Summary
The diversity of circulating human B cells is unknown. We use single-cell RNA sequencing (RNA-seq) to examine the diversity of both antigen-specific and total B cells in healthy subjects and malaria-exposed individuals. This reveals two B cell lineages: a classical lineage of activated and resting memory B cells and an alternative lineage, which includes previously described atypical B cells. Although atypical B cells have previously been associated with disease states, the alternative lineage is common in healthy controls, as well as malaria-exposed individuals. We further track Plasmodium-specific B cells after malaria vaccination in naive volunteers. We find that alternative lineage cells are primed after the initial immunization and respond to booster doses. However, alternative lineage cells develop an atypical phenotype with repeated boosts. The data highlight that atypical cells are part of a wider alternative lineage of B cells that are a normal component of healthy immune responses.
Keywords: B cell memory, atypical B cells, alternative B cell lineage, malaria, sporozoite, vaccination, single cell RNA-seq, CITE-seq
Graphical Abstract

Highlights
-
•
Single-cell RNA-seq reveals two distinct B cell lineages
-
•
An alternative lineage contains CXCR3+ and atypical B cells
-
•
Alternative B cells are primed after primary vaccination and respond to boosters
-
•
Alternative B cells adopt a more atypical phenotype following repeated antigen exposure
Using single-cell RNA sequencing, Sutton et al. show that a population of “atypical” B cells, normally associated with disease, are part of a wider landscape of alternative B cells that participate in normal responses to vaccination.
Introduction
Memory B cells (MBCs) are antigen experienced cells that are primed to produce a faster, larger, and more effective response upon secondary exposure (Tangye et al., 2003). In humans, circulating human B cells have been classified based on the expression of the surface proteins CD38, CD21, and CD27. Antibody-secreting plasma cells (PCs) express high levels of CD38 and CD27 (Horst et al., 2002) while MBCs are generally defined by the expression of CD21 and CD27 (Klein et al., 1997; Tangye et al., 1998). These CD21+ CD27+ B cells show high levels of affinity maturation and readily differentiate into antibody-secreting PCs after stimulation compared to CD21+ CD27− naive cells (Good et al., 2009; Tangye et al., 2003). Populations of activated CD21−, CD27+ B cells that are predisposed to differentiate into PCs have also been described (Avery et al., 2005; Lau et al., 2017). Finally, a subset of B cells that are CD21− CD27− was originally observed in tonsils and later in peripheral blood (Ehrhardt et al., 2005; Fecteau et al., 2006). These cells, commonly referred to as atypical B cells (atBC), are observed at high frequencies in conditions of chronic antigen stimulation, such as infection with Human Immunodeficiency Virus (HIV), Hepatitis C Virus, or malaria (Charles et al., 2011; Moir et al., 2008; Weiss et al., 2009), or autoimmune disease (Isnardi et al., 2010; Wei et al., 2007). atBCs have also commonly been associated with the transcription factor Tbet (Johnson et al., 2020; Knox et al., 2017; Obeng-Adjei et al., 2017; Portugal et al., 2015).
Because they are often found in chronic infection and autoimmune disease, atBCs have historically been considered to be a population of anergic or exhausted B cells that arise due to chronic antigenic stimulation. In support of this, atBCs typically express high levels of inhibitory receptors, such as those belonging to the family of Fc-receptor-like (FCRL) molecules, as well as having muted B cell receptor (BCR) signaling and limited capacity to differentiate into PCs following BCR stimulation in vitro (Moir et al., 2008; Portugal et al., 2015; Sullivan et al., 2015). However, not all data indicate that atBCs are an exhausted population. Although Systemic Lupus Erythematosus (SLE) patients with high disease scores carry high numbers of atBCs, a recent study suggested that these are short-lived activated cells, in the process of differentiating into PCs (Jenks et al., 2018). Interesting, it has also been noted that atBCs may be responsible for the secretion of autoimmune antibodies that target proteins on the membrane of uninfected erythrocytes, leading to anemia (Rivera-Correa et al., 2019). Similarly, it has been shown that BCRs used by atBCs specific to Plasmodium falciparum could also be found contributing to the anti-P. falciparum antibody response (Muellenbeck et al., 2013). Studies following B cells after influenza vaccination found that somatically hypermutated CD21− Tbethi influenza-specific B cells arise following vaccination, suggesting a germinal center (GC) origin (Andrews et al., 2019; Lau et al., 2017). Similarly, studies have observed that Tbethi populations among bulk B cells expand following yellow fever and vaccinia immunization and primary HIV infection (Knox et al., 2017), suggesting these cells are part of functional immune responses.
Part of the confusion around whether atBCs are a friend or foe in the immune system stems from the different definitions used in different contexts. Thus, the degree of relatedness between cells identified as “atypical” in different studies is unclear. To develop an unbiased understanding of the heterogeneity of circulating B cells in humans and gain insight into the role of atBCs, we performed single-cell RNA sequencing (RNA-seq) on antigen-specific B cells from malaria-exposed adults and compared these data to single-cell RNA-seq on non-antigen-specific B cells from both malaria-exposed and non-exposed individuals.
Results
Single-cell RNA-seq reveals three distinct populations of antigen-experienced circulating B cells
The initial studies focused on the transcriptional diversity of the circulating B cell populations in malaria-vaccinated and exposed humans by single-cell RNA-seq. Specifically, we isolated CD19+, CD20+ B cells that (1) were class switched, i.e., immunoglobulin D (IgD)−, and (2) bound specific antigens. P. falciparum circumsporozoite (PfCSP)-specific cells were isolated from the peripheral blood of five Kenyan children 6.5 and 74 months after receiving the CSP-based RTS,S vaccine. To further examine the response to natural exposure to malaria, PfCSP-specific B cells, as well as B cells specific for the P. falciparum merozoite surface protein-1 (PfMSP1), were also sorted from 6 adult Kenyans from an area of moderate to high malaria transmission (Figure 1A; Table S1). Individuals in this population carry high numbers of circulating atBCs characterized by the absence of CD21 and CD27 (Aye et al., 2020). We also sorted tetanus toxoid (TT)-specific B cells from the adult subjects, which we have previously shown to have a more classical MBC phenotype (Aye et al., 2020). Because B cells specific for a given antigen are rare within an individual, only ∼10–50 antigen-specific cells could be sorted per sample. Accordingly, we used a modified version of the relatively low-throughput Smart-seq2 protocol (Picelli et al., 2014) to obtain 163 transcriptomes from 11 individuals.
Figure 1.
Three distinct populations of antigen-experienced B cells revealed by single-cell RNA-seq
CSP-, MSP1-, and TT-specific B cells were index, single-cell sorted from malaria vaccinated and exposed donors; transcriptomic information for each cell was generated using Smart-seq2 methodology.
(A) Representative flow cytometry plots showing the gating strategy used to sort mature IgD− antigen-specific B cells.
(B) Heatmap showing the expression of the top 25 DEGs (row) per cluster for each cell (column).
(C) Unsupervised clustering of circulating antigen-specific B cells visualized using uniform manifold approximation and projection (UMAP). Each point represents a cell and is colored by cluster.
(D) Heatmap displaying the normalized enrichment scores of multiple GSEAs comparing each cluster versus previously published gene sets from atBCs, naive B cells, and actBCs.
(E) Percentage of mutations found in the heavy-chain V(D)J region of each antigen-specific cell per cluster; analysis was by two-way ANOVA, including each subject as a blocking factor; bars represent mean ± SD.
(F) Percentage of antibody isotype usage by each cluster; analysis was by chi-square test on the absolute values, which are given above each bar.
(G) Percentage of cells specific for each antigen that were found in each cluster; analysis was by chi-square test on the absolute values, which are given above each bar.
Where the exact p value is not quoted, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Unsupervised hierarchical clustering grouped the cells into 3 clusters (Figures 1B and 1C). To attempt to link these clusters to previously identified classifications of B cells we used gene set enrichment analysis (GSEA). GSEA showed that cluster 1 had many differentially expressed genes (DEGs) associated with the atBC phenotype (Portugal et al., 2015), such as FCRL5, ITGAX, TNFRSF1B, LILRB1, CD19, and MS4A1 (Figures 1D, S1A, and S1B). Cluster 2 expressed the lymphoid homing gene CCR7 and the antiproliferative BTG1 gene (Figures 1D and S1B), suggesting that this may be a quiescent resting/central B cell memory subset and therefore were classified as MBCs. GSEA comparing to previously published datasets (Ellebedy et al., 2016) showed that this population shared some differentially expressed genes with naive B cells (Figures 1D and S1A). However, examination of the V(D)J sequences of these cells revealed them to be somatically mutated and predominantly class switched, supporting their designation as MBCs (Figures 1E and 1F). Cluster 3 expressed high levels of the chemokine receptor CXCR3 but could not be definitively linked to a previously described B cell phenotype; accordingly, we designated these cells CXCR3+ memory cells. Both CXCR3+ memory cells and atBCs also appeared to be marginally enriched for genes associated with activated B cells, though no distinct activated B cell population was identified in this analysis (Ellebedy et al., 2016).
Because cells were index sorted prior to sequencing, we could also measure the surface protein expression on each cell, allowing us to investigate the expression of CD21 and CD27. 44.7% of cells in the atBC cluster had the CD21− CD27− phenotype typically used to describe atBCs, though 85.1% were CD21 negative. Similarly, only 20.6% of CXCR3+ B and 37.5% of MBCs were CD21+ CD27+, suggesting that there may be distinct transcriptional signatures that suggest greater heterogeneity than using the canonical cell surface markers used to delineate MBC subsets (Figures S1C and S1D).
50% of PfCSP-specific B cells were atBCs, which may be consistent with continuous exposure to low levels of this antigen via repeated P. falciparum infections (Figures 1G and S1E). PfCSP-specific B cells also had the lowest mutational burden (Figure 1E), as has been reported previously (McNamara et al., 2020; Murugan et al., 2018). Interestingly, most PfMSP1-specific cells mapped to the CXCR3+ B cell population rather than the atBC population. Finally, TT-specific cells were mostly MBCs consistent with the absence of ongoing antigenic exposure (Figures 1G and S1E). Overall, it is likely that differences in the response to each antigen are indicative of different exposure history rather than intrinsic properties of the antigens themselves.
High-throughput, single-cell analysis identifies atBC, MBC, and activated B cell populations in both malaria-exposed and non-exposed donors
We next wanted to know whether the 3 subsets of circulating B cells we identified via our Smart-seq analysis of a small number of antigen-specific cells were specific to malaria or reflected B cell memory in general. Moreover, we were concerned that the association of antigen with cell populations, although striking, could be confounded because cells of different specificities came from different donors (Figures S1E and S1F). We therefore used the 10× Chromium platform to sequence single CD20+ CD19+ IgD− MBCs, regardless of antigen specificity, sorted from the peripheral blood mononuclear cells (PBMCs) of two non-exposed donors (non-exp) and two malaria-exposed (malaria-exp) donors (Table S1). We also included barcoded antibodies specific for CD11c, CXCR3, CD21, and CD27 to perform cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) analysis, linking surface protein expression to transcriptomic data (Stoeckius et al., 2017). We chose these markers based on our Smart-seq2 experiment and to reconcile our data with established markers. Finally, we used immune profiling to obtain paired heavy- and light-chain V(D)J chain sequences for the BCR of each individual B cell.
We sequenced 7,167 cells from malaria-exposed donors (malaria-exp 1: 1,448; malaria-exp 2: 5,719). We further sequenced 5,813 cells from non-exposed individuals (non-exp 1: 2,252; non-exp 2: 3,561). Unsupervised clustering was performed on each sample to identify and remove any non-B cell clusters (Figure S2A). Strikingly, in one of the exposed individuals (malaria-exp 1), we identified a cluster enriched for CD5 and BCL2 in which all cells expressed the same heavy- and light-chain immunoglobulin genes (IGHV7-81 and IGKV1-8). We concluded that these cells might be from a premalignant B cell clonal expansion and were subsequently removed from further analysis (Figure S2A).
Following removal of non-B cell populations, we combined all 4 samples into one integrated dataset (Figure 2A). Unsupervised clustering was then performed on this combined dataset of 12 621 cells, which revealed 11 conserved clusters (Figures 2A and S2B). Three of these clusters were more distantly related to the others. The first outlier cluster consisted of naive B cells, characterized by the expression of IGHD, as well as BACH2 and BTG1 (Figure S3A). The second group of outliers were PCs, discerned by the high expression of the transcription factors XBP1, IRF4, and PRDM1 (Figure S3A), which are all associated with controlling PC differentiation and maintenance (Klein et al., 2006; Reimold et al., 2001; Shaffer et al., 2002). The small populations of naive B cells and PCs likely may come in part from contaminating cells in our sort. Finally, we identified a population of cells expressing high levels of proliferation markers CD69, IRF4, MYC, and CD83 (Figure S3A), which we designated proliferating (Prol) cells.
Figure 2.
High-throughput, single-cell analysis reveals the full diversity of circulating B cell populations
Single B cells were sorted from 2 malaria exposed and 2 non-exposed individuals, and gene expression was assessed using 10× Chromium methodology.
(A) Unsupervised clustering of circulating mature IgD− B cells pooled from all individuals visualized using UMAP. Each cell is represented by a point and colored by cluster.
(B) Phylogenetic tree based on the “average cell” from each cluster, showing relationships in gene expression patterns between clusters.
(C) Heatmap displaying the normalized enrichment scores of multiple GSEAs comparing each cluster against previously published gene sets.
(D) Unsupervised clustering of B cells from an integrated 10× and Smart-seq2 dataset visualized using UMAP split by sequencing technique. Each cell is represented by a point and colored by either 10× or Smart-seq2 clusters.
(E) Percentage of cells from each Smart-seq2 cluster found in each 10× Chromium cluster.
(F) Pseudotime analysis of circulating B cells generated visualized using UMAP. Each point represents a cell and is colored by cluster or progression along pseudotime. Pseudotime begins at “1,” rooted on the naive cluster.
(G) Percentage of IgD− B cells from each individual found in each cluster.
Where the exact p value is not quoted, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Visual inspection of a heatmap showing the top DEGs combined with phylogenetic analysis (Figures 2B and S2B) suggested that the 8 remaining clusters could be grouped into 3 distinct “superclusters.” GSEA revealed that one of superclusters, made up of 3 smaller clusters, was enriched for previously identified atBC genes (Portugal et al., 2015). We therefore designated these cells as atBCs (Figures 2C and S3B). This designation was further supported by the high expression of common atypical genes, such as ITGAX, FCRL5, TBET, LILRB1, and CD19 (Figure S3C). The second supercluster was found to be enriched for genes associated with previously described activated B cells (actBCs) (Ellebedy et al., 2016); therefore, cells within this supercluster were designated actBCs (Figures 2C and S3B). The final supercluster did not appear to be enriched for any previously described B cell gene sets (Figures 2C and S3B), suggesting that these populations may represent more quiescent cell types; we therefore designated these cell as true MBCs. MBC2 in particular upregulated the common memory surface marker CCR7 (Figure S2B).
To determine the relationship between our Smart-seq2 clusters and those found using 10× Chromium, we combined both datasets (Figures 2D and 2E). This integrated dataset revealed that the majority of atBCs identified in the Smart-seq2 dataset mapped to atBC clusters (Figures 2D and 2E). Similarly, most MBCs identified by Smart-seq2 belonged to the 10× Chromium MBC supercluster (principally MBC2). Finally, CXCR3+ B cells identified by Smart-seq2 mapped predominantly to the 10× Chromium MBC1 cluster. In agreement with this observation, CXCR3 was found to be most highly expressed on the MBC1 population (Figure S3C). Thus, the CXCR3+ memory population identified in malaria-specific cells could be identified in polyclonal cells from both malaria and non-exposed donors. The principal discrepancy between the datasets was that the 10× dataset distinguished a set of actBCs that was not resolved by the smaller Smart-seq2 analysis. Instead, in the Smart-seq dataset, actBCs were grouped with either cluster 1 (atypical cells) or cluster 3 (CXCR3+ B cells), which may explain the weak actBC gene signature of these clusters (Figure 1D).
To determine the lineage relationships between the distinct populations identified by unbiased hierarchical clustering, we conducted pseudotime analysis, which allows for the identification of trajectories that measure the progress of cells through biological progresses, such as cell differentiation. This analysis revealed 2 distinct, major branches of circulating B cells (Figure 2F; Video S1). The first branch, consisting of the more “classical” MBC2, MBC3, and actBCs, had low progression along pseudotime (Figure 2E), suggesting they were more closely related to naive cells, which marked the beginning of pseudotime. A second “alternative” branch consisted of the atBCs and, strikingly, the MBC1 population with a small number of MBC3s. This alternative branch had progressed further along pseudotime, indicating they had differentiated further away from naive precursors than the classical branch. The proliferating cells appeared to form their own distinct branch separate from either actBCs or atBC. Interestingly, the PCs appeared entirely detached from the pseudotime pathway, indicating that an intermediate PC population could not be found among circulating B cells.
Movie showing a 3D UMAP projection of the monocle pseudotime analysis. The naive population that marks the start of pseudotime is highlighted at the beginning. The PC cluster as well as the alternative and classical lineages are also highlighted.
Importantly, all populations could be found in all individuals (Figure 2G). As expected, malaria-exposed donors had high numbers of atBC1 and atBC2 populations, but the non-exposed donors also had significant numbers of atBCs (∼20%) that were largely atBC3 (Figure 2F). This number of atBCs, as identified by transcriptomic techniques, contrasts with previous flow cytometry analysis, which shows that non-malaria-exposed healthy donors typically carry few (generally <5%) CD27−, CD21− atypical cells (Illingworth et al., 2013; Weiss et al., 2009). Non-exposed donors and malaria-exposed donors alike also carried high numbers of the alternative lineage MBC1 population (∼20%; Figure 2F). Thus, alternative lineage cells are a major component of circulating B cell populations in both malaria-exposed and non-exposed healthy individuals.
Subsets of circulating B cells do not segregate with Ig subclass or V region usage
In murine models, different MBC subsets may be defined by their Ig subclass, including in malaria infection (Krishnamurty et al., 2016; Pape et al., 2011). We therefore performed analysis of the VDJ and constant region usage of the heavy chains of our different B cell populations. The switched (non-IgD) subclasses were found in all non-naive B cell populations, with the exception of IgE, of which only 2 cells were found across all donors (Figure 3A). The population we identified as naive was largely IgM+, with some cells expressing IgD, further supporting this designation. In both malaria-exposed donors, IgG3 was overrepresented in alternative lineage B cells, which is consistent with previous reports (Knox et al., 2017; Obeng-Adjei et al., 2017). Overall, these data suggest that the transcriptional signatures are distributed across all Ig subclasses.
Figure 3.
Lack of association between BCR variable and constant regions with different B cell subsets
V(D)J and constant region sequences for each cell from each donor described in Figure 2 were mapped to the individual transcriptomes and relationships analyzed.
(A) Percentage of isotype usage for each cluster per individual.
(B) Percentage of mutations found in the heavy-chain V(D)J region of cells for each cluster in each donor; mean ± SD shown.
(C) Heatmaps displaying the pairwise p values from Tukey’s post-tests based on one-way ANOVA of the data in (B) to determine the association between cell type and mutation frequency with subclass and individual also included in the model as fixed factors.
(D) Circos plots showing clonal B cell populations per individual; the thickness of the lines between or within clusters denotes the number of cells that belong to shared/expanded clones.
We further examined the variable region sequence of each cell to determine the level of somatic hypermutation (SHM) and clonal relationships based on identical IGHV Complementarity Determining Region 3s with matching IGLV or IGKV. Similar to the constant region, V region usage was similar between all donors and B cell clusters (Figures S3A and S3B). All clusters, with the exception of the naive cells, showed significant SHM, confirming that these cells were antigen experienced, post-GC B cells (Figure 3B). The degree of SHM differed significantly based on the cell population and donor (Figure 3B). Notably, across all populations, non-exposed donors apparently carried lower levels of SHM than malaria-exposed donors, perhaps indicating lower lifetime pathogen burden (Figure 3B). However, V(D)J databases are based on European donors, so the apparent increased mutation burden may in fact be due to allelic differences between populations of African and European ancestry. In 3/4 donors, actBC2 and PCs had higher levels of SHM compared to either atBC populations or MBC populations (Figures 3B and 3C). Finally, even though we did not sort antigen-specific cells, 1%–5% of all BCRs sequenced were shared between 2 or more cells in each sample (Figure 3D). In most cases, these expanded clones were found within clusters; however, some clones were shared between clusters, indicating that a single clone can potentially adopt multiple cell fates (Figure 3D).
Atypical B cells are represented at high frequencies in all individuals but are not necessarily CD27− CD21−
Our finding that the alternative lineage cells, including atBCs, were frequent in non-exposed donors contrasts with traditional flow cytometry analysis that shows that CD21− CD27− cells are generally rare in healthy western individuals. To address this discrepancy, we used CITE-seq to correlate the cell surface levels of CD21 and CD27 with our transcriptomic data for each cell and cluster (Figure 4A). Analysis of CD21 and CD27 expression showed that atBC1s, which are most abundant in malaria-exposed individuals, were almost exclusively CD21− CD27− or CD21− CD27+ (Figures 4B and 4C). However, the other related alternative lineage populations had more heterogeneous CD21/CD27 expression (Figures 4C and 4E). Combining this dataset with our pseudotime analysis, we hypothesize that atBC1 cells represent “true” atBCs, which may be an exhausted population akin to that previously described (Illingworth et al., 2013; Lau et al., 2017; Portugal et al., 2015; Weiss et al., 2009, 2010). On the other hand, the CD21− CD27− phenotype fails to capture other cells within in the alternative lineage, which may retain functional capacity.
Figure 4.
CITE-seq analysis reveals a cryptic population of atBCs found predominantly in non-exposed individuals
CITE-seq analysis to correlate expression of cell surface markers with gene expression was performed on cells from the four donors described in Figure 2.
(A) Surface protein expression measured by CITE-seq projected onto UMAP plots. Color was scaled for each marker with highest and lowest centered log ratio (CLR)-normalized expression level noted.
(B) Contour plots showing the CLR-normalized expression of CD27 and CD21 as measured by CITE-seq; data are concatenated from all individuals.
(C) Quantification of (B); data show the mean proportion ± SD of the CD21/CD27 CLR-normalized expression per individual.
(D) Histogram plots showing the CLR-normalized expression of CD11c on the different atBC, MBC, and actBC clusters; the gray histogram represents expression on naive B cells; data are concatenated from all individuals. Bar graph shows mean ± SD of the surface protein expression of CD11c for each individual, with triangles representing non-exposed and squares representing malaria-exposed individuals; analysis was performed using one-way ANOVA with each individual as a blocking factor, only comparisons to the naive population are shown.
(E) Histogram plots showing the CLR-normalized expression of CXCR3 on the different atBC, MBC, and actBC clusters; the gray histogram represents expression on naive B cells; data are concatenated from all individuals. Bar graph shows the mean ± SD of the surface protein expression of CXCR3 for each individual, with triangles representing non-exposed and squares representing malaria-exposed individuals; analysis was performed using one-way ANOVA with each individual as a blocking factor, only comparisons to the naive population are shown.
We also examined the markers CD11c and CXCR3, as these were differentially expressed in the atBC and CXCR3+ populations identified in our original Smart-seq experiment (Figures 4D and 4E). CD11c was found to be generally elevated on alternative lineage cells, with the exception of atBC3 cells from non-exposed donors (Figure 4D). CXCR3 expression was less useful for distinguishing the different lineages (Figure 4E). However, co-expression of CD11c and CXCR3 marked both the atBC2 and MBC1 populations (Figures S5B and S5C) although double-negative cells were mainly of the classical lineage. CXCR3 single-positive cells were relatively rare in our dataset and appear to be a heterogenous mixture of different memory populations (Figures S5B and S5C). Although the concordance between surface marker expression and populations identified transcriptomically is imperfect, it is important to note that CITE-seq data are generally over-dispersed compared to flow cytometry data (Stoeckius et al., 2017). Thus, individual memory populations do not always separate clearly in CITE-seq data. Nonetheless, these data also highlight some of the limitations of flow-cytometry-based approaches for identifying memory cell subsets. Notably, they also explain why significant populations of alternative lineage cells in healthy donors have been missed previously, given they express low levels of canonical atypical markers and are often CD21+ CD27+.
Flow cytometry analysis reveals heterogeneity in atBC populations from malaria-exposed individuals
To extend the analysis of circulating B cells beyond the original 4 donors, we performed flow cytometry analysis of B cells from the PBMCs of 11 malaria-exposed and 7 non-exposed individuals (Table S1). From our CITE-seq and transcriptomic data, we developed a panel to identify alternative lineage B cells from classical lineage cells based on CD11c, CXCR3, CD21, and CD27. CD11c+ and CD11c+ CXCR3+ double-positive (DP) were identified as alternative lineage cells although CD11c− CXCR3− double-negative (DN) cells were assumed to belong to the classical lineage (Figures S5D and S5E). CXCR3+ single-positive cells were harder to classify but are likely to be a mix of MBC1 (alternative lineage) and MBC2 (classical lineage) cells. Because actBCs are not well distinguished by this panel, we used CD71 as a marker, as this marker was the one originally used to identify the actBC population for the transcriptomic studies we reference in our GSEA (Ellebedy et al., 2016). It is important to note that this strategy does fail to identify the atBC3 population as they did not highly express any obvious candidate surface markers based on our transcriptomic and CITE-seq analysis.
Consistent with our own transcriptomic and CITE-seq data as well as the data of others (Portugal et al., 2015; Weiss et al., 2009), we found that CD11c+ and DP B cells were considerably enriched in malaria-exposed donors (Figures 5A and 5B). The alternative-lineage CD11c+ and DP populations, regardless of donor origin, were found to express higher levels of CD19 and CD20—a hallmark of atBCs (Figure 5C). In malaria-exposed individuals, these cells further expressed Tbet and FCRL5. Consistent with our CITE-seq analysis, many CD11c+ B cells in malaria-exposed individuals were CD21− CD27−, suggesting these cells belong to the atBC1 population. In non-exposed donors, a much smaller proportion of CD11c+ B cells were CD21− CD27−, showing that this marker captures a broader and more heterogeneous population of cells than the traditional CD21− CD27− strategy for identifying atBCs. Finally, in both non-exposed and malaria-exposed donors, around 20% of B cells were CD71+ DN B cells (Figures 5A and 5B), consistent with the proportion of actBCs identified by single-cell RNA-seq (Figure 2F).
Figure 5.
CD11c, CXCR3, and CD71 identify atBC, actBCs, and MBCs via flow cytometry
PBMCs from 7 non-exp and 11 malaria-exp donors were isolated and analyzed by flow cytometry for expression of markers associated with different B cell populations.
(A) Flow cytometry plots from representative individuals showing the CD11c, CXCR3, and CD71 expression on mature IgD− B cells.
(B) Quantification of (A) showing the percentage of cells found in each cell type by country; bars represent mean ± SD.
(C) The expression of surface markers on each cell type, measured by mean fluorescence intensity (MFI); analysis was performed using two-way ANOVA; bars represent mean ± SD.
(D) Representative flow cytometry plots showing the expression of CD27 and CD21 per cell type.
(E) Quantification of (D) showing the percentage of cells separated by expression of CD27 and CD21 found in each cell type; bars represent the mean proportion ± SD.
Where the exact p value is not quoted, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
The dynamics of antigen-specific alternative lineage populations following influenza vaccination
We next hypothesized that different atBC populations represent different activation states of alternative lineage cells. We therefore examined how these cells responded in a recall response to the inactivated influenza vaccine (IIV) (Koutsakos et al., 2018). Using recombinant Hemagglutinin (HA) probes to two IIV antigens, A/California/07/09−H1N1 (H1) or B/Phuket/3073/2013 (B-Phu), we were able to identify influenza-specific B cells from the peripheral blood of subjects prior to immunization (Figures 6A and S6A). CD11c+, CXCR3+, CD71− DN, and CD71+ DN specific to both influenza antigens could be found at baseline in most individuals, reflecting past exposure to influenza virus infection or vaccination (Figures 6A–6D). Following immunization, all B cell populations expanded 14 days post-immunization, in particular, the DP B cell population (Figures 6B and 6C). This suggests that at least some of the DP population might represent recently reactivated atBCs. This was supported by the fact that the DP population was particularly enriched for CD21− CD27+ B cells, a phenotype associated with recently activated cells (Lau et al., 2017). CD11c+ CXCR3− B cells were primarily CD21− CD27− or CD21− CD27+ at all time points, suggesting that these are mainly atBC1 (Figure S6B). Influenza-specific CD11c+ and DP B cells also expressed higher levels of CD19, CD20, and FCRL5, indicative of an atypical-like phenotype (Figure S6C). In contrast, the CD11c− cells had a greater proportion of CD21+ CD27+ cells and lower expression of atypical markers, indicating the more classical nature of these cells (Figure S6B). These data show that alternative lineage populations can expand in response to vaccination and suggest that recently reactivated alternative lineage cells express both CXCR3 and CD11c. Our previous data show that MBC1 cells and atBC2 share the CD11c+ CXCR3+ DP surface phenotype. Given that MBC1 cells also show markers of quiescence, it is therefore likely that the atBC2 population represents “activated” alternative lineage cells.
Figure 6.
Influenza-specific atBCs expand following vaccination
9 individuals were vaccinated with inactivated influenza vaccine (IIV) with blood drawn at baseline and 14 or 28 days later.
(A) Panel shows representative flow cytometry plots showing the number of either B/Phuket or H1-specific IgD− B cells and the CD11c, CXCR3, and CD71 expression found in these cells over time.
(B) Kinetics of the influenza-specific B cell response quantified by the number of cells of each population per mL of blood; analysis performed with a two-way ANOVA.
(C) The log10 fold change of cell numbers between days 0 and 14 for each cell type; analysis performed using two-way ANOVA; bars represent mean ± SD.
(D) The percentage of influenza-specific memory cells divided by CD11c, CXCR3, and CD71 expression (mean ± SD shown).
Where the exact p value is not quoted, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Alternative lineage B cells arise in the primary response to a malaria vaccine and respond to booster immunization
Our data above show that atypical-like B cells can be seen after influenza vaccination in humans. However, as individuals are likely to be have been exposed to influenza infection multiple times, this may not be reflective of a primary immune response. To determine whether alternative lineage cells differentiate during a primary response in humans, we tracked B cells specific for PfCSP in a cohort of individuals who were given three doses of a whole P. falciparum sporozoite vaccine (PfSPZ) at 8-week intervals (Ishizuka et al., 2016; Lyke et al., 2017). Importantly, individuals enrolled in this study had not been previously exposed to malaria; therefore, the majority of PfCSP-specific cells would be naive at baseline. We were also able to follow those cells upon boosting to examine how they behaved during a recall response. Blood samples were obtained at the time of each vaccination as well as 1 and 2 weeks after each vaccination. Flow cytometric analysis was performed to identify and characterize PfCSP-specific B cells as well as PfCSP-specific PCs (Figures 7A and S7A).
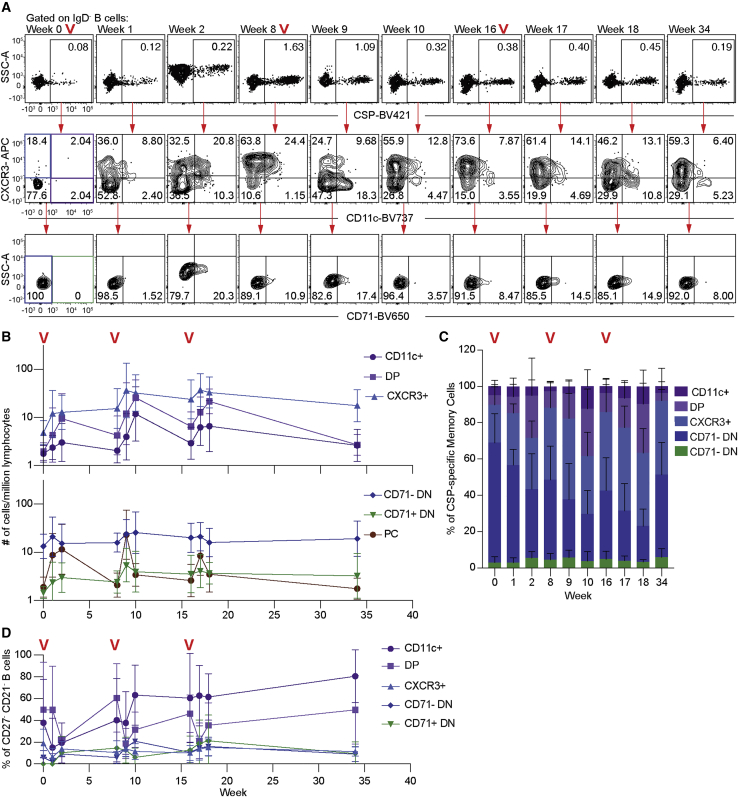
Figure 7.
Antigen-specific atBCs arise during the primary response to the PfSPZ vaccine and can be effectively recalled
15 individuals were vaccinated with 3 doses of 9 × 105 PfSPZ at 8-week intervals, with blood drawn at the indicated time points.
(A) Panel shows representative flow cytometry plots from a single individual of the gating of CSP-specific IgD− B cells and the CD11c, CXCR3, and CD71 expression found in these cells over time. Red “V”s indicate time points where booster immunizations were given.
(B) Kinetics of the CSP-specific B cell response quantified by the number of cells of each population per million lymphocytes; mean ± SD shown.
(C) The percentage of CSP-specific memory cells divided by CD11c, CXCR3, and CD71 expression over time; mean ± SD shown.
(D) Proportion of CD21− CD27− cells per cell population over time; mean ± SD shown.
By 2 weeks post-priming, a large population of CD11c-expressing B cells had developed, indicating that the priming of the alternative lineage occurs after a single immunization (Figures 7A–7C). Many of these CD11c+ cells co-expressed CXCR3, further supporting the idea that many double-positive cells are recently activated atBC2 cells. Interestingly, many B cells were CXCR3+ 8 weeks post-immunization. Our CITE-seq data suggest this population represents a heterogeneous population, including both MBC1 and MBC2 cells. In the absence of better markers, however, distinguishing these populations may require high-resolution, single-cell RNA-seq.
To examine the evolution of the alternative lineage, we tracked these B cell populations following boosts after 8 and 16 weeks (Figures 7A–7C). There was a significant expansion of the all-B-cell populations at the first boost, although magnitude of the response did not change at the second boost as described by us previously (McNamara et al., 2020). As expected, DN CD71+ cells, which we predicted to represent a transiently activated population, were observed to expand 1 week after each boost but rapidly contracted. Similar kinetics were observed in the plasmablast population (Figure 7C). We also observed an expansion of CD11c+ and DP B cells, albeit with different kinetics from the CD11c− cells, peaking 2 weeks after each boost. This suggests that either cells are recruited into the alternative lineage at each boost, or alternative lineage memory cells can be recalled after each boost. Circumstantial evidence for these cells expanding from a pool of alternative lineage memory cells comes from the fact that the proportion of CD11c+ and DP B cells that had the traditional CD27− CD21− atypical marker pattern increased with each immunization (Figure 7D), implying continued differentiation of this lineage over time. Across all time points, CD11c+, DP, and CXCR3+ B cells expressed higher levels of CD19 and CD20 although expressing lower levels of CCR7 than the CD71− DN population (Figure S7C). Collectively, our data suggest that alternative lineage cells are primed by vaccination and can be recalled on booster responses. Alternative lineage cells develop an increasingly atBC1 phenotype with each boost and continued antigen exposure.
Discussion
Here, we use single-cell RNA-seq to investigate the diversity of B cell populations in malaria-exposed and non-exposed individuals. As expected, many B cells from malaria-exposed individuals had an atypical phenotype, but surprisingly, these cells appeared to belong to a larger constellation of related cell types that were also abundant in non-exposed individuals. These cells, which we designate the alternative lineage, arise during the primary immune response to vaccination and can be recalled upon re-exposure. With repeated immunizations, a greater proportion of the alternative lineage adopted at atBC phenotype.
Our results are consistent with previous findings that atypical-like B cells can be found following influenza (Andrews et al., 2019; Lau et al., 2017) and tetanus (Kim et al., 2019) vaccinations. However, most people will generally be exposed to influenza and tetanus multiple times through immunization or infection, so whether or not these cells are generated following primary exposure is unknown. Utilizing a cohort of malaria-naive individuals immunized with PfSPZ, we were able to examine the primary response of antigen-specific B cells. We observed that small numbers of CD11c-expressing alternative lineage cells are primed after initial PfSPZ vaccination and can be recalled following booster immunizations. This would imply that chronic stimulation is not necessary for the priming of alternative lineage cells. However, our time course also shows that the canonical CD21− CD27− phenotype, which marks our atBC1 population, becomes more prominent following successive boosts. This is consistent with previous data showing that atBCs (defined using CD21− and CD27−) develop with chronic antigen exposure (Aye et al., 2020; Charles et al., 2011; Illingworth et al., 2013; Weiss et al., 2009).
It has been proposed based on mouse studies that atBCs are merely recently activated B cells (Pérez-Mazliah et al., 2018). However, our transcriptomic data clearly differentiated atBCs from previously described actBC subsets. Nonetheless, there is evidence that atBCs themselves can be activated. In particular, CD11c+ CXCR3+ DP B cells were seen to expand early in the influenza response. This population was also consistently found to be enriched for CD27+ CD21− B cells, a phenotype associated with recent activation (Avery et al., 2005). In contrast, CD21− CD27− CD11c+ B cells may represent a terminally differentiated effector population that wanes over time. Antigen-specific population CD11c+ B cells do diminish overtime in our PfSPZ-vaccinated individuals, suggesting that atBCs themselves are not long-lived memory cells. Alternatively, atBCs may migrate to other tissues: a recent study observed that influenza-specific CD11c+ B cells could be found in the spleens of mice 100 days post-infection and also showed that CD21− Tbethi B cells in humans preferentially take up residence within the spleen (Johnson et al., 2020).
The finding of an expanded population of atBCs and related alternative lineage cells in healthy donors was a surprising finding in our study. These cells have likely been undercounted in healthy individuals for two reasons: first because many cells belong to a cryptic atBC3 population that lacks expression of canonical atypical markers, and second because the relationship between “alternative memory B cells” (MBC1) and atBCs has not previously been demonstrated. Our inability to identify definitive surface markers of this MBC1 population is a limitation of this study. CITE-seq data suggest these cells would be a mix of CD11c+ CXCR3+ DP cells; however, this signature is shared with the activated atBC2 population. MBC1 cells may be included within the CXCR3+ single-positive population, but again this population likely includes other memory cells. A previous study also observed a CD21− Tbetlo CXCR3+ B cell population that continued to recirculate (Johnson et al., 2020), which may be analogous to our MBC1 population. Resolution of this issue may require a more extensive CITE-seq panel to build on recent work using time of flight cytometry (CyTOF) to investigate the heterogeneity of B cell populations via surface markers (Glass et al., 2020).
Although our data show that atBCs are part of a normal B cell response, the function of these cells remains elusive. Bulk RNA-seq analysis of atBC-like cells from SLE patients showed higher expression of genes associated with PC maintenance in this population leads to the hypothesis that atBCs represent a precursor PC population (Jenks et al., 2018). In contrast, we could not find any evidence of these genes being upregulated in any of our atBC populations in our single-cell RNA-seq datasets. We also found that atBCs were generally mutated, conflicting with suggestions that these cells represent an extrafollicular pre-plasmablast population, instead suggesting a GC origin for these cells. Finally, we also observe that atBC populations in sporozoite vaccinated individuals continue to expand even 2 weeks after booster immunization, although PC populations peaked after just 1 week. A reconciliation of these conflicting results may be that, in pathogenic conditions, such as SLE, atBCs can be driven to become pathogenic antibody-secreting cells. Consistent with this, the Toll-like receptor 7 (TLR7) pathway is implicated in SLE development, and it has been found that including TLR7 agonists within the stimulating condition can help differentiate atBCs into PCs (Jenks et al., 2018; Pérez-Mazliah et al., 2018; Rivera-Correa et al., 2019; Rubtsova et al., 2013).
Here, we provide an atlas of the B cell subsets circulating in human blood derived from both healthy and malaria-exposed individuals. Due to the high dimensionality of single-cell transcriptomic data and the power of our unsupervised clustering of >12,000 transcriptomes from malaria-exposed and healthy individuals, we refine previous analyses, including those based on other high-dimensional techniques, including CyTOF (Glass et al., 2020). By combining our analysis with CITE-seq technologies and VDJ profiling, we can further reconcile our data with existing classifications of B cell memory based on flow cytometry markers or Ig subclass. We find that atBCs are part of a broader alternative lineage that is abundant even in healthy donors. We follow this up by showing that these alternative lineage cells can be induced by primary exposure to acute antigen. Thus, our data suggest that alternative lineage B cells are a critical and typical component of the humoral immune response.
STAR★methods
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CCR7 BV650 | BioLegend | Clone G043h7; Cat#353234; RRID:AB_2563867 |
| Anti-human CD10 BV421 | BioLegend | Clone HI10a; Cat#312218; RRID:AB_2561833 |
| Anti-human CD10 BV510 | BioLegend | Clone HI10a; Cat#312220; RRID:AB_2563835 |
| Anti-human CD11c BUV737 | BD Biosciences | Clone B-ly6; Cat#741827; RRID:AB_2871162 |
| Anti-human CD11c BV421 | BioLegend | Clone Bu15; Cat#337226; RRID:AB_2564485 |
| Anti-human CD11c PE | BioLegend | Clone Bu15; Cat#337216; RRID:AB_2129790 |
| Anti-human CD11c PE Cy7 | BioLegend | Clone Bu15; Cat#337216; RRID:AB_2129790 |
| Anti-human CD14 BV510 | BioLegend | Clone M5E2; Cat#301842; RRID:AB_2561946 |
| Anti-human CD14 FITC | BioLegend | Clone M5E2; Cat#301804; RRID:AB_314186 |
| Anti-human CD15 (SEA-1) FITC | BioLegend | Clone H198; Cat#394706; RRID:AB_2750482 |
| Anti-human CD16 BV510 | BioLegend | Clone 3G8; Cat#302048; RRID:AB_2562085 |
| Anti-human CD183 (CXCR3) BV711 | BioLegend | Clone G025H7; Cat#353732; RRID:AB_2563533 |
| Anti-human CD184 (CXCR4) BuV395 | BD Bioscience | Clone 12G5; Cat#563924; RRID:AB_2738490 |
| Anti-human CD19 BV750 | BioLegend | Clone HIB19; Cat#302261; RRID:AB_2734257 |
| Anti-human CD19 ECD | Beckman Coulter | Clone J4-119; Cat#IM2708U; RRID:AB_130854 |
| Anti-human CD19 BV605 | BD Bioscience | Clone SJ25C1; Cat#562653; RRID:AB_2722592 |
| Anti-human CD197 (CCR7) BV785 | BioLegend | Clone G043H7; Cat#353230; RRID:AB_2563630 |
| Anti-human CD20 APC Cy7 | BD Bioscience | Clone 2H7; Cat#560853; RRID:AB_10561681 |
| Anti-human CD20 APC-Cy7 | BioLegend | Clone 2H7; Cat#302314; RRID:AB_314262 |
| Anti-human CD21 BV711 | BD Biosciences | Clone B-ly4; Cat#563163; RRID:AB_2738040 |
| Anti-human CD21 PeCy7 | BioLegend | Clone Bu32; Cat#354912; RRID:AB_2561577 |
| Anti-human CD27 BV605 | BioLegend | Clone O323; Cat#302830; RRID:AB_2561450 |
| Anti-human CD27 PerCP Cy5.6 | BioLegend | Clone M-T271; Cat#356408; RRID:AB_2561906 |
| Anti-human CD3 FITC | BioLegend | Clone HIT3A; Cat#300306; RRID:AB_314042 |
| Anti-human CD3 BV510 | BioLegend | Clone OKT3; Cat#317332; RRID:AB_2561943 |
| Anti-human CD307e (FCRL5) APC | BioLegend | Clone 509f6; Cat#340306; RRID:AB_256432 |
| Anti-human CD38 Ax700 | BD Biosciences | Clone HIT2; Cat#560676; RRID:AB_1727472 |
| Anti-human CD56 (NCAM) BV510 | BioLegend | Clone HCD56; Cat#318340; RRID:AB_2561944 |
| Anti-human CD56 (NCAM) FITC | BioLegend | Clone HCD56 ; Cat#318304; RRID:AB_604100 |
| Anti-human CD71 BV650 | BioLegend | Clone CU1G4; Cat#334108; RRID:AB_10915138 |
| Anti-human CD71 FITC | BD Biosciences | Clone M-A712; Cat#555536; RRID:AB_395920 |
| Anti-human CD8 BV510 | BioLegend | Clone RPA-T8; Cat#301048; RRID:AB_2561942 |
| Anti-human CD83 PE | BioLegend | Clone Hb15e; Cat#305308; RRID:AB_314516 |
| Anti-human CXCR3 APC | BioLegend | Clone G025H7; Cat#353708; RRID:AB_10983064 |
| Anti-human CXCR4 BUV395 | BD Biosciences | Clone 12G5; Cat#563924; RRID:AB_2738490 |
| Anti-human IgD PE-CF594 | BD Bioscience | Clone IA6-2; Cat#562540; RRID:AB_11153129 |
| Anti-human IgD PE-Cy7 | BD Biosciences | Clone IA6-2; Cat#561314; RRID:AB_10642457 |
| Anti-human IgG PE-CF594 | BD Biosciences | Clone G18-145; Cat#562538; RRID:AB_2737640 |
| Anti-human IgG A700 | BD Biosciences | Clone G18-145; Cat#555784; RRID:AB_396119 |
| Anti-human IgM BB700 | BD Biosciences | Clone UCA-B1; Cat#747879; RRID:AB_2872341 |
| Anti-human Tbet PerCP Cy5.5 | BioLegend | Clone 4B10; Cat#644806; RRID:AB_1595488 |
| Biological samples | ||
| Malaria Exposed and Non Exposed PMBCs | This paper (Table S1) | N/A |
| Human PBMCs from the VRC 314 Clinical Trial | Cellular Immunology Section, The National Institute of Health (Lyke et al., 2017). | https://clinicaltrials.gov/ct2/show/NCT02015091?term=NCT02015091&rank=1 |
| Human PBMCs following IIV immunization | Human T cell Laboratory, Doherty Institute(Koutsakos et al., 2018) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Agencourt AMPure XP beads | Beckman Coulter | Cat#A63880 |
| Betaine 5M | Sigma-Aldrich | Cat#B300-1VL |
| CloneAmp HiFi PCR Premix | Takara | Cat#639298 |
| ERCC Spike-In Mix | ThermoFisher | Cat#4456740 |
| RNase inhibitor | Clontech | Cat#2313A |
| SuperScript II RT | Life Technologies | Cat#18064-071 |
| Triton X-100 | Sigma-Aldrich | Cat#T9284 |
| Critical commercial assays | ||
| Nextera XT DNA Library Preparation kit | Illumina | Cat#FC-131-1096 |
| Nextera XT Index Kit v2 Set A (96 indices, 384 samples) | Illumina | Cat#FC-131-2001 |
| Nextera XT Index Kit v2 Set D (96 indices, 384 samples) | Illumina | Cat#FC-131-2004 |
| Quant-iT PicoGreen dsDNA Assay Kit | ThermoFisher | Cat#P11496 |
| Chromium Next GEM chip G Single cell Kit | 10x Genomics | Cat#1000127 |
| Chromium Next GEM Single5′ Library and Gel Bead Kit v1.1 | 10x Genomics | Cat#1000080 |
| Chromium Single cell 5′ Feature Barcode Library Kit | 10x Genomics | Cat#1000020 |
| Chromium Single cell 5′ Library Contruction Kit | 10x Genomics | Cat#1000016 |
| Chromium Single cell V(D)J Enrichment Kit Human B cell | 10x Genomics | Cat#1000005 |
| Single Index Kit N Set A | 10x Genomics | Cat#1000212 |
| Single Index Kit T Set A | 10x Genomics | Cat#1000213 |
| Deposited data | ||
| Atypical B cell Gene set | Portugal et al., 2015 | https://elifesciences.org/articles/07218#digest |
| Activated B cell Gene set | Ellebedy et al., 2016 | http://www.nature.com/articles/ni.3533 |
| Naive B cell Gene set | Ellebedy et al., 2016 | http://www.nature.com/articles/ni.3533 |
| Raw Sequence Data | This Paper | https://dataview.ncbi.nlm.nih.gov/object/PRJNA612353?reviewer=bf7ee45b186vstua0d23qud1nk |
| Oligonucleotides | ||
| ISPCR5′-AAGCAGTGGTATCAACGC AGAGT-3′ |
ThermoFisher | N/A |
| Oligo-DT5′-AAGCAGTGGTATCAA CGCAGAGTACT30VN-3′ |
ThermoFisher | N/A |
| Template Switching Oligo-5′-AAGC AGTGGTATCAACGCAGAGTACA TrGrG+G-3′ |
Exiquo | N/A |
| Software and algorithms | ||
| FlowJo | https://www.flowjo.com | |
| VDJPuzzle | (Rizzetto et al., 2018) | https://bitbucket.org/kirbyvisp/vdjpuzzle2/src/master/ |
| Cellranger | N/A | https://kb.10xgenomics.com/hc/en-us |
| R | R Development Core Team, 2020 | https://www.r-project.org |
| Monocle3 | Trapnell et al., 2014 | https://cole-trapnell-lab.github.io/monocle3/ |
| Seurat | (Butler et al., 2018) | https://satijalab.org/seurat/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ian Cockburn (ian.cockburn@anu.edu.au).
Materials availability
This study did not generate new unique reagents
Data and code availability
The accession number for single-cell RNA-seq is PRJNA612353 at the NCBI BioProject database:
Experimental model and subject details
All research was conducted according to the principles of the Declaration of Helsinki, which included the administration of informed consenting in the participant’s local language. Studies in Australia were further performed in accordance with the Australian National Health and Medical Research Council (NHMRC) Code of Practice.
Patient cohorts
The malaria-immunology cohort and vaccination studies, under which the samples described were collected in Kenya, were approved by the Kenyan Medical Research Institute Scientific and Ethics Review Unit, Nairobi, and the use of these samples at the Australian National University was further approved by the Australian National University Human Research Ethics Committee (protocol number 2014/102). The Kenyan adults are members of the KEMRI/Wellcome Research Programme’s longitudinal malaria immunology cohort studies in Junju and Ngerenya villages, 20 km apart from each other in Kilifi, Kenya. In addition, we included samples from the RTS,S/AS01 phase 3 clinical trial. Blood was also drawn from healthy control Australian donors who were recruited at the Australian National University. Information on the age and gender of human subjects in this cohort can be found is Table S1
VRC 314 clinical trial (https://clinicaltrials.gov/; NCT02015091) was an open-label evaluation of the safety, tolerability, immunogenicity and protective efficacy of PfSPZ Vaccine. Subjects in the high dose cohort received a total of three doses of 9 x105 PfSPZ intravenously at week 0, 8 and 16. Blood was drawn at the time of each immunization, as well as 7d and 14 d after each immunization. Plasma and PBMCs were isolated from all samples at these time points. Full details of the study are described in (Lyke et al., 2017).
The investigation of B cell responses after IIV immunization was approved by the University of Melbourne Human Ethics Committee (ID 1443389.3) and the Australian Red Cross Blood Service (ARCBS) Ethics Committee (ID 2015#8). PBMCs were used from 8 donors taken on the day of IIV immunization, as well as 14 and 28 days later. Full details of the study are described previously (Koutsakos et al., 2018).
Method details
Flow cytometry
For samples from Kenya and the IIV cohort PBMCs were thawed and washed in PBS with 2% heat-inactivated FBS. Cells were then stained with Live/Dead dye for 5 min in PBS before incubation with fluorescently labeled antibodies for a further 30 min. Details of all antibodies used are given in the Key resources table. Flow-cytometric data was collected on a BD Fortessa or X20 flow cytometer (Becton Dickinson) and analyzed using FlowJo (FlowJo Software) (). A BD FACs Aria I or II (Becton Dickinson) was used for sorting cells.
For VRC314 clinical trial specimens PBMCs were thawed into prewarmed RPMI media then washed with PBS. Cells were stained with Live/Dead dye for 15 minutes, washed in PBS with 2% heat inactivated FBS, and labeled with antibodies for an additional 30 minutes. Labeled cells were washed with PBS 2% FBS and fixed for 15 minutes in 0.5% PFA before and final wash and resuspension in PBS 2% FBS. Flow-cytometric data was collected on a BD X50 flow cytometer (Becton Dickinson) and analyzed using FlowJo ().
Tetramer Preparation
Pf MSP1, AMA1 and TT were biotinylated with the Sulfo-NHS-LC-Biotinylation Kit (ThermoFisher) at a ratio of 1:1 according to the manufacturer’s instructions, biotinylated (NANP)9 repeat region of Pf CSP was sourced from Biomatik (Ontario, Canada). Biotinylated antigens were incubated with premium-grade SA-PE and SA-APC (Molecular Probes) or SA-BV421 and SA-BB660 (Biolegend and BD Horizon) at a molar ratio of 4:1, added four times with 15 min incubation at room temperature.
Single-cell RNA-seq using Smart-seq2
Antigen-specific single-cell RNA sequencing was performed using a Smart-seq 2 protocol (Picelli et al., 2014) with the following modifications. Cells were sorted into plates with wells containing 1 μl of the cell lysis buffer, 0.5 μl dNTP mix (10 mM) and 0.5 μl of the oligo-dT primer at 5 μM. We then reduced the amount reagent used in the following reverse-transcription and PCR amplification step by half. The concentration of the ISPCR primer was also further reduced to 50 nM. Due to the low transcriptional activity of memory B cells, we increased the number of PCR cycles to 28. cDNA was then purified with AMPure XP beads at a bead to sample ratio of 0.8:1. Sequencing libraries were prepared using the Nextera XT Library Preparation Kit with the protocol modified by reducing the original volumes of all reagents in the kit by 1/5th. Another round of bead cDNA bead purification was preformed using a bead to sample ratio of 0.6:1. Sequencing was performed on the Illumina NextSeq sequencing platform. Following sequencing, fastq files were passed through the program VDJPuzzle (Rizzetto et al., 2018) where reads were trimmed using Trimmomatic, then aligned to the human reference genome GRCh37 using tophat2. Gene expression profiles were then generated using cufflinks 2. As a further QC step, cells where reads were mapped to less than 30% of the reference genome were removed. All following downstream analysis for transcriptomic data was performed using Seurat (Butler et al., 2018). Details of all key reagents for single-cell RNA-seq are given in the Key resources table.
Single-cell RNA-seq and CITE-seq using 10x Chromium
Post-sorting, CD19+ CD20+ IgD- B cells were incubated with Total-seq C antibodies (Biolegend) for 30 min and washed 3 times. The number of cells were then counted and 14 000 cells per sample were run on the 10X Chromium (10X Genomics). Library preparation was completed by Biomedical Research Facility (BRF) at the JCSMR following the recommended protocols for the Chromium Single-cell 5′ Reagent Kit as well as 5′ Feature Barcode and V(D)J Enrichment Kit for Human B cells. Libraries were sequenced using the Illumina NovaSeq6000 (Illumina). The 10X Cell Ranger package (v1.2.0, 10X Genomics) was used to process transcript, CITE-seq and VDJ libraries and prepare them for downstream analysis. Details of all key reagents for single-cell RNA-seq are given in the Key resources table.
Quantification and statistical analysis
Statistical Analysis
Statistical analysis of flow cytometry data was performed in GraphPad Prism for simple analyses without blocking factors; all other analyses was performed in R (R Development Core Team, 2020) with details of statistical tests in the relevant figure legends. Abbreviations for p values are as follows: p < 0.05 = ∗, p < 0.01 = ∗∗, p < 0.001 = ∗∗∗, p < 0.0001 = ∗∗∗∗; with only significant p values shown.
Single-cell RNA-seq analysis
The package Seurat (version 3.1) (Butler et al., 2018) was used for graph-based clustering and visualizations. All functions described are from Seurat or the standard R package (version 3.60) (R Development Core Team, 2020) using the default parameters unless otherwise stated. Each sample was initially analyzed separately using the following procedures. Cells that expressed less than 200 genes and genes that were expressed in less than 3 cells were excluded, along with cells that had greater than 10% mitochondrial genes. Gene expression was normalized for both mRNA and CITE-seq assays using the NormalizeData function, then the 2000 most variable genes for each sample were identified using FindVaraibleFeatures. Next expression of all genes was scaled using ScaleData to linearly regress out sources of variation. Principal component analysis on the variable genes identified above was then run with RunPCA. Based on ElbowPlot results we decided to use 13, 20, 12 and 20 principal components (PCs) for the clustering of samples Non-Exp 1, Non-Exp 2, Exp 1 and Exp 2 respectively using FindNeighbors. FindClusters was then run to identify clusters for each sample, using the resolutions 0.3, 0.4, 0.5 and 0.4 respectively. FindAllMarkers was then used to identify clusters of non-B cells. The remaining cells in each sample were then normalized and scaled again as above. Australian and Kenyan samples were combined together first using FindIntegrationAnchors and then Intergratedata to create two combined datasets, 1 with both non-exposed samples and one with both malaria-exposed samples. These two combined samples were further combined using the commands to from one combined dataset containing all 4 samples. The combined dataset was then scaled again as above and a PCA was run. Using FindClusters with a resolution of 0.8, we identified our 11 clusters. A resolution of 0.8 allowed us to identify expected clusters related to Atypical, Activated, Memory and PC B cell populations, while increasing the resolution above 0.8 led to exponential increases in the numbers of clusters. DEGs were identified using FindAllMarkers. The clustering was visualized with Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction using RunUMAP and plotted using DimPlot with umap as the reduction. Phylogentic analysis was done using BuildClusterTree to report the hierarchical distance matrix relating an ‘average’ cell from each cluster. Log-normalized gene expression data was visualized using violin plots (VlnPlots) as well as onto -UMAP plots (FeaturePlot). Heatmaps were generated using DoHeatmap.
For Smart-seq2 analysis, cells with greater than 10% mitochondrial genes were not excluded. 8 PCs were used as determine by ElbowPlot. For clustering a resolution of 0.8 was used.
Pseudotime Analysis
Pseudotime analysis was preformed using the R package Monocle 3 (Trapnell et al., 2014). Our Seurat Object was converted into monocle3 main data calls cell_data_set. The default Monocle 3 workflow was then followed.
Gene Set Enrichment Analysis (GSEA)
GSEA was done using javaGSEA through the Broad Institute. For each comparison, DEGs were ranked by log-fold change and pre-ranked analysis using 1000 permutations was used to examine enrichment in the referenced gene sets.
CITE-seq analysis
CITE-seq expression data was normalized using centered log-ratio normalization and exported into the flow cytometry analysis software FlowJo for further processing and presentation.
VDJ Analysis
To determine the antigen-specific BCR repertoire, we made use of VDJpuzzle (Rizzetto et al., 2018) to reconstruct full-length heavy and light chains from each cell from out Smart-seq2 dataset. From this we were able to determine V region usage and mutation frequency.
VDJ sequences from the 10x dataset were obtained using the cellranger vdj command. From this output, V region usage and mutation frequency could be determined.
Acknowledgments
We would like to thank the University of Maryland study volunteers from malaria clinical trial VRC314. We are grateful to the KEMRI/CGMRC field team for their dedication in the recruitments, malaria surveillance data, and sample collection and the laboratory team that processed the samples. We are also indebted to the study participants. We would like to thank Harpreet Vohra and Michael Devoy of the Imaging and Cytometry Facility at the Australian National University for assistance with flow cytometry and sorting. This work was supported by National Health and Medical Research Council project grant support to I.A.C. (GNT1158404). Production and characterization of PfSPZ vaccine were supported in part by National Institute of Allergy and Infectious Diseases Small Business Innovation Research Grants 5R44AI055229-11 (to S.L.H.), 5R44AI058499-08 (to S.L.H.), and 5R44AI058375-08 (to S.L.H.). F.M.N. was supported by an MRC/DFID African Research Leadership Award (MR/P020321/1) and a Senior Fellowship from the European and Developing Countries Clinical Trial Partnership (TMA2016SF-1513). R.A. was supported by the African Academy of Sciences through the DELTAS Africa Initiative (DEL-15-003).
Author contributions
Conceptualization, H.J.S., R.A., A.H.I., F.M.N., and I.A.C.; Data Curation, H.J.S., X.L., T.D.A., F.M.N., and I.A.C.; Formal Analysis, H.J.S., R.A., X.L., X.G., T.D.A., and F.L.; Funding Acquisition, K.K., R.A.S., F.M.N., and I.A.C.; Project Administration, K.E.L. and S.L.H.; Investigation, H.J.S., R.A., A.H.I., R.V., E.N., O.K., J.M., M.K., T.H.O.N., M.N., P.M., A.A.B., N.K., and S.C.; Methodology, A.E. and F.L.; Resources, A.K.W., S.J.K., F.L., K.K., R.A.S., and F.M.N.; Software, F.L.; Supervision, S.J.K., F.L., K.K., R.A.S., F.M.N., and I.A.C.; Visualization, H.J.S., X.L., and I.A.C.; Writing – Original Draft, H.J.S. and I.A.C.; Writing – Review & Editing, F.L., K.K., R.A.S., and F.M.N.
Declaration of interests
S.C., N.K., B.K.L.S., and S.L.H. are salaried employees of Sanaria, the developer and owner of PfSPZ Vaccine and the investigational new drug (IND) application sponsor of the clinical trials. S.L.H. and B.K.L.S. have a financial interest in Sanaria. All other authors declare no competing interests.
Published: February 9, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2020.108684.
Supplemental information
References
- Andrews S.F., Chambers M.J., Schramm C.A., Plyler J., Raab J.E., Kanekiyo M., Gillespie R.A., Ransier A., Darko S., Hu J. Activation dynamics and immunoglobulin evolution of pre-existing and newly generated human memory B cell responses to influenza hemagglutinin. Immunity. 2019;51:398–410.e5. doi: 10.1016/j.immuni.2019.06.024. [DOI] [PubMed] [Google Scholar]
- Avery D.T., Ellyard J.I., Mackay F., Corcoran L.M., Hodgkin P.D., Tangye S.G. Increased expression of CD27 on activated human memory B cells correlates with their commitment to the plasma cell lineage. J. Immunol. 2005;174:4034–4042. doi: 10.4049/jimmunol.174.7.4034. [DOI] [PubMed] [Google Scholar]
- Aye R., Sutton H.J., Nduati E.W., Kai O., Mwacharo J., Musyoki J., Otieno E., Wambua J., Bejon P., Cockburn I.A., Ndungu F.M. Malaria exposure drives both cognate and bystander human B cells to adopt an atypical phenotype. Eur. J. Immunol. 2020;50:1187–1194. doi: 10.1002/eji.201948473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species Nat. Biotech. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles E.D., Brunetti C., Marukian S., Ritola K.D., Talal A.H., Marks K., Jacobson I.M., Rice C.M., Dustin L.B. Clonal B cells in patients with hepatitis C virus-associated mixed cryoglobulinemia contain an expanded anergic CD21low B-cell subset. Blood. 2011;117:5425–5437. doi: 10.1182/blood-2010-10-312942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt G.R.A., Hsu J.T., Gartland L., Leu C.M., Zhang S., Davis R.S., Cooper M.D. Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J. Exp. Med. 2005;202:783–791. doi: 10.1084/jem.20050879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellebedy A.H., Jackson K.J., Kissick H.T., Nakaya H.I., Davis C.W., Roskin K.M., McElroy A.K., Oshansky C.M., Elbein R., Thomas S. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat. Immunol. 2016;17:1226–1234. doi: 10.1038/ni.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecteau J.F., Côté G., Néron S. A new memory CD27-IgG+ B cell population in peripheral blood expressing VH genes with low frequency of somatic mutation. J. Immunol. 2006;177:3728–3736. doi: 10.4049/jimmunol.177.6.3728. [DOI] [PubMed] [Google Scholar]
- Glass D.R., Tsai A.G., Oliveria J.P., Hartmann F.J., Kimmey S.C., Calderon A.A., Borges L., Glass M.C., Wagar L.E., Davis M.M., Bendall S.C. An integrated multi-omic single-cell atlas of human B cell identity. Immunity. 2020;53:217–232.e5. doi: 10.1016/j.immuni.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good K.L., Avery D.T., Tangye S.G. Resting human memory B cells are intrinsically programmed for enhanced survival and responsiveness to diverse stimuli compared to naive B cells. J. Immunol. 2009;182:890–901. doi: 10.4049/jimmunol.182.2.890. [DOI] [PubMed] [Google Scholar]
- Horst A., Hunzelmann N., Arce S., Herber M., Manz R.A., Radbruch A., Nischt R., Schmitz J., Assenmacher M. Detection and characterization of plasma cells in peripheral blood: correlation of IgE+ plasma cell frequency with IgE serum titre. Clin. Exp. Immunol. 2002;130:370–378. doi: 10.1046/j.1365-2249.2002.02025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth J., Butler N.S., Roetynck S., Mwacharo J., Pierce S.K., Bejon P., Crompton P.D., Marsh K., Ndungu F.M. Chronic exposure to Plasmodium falciparum is associated with phenotypic evidence of B and T cell exhaustion. J. Immunol. 2013;190:1038–1047. doi: 10.4049/jimmunol.1202438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka A.S., Lyke K.E., DeZure A., Berry A.A., Richie T.L., Mendoza F.H., Enama M.E., Gordon I.J., Chang L.J., Sarwar U.N. Protection against malaria at 1 year and immune correlates following PfSPZ vaccination. Nat. Med. 2016;22:614–623. doi: 10.1038/nm.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isnardi I., Ng Y.S., Menard L., Meyers G., Saadoun D., Srdanovic I., Samuels J., Berman J., Buckner J.H., Cunningham-Rundles C., Meffre E. Complement receptor 2/CD21- human naive B cells contain mostly autoreactive unresponsive clones. Blood. 2010;115:5026–5036. doi: 10.1182/blood-2009-09-243071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks S.A., Cashman K.S., Zumaquero E., Marigorta U.M., Patel A.V., Wang X., Tomar D., Woodruff M.C., Simon Z., Bugrovsky R. Distinct effector B cells induced by unregulated Toll-like receptor 7 contribute to pathogenic responses in systemic lupus erythematosus. Immunity. 2018;49:725–739.e6. doi: 10.1016/j.immuni.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.L., Rosenthal R.L., Knox J.J., Myles A., Naradikian M.S., Madej J., Kostiv M., Rosenfeld A.M., Meng W., Christensen S.R. The transcription factor T-bet resolves memory B cell subsets with distinct tissue distributions and antibody specificities in mice and humans. Immunity. 2020;52:842–855.e6. doi: 10.1016/j.immuni.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.C., Baccarella A.M., Bayat A., Pepper M., Fontana M.F. FCRL5+ memory B cells exhibit robust recall responses. Cell Rep. 2019;27:1446–1460.e4. doi: 10.1016/j.celrep.2019.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U., Küppers R., Rajewsky K. Evidence for a large compartment of IgM-expressing memory B cells in humans. Blood. 1997;89:1288–1298. [PubMed] [Google Scholar]
- Klein U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Knox J.J., Buggert M., Kardava L., Seaton K.E., Eller M.A., Canaday D.H., Robb M.L., Ostrowski M.A., Deeks S.G., Slifka M.K. T-bet+ B cells are induced by human viral infections and dominate the HIV gp140 response. JCI Insight. 2017;2:e92943. doi: 10.1172/jci.insight.92943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsakos M., Wheatley A.K., Loh L., Clemens E.B., Sant S., Nüssing S., Fox A., Chung A.W., Laurie K.L., Hurt A.C. Circulating TFH cells, serological memory, and tissue compartmentalization shape human influenza-specific B cell immunity. Sci. Transl. Med. 2018;10:eaan8405. doi: 10.1126/scitranslmed.aan8405. [DOI] [PubMed] [Google Scholar]
- Krishnamurty A.T., Thouvenel C.D., Portugal S., Keitany G.J., Kim K.S., Holder A., Crompton P.D., Rawlings D.J., Pepper M. Somatically hypermutated plasmodium-specific IgM(+) memory B cells are rapid, plastic, early responders upon malaria rechallenge. Immunity. 2016;45:402–414. doi: 10.1016/j.immuni.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau D., Lan L.Y., Andrews S.F., Henry C., Rojas K.T., Neu K.E., Huang M., Huang Y., DeKosky B., Palm A.E. Low CD21 expression defines a population of recent germinal center graduates primed for plasma cell differentiation. Sci. Immunol. 2017;2:eaai8153. doi: 10.1126/sciimmunol.aai8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyke K.E., Ishizuka A.S., Berry A.A., Chakravarty S., DeZure A., Enama M.E., James E.R., Billingsley P.F., Gunasekera A., Manoj A. Attenuated PfSPZ vaccine induces strain-transcending T cells and durable protection against heterologous controlled human malaria infection. Proc. Natl. Acad. Sci. USA. 2017;114:2711–2716. doi: 10.1073/pnas.1615324114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara H.A., Idris A.H., Sutton H.J., Vistein R., Flynn B.J., Cai Y., Wiehe K., Lyke K.E., Chatterjee D., Kc N. Antibody feedback limits the expansion of B cell responses to malaria vaccination but drives diversification of the humoral response. Cell Host Microbe. 2020;28:572–585.e7. doi: 10.1016/j.chom.2020.07.001. [DOI] [PubMed] [Google Scholar]
- Moir S., Ho J., Malaspina A., Wang W., DiPoto A.C., O’Shea M.A., Roby G., Kottilil S., Arthos J., Proschan M.A. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J. Exp. Med. 2008;205:1797–1805. doi: 10.1084/jem.20072683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muellenbeck M.F., Ueberheide B., Amulic B., Epp A., Fenyo D., Busse C.E., Esen M., Theisen M., Mordmüller B., Wardemann H. Atypical and classical memory B cells produce Plasmodium falciparum neutralizing antibodies. J. Exp. Med. 2013;210:389–399. doi: 10.1084/jem.20121970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan R., Buchauer L., Triller G., Kreschel C., Costa G., Pidelaserra Martí G., Imkeller K., Busse C.E., Chakravarty S., Sim B.K.L. Clonal selection drives protective memory B cell responses in controlled human malaria infection. Sci. Immunol. 2018;3:eaap8029. doi: 10.1126/sciimmunol.aap8029. [DOI] [PubMed] [Google Scholar]
- Obeng-Adjei N., Portugal S., Holla P., Li S., Sohn H., Ambegaonkar A., Skinner J., Bowyer G., Doumbo O.K., Traore B. Malaria-induced interferon-γ drives the expansion of Tbethi atypical memory B cells. PLoS Pathog. 2017;13:e1006576. doi: 10.1371/journal.ppat.1006576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape K.A., Taylor J.J., Maul R.W., Gearhart P.J., Jenkins M.K. Different B cell populations mediate early and late memory during an endogenous immune response. Science. 2011;331:1203–1207. doi: 10.1126/science.1201730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Mazliah D., Gardner P.J., Schweighoffer E., McLaughlin S., Hosking C., Tumwine I., Davis R.S., Potocnik A.J., Tybulewicz V.L., Langhorne J. Plasmodium-specific atypical memory B cells are short-lived activated B cells. eLife. 2018;7:e39800. doi: 10.7554/eLife.39800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Portugal S., Tipton C.M., Sohn H., Kone Y., Wang J., Li S., Skinner J., Virtaneva K., Sturdevant D.E., Porcella S.F. Malaria-associated atypical memory B cells exhibit markedly reduced B cell receptor signaling and effector function. eLife. 2015;4:e07218. doi: 10.7554/eLife.07218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; 2020. R: A language and environment for statistical computing. [Google Scholar]
- Reimold A.M., Iwakoshi N.N., Manis J., Vallabhajosyula P., Szomolanyi-Tsuda E., Gravallese E.M., Friend D., Grusby M.J., Alt F., Glimcher L.H. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Rivera-Correa J., Mackroth M.S., Jacobs T., Schulze Zur Wiesch J., Rolling T., Rodriguez A. Atypical memory B-cells are associated with Plasmodium falciparum anemia through anti-phosphatidylserine antibodies. eLife. 2019;8:e48309. doi: 10.7554/eLife.48309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzetto S., Koppstein D.N.P., Samir J., Singh M., Reed J.H., Cai C.H., Lloyd A.R., Eltahla A.A., Goodnow C.C., Luciani F. B-cell receptor reconstruction from single-cell RNA-seq with VDJPuzzle. Bioinformatics. 2018;34:2846–2847. doi: 10.1093/bioinformatics/bty203. [DOI] [PubMed] [Google Scholar]
- Rubtsova K., Rubtsov A.V., van Dyk L.F., Kappler J.W., Marrack P. T-box transcription factor T-bet, a key player in a unique type of B-cell activation essential for effective viral clearance. Proc. Natl. Acad. Sci. USA. 2013;110:E3216–E3224. doi: 10.1073/pnas.1312348110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer A.L., Lin K.I., Kuo T.C., Yu X., Hurt E.M., Rosenwald A., Giltnane J.M., Yang L., Zhao H., Calame K., Staudt L.M. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan R.T., Kim C.C., Fontana M.F., Feeney M.E., Jagannathan P., Boyle M.J., Drakeley C.J., Ssewanyana I., Nankya F., Mayanja-Kizza H. FCRL5 delineates functionally impaired memory B cells associated with Plasmodium falciparum exposure. PLoS Pathog. 2015;11:e1004894. doi: 10.1371/journal.ppat.1004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye S.G., Liu Y.J., Aversa G., Phillips J.H., de Vries J.E. Identification of functional human splenic memory B cells by expression of CD148 and CD27. J. Exp. Med. 1998;188:1691–1703. doi: 10.1084/jem.188.9.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye S.G., Avery D.T., Deenick E.K., Hodgkin P.D. Intrinsic differences in the proliferation of naive and memory human B cells as a mechanism for enhanced secondary immune responses. J. Immunol. 2003;170:686–694. doi: 10.4049/jimmunol.170.2.686. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Anolik J., Cappione A., Zheng B., Pugh-Bernard A., Brooks J., Lee E.H., Milner E.C., Sanz I. A new population of cells lacking expression of CD27 represents a notable component of the B cell memory compartment in systemic lupus erythematosus. J. Immunol. 2007;178:6624–6633. doi: 10.4049/jimmunol.178.10.6624. [DOI] [PubMed] [Google Scholar]
- Weiss G.E., Crompton P.D., Li S., Walsh L.A., Moir S., Traore B., Kayentao K., Ongoiba A., Doumbo O.K., Pierce S.K. Atypical memory B cells are greatly expanded in individuals living in a malaria-endemic area. J. Immunol. 2009;183:2176–2182. doi: 10.4049/jimmunol.0901297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G.E., Traore B., Kayentao K., Ongoiba A., Doumbo S., Doumtabe D., Kone Y., Dia S., Guindo A., Traore A. The Plasmodium falciparum-specific human memory B cell compartment expands gradually with repeated malaria infections. PLoS Pathog. 2010;6:e1000912. doi: 10.1371/journal.ppat.1000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie showing a 3D UMAP projection of the monocle pseudotime analysis. The naive population that marks the start of pseudotime is highlighted at the beginning. The PC cluster as well as the alternative and classical lineages are also highlighted.
Data Availability Statement
The accession number for single-cell RNA-seq is PRJNA612353 at the NCBI BioProject database: