Abstract
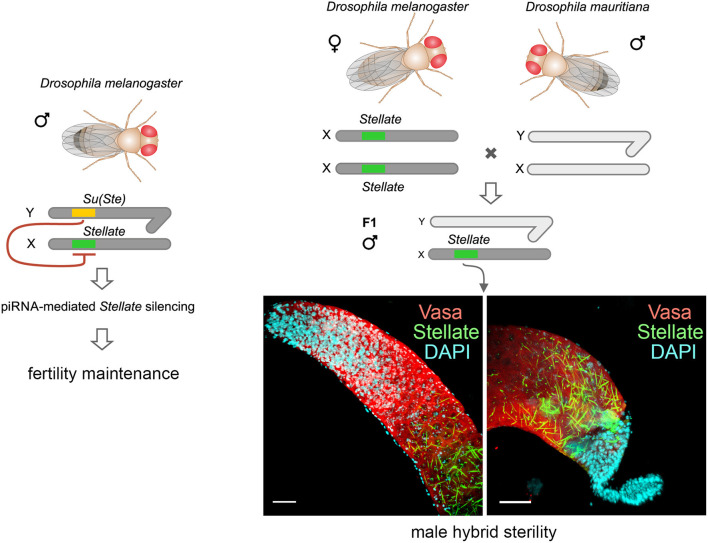
One of the main conditions of the species splitting from a common precursor lineage is the prevention of a gene flow between diverging populations. The study of Drosophila interspecific hybrids allows to reconstruct the speciation mechanisms and to identify hybrid incompatibility factors that maintain post-zygotic reproductive isolation between closely related species. The regulation, evolution, and maintenance of the testis-specific Ste-Su(Ste) genetic system in Drosophila melanogaster is the subject of investigation worldwide. X-linked tandem testis-specific Stellate genes encode proteins homologous to the regulatory β-subunit of protein kinase CK2, but they are permanently repressed in wild-type flies by the piRNA pathway via piRNAs originating from the homologous Y-linked Su(Ste) locus. Derepression of Stellate genes caused by Su(Ste) piRNA biogenesis disruption leads to the accumulation of crystalline aggregates in spermatocytes, meiotic defects and male sterility. In this review we summarize current data about the origin, organization, evolution of the Ste-Su(Ste) system, and piRNA-dependent regulation of Stellate expression. The Ste-Su(Ste) system is fixed only in the D. melanogaster genome. According to our hypothesis, the acquisition of the Ste-Su(Ste) system by a part of the ancient fly population appears to be the causative factor of hybrid sterility in crosses of female flies with males that do not carry Y-linked Su(Ste) repeats. To support this scenario, we have directly demonstrated Stellate derepression and the corresponding meiotic disorders in the testes of interspecies hybrids between D. melanogaster and D. mauritiana. This finding embraces our hypothesis about the contribution of the Ste-Su(Ste) system and the piRNA pathway to the emergence of reproductive isolation of D. melanogaster lineage from initial species.
Keywords: Drosophila, Stellate genes, piRNA pathway, reproductive isolation, hybrid sterility
Graphical Abstract.
The Stellate-Su(Ste) genetic system is maintained only in the D. melanogaster genome. X-linked testis-specific Stellate genes are repressed in the testes of wild-type flies with the aid of piRNAs derived from the homologous Y-linked Su(Ste) locus that is essential for male fertility. Derepression of Stellate genes in the testes of hybrids of D. melanogaster with closely related D. mauritiana leads to the crystalline aggregate formation and causes strong meiotic failures and male hybrid sterility. Thus, the Stellate-Su(Ste) system and the piRNA pathway contribute to the emergence of the reproductive isolation of D. melanogaster from initial species.
Introduction
At the beginning of the XXI century, a new class of small regulatory RNAs, piRNAs, was discovered in the testes of Drosophila melanogaster (Aravin et al., 2001, 2004; Vagin et al., 2006). X-linked Stellate genes, encoding proteins with homology to the regulatory β-subunit of protein kinase CK2, are currently known as the main targets of piRNA silencing in D. melanogaster testes (Nishida et al., 2007; Ryazansky et al., 2016; Kotov et al., 2019) and the maintenance of their repressed state is essential for male fertility (Palumbo et al., 1994; Bozzetti et al., 1995). Stellate genes are repressed via complementary piRNAs derived from transcripts of the homologous Y-linked Suppressor of Stellate [Su(Ste)] locus. The Ste-Su(Ste) family is present in the genome as abundant homologous tandem repeats found exclusively in D. melanogaster (Livak, 1984, 1990; Balakireva et al., 1992; Tulin et al., 1997). Loss of the Su(Ste) locus leads to Stellate derepression, accumulation of abundant crystalline aggregates in spermatocytes and to meiotic disorders (Hardy et al., 1984; Palumbo et al., 1994; Bozzetti et al., 1995). However, the biological functions of the Ste-Su(Ste) system have remained enigmatic for a long time. Currently, numerous data have been accumulated allowing the reconstruction of the essential stages of Ste-Su(Ste) evolution and piRNA-mediated regulation of Stellate gene expression. Our analysis involving interspecies hybrids reveals the contribution of the Ste-Su(Ste) system to hybrid male sterility. This is the first finding that the disruption of piRNA silencing of a protein-coding gene is able to cause the reproductive isolation of closely related species. In this review, we focus on analyzing the recent advances in understanding of the functional significance of the Ste-Su(Ste) genetic system and novel functions of piRNAs in reproductive isolation.
Discovery of the Crystal-Stellate System and Their Structural Organization
The crystal-Stellate genetic system was discovered by studying the testes of D. melanogaster males with a missing Y chromosome (X/0) using the phase contrast microscopy. Crystalline aggregates of star-like and needle-like shape were found in the nuclei and cytoplasm of primary spermatocytes in these testes (Meyer et al., 1961). It was later shown that the testes of X/0 males also exhibited defects in the condensation and segregation of meiotic chromosomes, such as frequent chromosome non-disjunctions, and X/0 males were sterile (Lifschytz and Hareven, 1977; Hardy et al., 1984).
Now it is established that the crystal-Stellate genetic system contains several interacting loci mapping to the X and Y chromosomes. The Y chromosome of D. melanogaster is completely heterochromatic and contains only a few genes, mainly responsible for male fertility (Charlesworth, 2001; Hoskins et al., 2002; Chang and Larracuente, 2019). The first uncovered locus of the crystal-Stellate system was mapped to the h11 region of the mitotic prometaphase map of the Y chromosome. The loss of this locus or even its partial deletion was found to be sufficient for the crystal accumulation in spermatocytes (Hardy et al., 1984). Thus, the locus was named crystal (cry), but later it was renamed to Suppressor of Stellate [Su(Ste)] (Hardy et al., 1984). Along with the generation of crystalline aggregates in the testes of males with a deficiency in the cry locus (X/Y cry1), similar defects of chromosome condensation and segregation with the X/0 male testes were found (Palumbo et al., 1994).
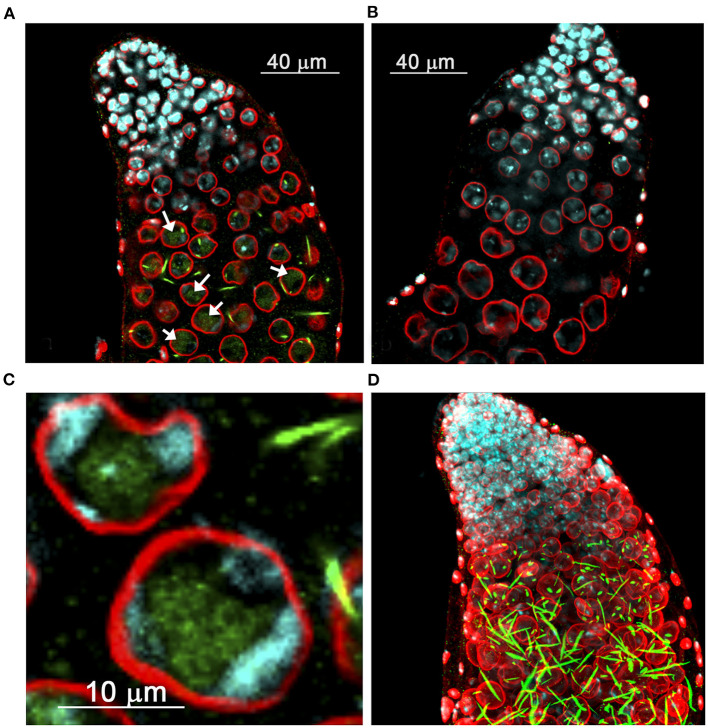
The components of the system include two Stellate (Ste) loci, one of which resides in euchromatic cytolocation 12E1-2 of the X chromosome, whereas the other is mapped to pericentromeric heterochromatin of the X chromosome (the h26 region of the mitotic prometaphase map) (Hardy et al., 1984; Livak, 1984; Palumbo et al., 1994; Tulin et al., 1997). Molecular analysis revealed that the crystal and Stellate loci consist of multiple homologous tandemly repeated sequences (Livak, 1984, Figure 1). The severity of meiotic abnormalities, abundance and shape of crystals in the cry1 testes have been shown to depend on the Ste allele (Livak, 1984; Palumbo et al., 1994). The low-copy Ste + allele contains a small number of Stellate repeats (15–50 copies) and leads only to the appearance of little needle-like aggregates, weak meiotic disturbances and reduced male fertility, whereas the high-copy Ste allele (150–400 copies) leads to the formation of a multitude of crystals, visible under phase contrast as star-shaped entities, severe meiotic defects and complete sterility. Non-disjunction of the XY- and 2nd chromosomes, fragmented chromatin, and chromatin bridges have been found among the intrinsic meiotic defects in the testes of cry1 males. However, in the examined natural and laboratory lineages of D. melanogaster, the Ste+ alleles significantly predominate over the Ste ones (Palumbo et al., 1994). The severity of male fertility defects and the degree of meiotic disorders are associated with the number of Stellate copies and independent from the ratio of euchromatic and heterochromatic Stellate repeats. The boundary for fertility is considered to be 50–60 Stellate copies; the presence of more copies in the genome leads to complete male sterility (Palumbo et al., 1994). Stellate genes are expressed in the testes of cry1 males as 750 nt polyadenylated transcripts (Livak, 1990), and their abundance is proportional to the number of repeats in both Stellate loci (Palumbo et al., 1994). In the cry1 testes Stellate transcripts from both loci are translated generating small proteins of about 17–18 kDa, which have homology with the regulatory β-subunit of protein kinase CK2, CK2β (Livak, 1990; Bozzetti et al., 1995; Egorova et al., 2009; Olenkina et al., 2012b). Stellate proteins, products of the heterochromatic and euchromatic clusters, possess high intra-cluster homogeneity, having minor differences in amino acid sequence between themselves and slightly different electrophoretic mobility (Olenkina et al., 2012b). Immunostaining of the cry1 testes with anti-Stellate antibodies reveals that Stellate is main or the only component of crystalline aggregates (Bozzetti et al., 1995; Egorova et al., 2009, Figures 2A,D). In wild-type flies, Stellate gene expression is strongly suppressed and no Stellate proteins are detected (Figure 2B).
Figure 1.
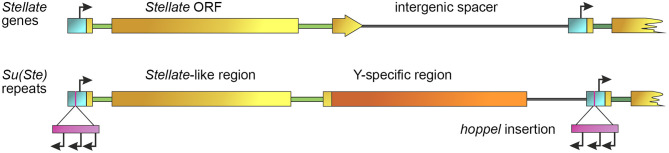
General scheme of Stellate and Su(Ste) repeats. Promoters are indicated by a blue color bar, introns are depicted by green lines, intergenic spacers are depicted by gray lines. Stellate gene contains an ORF (brown color bar) and two introns (green lines). An individual Su(Ste) repeat carries the region homologous to the Stellate ORF (brown color bar), Y-specific region (orange bar) and an insertion of the defective hoppel transposon (violet bar) flanked by inverted repeats (not shown) in the promoter. Start sites of sense transcription of Stellates and Su(Ste) and multiple starts of antisense Su(Ste) transcription within the body of hoppel are indicated by black arrows [modified from Aravin et al. (2001)].
Figure 2.
Distribution of derepressed Stellate protein in the testes of D. melanogaster. (A–C) Internal confocal slices of stained testis preparation of cry1 males (A,C) and wild-type control (B). Testes were immunofluorescently stained with anti-Stellate (green) and anti-lamin (red) antibodies, chromatin was stained with DAPI (cyan). Anti-lamin staining indicates nuclear membrane position. (A,C) Diffuse Stellate signals in the nuclei (arrows in A) and bright needle-like and dot-like crystalline Stellate aggregates mainly in the cytoplasm are seen in spermatocytes of cry1 males. (C) The nuclei of mature spermatocytes. (D) 3D reconstruction of the stained testis preparation of cry1 males. (A–C) are reproduced from Figure 2 in Egorova et al. (2009). (D) is reproduced from Figure 2 in Kibanov et al. (2013) by permission of Elsevier (Licenses ## 4913121387410 and 4913131090753).
The organization of the Su(Ste) locus has also been studied in detail. According to previously published data in most laboratory strains of D. melanogaster the number of Su(Ste) repeats comprises about 80 copies, whereas in natural populations, strains with 240 repeats were found (Lyckegaard and Clark, 1989; Balakireva et al., 1992; McKee and Satter, 1996). However, recent Y chromosome assembly using the Iso1 strain of D. melanogaster with improved annotation of both protein-coding genes and repeats contains more than 500 Su(Ste) copies (Chang and Larracuente, 2019). The size of a typical complete Su(Ste) repeat is about 28 000 nt. It consists of three main parts: the region homologous to Stellate gene, the AT-rich region specific for the Y chromosome, and the insertion of transposable element hoppel (1360) into the promoter sequence (Figure 1). Su(Ste) repeats are transcribed and processed to polyadenylated mRNAs, however, unlike Stellate transcripts, they contain numerous frameshift mutations due to point mutations and deletions (Balakireva et al., 1992; Shevelyov, 1992). Translation products of Su(Ste) repeats are not detected. The insertion of a defective transposon hoppel copy is responsible for antisense transcription of Su(Ste) repeats (Aravin et al., 2001).
Reconstruction of Ste-Su(Ste) System Origin in the D. Melanogaster Genome
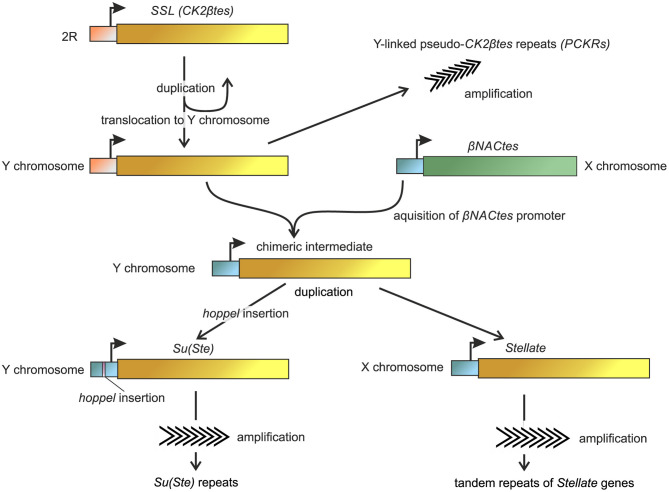
Analysis of published data allows to partially reconstruct the origin and evolution of the Ste-Su(Ste) system in the D. melanogaster genome (Figure 3). A unique gene mapping to the 60D1-2 euchromatic region of the 2nd chromosome is considered a precursor of the Ste-Su(Ste) family (Kalmykova et al., 1997). This gene, called SSL (Su(Ste)-like) or CK2βtes, encodes a functional testis-specific β-subunit of the protein kinase CK2 (Kalmykova et al., 1997, 2002). Identical exon-intron structure of the CK2βtes and Stellate genes is observed. The introns of CK2βtes and Stellate are of the same length and are highly homologous. The amino acid sequence of the CK2βtes shows 45% identity to CK2β and 53% identity to Stellate protein. However, the C-terminus of CK2βtes is 40 amino acids longer than that of Stellate. It has been assumed that CK2βtes gene itself originated from a retroposition of a semi-processed transcript of the canonical β-subunit of protein kinase CK2 (CK2β). The retroposition appears to be accompanied by the loss of all introns except the fourth one, which remains conserved for both CK2β and CK2βtes. At the same time, a new intron was acquired in the non-conserved 5′-region of the CK2βtes ORF. High homology between CK2βtes and Stellate, along with the results of the analysis of the ratio between non-synonymous and synonymous substitutions, suggests that Ste-Su(Ste) repeats originated from CK2βtes via translocation from the autosome to the sex chromosomes and subsequent amplification (Kalmykova et al., 1997).
Figure 3.
Reconstruction of the basic steps of the origin and evolution of Ste-Su(Ste) repeats in the D. melanogaster genome. Non-homologous recombination between the promoter of βNACtes genes and Y-linked βCK2tes copy led to the formation of a chimeric intermediate. Subsequently, the ancestral copies with the acquired promoters were amplified in the sex chromosomes. Insertion of the hoppel transposon in the Su(Ste) promoter region allowed the acquisition of Stellate repressor functions. The homologous sequences are marked by the same color. The model is developed with the usage and modification of ones from Usakin et al. (2005), Chang and Larracuente (2019).
However, the promoter regions of CK2βtes gene and Ste-Su(Ste) family do not show sequence similarity to each other. BLAST search revealed extremely high homology of the promoter sequences of Ste-Su(Ste) repeats with the promoter of an unrelated gene family located as a single cluster in 12DE region of the X chromosome (Figure 3). These genes maintain testes-specific expression and were called βNACtes owing to their homology with bicaudal gene, which encoding β-subunit of NAC (Nascent polypeptide-Associated Complex) (Usakin et al., 2005). Alignment of the promoters of the Ste-Su(Ste) and βNACtes families demonstrates a striking similarity of 180 nt nucleotide regions before the transcription start site—up to 95% identity. Taken together with the data about testis-specific expression of a transgene construct containing a 134 bp part of the promoter region of Stellate gene (Aravin et al., 2004), these results confirm the defining role of the new promoter region for testis-specific transcription of both gene families, Ste-Su(Ste) and βNACtes. The common promoter of Ste-Su(Ste) and βNACtes genes has been functionally mapped and three cis-regulatory E-box sites have been determined within this region. Transgene constructs carrying the promoter with the disruption of all three E-boxes do not express the reporter in the testes (Olenkina et al., 2012a,b). E-boxes are known as cis-acting elements for the recognition by transcription factors of the basic helix-loop-helix (bHLH) family (Ledent and Vervoort, 2001), and one of them, dUSF, from testis nuclear extract, is able to interact with oligonucleotide fragments of the Stellate promoter in vitro (Olenkina et al., 2012a). βNACtes genes are present in all closely related Drosophila species and are considered to be more ancient than the Ste-Su(Ste) system (Usakin et al., 2005; Kogan et al., 2012). Thus, a precursor of the Ste-Su(Ste) family captured an alien promoter during evolution of D. melanogaster to acquire testis-specific expression.
The first step of precursor generation of the Ste-Su(Ste) repeats appears to occur through a duplication of SSL (CK2βtes) gene and translocation of the latter to the Y chromosome (Danilevskaya et al., 1991). Indeed, recently a set of 122 pseudogenes that are similar to the initial duplication of SSL and carry the ancestral promoter was mapped on the Y chromosome of the new assembly (Chang and Larracuente, 2019). They were designated as pseudo-CK2βtes repeats on the Y chromosome (PCKRs) (Figure 3). PCKRs are located proximal to Su(Ste) repeats, between WDY and Pp1-Y1 genes (Chang and Larracuente, 2019). A presumed subsequent Ste-Su(Ste) evolution step is the creation of a chimeric intermediate, as a result of putative non-homologous recombination between the X-linked βNACtes promoter and the Y-linked SSL gene. In support of this hypothesis, sequenced scaffold AE003039 has been found earlier in the non-annotated region of the Y chromosome that possesses a pseudogene that could be considered as a chimeric precursor of the Ste-Su(Ste) family (Usakin et al., 2005). This pseudogene carries the βNACtes-derived promoter fused with a damaged ORF having exon-intron structure which is identical to Ste-Su(Ste)-SSL. The 5′-region upstream the transcription start site of this pseudogene contains a fragment of about 200 nt with high homology to the sequence adjacent to the promoter of the βNACtes3 gene and other sequences specific for the βNACtes cluster (Usakin et al., 2005), implying that a recombination event may occur between the X and Y chromosomes. Following the initial duplication of SSL in the Y chromosome, the ancestral copies with the acquired testes-specific promoters were amplified independently in the sex chromosomes with the possible participation of the Helitron family transposable elements (Kogan et al., 2012), which led to the formation of both euchromatic and heterochromatic clusters of Stellate genes on the X chromosome and Su(Ste) repeats on the Y. The X-linked Stellate genes and Y-linked Su(Ste) copies were amplified independently. The insertion of the hoppel transposon in the Su(Ste) promoter region is considered as a key event responsible for the acquisition of repressor functions by Su(Ste) repeats. This insertion defined the antisense transcription of Su(Ste) repeats and subsequent generation of Su(Ste) piRNAs (Aravin et al., 2001, 2004). Note that the promoter region of heterochromatic Stellate genes contains no traces of hoppel insertion and is closest to the parental βNACtes promoter. However, euchromatic Stellate genes carry a 16 bp deletion in the corresponding region of the promoter, which could be a sign of the insertion and subsequent imperfect excision of hoppel (Olenkina et al., 2012a). This data argues for the hypothesis according to which ancestral copies of euchromatic and heterochromatic Stellate clusters translocate on the X chromosome independently and at different stages of the evolutionary history of the Ste-Su(Ste) family. Phylogenetic analysis provided recently (Chang and Larracuente, 2019) supports that euchromatic and heterochromatic Stellate clusters are amplified independently of each other. Both euchromatic and heterochromatic Stellate repeats are fixed in the D. melanogaster genome and contain intact ORFs (Tulin et al., 1997; Kogan et al., 2000). Polymorphism in their coding regions is mainly determined by synonymous substitutions. Apparently, translational selection supports high homogeneity of Stellate genes. Intra-locus divergence between adjacent heterochromatic Stellate genes is nearby 0.1–0.2% (Tulin et al., 1997), and divergence between randomly selected Stellate pairs does not exceed 2.5% (McKee and Satter, 1996).
It should be noted that the Ste-Su(Ste) genetic system is found only in D. melanogaster. The genomes of three closely related Drosophila species have diverged from D. melanogaster. Analysis of recent genome assemblies of the simulans clade species reveals that pseudo-βCK2tes repeat (PCKR) duplications are present in the Y chromosome of all three sibling species, D. simulans, D. mauritiana, and D. sechellia, in the range from 22 to 117 copies (Chakraborty et al., 2020). These findings allow suggesting that the first stages of the Ste-Su(Ste) system formation occurred before the splitting of D. melanogaster from the common precursor, but subsequent dynamics of the evolution process led to species diversification.
Properties of Stellate Proteins
The derepression of Stellate genes causes the emergence of protein crystals both in the cytoplasm and nuclei of spermatocytes of cry1 males. Stellate appears to be the predominant or the exclusive component of these crystals (Bozzetti et al., 1995, Egorova et al., 2009). The needle-like shape of crystalline aggregates indicates their generation in a head-to-tail manner (Figures 2A,D). Mass-spectrometry analysis points to the presence in their content of protein products of both euchromatic and heterochromatic Stellate clusters (Olenkina et al., 2012b). Since Stellate protein possesses 38% homology with the canonical regulatory β-subunit of the protein kinase CK2 (CK2β) (Livak, 1990; Bozzetti et al., 1995), it is suggested that Stellate is able to interact with the catalytic α-subunit of CK2 (CK2α) in certain conditions. Protein kinase CK2 (also known as casein kinase 2) is a multi-functional ubiquitous heterotetrameric α2β2 complex, which takes part in numerous signaling cascades, cell differentiation, proliferation and surviving (Pinna, 2002; Bibby and Litchfield, 2005; Bandyopadhyay et al., 2016). CK2 phosphorylates protein substrates at serine or threonine residues embedded in an S/TXXD/E motif, where X could be any amino acid residue except the basic ones (Allende and Allende, 1995). Among the established CK2 targets in D. melanogaster there are nuclear proteins such as HP1 (Heterochromatin Protein 1) (Zhao and Eissenberg, 1999), topoisomerase II (Ackerman et al., 1985, 1988), Mi-2 (Bouazoune and Brehm, 2005), transcription factor GAGA (Bonet et al., 2005), and other proteins with a predominance of transcription factors (Bandyopadhyay et al., 2016). The regulatory subunits CK2β modulate the substrate specificity of CK2α and its phosphotransferase activity and also ensure the stability of the holoenzyme (Bibby and Litchfield, 2005).
Bozzetti and colleagues showed that recombinant Stellate protein was able to interact in vitro with recombinant CK2α, and its 17-fold excess somewhat stimulated the basal activity of CK2α (Bozzetti et al., 1995). However, the functional interaction of Stellate present in the form of insoluble aggregates in spermatocytes with CK2α seems to be problematic and doubtful. Accordingly, the two-hybrid assay did not detect any interaction of Stellate with CK2α, in contrast to canonical CK2β subunits of D. melanogaster, testis-specific subunits CK2β′, and CK2βtes (SSL) (Karandikar et al., 2003). Stellate protein lacks the conserved C-terminal domain with the aid of which CK2β interacts with CK2α. Unlike the other β-subunits of Drosophila, Stellate was not able to compensate for the absence of CK2β and did not rescue the defects of ionic homeostasis when it was expressed in Saccharomyces cerevisiae strains with deletion of yeast genes encoding their own β-subunits (Karandikar et al., 2003). However, biochemical and immunofluorescence studies have shown that hyperexpression of Stellate protein causes the accumulation Stellate crystalline aggregates mainly in the cytoplasm of premeiotic spermatocytes, whereas soluble Stellate protein is clearly detected in the spermatocyte nuclei of the cry1 testes (Egorova et al., 2009) (Figures 2A,C). The soluble Stellate appears in the spermatocytes in the mid-G2 phase and persists throughout meiotic interphase. It is coimmunoprecipitated with CK2α, despite the lack of canonical CK2β C-terminal domain (Egorova et al., 2009), which indicates possible formation of heterotetrameric holoenzyme CK2 via alternative contacts. The biological significance of this interaction is not clear. It is possible that at some stage of D. melanogaster evolution the appearance of Stellate gene encoding the alternative testis-specific β-subunit of CK2 was supported by positive selection, but later its functions turned out to be dispensable or harmful.
One more surprising circumstance is in agreement with this assumption. Immunochemical and mass-spectrometry data confirmed that both forms of Stellate proteins, soluble and crystalline, undergo trimethylation of lysine residue K92. Endogenous Stellate proteins are recognized with a high affinity by antibodies against histone H3 trimethylated at K9 lysine residue (Egorova et al., 2009). Thus, the methylated site of Stellate proteins mimics the epigenetic modification of histone H3, H3K9me3, that is a hallmark of transcriptionally repressed heterochromatin (Kouzarides, 2007; Allis and Jenuwein, 2016; Ninova et al., 2019). Three histone methyltransferase, Su(var)3-9, dG9a, and dSETDB1, are involved in the H3K9 methylation in Drosophila (Tachibana et al., 2001; Schotta et al., 2002; Schultz et al., 2002). While Stellate does not exhibit similarity to the corresponding histone H3 sequence (–TARK9ST–), the methylated site of Stellate, –MHRK92YL/M–, contains an –RK– dipeptide motif that has been shown to be the principal determinant for lysine methylation provided by methyltransferase G9a (Rathert et al., 2008). The site for lysine methylation has emerged and has been fixed only in Stellate sequence because sequences of other Drosophila CK2β subunits, including CK2βtes, do not contain it (Egorova et al., 2009). The functional significance of the trimethylation of Stellate is still obscure. Proteins of the highly conserved Heterochromatin Protein 1 family are known as H3K9me3 readers aided by their N-terminal chromo domain (Vermaak and Malik, 2009; Ninova et al., 2019). The H3K9me3 modification is recognized by HP1 that is essential for the formation of heterochromatin genomic regions (Fischle et al., 2005; Kouzarides, 2007; Vermaak and Malik, 2009). Both hypo- and hyper-expression of HP1 lead to disturbances in the compaction of pericentromeric heterochromatin and abnormalities in centromeric cohesion and segregation of chromosomes (Inoue et al., 2008; Vermaak and Malik, 2009). Besides the K92me3 site mimicking the H3K9me3 mark, Stellate also possesses two -P(L)XVXL- motifs, known as potential sites for interaction with the chromo shadow domain of HP1 (Lechner et al., 2005). Taking into account the above-mentioned observations, it can be proposed that thrimethylated Stellate is able to interact with HP1 or other chromo domain-containing proteins in premeiotic spermatocytes. However, the functional significance of these possible interactions is unexplored to date.
Stellate Genes as a Major Target of Pirna Silencing in the Testes of D. Melanogaster
At the beginning of the XXI century, in 2001, a new class of small non-coding RNAs, piRNAs, 23–30 nt in length, associated with silencing of Stellate genes has been discovered in the testes of Drosophila melanogaster by Aravin and colleagues (Aravin et al., 2001). The piRNA pathway is conserved in a wide range of animals from fungi to mammals (Aravin et al., 2007; Malone et al., 2009; Gainetdinov et al., 2018). The piRNA pathway provides both innate and adaptive immune defense against the activity of transposable elements protecting genome integrity in germinal tissues. It also contributes to the maintenance of germline stem cells, ensures expression regulation of protein-coding genes, functions in the establishment of embryonic patterning (in Drosophila), and provides trans-generational epigenetic inheritance (Malone et al., 2009; Le Thomas et al., 2014; Rojas-Ríos et al., 2017; Rojas-Ríos and Simonelig, 2018; Ozata et al., 2019). The piRNAs mainly function in the gonads and are characterized by extreme diversity existing as millions of unique piRNA sequences (Huang et al., 2017), unlike other main classes of small non-coding RNAs, siRNAs and microRNAs. RNA-induced silencing complexes (RISCs) operate in all pathways with the participation of small RNAs, and ARGONAUTE proteins are the key players of different RISCs (Czech and Hannon, 2016; Huang et al., 2017). piRNAs load to PIWI subfamily of ARGONAUTE family proteins: Piwi, Aubergine (Aub) and ARGONAUTE 3 (AGO3) in Drosophila (Aravin et al., 2001; Vagin et al., 2006; Li et al., 2009; Malone et al., 2009). AGO3 and Aub are expressed only in the germline, while Piwi is also found in somatic cells of gonads. Complementary base pairing with a small RNA guides the piRISC to a specific RNA target, generally providing silencing via its direct cleavage or translational repression. Mature piRISC complexes have a different destiny depending on the protein component. Piwi loaded by piRNA enters the nucleus, where it participates in co-transcriptional silencing, whereas Aub and AGO3 reside in cytoplasmic granules and function in post-transcriptional silencing.
Early classification of piRNAs based on the peculiarities of their biogenesis divided piRNAs into two main groups: primary and secondary piRNAs. The main sources of primary piRNAs are long transcripts originating from piRNA clusters, specific genome loci residing mainly in heterochromatin and generally representing transposon “graveyards” (Brennecke et al., 2007; Czech and Hannon, 2016; Huang et al., 2017). In germ cells of Drosophila gonads primary or maternally inherited piRNAs initiate the formation of secondary piRNAs. The cyclic mechanism called “ping-pong” provides amplification of the piRNA pool for the rigorous repression of selfish element activity. Ping-pong was discovered in Drosophila ovarian nurse cells (Brennecke et al., 2007) and was subsequently defined as the conserved property of the piRNA pathway (Aravin et al., 2007; Gainetdinov et al., 2018). Piwi- and Aub-associated piRNAs are found to be enriched with uridine at the 1st position from the 5′-end. AGO3-related piRNAs are enriched with adenosine at the 10th position and do not have a clear preference for the 1st position. Consistent with the initially proposed model, Aub complexed with a primary antisense piRNA, derived from a piRNA cluster transcript, recognizes and trims the transcript of a transposable element, creating the 5′-end of a secondary sense piRNA. All PIWI proteins cleave their target transcripts between nucleotides 10 and 11 of the paired piRNA guide. Thus, the first 10 nucleotides of a secondary piRNA possess complementarity to the first 10 nucleotides of a primary piRNA directing the cleavage act. This secondary piRNA precursor is loaded into AGO3 and matures via trimming at the 3′-end. The sense piRNA-AGO3 complexes in turn, recognize and process a long transcript of a piRNA cluster to form a new antisense piRNA with the sequences that are identical or very similar to the original primary one. This process is repeated cyclically (Figure 4A). Ping-pong processing increases the piRNA pool quantitatively, but it does not create new piRNA sequences. The maturation of piRNA intermediates, associated with Aub or Piwi, can also take place through the production of phased piRNAs from adjacent regions of the transcript (Han et al., 2015; Mohn et al., 2015; Senti et al., 2015; Wang et al., 2015). It can be considered as a mechanism of adaptation to the targets through diversification of piRNA sequences. Thus, at least in the germline, the separation of primary and secondary piRNA biogenesis is rather provisional, since both variants of processing are more closely related than previously thought (Czech and Hannon, 2016; Huang et al., 2017; Gainetdinov et al., 2018).
Figure 4.
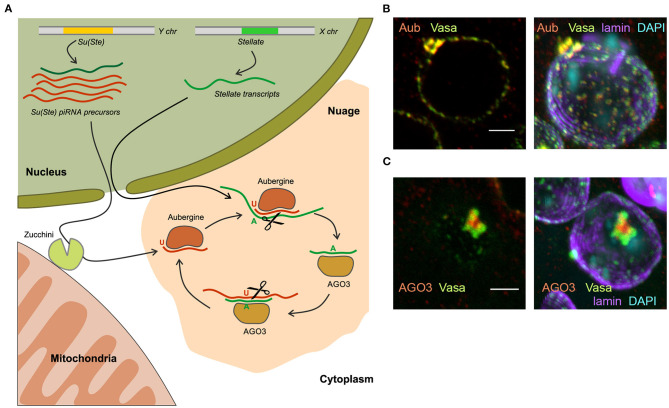
The piRNA biogenesis in the nuage granules of Drosophila spermatocytes. (A) Long transcripts of Su(Ste) piRNA precursors are exported from the nucleus and are presumably cleaved by endonuclease Zucchini located on the outer membrane of mitochondria, forming the 5′-end of the future piRNA. The cleaved transcript is loaded into Aubergine, and then trimmed from the 3′-end by an unknown trimmer nuclease. Aubergine loaded with guide antisense piRNAs recognizes and cleaves the complementary Stellate mRNAs producing the 5′-end of a new sense piRNA. The new piRNA is loaded into AGO3 and in turn performs cleavage of the complementary Su(Ste) transcript. This step generates a new antisense piRNA that is identical or very similar to the initiating piRNA (ping-pong cycle). (B) Vasa and Aubergine are colocalized in the periphery lobes of the piNG-body. (C) AGO3 is located in the central lobe of the piNG-body and does not colocalized with Vasa. (B,C) Confocal slices (left) and 3D images (right) of premeiotic spermatocytes in testis preparations are reproduced from Figure 3 in Kibanov et al. (2011) by permission of MBoC. Under the License and Publishing Agreement, authors grant to the general public, the non-exclusive right to copy, distribute, or display the manuscript subject to the terms of the Creative Commons-Non-commercial-Share Alike 3.0 Unported license (http://creativecommons.org/licenses/by-nc-sa/3.0). Scale bars are 3 μm.
Cytoplasmic electron-dense perinuclear granules called nuage are considered to be centers of piRNA biogenesis and piRNA-mediated post-transcriptional silencing in the germline of Drosophila. To date, the organization and functions of the nuage are described in some details (Findley et al., 2003; Snee and Macdonald, 2004; Lim and Kai, 2007; Lim et al., 2009; Patil and Kai, 2010; Kibanov et al., 2011; Nosov et al., 2014; Webster et al., 2015). Among the known components that are concentrated in the nuage, three classes of proteins prevail: PIWI proteins (Aub and AGO3), RNA helicases, and Tudor-domain-containing proteins. The nuage contains RNA as an essential component. A growing body of evidence suggests that nuage formation is based on the principles of liquid–liquid phase separation (Brangwynne et al., 2009; Nott et al., 2015). A number of proteins are necessary for piRNA-mediated repression of Stellates in the testes and retrotransposons in the ovarian germline are located in or associated with the nuage: Aub, AGO3, Vasa, Spindle-E, Armitage, Tudor, Tejas, Krimper, Maelstrom, Squash, Qin/Kumo, Zhuccini, etc. (Aravin et al., 2004; Vagin et al., 2006; Lim and Kai, 2007; Pane et al., 2007; Malone et al., 2009; Patil and Kai, 2010; Kibanov et al., 2011; Zhang et al., 2011; Anand and Kai, 2012). Vasa and Spindle-E are ATP-dependent RNA helicases. Vasa takes part in the different stages of the piRNA silencing, it presumably mediates nuclear export of piRNA precursors (Zhang et al., 2012) and has basic architectural functions in nuage formation, being the top component of the nuage hierarchy. vasa mutations lead to a loss of nuage granules, mislocalization of Aub, AGO3 and other components in the cytoplasm, and to the disruption of piRNA silencing both in the ovaries and testes (Lim and Kai, 2007; Li et al., 2009; Malone et al., 2009; Patil and Kai, 2010; Kibanov et al., 2011). Tudor, Spindle-E, Qin/Kumo, Tejas and Krimper proteins possess the TUDOR-domains, which recognizes post-translationally methylated lysine residues in PIWI proteins, which are essential for the nuage assembly (Kirino et al., 2009; Nishida et al., 2009; Kibanov et al., 2011).
In Drosophila spermatocytes, among small nuage granules, a large granule, the piNG-body was uncovered, that was more than 50 times larger than the smaller ones (Kibanov et al., 2011). The piNG-body is present in primary spermatocytes as one giant nuage granule per cell. piNG-bodies move around the outer surface of the nuclei (Nosov et al., 2014) and persist in spermatocytes throughout the entire meiotic interphase. The large size of the piNG-body allowed to determine a specific pattern of piRNA pathway component distribution: Spindle-E, Aub, and Tudor are colocalized with Vasa in the periphery lobes of the piNG-body, while AGO3 is found only in the central lobe (Kibanov et al., 2011, 2013; Ryazansky et al., 2016) (Figures 4B,C). These results are consistent with data indicating different subcellular localization of Aub and AGO3 and independent mechanisms of their recruitment to the nuage in the ovaries (Webster et al., 2015). With the aid of fluorescence recovery after photobleaching (FRAP) experiments, the existence of a dynamic exchange of GFP-tagged Vasa between the piNG-body and the cytoplasm in spermatocytes under normal conditions was shown (Nosov et al., 2014). A similar exchange of Vasa, Aub, AGO3, Tudor, Tejas, and Spindle-E molecules was observed between the nuage and their cytoplasmic pool in the ovaries (Snee and Macdonald, 2004; Xiol et al., 2014; Webster et al., 2015). Only 71% recovery after photobleaching of the Vasa-GFP signal in the piNG-bodies indicates the existence of at least 29% stationary fraction that is not involved in the rapid exchange with the cytosolic mobile fraction (Nosov et al., 2014). Liquid–liquid phase separation is proposed to slow this exchange down owing to the higher viscosity of nuage granules supported by specific protein-protein interactions (Ozata et al., 2019).
Historically, the first example of piRNA-mediated gene repression is Stellate gene silencing in the Drosophila testes with the aid of piRNAs derived from Y-linked Su(Ste) repeats (Aravin et al., 2001). To date, the mechanism of Sellate repression has been clarified in some details. Su(Ste) represents the major testis piRNA cluster producing about 43% of total piRNAs (Kotov et al., 2019; Chen et al., 2020) unlike the situation in the ovarian germline, where 42AB in pericentromeric region of the 2nd chromosome is found to be the most active piRNA cluster (Malone et al., 2009). Chronologically, antisense Su(Ste) transcripts appear in the nuclei of early spermatocytes before the sense ones. Antisense transcription is initiated from several sites within the hoppel transposon inserted in the Su(Ste) promoter (Aravin et al., 2001, Figure 1). In the nuclei of mature primary spermatocytes sense and antisense Su(Ste) transcripts are found to be colocalized (Aravin et al., 2004). However, antisense Su(Ste) transcripts are much more abundant (more than 20-fold) than sense ones and provide a great amount of piRNAs with high complementarity to Stellate transcripts (Kotov et al., 2019). Stellate gene transcription starts in early spermatocytes. Deletion of the bulk part of Su(Ste) caused increased accumulation of spliced, but not non-spliced Stellate transcripts in the nuclei and cytoplasm of spermatocytes (Kotelnikov et al., 2009), indicating the post-transcriptional mode of Stellate silencing. Analysis of Su(Ste)-Stellate piRNAs reveals strong U1 (78%) and A10 (54%) nucleotide biases for antisense and sense piRNAs, respectively. This points to the existence of piRNA pairs with a 10-nt overlap generated through the ping-pong mechanism (Ryazansky et al., 2016; Kotov et al., 2019, Figure 4A). Expression and silencing of Stellate genes coincide with the organization of the piNG-bodies, that contain the components of the piRNA machinery in a high concentration. Mutations of vasa, aub and ago3 cause the destruction of the piNG-bodies that results in Stellate derepression. Note that the ago3 mutation does not lead to the disappearance of the small nuage granules, indicating that a high concentration and compartmentalization of the piRNA pathway components achieved in the piNG-body are essential for successful silencing (Kibanov et al., 2011).
The piRNA pathway has evolved to keep a high level of sequence complementarity between a piRNA and its target in the germline (Kotov et al., 2019; Chen et al., 2020). Expression of Stellate genes is perfectly repressed by Su(Ste) piRNAs in the testes of wild-type males. Other gene encoding testis-specific β-subunit of CK2, CK2βtes, possesses a significant homology with Su(Ste) and Stellates, about 70% identity of nucleotide sequence. However, CK2βtes is expressed in the male germline at a high level (Kalmykova et al., 2002, 2005) and does not undergo targeting by Su(Ste) piRNAs. The second major piRNA cluster in the testes, AT-chX, resides in the pericentromeric region of the X chromosome and also contains internal tandem repeats (Kotov et al., 2019; Chen et al., 2020). This cluster produces abundant and diverse piRNAs with 76% homology to vasa transcripts mainly in antisense orientation. However, in contrast to the previously published data (Nishida et al., 2007) we found that sequence similarity between AT-chX-derived piRNAs and vasa transcripts is not enough for their repression in the testes and ovaries of D. melanogaster (Kotov et al., 2019). On the whole, these data suggest that effective piRNA silencing at least in premeiotic germ cells of Drosophila testes requires a high level of sequence complementarity between piRNAs and their targets to prevent harmful off-target effects.
Contribution of the Ste-Su(Ste) System to Hybrid Sterility and Reproductive Isolation Between D. MELANOGASTER and Closely Related Species
Despite the deep insight into the evolution and regulation of Stellate genes expression, functions and biological significance of the Ste-Su(Ste) system have been obscure for a long time. According to one early hypothesis, the Ste-Su(Ste) system exists as a parasitic self-maintaining genetic system that only mimics functions crucial for meiosis (Bozzetti et al., 1995). According to another assumptions, the Ste-Su(Ste) system is similar to toxin-antitoxin systems, which are widely found in prokaryotes (Aravin, 2020). However, the most common fate for insignificant duplicated genes is their rapid degeneration and loss of function (Conant and Wolfe, 2008). The energy-consuming maintenance of multiple highly homogenous copies of Stellate genes in the genome and their permanent silencing indicate the existence of selective forces that prevent the loss of long-lived Stellate genes. Thus, the Ste-Su(Ste) system is fixed in the D. melanogaster genome and maintained under positive selection. Stellate genes are normally completely repressed in the male germline by piRNAs generated from Y-linked Su(Ste) repeats. A deletion of the cry1 locus or a loss of the entire Y chromosome leads to Stellate derepression in spermatocytes and to subsequent meiotic disorders causing complete or partial male infertility. Tandem Stellate genes are only found in the D. melanogaster genome, but not in other Drosophila species, as mentioned above. We have hypothesized that Stellate genes may function in the reproductive isolation of D. melanogaster from closely related species lacking Su(Ste) repeats in the Y chromosomes.
Highly homogenous AT-chX repeats are also fixed in D. melanogaster and not found in other Drosophila species. Strikingly, we revealed that vasa sequences from closely related species, D. simulans, D. sechellia and D. mauritiana, that have diverged from an ancestor common with D. melanogaster 2.0–5.4 million years ago (Russo et al., 1995; Tamura et al., 2004), maintain more than 90% complementarity with piRNAs from AT-chX repeats of D. melanogaster compared with only 76% for vasa of D. melanogaster itself (Kotov et al., 2019). In line with this observation, we have proposed that AT-chX piRNAs could repress alien vasa transcripts in the gonads of interspecies hybrids.
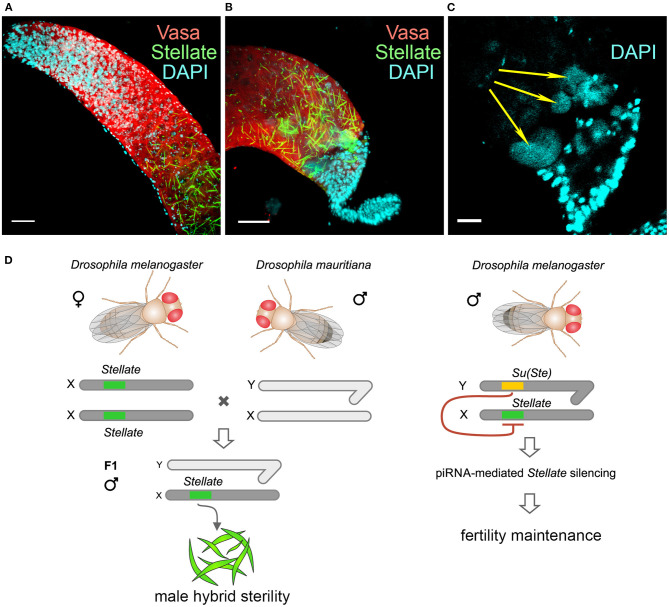
The studying of viable interspecific hybrids is a way to identify hybrid incompatibility factors that cause reproductive isolation between species. It has been shown earlier that the Hmr mutation significantly rescues survival of F1 male hybrid progeny of crosses between D. melanogaster females and males of the simulans clade (Barbash et al., 2000; Barbash and Ashburner, 2003). We raised hybrid males by crossing Hmr1 D. melanogaster females and D. mauritiana males to test our hypotheses (Kotov et al., 2019). The hybrid males inherit the maternal X chromosome carrying Stellate genes and the AT-chX piRNA cluster, and the paternal Y chromosome that does not contain Su(Ste) repeats. We uncovered that the piRNA biogenesis was not disrupted in the testes of hybrids, because the analysis of small RNAs reveals the presence of 23–30 nt piRNAs mapped to various transposable elements (Kotov et al., 2019). These piRNAs exhibit U1-bias for antisense piRNAs, whereas the sense ones possess A10-bias, indicating that the piRNA pathway is still functional. We have observed a strong silencing of D. mauritiana vasa by trans-acting piRNAs from the AT-chX cluster in hybrid testes, whereas vasa of D. melanogaster does not undergo repression. The majority of hybrid testes has reduced size and contains just a few or no germ cells. We assumed that selective repression of one allele of the vasa gene in diploid hybrids might cause haploinsufficiency of vasa functions and disruption of germ cell development at early stages, appearing to contribute to hybrid male sterility. However, the molecular basis of hybrid dysfunction is not characterized yet in this case. For the analysis of Stellate gene regulation we selected only perfectly developed hybrid testes with maximal germ cell content (Kotov et al., 2019). These wild-type size testes are filled a large number of germ cells at different stages of spermatogenesis. Expectedly, Su(Ste) piRNAs are not generated in the testes of the hybrids. Immunostaining of the testis preparations reveals the presence of abundant Stellate aggregates in spermatocytes of the hybrid testes (Figures 5A,B), but not in the testes of parent flies. Spermatocytes in the hybrid testes do not progress through meiosis, haploid spermatids are absent at the distal end of the testes and the seminal vesicles do not contain mature sperm. Giant conglomerations of decondensed chromatin at the distal end of these testes indicate a severe meiotic catastrophe (Kotov et al., 2019, Figure 5C). Thus, strong Stellate derepression in the testes of hybrid males definitely leads to their infertility.
Figure 5.
Derepression of Stellate genes in the testes of interspecies hybrids D. melanogaster with D. mauritiana. (A–C) Hybrid testes were stained with anti-Vasa (red) and anti-Stellate (green) antibodies, chromatin was stained with DAPI (cyan). 3D images are present for apical (A) and distal (B) testes ends; an internal confocal slice is present for the distal testis end stained with DAPI (C). Scale bars are 30 μm for (A) and (B), 15 μm for C. Stellate genes are derepressed in spermatocytes of hybrid testes and generate abundant crystals (A,B). Spermatids are absent at the distal end of hybrid testes (B). Giant conglomerations of decondensed chromatin (yellow arrows) are found at the distal ends of hybrid testis (C) indicating a severe meiotic catastrophe. (D) Derepession of Stellate genes in the testes of hybrids causes meiotic failures and male hybrid sterility. (A–D) Images are reproduced from Figure 6B in Kotov et al. (2019), and the scheme is adapted from Figure 7B in Kotov et al. (2019) by permission of Oxford University Press (http://global.oup.com/academic) without the need to obtain written permission from OUP and payment as authors of this publication.
Conclusions and Perspectives
One of main condition of species splitting from a common precursor lineage is the prevention of gene flow between diverging populations. Thus, reproductive isolation caused by hybrid lethality or sterility is a necessary condition for speciation (Orr, 2005; Castillo and Barbash, 2017). Postzygotic reproductive isolation emerges as a consequence of gradual accumulation of genetic differences between isolated populations of the ancient precursor. These differences, emerging due to selection or genetic drift, lead to lower fitness or sterility of hybrid progeny. The well-known Dobzhansky–Muller model suggests that a common cause of genetic incompatibility between closely related species is the divergence among alleles at two or more loci (Dobzhansky, 1936; Muller, 1940; Muller and Pontecorvo, 1942). We did not identify new Dobzhansky–Muller gene pairs in parental genomes. However, we showed that abnormal regulatory divergence triggered by the piRNA pathway provides two opposite harmful effects in the testes of interspecies hybrids: loss of Su(Ste) piRNAs leads to derepression of Stellate genes causing defects in male meiosis (Figure 5D), whereas silencing of D. mauritiana vasa by AT-chX piRNAs originating from the D. melanogaster genome appears to contribute to defects of early spermatogenesis stages (Kotov et al., 2019). In accordance with these findings a recently published study leads to the speculation that rapid intrinsic divergence of human pachytene piRNA loci among placental mammals also may be an undescribed driver of reproductive isolation (Özata et al., 2020). It would be attractive to test in future the derepression of Stellate genes in the interspecific hybrids of D. melanogaster with D. simulans or D. sechellia as well. However, to date our attempts to cross D. melanogaster females with males of these species have resulted in hybrid males with severely reduced testes that did not contain germ cells. This indicates a failure in germ cell maintenance at earlier stages of testis development. It can be proposed that the repression of one of the vasa alleles affects this phenotype. But this hypothesis requires a further investigation that is strongly hampered owing to a loss of germ cells. Potential advances in this field can be provided by studies of the ovaries of hybrid females, since the AT-chX piRNA cluster is shown to be active in the ovaries of D. melanogaster and produce abundant piRNAs (Chen et al., 2020). The generation and usage of female hybrid offspring with developed ovaries may allow to study the impact of vasa misregulation on the maintenance of early germ cells and fertility in the hybrid genomic background.
The studies of hybrid male sterility in Drosophila allowed to uncover that reproductive isolation can occur due to a small number of genes misexpressed in hybrids (Michalak and Noor, 2003; Haerty and Singh, 2006; Moehring et al., 2007; Wei et al., 2014). The acquisition of the Ste-Su(Ste) system by a part of the ancient fly population appears to be one of the causative factors of hybrid sterility in crosses of female flies with males that do not carry Y-linked Su(Ste) repeats. In our experiments, we reproduced this putative scenario by directly demonstrating Stellate derepression in the background of interspecies hybrids between D. melanogaster and D. mauritiana (Figure 5). The results embrace our hypothesis about the contribution of the Ste-Su(Ste) system and its epigenetic regulation by the piRNA pathway in reproductive isolation of the D. melanogaster lineage. By studying the molecular and genetic events that lead to the emergence of reproductive isolation between lineages originating from a single ancestral population, we have pieced together some of the processes that cause D. melanogaster divergence from closely related species to improve our understanding of the process of speciation.
Author Contributions
VA and LO prepared the initial version of the manuscript and created the figures. AK, SB, AS, and AA rigorously revised and improved the manuscript. VA, LO, AK, and AA polished the final version of the manuscript. All authors provided intellectual contribution, edited, and approved the manuscript for publication in its present version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank A. D. Stolyarenko for discussions on this manuscript.
Footnotes
Funding. This work was funded by RFBR, project number #20-04-00562.
References
- Ackerman P., Glover C. V., Osheroff N. (1985). Phosphorylation of DNA topoisomerase II by casein kinase II: modulation of eukaryotic topoisomerase II activity in vitro. Proc. Natl. Acad. Sci. U. S. A. 82, 3164–3168. 10.1073/pnas.82.10.3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman P., Glover C. V., Osheroff N. (1988). Phosphorylation of DNA topoisomerase II in vivo and in total homogenates of Drosophila Kc cells. The role of casein kinase II. J. Biol. Chem. 263, 12653–12660. [PubMed] [Google Scholar]
- Allende J. E., Allende C. C. (1995). Protein kinases. 4. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 9, 313–323. 10.1096/fasebj.9.5.7896000 [DOI] [PubMed] [Google Scholar]
- Allis C. D., Jenuwein T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500. 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- Anand A., Kai T. (2012). The tudor domain protein kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. EMBO J. 31, 870–882. 10.1038/emboj.2011.449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A. A. (2020). Pachytene piRNAs as beneficial regulators or a defense system gone rogue. Nat. Genet. 52, 644–645. 10.1038/s41588-020-0656-8 [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Klenov M. S., Vagin V. V., Bantignies F., Cavalli G., Gvozdev V. A. (2004). Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell. Biol. 24, 6742–6750. 10.1128/MCB.24.15.6742-6750.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A. A., Naumova N. M., Tulin A. V., Vagin V. V., Rozovsky Y. M., Gvozdev V. A. (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11, 1017–1027. 10.1016/S0960-9822(01)00299-8 [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G. J. (2007). Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747. 10.1126/science.1142612 [DOI] [PubMed] [Google Scholar]
- Balakireva M. D., Shevelyov Y. Y., Nurminsky D. I., Livak K. J., Gvozdev V. A. (1992). Structural organization and diversification of Y-linked sequences comprising Su(Ste) genes in Drosophila melanogaster. Nucleic Acids Res. 20, 3731–3736. 10.1093/nar/20.14.3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay M., Arbet S., Bishop C. P., Bidwai A. P. (2016). Drosophila protein kinase CK2: genetics, regulatory complexity and emerging roles during development. Pharmaceuticals 10:4. 10.3390/ph10010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash D. A., Ashburner M. (2003). A novel system of fertility rescue in Drosophila hybrids reveals a link between hybrid lethality and female sterility. Genetics 163, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash D. A., Roote J., Ashburner M. (2000). The Drosophila melanogaster hybrid male rescue gene causes inviability in male and female species hybrids. Genetics 154, 1747–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby A. C., Litchfield D. W. (2005). The multiple personalities of the regulatory subunit of protein kinase CK2: CK2 dependent and CK2 independent roles reveal a secret identity for CK2beta. Int. J. Biol. Sci. 1, 67–79. 10.7150/ijbs.1.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonet C., Fernández I., Aran X., Bernués J., Giralt E., Azorín F. (2005). The GAGA protein of Drosophila is phosphorylated by CK2. J. Mol. Biol. 351, 562–572. 10.1016/j.jmb.2005.06.039 [DOI] [PubMed] [Google Scholar]
- Bouazoune K., Brehm A. (2005). dMi-2 chromatin binding and remodeling activities are regulated by dCK2 phosphorylation. J. Biol. Chem. 280, 41912–41920. 10.1074/jbc.M507084200 [DOI] [PubMed] [Google Scholar]
- Bozzetti M. P., Massari S., Finelli P., Meggio F., Pinna L. A., Boldyreff B., et al. (1995). The Ste locus, a component of the parasitic cry-Ste system of Drosophila melanogaster, encodes a protein that forms crystals in primary spermatocytes and mimics properties of the α-subunit of casein kinase 2. Proc. Natl. Acad. Sci. U. S. A. 92, 6067–6071. 10.1073/pnas.92.13.6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., et al. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., Sachidanandam R., et al. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103. 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- Castillo D. M., Barbash D. A. (2017). Moving speciation genetics forward: modern techniques build on foundational studies in Drosophila. Genetics 207, 825–842. 10.1534/genetics.116.187120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M., Chang C., Khost D. E., Vedanayagam J., Adrion J. R., Liao Y., et al. (2020). Evolution of genome structure in the Drosophila simulans species complex. bioRxiv [Preprint]. 10.1101/2020.02.27.968743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. H., Larracuente A. M. (2019). Heterochromatin-enriched assemblies reveal the sequence and organization of the Drosophila melanogaster Y chromosome. Genetics 211, 333–348. 10.1534/genetics.118.301765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. (2001). Genome analysis: more Drosophila Y chromosome genes. Curr. Biol. 11, R182–R184. 10.1016/S0960-9822(01)00089-6 [DOI] [PubMed] [Google Scholar]
- Chen P., Kotov A. A., Godneeva B. K., Bazylev S. S., Olenina L. V., Aravin A. A. (2020). piRNA-mediated gene regulation and adaptation to sex-specific transposon expression in D. melanogaster male germline. bioRxiv [Preprint]. 10.1101/2020.08.25.266585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant G. C., Wolfe K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. 10.1038/nrg2482 [DOI] [PubMed] [Google Scholar]
- Czech B., Hannon G. J. (2016). One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem. Sci. 41, 324–337. 10.1016/j.tibs.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya O. N., Kurenova E. V., Pavlova M. N., Bebehov D. V., Link A. J., Koga A., et al. (1991). He-T family DNA sequences in the Y chromosome of Drosophila melanogaster share homology with the X-linked stellate genes. Chromosoma 100, 118–124. 10.1007/BF00418245 [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. (1936). Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21, 113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorova K. S., Olenkina O. M., Kibanov M. V., Kalmykova A. I., Gvozdev V. A., Olenina L. V. (2009). Genetically derepressed nucleoplasmic stellate protein in spermatocytes of D. melanogaster interacts with the catalytic subunit of protein kinase 2 and carries histone-like lysine-methylated mark. J. Mol. Biol. 389, 895–906. 10.1016/j.jmb.2009.04.064 [DOI] [PubMed] [Google Scholar]
- Findley S. D., Tamanaha M., Clegg N. J., Ruohola-Baker H. (2003). Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 130, 859–871. 10.1242/dev.00310 [DOI] [PubMed] [Google Scholar]
- Fischle W., Tseng B. S., Dormann H. L., Ueberheide B. M., Garcia B. A., et al. (2005). Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122. 10.1038/nature04219 [DOI] [PubMed] [Google Scholar]
- Gainetdinov I., Colpan C., Arif A., Cecchini K., Zamore P. D. (2018). A single mechanism of biogenesis, initiated and directed by PIWI proteins, explains piRNA production in most animals. Mol. Cell 71, 775–790. 10.1016/j.molcel.2018.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty W., Singh R. S. (2006). Gene regulation divergence is a major contributor to the evolution of Dobzhansky-Muller incompatibilities between species of Drosophila. Mol. Biol. Evol. 23, 1707–1714. 10.1093/molbev/msl033 [DOI] [PubMed] [Google Scholar]
- Han B. W., Wang W., Li C., Weng Z., Zamore P. D. (2015). Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348, 817–821. 10.1126/science.aaa1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R. W., Lindsley D. L., Livak K. J., Lewis B., Siversten A. V., Joslyn G. L., et al. (1984). Cytogenetic analysis of a segment of the Y chromosome of Drosophila melanogaster. Genetics 107, 591–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R. A., Smith C. D., Carlson J. W., Carvalho A. B., Halpern A., Kaminker J. S., et al. (2002). Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 3:RESEARCH0085. 10.1186/gb-2002-3-12-research0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Fejes Tóth K., Aravin A. A. (2017). piRNA biogenesis in Drosophila melanogaster. Trends Genet. 33, 882–894. 10.1016/j.tig.2017.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A., Hyle J., Lechner M. S., Lahti J. M. (2008). Perturbation of HP1 localization and chromatin binding ability causes defects in sister-chromatid cohesion. Mutat. Res. 657, 48–55. 10.1016/j.mrgentox.2008.08.010 [DOI] [PubMed] [Google Scholar]
- Kalmykova A. I., Nurminsky D. I., Ryzhov D. V., Shevelyov Y. Y. (2005). Regulated chromatin domain comprising cluster of co-expressed genes in Drosophila melanogaster. Nucleic Acids Res. 33, 1435–1444. 10.1093/nar/gki281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova A. I., Shevelyov Y. Y., Dobritsa A. A., Gvozdev V. A. (1997). Acquisition and amplification of a testis-expressed autosomal gene, SSL, by the Drosophila Y chromosome. Proc. Natl. Acad. Sci. U. S. A. 94, 6297–6302. 10.1073/pnas.94.12.6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova A. I., Shevelyov Y. Y., Polesskaya O. O., Dobritsa A. A., Evstafieva A. G., Boldyreff B., et al. (2002). CK2(beta)tes gene encodes a testis-specific isoform of the regulatory subunit of casein kinase 2 in Drosophila melanogaster. Eur. J. Biochem. 269, 1418–1427. 10.1046/j.1432-1033.2002.02785.x [DOI] [PubMed] [Google Scholar]
- Karandikar U., Anderson S., Mason N., Trott R. L., Bishop C. P., Bidwai A. P. (2003). The Drosophila SSL gene is expressed in larvae, pupae, and adults, exhibits sexual dimorphism, and mimics properties of the beta subunit of casein kinase II. Biochem. Biophys. Res. Commun. 301, 941–947. 10.1016/S0006-291X(03)00073-1 [DOI] [PubMed] [Google Scholar]
- Kibanov M. V., Egorova K. S., Ryazansky S. S., Sokolova O. A., Kotov A. A., Olenkina O. M., et al. (2011). A novel organelle, the piNG-body, in the nuage of Drosophila male germ cells is associated with piRNA-mediated gene silencing. Mol. Biol. Cell. 22, 3410–3419. 10.1091/mbc.e11-02-0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibanov M. V., Kotov A. A., Olenina L. V. (2013). Multicolor fluorescence imaging of whole-mount Drosophila testes for studying spermatogenesis. Anal. Biochem. 436, 55–64. 10.1016/j.ab.2013.01.009 [DOI] [PubMed] [Google Scholar]
- Kirino Y., Kim N., de Planell-Saguer M., Khandros E., Chiorean S., Klein P. S., et al. (2009). Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat. Cell Biol. 11, 652–658. 10.1038/ncb1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan G. L., Epstein V. N., Aravin A. A., Gvozdev V. A. (2000). Molecular evolution of two paralogous tandemly repeated heterochromatic gene clusters linked to the X and Y chromosomes of Drosophila melanogaster. Mol. Biol. Evol. 17, 697–702. 10.1093/oxfordjournals.molbev.a026348 [DOI] [PubMed] [Google Scholar]
- Kogan G. L., Usakin L. A., Ryazansky S. S., Gvozdev V. A. (2012). Expansion and evolution of the X-linked testis specific multigene families in the melanogaster species subgroup. PLoS ONE 7:e37738. 10.1371/journal.pone.0037738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotelnikov R. N., Klenov M. S., Rozovsky Y. M., Olenina L. V., Kibanov M. V., Gvozdev V. A. (2009). Peculiarities of piRNA-mediated post-transcriptional silencing of Stellate repeats in testes of Drosophila melanogaster. Nucleic Acids Res. 37, 3254–3263. 10.1093/nar/gkp167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotov A. A., Adashev V. E., Godneeva B. K., Ninova M., Shatskikh A. S., Bazylev S. S., et al. (2019). piRNA silencing contributes to interspecies hybrid sterility and reproductive isolation in Drosophila melanogaster. Nucleic Acids Res. 47, 4255–4271. 10.1093/nar/gkz130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128, 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Le Thomas A., Stuwe E., Li S., Du J., Marinov G., Rozhkov N., et al. (2014). Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 28, 1667–1680. 10.1101/gad.245514.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M. S., Schultz D. C., Negorev D., Maul G. G., Rauscher F. J., III. (2005). The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem. Biophys. Res. Commun. 331, 929–937. 10.1016/j.bbrc.2005.04.016 [DOI] [PubMed] [Google Scholar]
- Ledent V., Vervoort M. (2001). The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 11, 754–770. 10.1101/gr.177001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Vagin V. V., Lee S., Xu J., Ma S., Xi H., et al. (2009). Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137, 509–521. 10.1016/j.cell.2009.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschytz E., Hareven D. (1977). Gene expression and the control of spermatid morphogenesis in Drosophila melanogaster. Dev. Biol. 58, 276–294. 10.1016/0012-1606(77)90092-6 [DOI] [PubMed] [Google Scholar]
- Lim A. K., Kai T. (2007). Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 104, 6714–6719. 10.1073/pnas.0701920104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A. K., Tao L., Kai T. (2009). piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J. Cell Biol. 186, 333–342. 10.1083/jcb.200904063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. (1984). Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics 107, 611–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. (1990). Detailed structure of the Drosophila melanogaster Stellate genes and their transcripts. Genetics 124, 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyckegaard E. M., Clark A. G. (1989). Ribosomal DNA and Stellate gene copy number variation on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 86, 1944–1948. 10.1073/pnas.86.6.1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C. D., Brennecke J., Dus M., Stark A., Mccombie W. R., Sachidanandam R., et al. (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535. 10.1016/j.cell.2009.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee B. D., Satter M. T. (1996). Structure of the Y chromosomal Su(Ste) locus in Drosophila melanogaster and evidence for localized recombination among repeats. Genetics 142, 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G. F., Hess O., Beermann W. (1961). Phase-specific function structure in spermatocyte nuclei of Drosophila melanogaster and their dependence of Y chromosomes. Chromosoma 12, 676–716. 10.1007/BF00328946 [DOI] [PubMed] [Google Scholar]
- Michalak P., Noor M. A. (2003). Genome-wide patterns of expression in Drosophila pure species and hybrid males. Mol. Biol. Evol. 20, 1070–1076. 10.1093/molbev/msg119 [DOI] [PubMed] [Google Scholar]
- Moehring A. J., Teeter K. C., Noor M. A. (2007). Genome-wide patterns of expression in Drosophila pure species and hybrid males. II. Examination of multiple-species hybridizations, platforms, and life cycle stages. Mol. Biol. Evol. 24, 137–145. 10.1093/molbev/msl142 [DOI] [PubMed] [Google Scholar]
- Mohn F., Handler D., Brennecke J. (2015). Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348, 812–817. 10.1126/science.aaa1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J. (1940). Bearings of the Drosophila work on systematics, in The New Systematics, ed J. Huxley (Oxford: Clarendon Press; ), 185–268. [Google Scholar]
- Muller H. J., Pontecorvo G. (1942). Recessive genes causing interspecific sterility and other disharmonies between Drosophila melanogaster and simulans. Genetics 27:157. [Google Scholar]
- Ninova M., Fejes Tóth K., Aravin A. A. (2019). The control of gene expression and cell identity by H3K9 trimethylation. Development 146:dev181180. 10.1242/dev.181180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K. M., Okada T. N., Kawamura T., Mituyama T., Kawamura Y., Inagaki S., et al. (2009). Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 28, 3820–3831. 10.1038/emboj.2009.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K. M., Saito K., Mori T., Kawamura Y., Nagami-Okada T., Inagaki S., et al. (2007). Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13, 1911–1922. 10.1261/rna.744307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosov G. A., Kibanov M. V., Olenina L. V. (2014). Dynamic properties of germinal granule ping-body in the testes of Drosophila melanogaster. Mol. Biol. 48, 805–813. 10.1134/S0026893314050112 [DOI] [PubMed] [Google Scholar]
- Nott T. J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., et al. (2015). Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947. 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olenkina O. M., Egorova K. S., Aravin A. A., Naumova N. M., Gvozdev V. A., Olenina L. V. (2012b). Mapping of cis-regulatory sites in the promoter of testis-specific Stellate genes of Drosophila melanogaster. Biochem. Mosc. 77, 1285–1293. 10.1134/S0006297912110077 [DOI] [PubMed] [Google Scholar]
- Olenkina O. M., Egorova K. S., Kibanov M. V., Gervaziev Y. V., Gvozdev V. A., Olenina L. V. (2012a). Promoter contribution to the testis-specific expression of Stellate gene family in Drosophila melanogaster. Gene 499, 143–153. 10.1016/j.gene.2012.03.023 [DOI] [PubMed] [Google Scholar]
- Orr H. A. (2005). The genetic basis of reproductive isolation: insights from Drosophila. Proc. Natl. Acad. Sci. U. S. A. 102, 6522–6526. 10.1073/pnas.0501893102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata D. M., Gainetdinov I., Zoch A., O'Carroll D., Zamore P. D. (2019). PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 20, 89–108. 10.1038/s41576-018-0073-3 [DOI] [PubMed] [Google Scholar]
- Özata D. M., Yu T., Mou H., Gainetdinov I., Colpan C., Cecchini K., et al. (2020). Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat. Ecol. Evol. 4, 156–168. 10.1038/s41559-019-1065-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo G., Bonaccorsi S., Robbins L. G., Pimpinelli S. (1994). Genetic analysis of Stellate elements of Drosophila melanogaster. Genetics 138, 1181–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A., Wehr K., Schüpbach T. (2007). zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell 12, 851–862. 10.1016/j.devcel.2007.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil V. S., Kai T. (2010). Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr. Biol. 20, 724–730. 10.1016/j.cub.2010.02.046 [DOI] [PubMed] [Google Scholar]
- Pinna L. A. (2002). Protein kinase CK2: a challenge to canons. J. Cell Sci. 115, 3873–3878. 10.1242/jcs.00074 [DOI] [PubMed] [Google Scholar]
- Rathert P., Dhayalan A., Murakami M., Zhang X., Tamas R., Jurkowska R., et al. (2008). Protein lysine methyltransferase G9a acts on non-histone targets. Nat. Chem. Biol. 4, 344–346. 10.1038/nchembio.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Ríos P., Chartier A., Pierson S., Simonelig M. (2017). Aubergine and piRNAs promote germline stem cell self-renewal by repressing the proto-oncogene Cbl. EMBO J. 36, 3194–3211. 10.15252/embj.201797259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Ríos P., Simonelig M. (2018). piRNAs and PIWI proteins: regulators of gene expression in development and stem cells. Development 145:dev161786. 10.1242/dev.161786 [DOI] [PubMed] [Google Scholar]
- Russo C. A., Takezaki N., Nei M. (1995). Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 12, 391–404. [DOI] [PubMed] [Google Scholar]
- Ryazansky S. S., Kotov A. A., Kibanov M. V., Akulenko N. V., Korbut A. P., Lavrov S. A., et al. (2016). RNA helicase Spn-E is required to maintain Aub and AGO3 protein levels for piRNA silencing in the germline of Drosophila. Eur. J. Cell Biol. 95, 311–322. 10.1016/j.ejcb.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Schotta G., Ebert A., Krauss V., Fischer A., Hoffmann J., Rea S., et al. (2002). Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21, 1121–1131. 10.1093/emboj/21.5.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D. C., Ayyanathan K., Negorev D., Maul G. G., Rauscher F. J., III. (2002). SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16, 919–932. 10.1101/gad.973302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti K. A., Jurczak D., Sachidanandam R., Brennecke J. (2015). piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev. 29, 1747–1762. 10.1101/gad.267252.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevelyov Y. Y. (1992). Copies of a Stellate gene variant are located in the X heterochromatin of Drosophila melanogaster and are probably expressed. Genetics 132, 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snee M. J., Macdonald P. M. (2004). Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J. Cell Sci. 117, 2109–2120. 10.1242/jcs.01059 [DOI] [PubMed] [Google Scholar]
- Tachibana M., Sugimoto K., Fukushima T., Shinkai Y. (2001). SET Domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276, 25309–25317. 10.1074/jbc.M101914200 [DOI] [PubMed] [Google Scholar]
- Tamura K., Subramanian S., Kumar S. (2004). Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21, 36–44. 10.1093/molbev/msg236 [DOI] [PubMed] [Google Scholar]
- Tulin A. V., Kogan G. L., Filipp D., Balakireva M. D., Gvozdev V. A. (1997). Heterochromatic Stellate gene cluster in Drosophila melanogaster: structure and molecular evolution. Genetics 146, 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usakin L. A., Kogan G. L., Kalmykova A. I., Gvozdev V. A. (2005). An alien promoter capture as a primary step of the evolution of testes-expressed repeats in the Drosophila melanogaster genome. Mol. Biol. Evol. 22, 1555–1560. 10.1093/molbev/msi147 [DOI] [PubMed] [Google Scholar]
- Vagin V. V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P. D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324. 10.1126/science.1129333 [DOI] [PubMed] [Google Scholar]
- Vermaak D., Malik H. S. (2009). Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu. Rev. Genet. 43, 467–492. 10.1146/annurev-genet-102108-134802 [DOI] [PubMed] [Google Scholar]
- Wang W., Han B. W., Tipping C., Ge D. T., Zhang Z., Weng Z., et al. (2015). Slicing and binding by Ago3 or Aub trigger piwi-bound piRNA production by distinct mechanisms. Mol. Cell 59, 819–830. 10.1016/j.molcel.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster A., Li S., Hur J. K., Wachsmuth M., Bois J. S., Perkins E. M., et al. (2015). Aub and Ago3 are recruited to nuage through two mechanisms to form a ping-pong complex assembled by Krimper. Mol. Cell 59, 564–575. 10.1016/j.molcel.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K. H., Clark A. G., Barbash D. A. (2014). Limited gene misregulation is exacerbated by allele-specific upregulation in lethal hybrids between Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 31, 1767–1778. 10.1093/molbev/msu127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J., Spinelli P., Laussmann M. A., Homolka D., Yang Z., Cora E., et al. (2014). RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157, 1698–1711. 10.1016/j.cell.2014.05.018 [DOI] [PubMed] [Google Scholar]
- Zhang F., Wang J., Xu J., Zhang Z., Koppetsch B. S., Schultz N., et al. (2012). UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 151, 871–884. 10.1016/j.cell.2012.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Xu J., Koppetsch B. S., Wang J., Tipping C., Ma S., et al. (2011). Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol. Cell 44, 572–584. 10.1016/j.molcel.2011.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Eissenberg J. C. (1999). Phosphorylation of heterochromatin protein 1 by casein kinase II is required for efficient heterochromatin binding in Drosophila. J. Biol. Chem. 274, 15095–15100. 10.1074/jbc.274.21.15095 [DOI] [PubMed] [Google Scholar]