 2D and 3D cells. Gene expression was determined by q-RT PCR. GAPDH was used as an internal control. The results were analyzed using the comparative threshold cycle (ΔΔCT) and are presented as fold change to the 2D cells. (E). Cytotoxicity of naringenin in MCF-7 cell monolayer (2D) and mammospheres (3D). A total of 10,000 cells per well were seeded in 96-well plates, as described in the method section. Cells were treated with naringenin and incubated for 72 h. MTT assay is used to determine cell viability. (F). Naringenin inhibits mammosphere formation from MCF-7 cells. Cells were seeded in 6-well plates and then incubated to 80% confluence. Subsequently, cells were treated with samples for 72 h. After that, the media was replaced with new media, and the cells were incubated for a further 24 h. Furthermore, 10,000 cells per well were seeded in 96-well plates and incubated for five days. At the end of incubation, the number of mammospheres formed is then calculated manually and analyzed as MFP. (G). Naringenin inhibits colony formation in MCF-7 cells. Naringenin treatment was given at 500 cells/well for 72 h, then the culture medium was replaced, and cells were grown for 14 days and stained at the end of the incubation period using gentian violet. The number of colonies was quantified with ColonyArea. Results represent the average of three independent experiments (mean ± SD). Two-way ANOVA with post-hoc Bonferroni’s multiple comparisons test was used to analyze the CD44+/CD24− population enrichment assay and cytotoxic effect of naringenin, while one-way ANOVA with post-hoc Bonferroni’s multiple comparisons test was used to analyze the effect of naringenin on mammosphere and colony formation. *** or **** indicates p < 0.001 or p < 0.0001, respectively. In Fig. 2C **** indicates significant to MCF-7 2D (p < 0.0001), while ++ in indicates significant to day 3 (p < 0.01).
CD44+/CD24− was selected as a molecular marker to identify the population of BCSCs and examine the stem cell enrichment characteristics in the mammosphere. Q1-UL area in Fig. 2B showed cell population in CD44+/CD24−. Quantification of the cell population showed a significant increase of CD44+/CD24− population after 3 days, a further rise after 5 days, and was stable after 7 days (Fig. 2C, indicating that CD44+/CD24− population at day 5 was significant than that on day 3 but was not significant on day 7. In addition, Gene expression of BCL2 and ALDH1 were significantly upregulated in 3D cells (Fig. 2D), however, p53 and ESR1 were significantly downregulated. Taken together, BCSCs were enriched in mammospheres from MCF-7 cells. Therefore, the authors decided to use mammospheres for the subsequent naringenin treatment after day 5.
3.3. Naringenin effects on cell viability and colony and mammosphere formation
After successful formation and characterization, mammospheres were treated with naringenin. Measured using MTT, with a parameter of cell viability, naringenin showed cytotoxicity in MCF-7 3D cells (Fig. 2E). Based on the number of mammospheres per cell seeded or the MFP, the effect of naringenin on mammosphere formation inhibition was analyzed by the number of mammospheres formed after treatment. Results revealed that naringenin inhibited mammosphere formation from MCF-7 cells (Fig. 2F). Moreover, the MFP value of naringenin is more or less similar to metformin, which targets cancer stem cell.
The drugs’ effect on colony formation was examined using the colony formation assay. Preliminary experiment reveal that naringenin concentration for colony and mammosphere formation was respectively 200 and 100 μM, which produced>80% cell viability (Fig. 2E) to ensure that cell growth inhibition does not influence the colony's inhibitory effect and mammosphere formation. The effect of naringenin on cell proliferation or cancer colonization was observed for 14 days; and after>24 h of observation, naringenin inhibited colony formation similar to that of doxorubicin (Fig. 2G).
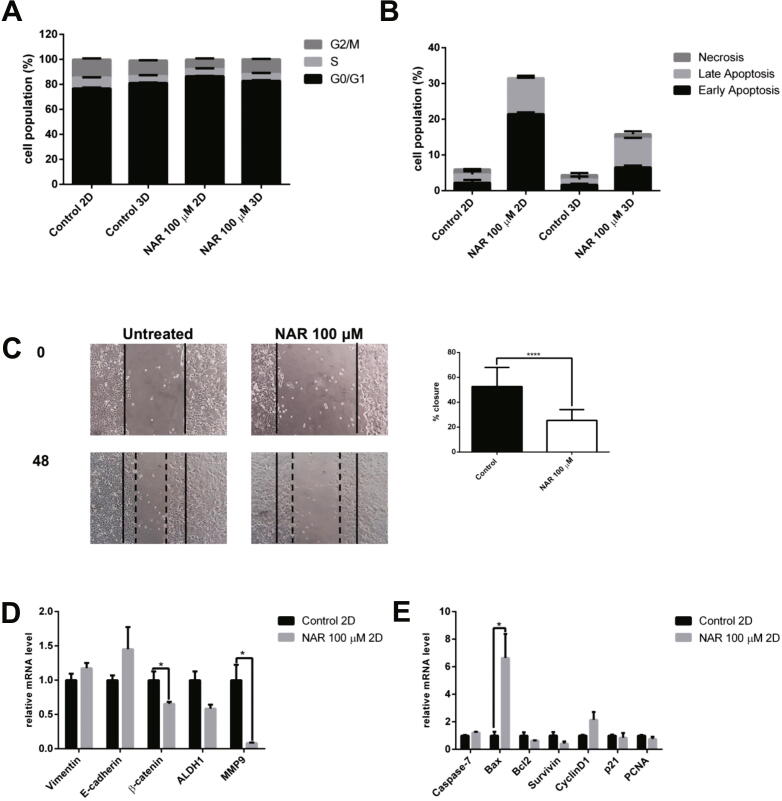
3.4. Effects of naringenin on cell cycle arrest, apoptosis, and migration
The effect of naringenin in modulating the cell cycle was tested using flow cytometry with propidium iodide (PI) staining on DNA. In G1 phase, an increase in 3D cells was observed compare with 2D cells (Fig. 3A); naringenin increased G1 cell cycle arrest in 2D cells. The cell cycle profile of naringenin-treated 3D cells is not significantly different from the untreated cells. Flow cytometry with annexin V and PI reagents was used to carry out confirmation of cell death upon naringenin treatment. Naringenin was shown to induce apoptosis in both 2D and 3D cell culture (Fig. 3B). Moreover, the effect of apoptosis induction in 2D cells was higher than in 3D cells. One indication of BCSC is migration, and a scratch wound-healing assay was performed to measure the potency of naringenin to inhibit the migration of mammosphere-derived MCF-7 cells. Fig. 2E shows a naringenin concentration of 100 μM produced>80% cell viability, ensuring that cell growth inhibition does not influence the inhibitory effect of migration. The results in Fig. 3C showed that naringenin inhibited cell migration, which agrees with the present study that naringenin inhibits proliferation and migration and induces apoptosis in BCSCs.
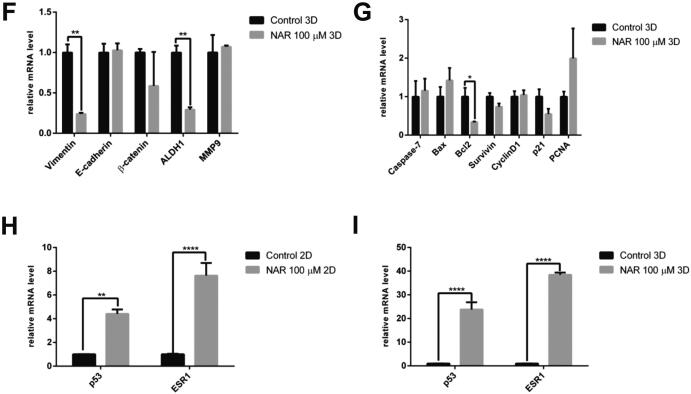
Fig. 3.
The effect of naringenin on the (A). Cell cycle profile. Cells were harvested after naringenin treatment for 72 h, stained with propidium iodide reagents, before analysis of DNA content using flow cytometry. (B). Apoptosis induction. Cells were harvested after naringenin treatment for 120 h, stained with annexin V and propidium iodide reagents, incubated, and analyzed using flow cytometry. The total percentage of cells consists of living cells, and cells undergoing early apoptosis, late apoptosis, and necrosis. (C). Naringenin inhibits migration. Mammosphere-derived MCF-7 cells were seeded and incubated for 24 h and starved with serum-free medium for another 24 h. After starvation, the cells were scratched using a sterile pipette tip and treated with naringenin. Images of the cells were captured at 0, 18, 24, 42, and 48 h after treatment. The results were analyzed using ImageJ and presented as percentage closure (n = 6). (D). Gene expression of EMT and stemness regulators in 2D cells. (E). Gene expression of apoptosis and cell cycle regulators upon naringenin treatment in 2D cells. (F). Gene expression of EMT and stemness regulators in 3D cells. (G). Gene expression of apoptosis and cell cycle regulators upon naringenin treatment in 3D cells. The effect of naringenin on p53 and ESR1 gene expression in 2D (H) and 3D cells (I). Gene expression was determined by q-RT PCR. GAPDH was used as an internal control. The results were analyzed using a comparative threshold cycle (ΔΔCT) and presented as fold change to the untreated control. Results represent the average of three independent experiments (mean ± SD). Statistical analyses were conducted using Student's t-test. *, **, and **** indicate p < 0.05, p < 0.01, and p < 0.0001, respectively.
3.5. Alterations of gene expression upon naringenin treatment
q-RT PCR was performed to examine the molecular mechanism of naringenin on BCSCs. The expression of stemness regulatory gene ß-catenin and ALDH1 was affected by naringenin treatment, as well as the epithelial-mesenchymal transition (EMT) regulators E-cadherin and Vimentin and migration regulator MMP9. The significant downregulation of ß-catenin and MMP9 and the reduced the expression of ALDH1 in 2D cells shown in Fig. 3D was due to naringenin treatment. Moreover, naringenin also increased Bax expression shown in Fig. 3E. In 3D cells, naringenin significantly reduced the expression of Vimentin and ALDH1 and BCL2 (Fig. 3F,G). In addition, naringenin reduced the expression of survivin and p21, but not statistically significant (Fig. 3G), and significantly increased p53 and ESR1 gene expression in 2D and 3D cells (Fig. 3I,J).
4. Discussion
This study aimed to explore the effect of naringenin in BCSC inhibition using bioinformatics approach and in vitro experiments. Integrated bioinformatics analysis retrieved DTPs, ITPs, and BCSCs-related genes, which resulted in the PTTNs in BCSCs. ERα is the only DTP having the highest degree score among hub proteins, indicating the pivotal role of ERα in the effect of naringenin on BCSCs. A similar bioinformatics work was published, where the association between gene-prostate cancer and naringenin by functional network analysis was identified (Fu et al., 2019).
The present study uses a similar approach in identifying key genes in naringenin effects against BCSCs. Hub proteins include EGFR, MAPK8, PIK3R1, Cyclin D1, and p53. This result is supported by previous studies, which showed that MAPK is one of the molecular targets of naringenin in MCF-7 cells (Eanes and Patel, 2016). A further study demonstrated that naringenin induces apoptosis in PC3 and LNCaP prostate cancer cells through inhibition of the PI3K/AKT and MAPK pathways (Lim et al., 2017).
A GO enrichment analysis revealed that PTTNs participated in the biological processes of apoptosis and intracellular signaling cascades. The PTTNs are located in the extracellular space and cytosol. Moreover, the PTTNs play a molecular function in growth factor and transcription factor activity. KEGG’s pathway enrichment of the PTTNs showed the regulation of approximately 53 pathways, including cancer, Wnt, and TGF-beta signaling pathways. Wnt signaling plays an important role in the proliferation, differentiation, invasion, migration, metastasis, and metabolism of cancer cells (Asem et al., 2016).
CSC formation can be achieved using 3D cell culture (Zhang et al., 2019), and to enrich BCSC properties, mammospheres were utilized. A serum-free medium in a polyHEMA-coated plate, with a diameter of around 100 µm, was used in culturing to generate the mammospheres. Based on our study, mammospheres were stable until day 7 and examination using CD44+/CD24- revealed that mammospheres BCSC properties were maintained. Mammospheres after day 5 were used for subsequent naringenin treatment because BCSC population at day 5 is not significantly different from the population at day 7. Results from this study, supported by a previous study, demonstrated that culturing spheroids to over 5–7 days will result to a growth of 100–500 µm in diameter (Liu et al., 2016). The effects of naringenin to the drug resistance phenotype of BCSCs, including drug uptake and MDR gene expression, needs further studies.
Naringenin exerts cytotoxicity in 2D and 3D cells, but its effects are stronger in 2D than in 3D cells. Previous studies showed that the IC50 value of naringenin is high. Naringenin showed cytotoxicity in MCF-7 (Fitriasari et al., 2010) and T47D (Junedi et al., 2010) breast cancer cells with IC50 values of 400 and 500 μM, respectively. The IC50 values of naringenin on MCF-7, HT-29, PC-12, and L-929 ranges from 780 to 880 µM (Kocyigit et al., 2016). According to Hakimuddin et al. (2004), Naringenin showed selectivity toward cancer cells (Hakimuddin et al., 2004). Naringenin treatment on CRL1554 human fibroblast cells, with a concentration of 4 mM for 24 h, induced cytotoxicity by about 30% (Abaza et al., 2015). In addition, Salehi et al. discussed clinical trials of naringenin on bioavailability and its effect on cardiovascular systems with a dosage between 600 and 800 μM/day (Salehi et al., 2019). Generally, the concentration of 100 and 200 µM is not toxic toward normal cells. A concentration of 100–500 µM was chosen for this study to determine cytotoxicity and a maximum dose of 200 µM was chosen for the other experiments on BCSCs (i.e., mammosphere forming analysis, clonogenic assay, cell cycle, apoptosis, anti-migratory assay, and q-RT PCR experiments).
Naringenin inhibited mammosphere formation based on MFP value. MFP is an accessible and suitable assay in estimating BSCS behavior (Lombardo et al., 2015) and it represents the population of CSCs responsible for chemoresistance and disease recurrence (Ji et al., 2016b). In this study, naringenin inhibited colony formation similar to doxorubicin (Fig. 2G). The colony formation or clonogenic assay is an in vitro technique that measures the ability of a single cell to grow into a larger colony, and it is a sensitive indicator of an undifferentiated CSC (Rajendran and Jain, 2018). To validate the results of this study, a study using methylcellulose based clonogenic assay is needed to replate the surviving colonies in the primary clonogenic assay and check the secondary colony formation potential.
The importance of p53 and ERα in the effect of naringenin on BCSCs were emphasized int the OncoPrint analysis, and additional mutual exclusivity analysis revealed the co-occurrence of p53 and ERα. Gene network analysis showed the importance of TP53 among neighboring genes and that p53 and ERα are the most druggable targets. Aside from naringenin, drugs that target p53 are PRIMA-1-Me, RITA, PRIMA-1, and those that target ERα are tamoxifen, raloxifene, and fulvestrant. Naringenin is clinically proven safe to use as a daily dietary supplement in healthy subjects (Rebello et al., 2020), and it can prevent osteosarcoma relapse in post-surgery osteosarcoma patients (Zhang et al., 2018), which is advantageous compared with other drugs.
In vitro results showed that mRNA of p53 and ERα were downregulated in 3D cells (Fig. 2D). Previous studies supported these findings, which demonstrated that ERα is downregulated in luminal A breast cancer that is resistant to chemotherapy and displays estrogen-independent signaling (Morimoto et al., 2009), and that loss of p53 promotes the maintenance of BCSCs (Chiche et al., 2017) and promotes Myc-activation and stem cell reprogramming of progenitors (Santoro et al., 2019). In this study, naringenin treatment significantly increased p53 and ERα mRNA expression in 3D cells (Fig. 3I). Bioinformatics finding was supported by the in vitro results, which emphasizes the pivotal role of p53 and ERα as PTTNs on BCSCs. A study using monolayer cell culture by Kanno et al. (2005) revealed no correlation between naringenin cytotoxicity and p53 status in cancer cells (Kanno et al., 2005). Nevertheless, the correlation between naringenin cytotoxicity and p53 status in BCSCs and the protein level regulation of p53 and ERα upon naringenin treatment remains unclear and needs to be confirmed in future studies.
The KEGG pathway enrichment analysis showed that cancer, β-catenin, and TGF-β signaling pathways may be regulated by naringenin in BCSC. Naringenin inhibition of Wnt/ß-catenin pathway in 2D cells are indicated in this study. These findings were supported by Li et al., (2013) showing that through the downregulation of survivin, naringin, the glycoside form of naringenin, inhibited the Wnt/ß-catenin pathway in triple-negative breast cancer cells through (Li et al., 2013). Activation of Wnt/β-catenin signaling plays a critical role in mesenchymal stem cell aging, which involves interactions with the p21 pathway (Zhang et al., 2011). Previous studies demonstrated the crosstalk between p53, ERα, and Wnt/ß-catenin signaling. Sadot et al., (2001) showed that activation of p53 leads to the downregulation of ß-catenin signaling (Sadot et al., 2001). In addition, the loss of wild-type p53 expression is driven by the excessive accumulation of ß-catenin (Sadot et al., 2001). Gupta et al., (2011) demonstrated that ß-catenin is a positive regulator of ERα function in breast cancer cells (Gupta et al., 2011). Moreover, the protein level of ERα is correlated with DKK1, a target gene of Wnt/ß-catenin signaling, highlighting the crosstalk between ERα and ß-catenin signaling in endometrial cancer (Kasoha et al., 2019). Generally, naringenin’s effect on the Wnt/ß-catenin pathway, p53, and ERα in inhibiting BCSCs is highlighted as an interesting topic for future research.
In the present study, TGF-β signaling is also inhibited by naringenin in 3D cells. Through increased expression of Vimentin, a marker of EMT, TGF-β activation triggers an EMT in breast cancer cells (Yoshida et al., 2013). EMT is important for the acquisition and maintenance of stem cell properties and CSCs. CSCs often exhibit EMT properties (Liu and Fan, 2015). In this study, Vimentin was upregulated in 3D cells compared with 2D cells, and this was also supported by previous studies that reported vimentin upregulation in BCSCs (Makki et al., 2015). The expression of Vimentin in 3D cells decreases with naringenin treatment. Results presented in this study demonstrated that naringenin inhibits cell migration. MMP9 was downregulated upon naringenin treatment in 2D cells; however, MMP9 expression was not downregulated upon naringenin treatment in 3D cells. This may be due to MMP9 activity inhibition rather than MMP9 expression. Previous studies supported this hypothesis which showed inhibition of MMP9 activity upon naringenin treatment in A549 lung cancer cells via regulation of the NFkB pathway (Chang et al., 2017) and in GBM 8901 glioblastoma cells through modulation of the ERK and p38 pathways (Chen et al., 2019). A study by Lou et al., (2012) demonstrated that by inhibiting TGF-β-induced EMT, naringenin reduced migration and invasion in pancreatic cancer cells (Lou et al., 2012).
TGF-ß signaling is also regulated by p53 and ERα. The transcriptional activity of TGF-ß target genes is suppressed by a p53 mutant gene (Elston and Inman, 2012); one of which is the p21 gene, which requires p53 for its transcriptional regulation (Cordenonsi et al., 2003). Moreover, p53 function inactivation modulates TGF-ß signaling (Kawarada et al., 2016). Stope et al. (2010) showed that ERα signaling activation inhibits TGF-ß signaling in breast cancer cells (Stope et al., 2010). In general, the mechanism of naringenin in BCSCs by regulating TGF-ß signaling, p53, and ERα is interesting topic to be evaluated.
The results of this study indicated that naringenin induces G1 cell cycle arrest in 2D cells, but cell cycle does not change compared with controls. In addition, it demonstrated that naringenin inhibits BCSCs by downregulating mRNA levels of ALDH1 and p21 in 3D cells. In correlation with Meng et al. (2014), it showed the importance of ALDH1 and p21 interaction in maintaining the CSC properties of ovarian cancer, in which downregulation of ALDH1 regulates cell cycle by decreasing p21 expression (Meng et al., 2014). Therefore, the interaction between ALDH1 and p21 and treatment with naringenin in mammospheres are interesting topics for further study.
The present study was done using the MCF-7 breast cancer cell line, which is a luminal subtype and has wild-type p53. In previous studies, naringenin exhibited cytotoxicity on different subtypes of breast cancer, including luminal, HER2+, and triple-negative breast cancer and it showed anticancer activity in triple-negative breast cancer cells 4 T1 (Qin et al., 2011) and MDA-MB-231 (Filho et al., 2014, Wang et al., 2019). Naringenin also exhibited cytotoxicity in MCF-7 breast cancer cells (Eanes and Patel, 2016, Xu et al., 2018) and HER2 positive breast cancer cells (Chandrika et al., 2016). In Kanno et al. (2005), cytotoxicity in various subtypes of monolayer breast cancer cells did not show any difference in naringenin treatment (Kanno et al., 2005). Breast cancer is a complex and heterogeneous disease; therefore, further study using the mammosphere from luminal breast cancer with mutant p53 or other types of breast cancer cell types is needed.
Several studies have shown the development of databases for predicting in vitro dose extrapolation to in vivo or clinical trials (Yoshida et al., 2017, Fabian et al., 2019). In addition, databases like ADMETlab (Dong et al., 2018) and SwissADME (Daina et al., 2017) can also be used for dose adjustments related to absorption, distribution, metabolism, excretion of a drug. A pharmacokinetic study of naringenin revealed the pharmacokinetic profile of naringenin in humans and rats (Bai et al., 2020). Taken together, the databases and results of the pharmacokinetics study can be used as basis in determining the clinical trial dose of naringenin in breast cancer patients.
One limitation of naringenin development is poor bioavailability. A previous study showed that the bioavailability of naringenin is 4% in oral administration (Hsiu et al., 2002). Naringenin undergoes further metabolism into several metabolites, including mainly sulfates and glucoronides (Zeng et al., 2019). Pharmacokinetics studies on rats, dogs, and humans revealed that apigenin, eriodictyol, hesperetin, naringenin-o-glucoronides, naringenin-o-glucosides, and 5,7-dihydroxychromone are naringenin metabolites (Bai et al., 2020). Another previous study showed that metabolites such as apigenin inhibited CSC properties in breast (Kim et al., 2016, Li et al., 2018) and prostate cancer (Erdogan et al., 2016) and eriodictyol in glioblastoma cells (Li et al., 2020). Therefore, the effect of naringenin on BCSCs inhibition in vivo might be due to its metabolites. However, this hypothesis need further investigation.
Studies were carried out to improve naringenin bioavailability. The naringenin nanoparticles formulation was shown to increase the physicochemical properties and hepatoprotective effects in vivo (Yen et al., 2009) and the cytotoxicity of naringenin in HeLa cervical cancer cells (Krishnakumar et al., 2011). Solid lipid nanoparticle naringenin was shown to increase the bioavailability of naringenin in the pulmonary delivery system in vitro and in vivo (Ji et al., 2016a). Therefore, further studies of the effects of naringenin nanoparticles on BCSCs are warranted.
In summary, this study highlighted the potential therapeutic use of naringenin for BCSC inhibition. The critical role of p53 and ERα in the effects of naringenin on BCSCs was emphasized through bioinformatics analysis and in vitro results. KEGG pathway enrichment analysis revealed TGF-ß and Wnt/ß-catenin pathways regulation by PTTN, p53 and ESR1 mRNA downregulation in mammospheres, and significant increased p53 and ESR1 mRNA levels with naringenin treatment. However, these results need to be confirmed at protein expression level. Naringenin has a more significant effect on mammosphere and colony formation. Validation of inhibiting those key genes could reverse the anti-proliferative, anti-migratory, or proapoptotic action of naringenin in BCSCs is needed. This study has a limitation as it is challenging to have a physiological significance because in vivo data is not available. Data presented in the manuscript is derived from a single cell line that needs to complement another cell line. Molecular mechanism of naringenin, including p53 and ER, in the inhibition of BCSCs requires further investigation for its development as a BCSC-targeted drug.
5. Conclusions
The results in this study showed that naringenin exhibits cytotoxicity in mammospheres and inhibits EMT and migration. More importantly, PTTNs that play a pivotal role in the effects of naringenin on BCSCs are p53 and ERα. Naringenin has the potential to eliminate BCSCs. Further in vitro and in vivo studies on naringenin’s molecular mechanism in BCSCs, as well as formulation for increasing bioavailability of naringenin, and clinical trials would be beneficial for its development as a BCSC-targeted drug.
Compliance with ethical standards
This study does not contain any studies with human participants or animals performed by any of the authors.
Availability of data and material
All data and material could be available from the corresponding author upon request.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported by the Penelitian Unggulan Perguruan Tinggi (PUPT) 2017 and 2018, Contract No. 2398/UN1.P.III/DIT-LIT/LT/2017) and No. 189/UN1/DITLIT/DIT-LIT/LT/2018.
Authors' contributions
AH—conception and design of the study, acquisition, analysis and interpretation of data, drafting, and revising the article, MI, RIJ, AK, IPN, HP, SMA, and HAM—acquisition and analysis of data. All authors had final approval of the submitted manuscript.
Acknowledgements
The authors thank Badan Penerbit dan Publikasi Universitas Gadjah Mada for their assistance in writing.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2020.12.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abaza, M.S.I., Orabi, K.Y., Al-Quattan, E. and Al-Attiyah, R.a.J., 2015. Growth inhibitory and chemo-sensitization effects of naringenin, a natural flavanone purified from Thymus vulgaris, on human breast and colorectal cancer. Cancer Cell International 15, 46. [DOI] [PMC free article] [PubMed]
- Aggarwal V., Tuli H.S., Varol A., Thakral F., Yerer M.B., Sak K., Varol M., Jain A., Khan M.A., Sethi G. Role of Reactive Oxygen Species in Cancer Progression: Molecular Mechanisms and Recent Advancements. Biomolecules. 2019;9 doi: 10.3390/biom9110735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed O.M., Ahmed A.A., Fahim H.I., Zaky M.Y. Quercetin and naringenin abate diethylnitrosamine/acetylaminofluorene-induced hepatocarcinogenesis in Wistar rats: the roles of oxidative stress, inflammation and cell apoptosis. Drug Chem. Toxicol. 2019:1–12. doi: 10.1080/01480545.2019.1683187. [DOI] [PubMed] [Google Scholar]
- Alvarez R.H., Booser D.J., Cristofanilli M., Sahin A.A., Strom E.A., Guerra L., Kau S.W., Gonzalez-Angulo A.M., Hortobagyi G.N., Valero V. Phase 2 trial of primary systemic therapy with doxorubicin and docetaxel followed by surgery, radiotherapy, and adjuvant chemotherapy with cyclophosphamide, methotrexate, and 5-fluorouracil based on clinical and pathologic response in patients with stage IIB to III breast cancer : long-term results from the University of Texas M. D. Anderson Cancer Center Study ID97-099. Cancer. 2010;116:1210–1217. doi: 10.1002/cncr.24901. [DOI] [PubMed] [Google Scholar]
- Asem M.S., Buechler S., Wates R.B., Miller D.L., Stack M.S. Wnt5a Signaling in Cancer. Cancers. 2016;8:79. doi: 10.3390/cancers8090079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badve S., Nakshatri H. Breast-cancer stem cells-beyond semantics. Lancet Oncol. 2012;13:e43–e48. doi: 10.1016/S1470-2045(11)70191-7. [DOI] [PubMed] [Google Scholar]
- Bai Y., Peng W., Yang C., Zou W., Liu M., Wu H., Fan L., Li P., Zeng X., Su W. Pharmacokinetics and Metabolism of Naringin and Active Metabolite Naringenin in Rats, Dogs, Humans, and the Differences Between Species. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Liu F., Guo H.B., Li Y., Tan B.B., Zhang W.X., Peng Y.H. Naringenin inhibits proliferation, migration, and invasion as well as induces apoptosis of gastric cancer SGC7901 cell line by downregulation of AKT pathway. Tumour Biol. 2016 doi: 10.1007/s13277-016-5013-2. [DOI] [PubMed] [Google Scholar]
- Bussmann A.J.C., Borghi S.M., Zaninelli T.H., Dos Santos T.S., Guazelli C.F.S., Fattori V., Domiciano T.P., Pinho-Ribeiro F.A., Ruiz-Miyazawa K.W., Casella A.M.B., Vignoli J.A., Camilios-Neto D., Casagrande R., Verri W.A., Jr. The citrus flavanone naringenin attenuates zymosan-induced mouse joint inflammation: induction of Nrf2 expression in recruited CD45(+) hematopoietic cells. Inflammopharmacology. 2019;27:1229–1242. doi: 10.1007/s10787-018-00561-6. [DOI] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrika B.B., Steephan M., Kumar T.R.S., Sabu A., Haridas M. Hesperetin and Naringenin sensitize HER2 positive cancer cells to death by serving as HER2 Tyrosine Kinase inhibitors. Life Sci. 2016;160:47–56. doi: 10.1016/j.lfs.2016.07.007. [DOI] [PubMed] [Google Scholar]
- Chang H.-L., Chang Y.-M., Lai S.-C., Chen K.-M., Wang K.-C., Chiu T.-T., Chang F.-H., Hsu L.-S. Naringenin inhibits migration of lung cancer cells via the inhibition of matrix metalloproteinases-2 and -9. Experimental Therapeutic Medicine. 2017;13:739–744. doi: 10.3892/etm.2016.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.Y., Chang Y.M., Wang K.Y., Chen P.N., Hseu Y.C., Chen K.M., Yeh K.T., Chen C.J., Hsu L.S. Naringenin inhibited migration and invasion of glioblastoma cells through multiple mechanisms. Environ. Toxicol. 2019;34:233–239. doi: 10.1002/tox.22677. [DOI] [PubMed] [Google Scholar]
- Chiche A., Moumen M., Romagnoli M., Petit V., Lasla H., Jezequel P., de la Grange P., Jonkers J., Deugnier M.A., Glukhova M.A., Faraldo M.M. p53 deficiency induces cancer stem cell pool expansion in a mouse model of triple-negative breast tumors. Oncogene. 2017;36:2355–2365. doi: 10.1038/onc.2016.396. [DOI] [PubMed] [Google Scholar]
- Chin C.H., Chen S.H., Wu H.H., Ho C.W., Ko M.T., Lin C.Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.Y., Sung M.K., Kim N.H., Jang J.O., Go E.J., Lee H.J. Inhibition of P-glycoprotein by natural products in human breast cancer cells. Arch Pharm Res. 2005;28:823–828. doi: 10.1007/BF02977349. [DOI] [PubMed] [Google Scholar]
- Cordenonsi M., Dupont S., Maretto S., Insinga A., Imbriano C., Piccolo S. Links between tumor suppressors: p53 is required for TGF-β gene responses by cooperating with Smads. Cell. 2003;113:301–314. doi: 10.1016/s0092-8674(03)00308-8. [DOI] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Wang N.-N., Yao Z.-J., Zhang L., Cheng Y., Ouyang D., Lu A.-P., Cao D.-S. ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminf. 2018;10:29. doi: 10.1186/s13321-018-0283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes L., Patel Y.M. Inhibition of the MAPK pathway alone is insufficient to account for all of the cytotoxic effects of naringenin in MCF-7 breast cancer cells. Biochim. Open. 2016;3:64–71. doi: 10.1016/j.biopen.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston, R. and Inman, G.J., 2012. Crosstalk between p53 and TGF-β Signalling. Journal of signal transduction 2012. [DOI] [PMC free article] [PubMed]
- Erdogan S., Doganlar O., Doganlar Z.B., Serttas R., Turkekul K., Dibirdik I., Bilir A. The flavonoid apigenin reduces prostate cancer CD44+ stem cell survival and migration through PI3K/Akt/NF-κB signaling. Life Sci. 2016;162:77–86. doi: 10.1016/j.lfs.2016.08.019. [DOI] [PubMed] [Google Scholar]
- Fabian E., Gomes C., Birk B., Williford T., Hernandez T.R., Haase C., Zbranek R., van Ravenzwaay B., Landsiedel R. In vitro-to-in vivo extrapolation (IVIVE) by PBTK modeling for animal-free risk assessment approaches of potential endocrine-disrupting compounds. Arch. Toxicol. 2019;93:401–416. doi: 10.1007/s00204-018-2372-z. [DOI] [PubMed] [Google Scholar]
- Filho J.C., Sarria A.L., Becceneri A.B., Fuzer A.M., Batalhao J.R., da Silva C.M., Carlos R.M., Vieira P.C., Fernandes J.B., Cominetti M.R. Copper (II) and 2,2'-bipyridine complexation improves chemopreventive effects of naringenin against breast tumor cells. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0107058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitriasari A., Susidarti R.A., Meiyanto E. Peningkatan efek sitotoksik doxorubicin oleh naringenin melalui pemacuan apoptosis sel kanker payudara MCF-7. Jurnal Bahan Alam. Indonesia. 2010;7 [Google Scholar]
- Fu S., Zhang Y., Shi J., Hao D., Zhang P. Identification of gene-phenotype connectivity associated with flavanone naringenin by functional network analysis. PeerJ. 2019;7 doi: 10.7717/peerj.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimshaw M.J., Cooper L., Papazisis K., Coleman J.A., Bohnenkamp H.R., Chiapero-Stanke L., Taylor-Papadimitriou J., Burchell J.M. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10:R52. doi: 10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Schmitt F., Grebhardt S., Mayer D. beta-Catenin Is a Positive Regulator of Estrogen Receptor-alpha Function in Breast Cancer Cells. Cancers (Basel) 2011;3:2990–3001. doi: 10.3390/cancers3032990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman C., Bagga M., Kaur A., Westermarck J., Abankwa D. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0092444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimuddin F., Paliyath G., Meckling K. Selective cytotoxicity of a red grape wine flavonoid fraction against MCF-7 cells. Breast Cancer Res. Treat. 2004;85:65–79. doi: 10.1023/B:BREA.0000021048.52430.c0. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hirsch H.A., Iliopoulos D., Tsichlis P.N., Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 2009;69:7507–7511. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiu S.L., Huang T.Y., Hou Y.C., Chin D.H., Chao P.D. Comparison of metabolic pharmacokinetics of naringin and naringenin in rabbits. Life Sci. 2002;70:1481–1489. doi: 10.1016/s0024-3205(01)01491-6. [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P., Yu T., Liu Y., Jiang J., Xu J., Zhao Y., Hao Y., Qiu Y., Zhao W., Wu C. Naringenin-loaded solid lipid nanoparticles: preparation, controlled delivery, cellular uptake, and pulmonary pharmacokinetics. Drug design, development and therapy. 2016:911–925. doi: 10.2147/DDDT.S97738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P., Zhang Y., Wang S.-J., Ge H.-L., Zhao G.-P., Xu Y.-C., Wang Y. CD44hiCD24lo mammosphere-forming cells from primary breast cancer display resistance to multiple chemotherapeutic drugs. Oncol. Rep. 2016;35:3293–3302. doi: 10.3892/or.2016.4739. [DOI] [PubMed] [Google Scholar]
- Jin C.Y., Park C., Hwang H.J., Kim G.Y., Choi B.T., Kim W.J., Choi Y.H. Naringenin up-regulates the expression of death receptor 5 and enhances TRAIL-induced apoptosis in human lung cancer A549 cells. Mol. Nutr. Food Res. 2011;55:300–309. doi: 10.1002/mnfr.201000024. [DOI] [PubMed] [Google Scholar]
- Junedi S., Susidarti R., Meiyanto E. Increased Cytotoxic Effect of Doxorubicin by Naringenin on T47D Cancer Cells through Apoptosis Induction. Jurnal Ilmu Kefarmasian Indonesia. 2010;8:85–90. [Google Scholar]
- Kanno S., Tomizawa A., Hiura T., Osanai Y., Shouji A., Ujibe M., Ohtake T., Kimura K., Ishikawa M. Inhibitory effects of naringenin on tumor growth in human cancer cell lines and sarcoma S-180-implanted mice. Biol. Pharm. Bull. 2005;28:527–530. doi: 10.1248/bpb.28.527. [DOI] [PubMed] [Google Scholar]
- Kasoha M., Dernektsi C., Seibold A., Bohle R.M., Takacs Z., Ioan-Iulian I., Solomayer E.F., Juhasz-Boss I. Crosstalk of estrogen receptors and Wnt/beta-catenin signaling in endometrial cancer. J. Cancer Res. Clin. Oncol. 2019 doi: 10.1007/s00432-019-03114-8. [DOI] [PubMed] [Google Scholar]
- Kawarada Y., Inoue Y., Kawasaki F., Fukuura K., Sato K., Tanaka T., Itoh Y., Hayashi H. TGF-β induces p53/Smads complex formation in the PAI-1 promoter to activate transcription. Sci. Rep. 2016;6:35483. doi: 10.1038/srep35483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Jung N., Lee S., Sohng J.K., Jung H.J. Apigenin Inhibits Cancer Stem Cell-Like Phenotypes in Human Glioblastoma Cells via Suppression of c-Met Signaling. Phytother. Res. 2016;30:1833–1840. doi: 10.1002/ptr.5689. [DOI] [PubMed] [Google Scholar]
- Kocyigit A., Koyuncu I., Dikilitas M., Bahadori F., Turkkan B. Cytotoxic, genotoxic and apoptotic effects of naringenin-oxime relative to naringenin on normal and cancer cell lines. Asian Pacific J. Tropical Biomedicine. 2016;6:872–880. [Google Scholar]
- Krishnakumar N., Sulfikkarali N., RajendraPrasad N., Karthikeyan S. Enhanced anticancer activity of naringenin-loaded nanoparticles in human cervical (HeLa) cancer cells. Biomedicine Preventive Nutrition. 2011;1:223–231. [Google Scholar]
- Kuhn M., von Mering C., Campillos M., Jensen L.J., Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2007;36:D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tiku A.B. Biochemical and Molecular Mechanisms of Radioprotective Effects of Naringenin, a Phytochemical from Citrus Fruits. J. Agric. Food Chem. 2016;64:1676–1685. doi: 10.1021/acs.jafc.5b05067. [DOI] [PubMed] [Google Scholar]
- Lefebvre C., Bachelot T., Filleron T., Pedrero M., Campone M., Soria J.C., Massard C., Levy C., Arnedos M., Lacroix-Triki M., Garrabey J., Boursin Y., Deloger M., Fu Y., Commo F., Scott V., Lacroix L., Dieci M.V., Kamal M., Dieras V., Goncalves A., Ferrerro J.M., Romieu G., Vanlemmens L., Mouret Reynier M.A., Thery J.C., Le Du F., Guiu S., Dalenc F., Clapisson G., Bonnefoi H., Jimenez M., Le Tourneau C., Andre F. Mutational Profile of Metastatic Breast Cancers: A Retrospective Analysis. PLoS Med. 2016;13(12) doi: 10.1371/journal.pmed.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Yang B., Huang J., Xiang T., Yin X., Wan J., Luo F., Zhang L., Li H., Ren G. Naringin inhibits growth potential of human triple-negative breast cancer cells by targeting beta-catenin signaling pathway. Toxicol. Lett. 2013;220:219–228. doi: 10.1016/j.toxlet.2013.05.006. [DOI] [PubMed] [Google Scholar]
- Li W., Du Q., Li X., Zheng X., Lv F., Xi X., Huang G., Yang J., Liu S. Eriodictyol Inhibits Proliferation, Metastasis and Induces Apoptosis of Glioma Cells via PI3K/Akt/NF-κB Signaling Pathway. Front. Pharmacol. 2020;11:114. doi: 10.3389/fphar.2020.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-W., Xu J., Zhu G.-Y., Huang Z.-J., Lu Y., Li X.-Q., Wang N., Zhang F.-X. Apigenin suppresses the stem cell-like properties of triple-negative breast cancer cells by inhibiting YAP/TAZ activity. Cell Death Discovery. 2018;4:105. doi: 10.1038/s41420-018-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim W., Park S., Bazer F.W., Song G. Naringenin-Induced Apoptotic Cell Death in Prostate Cancer Cells Is Mediated via the PI3K/AKT and MAPK Signaling Pathways. J. Cell. Biochem. 2017;118:1118–1131. doi: 10.1002/jcb.25729. [DOI] [PubMed] [Google Scholar]
- Liu F.S. Mechanisms of chemotherapeutic drug resistance in cancer therapy–a quick review. Taiwan J Obstet Gynecol. 2009;48:239–244. doi: 10.1016/S1028-4559(09)60296-5. [DOI] [PubMed] [Google Scholar]
- Liu H., Wang H., Li C., Zhang T., Meng X., Zhang Y., Qian H. Spheres from cervical cancer cells display stemness and cancer drug resistance. Oncol Lett. 2016;12:2184–2188. doi: 10.3892/ol.2016.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Fan D. The epithelial-mesenchymal transition and cancer stem cells: functional and mechanistic links. Curr. Pharm. Des. 2015;21:1279–1291. doi: 10.2174/1381612821666141211115611. [DOI] [PubMed] [Google Scholar]
- Lombardo Y., de Giorgio A., Coombes C.R., Stebbing J., Castellano L. Mammosphere formation assay from human breast cancer tissues and cell lines. J. Visualized Experiments : JoVE. 2015;52671 doi: 10.3791/52671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou C., Zhang F., Yang M., Zhao J., Zeng W., Fang X., Zhang Y., Zhang C., Liang W. Naringenin decreases invasiveness and metastasis by inhibiting TGF-beta-induced epithelial to mesenchymal transition in pancreatic cancer cells. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0050956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou P.-J., Lai P.-S., Shieh M.-J., MacRobert A.J., Berg K., Bown S.G. Reversal of doxorubicin resistance in breast cancer cells by photochemical internalization. Int. J. Cancer. 2006;119:2692–2698. doi: 10.1002/ijc.22098. [DOI] [PubMed] [Google Scholar]
- Makki J., Myint O., Wynn A.A., Samsudin A.T., John D.V. Expression distribution of cancer stem cells, epithelial to mesenchymal transition, and telomerase activity in breast cancer and their association with clinicopathologic characteristics. Clin Med Insights Pathol. 2015;8:1–16. doi: 10.4137/CPath.S19615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng E., Mitra A., Tripathi K., Finan M.A., Scalici J., McClellan S., da Silva L.M., Reed E., Shevde L.A., Palle K., Rocconi R.P. ALDH1A1 Maintains Ovarian Cancer Stem Cell-Like Properties by Altered Regulation of Cell Cycle Checkpoint and DNA Repair Network Signaling. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0107142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsunaga Y., Takanaga H., Matsuo H., Naito M., Tsuruo T., Ohtani H., Sawada Y. Effect of bioflavonoids on vincristine transport across blood-brain barrier. Eur. J. Pharmacol. 2000;395:193–201. doi: 10.1016/s0014-2999(00)00180-1. [DOI] [PubMed] [Google Scholar]
- Moitra K. Overcoming Multidrug Resistance in Cancer Stem Cells. Biomed Res. Int. 2015;2015:635745. doi: 10.1155/2015/635745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto K., Kim S.J., Tanei T., Shimazu K., Tanji Y., Taguchi T., Tamaki Y., Terada N., Noguchi S. Stem cell marker aldehyde dehydrogenase 1-positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression. Cancer Sci. 2009;100:1062–1068. doi: 10.1111/j.1349-7006.2009.01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Oak P.S., Kopp F., Thakur C., Ellwart J.W., Rapp U.R., Ullrich A., Wagner E., Knyazev P., Roidl A. Combinatorial treatment of mammospheres with trastuzumab and salinomycin efficiently targets HER2-positive cancer cells and cancer stem cells. Int. J. Cancer. 2012;131:2808–2819. doi: 10.1002/ijc.27595. [DOI] [PubMed] [Google Scholar]
- Park H.J., Choi Y.J., Lee J.H., Nam M.J. Naringenin causes ASK1-induced apoptosis via reactive oxygen species in human pancreatic cancer cells. Food Chem. Toxicol. 2017;99:1–8. doi: 10.1016/j.fct.2016.11.008. [DOI] [PubMed] [Google Scholar]
- Peitzsch C., Tyutyunnykova A., Pantel K., Dubrovska A. Cancer stem cells: The root of tumor recurrence and metastases. Semin. Cancer Biol. 2017;44:10–24. doi: 10.1016/j.semcancer.2017.02.011. [DOI] [PubMed] [Google Scholar]
- Pickl M., Ries C.H. Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab. Oncogene. 2009;28:461–468. doi: 10.1038/onc.2008.394. [DOI] [PubMed] [Google Scholar]
- Qin L., Jin L., Lu L., Lu X., Zhang C., Zhang F., Liang W. Naringenin reduces lung metastasis in a breast cancer resection model. Protein Cell. 2011;2:507–516. doi: 10.1007/s13238-011-1056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran V., Jain M.V. In vitro tumorigenic assay: colony forming assay for cancer stem cells. Cancer Stem Cells. Springer. 2018:89–95. doi: 10.1007/978-1-4939-7401-6_8. [DOI] [PubMed] [Google Scholar]
- Rebello C.J., Beyl R.A., Lertora J.J.L., Greenway F.L., Ravussin E., Ribnicky D.M., Poulev A., Kennedy B.J., Castro H.F., Campagna S.R., Coulter A.A., Redman L.M. Safety and pharmacokinetics of naringenin: A randomized, controlled, single-ascending-dose clinical trial. Diabetes Obes. Metab. 2020;22:91–98. doi: 10.1111/dom.13868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadot E., Geiger B., Oren M., Ben-Ze'ev A. Down-regulation of beta-catenin by activated p53. Mol. Cell. Biol. 2001;21:6768–6781. doi: 10.1128/MCB.21.20.6768-6781.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi B., Fokou P.V.T., Sharifi-Rad M., Zucca P., Pezzani R., Martins N., Sharifi-Rad J. The Therapeutic Potential of Naringenin: A Review of Clinical Trials. Pharmaceuticals (Basel) 2019;12 doi: 10.3390/ph12010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro A., Vlachou T., Luzi L., Melloni G., Mazzarella L., D’Elia E., Aobuli X., Pasi C.E., Reavie L., Bonetti P., Punzi S., Casoli L., Sabò A., Moroni M.C., Dellino G.I., Amati B., Nicassio F., Lanfrancone L., Pelicci P.G. p53 Loss in Breast Cancer Leads to Myc Activation, Increased Cell Plasticity, and Expression of a Mitotic Signature with Prognostic Value. Cell Reports. 2019;26:624–638.e8. doi: 10.1016/j.celrep.2018.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E.J., Wiench B., Hamm R., Paulsen M., Zu Y., Fu Y., Efferth T. Cytotoxicity of natural products and derivatives toward MCF-7 cell monolayers and cancer stem-like mammospheres. Phytomedicine. 2015;22:438–443. doi: 10.1016/j.phymed.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stope M.B., Popp S.L., Knabbe C., Buck M.B. Estrogen receptor α attenuates transforming growth factor-β signaling in breast cancer cells independent from agonistic and antagonistic ligands. Breast Cancer Res. Treat. 2010;120:357–367. doi: 10.1007/s10549-009-0393-2. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., Kuhn M., Bork P., Jensen L.J., von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng J., Li Y., Yu W., Zhao Y., Hu X., Tao N.P., Wang M. Naringenin, a common flavanone, inhibits the formation of AGEs in bread and attenuates AGEs-induced oxidative stress and inflammation in RAW264.7 cells. Food Chem. 2018;269:35–42. doi: 10.1016/j.foodchem.2018.06.126. [DOI] [PubMed] [Google Scholar]
- Wang R., Wang J., Dong T., Shen J., Gao X., Zhou J. Naringenin has a chemoprotective effect in MDA-MB-231 breast cancer cells via inhibition of caspase-3 and -9 activities. Oncol Lett. 2019;17:1217–1222. doi: 10.3892/ol.2018.9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2015. World Cancer Report 2014.
- Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., Maciejewski A., Gale N., Wilson A., Chin L., Cummings R., Le D., Pon A., Knox C., Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Huang B., Liu J., Wu X., Luo N., Wang X., Zheng X., Pan X. Combinatorial anti-proliferative effects of tamoxifen and naringenin: The role of four estrogen receptor subtypes. Toxicology. 2018;410:231–246. doi: 10.1016/j.tox.2018.08.013. [DOI] [PubMed] [Google Scholar]
- Yen F.-L., Wu T.-H., Lin L.-T., Cham T.-M., Lin C.-C. Naringenin-Loaded Nanoparticles Improve the Physicochemical Properties and the Hepatoprotective Effects of Naringenin in Orally-Administered Rats with CCl4-Induced Acute Liver Failure. Pharm. Res. 2009;26:893–902. doi: 10.1007/s11095-008-9791-0. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Saito T., Kamida A., Matsumoto K., Saeki K., Mochizuki M., Sasaki N., Nakagawa T. Transforming growth factor-beta transiently induces vimentin expression and invasive capacity in a canine mammary gland tumor cell line. Res. Vet. Sci. 2013;94:539–541. doi: 10.1016/j.rvsc.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Zhao P., Zhang L., Abernethy D.R., Rekić D., Reynolds K.S., Galetin A., Huang S.M. In Vitro-In Vivo Extrapolation of Metabolism- and Transporter-Mediated Drug-Drug Interactions-Overview of Basic Prediction Methods. J. Pharm. Sci. 2017;106:2209–2213. doi: 10.1016/j.xphs.2017.04.045. [DOI] [PubMed] [Google Scholar]
- Zeng X., Su W., Zheng Y., He Y., He Y., Rao H., Peng W., Yao H. Pharmacokinetics, Tissue Distribution, Metabolism, and Excretion of Naringin in Aged Rats. Front. Pharmacol. 2019;10:34. doi: 10.3389/fphar.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.-Y., Wang H.-J., Tan Y.-Z. Wnt/β-Catenin Signaling Induces the Aging of Mesenchymal Stem Cells through the DNA Damage Response and the p53/p21 Pathway. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0021397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhong X., Zhang X., Shang D., Zhou Y.I., Zhang C. Enhanced anticancer effect of ABT-737 in combination with naringenin on gastric cancer cells. Exp Ther Med. 2016;11:669–673. doi: 10.3892/etm.2015.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Xu X., Jiang T., Wu K., Ding C., Liu Z., Zhang X., Yu T., Song C. Citrus aurantium naringenin prevents osteosarcoma progression and recurrence in the patients who underwent osteosarcoma surgery by improving antioxidant capability. Oxidative medicine and cellular longevity 2018. 2018 doi: 10.1155/2018/8713263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li F., Zheng Y., Wang X., Wang K., Yu Y., Zhao H. Propofol Reduced Mammosphere Formation of Breast Cancer Stem Cells via PD-L1/Nanog In Vitro. Oxid. Med. Cell. Longevity. 2019;2019:9078209. doi: 10.1155/2019/9078209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and material could be available from the corresponding author upon request.