Abstract
TERT gene promoter mutations are known in multiple cancer types. Other TERT alterations remain poorly characterized. Sequencing data from 30,773 tumors analyzed by a hybridization capture next-generation sequencing assay (Memorial Sloan Kettering Cancer Center Integrated Mutation Profiling of Actionable Cancer Targets) were analyzed for the presence of TERT alterations. Promoter rearrangements (500 bases upstream of the transcriptional start site), hypermethylation (n = 57), and gene expression (n = 155) were evaluated for a subset of cases. Mutually exclusive and recurrent promoter mutations were identified at three hot spots upstream of the transcriptional start site in 11.3% of cases (−124: 74%; −146: 24%; and −138: <2%). Mutually exclusive amplification events were identified in another 2.3% of cases, whereas mutually exclusive rearrangements proximal to the TERT gene were seen in 24 cases. The highest incidence of TERT promoter mutations was seen in cutaneous melanoma (82%), whereas amplification events significantly outnumbered promoter mutations in well-differentiated/dedifferentiated liposarcoma (14.1% versus 2.4%) and adrenocortical carcinoma (13.6% versus 4.5%). Gene expression analysis suggests that the highest levels of gene expression are seen in cases with amplifications and rearrangements. Hypermethylation events upstream of the TERT coding sequence were not mutually exclusive with known pathogenic alterations. Studies aimed at defining the prevalence and prognostic impact of TERT alterations should incorporate other pathogenic TERT alterations as these may impact telomerase function.
Human telomeres are composed of telomeric TTAGGG DNA repeats that are protected by the shelterin complex and the telomerase complex.1,2 Unlike stem cells, in most differentiated cells, silencing of telomerase reverse transcriptase (TERT) occurs over time with consequent telomeric shortening.1 This leads to activation of DNA damage response signaling and replicative arrest.1 An early event in many neoplasms involves bypassing this replicative arrest through telomerase reactivation, thereby increasing both cancer cell viability and genomic instability.1 The most common mechanism of telomerase reactivation across several cancer types involves TERT reactivation through hot spot promoter mutations, which generates a de novo binding site for activating erythroblast transformation-specific (ETS) family transcription factors.2, 3, 4 This leads to increased recruitment of multimeric GA-binding protein transcription factors to the mutant TERT promoter sequence and transcriptional up-regulation of TERT.2,3,5
In addition to promoter mutations, the spectrum of activating TERT alterations includes genomic amplifications as well as structural alterations at the TERT locus or hypermethylation events upstream of the TERT transcriptional start site.6, 7, 8, 9, 10 The Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) assay used in this study is designed to detect TERT promoter mutations and genomic amplifications. Although a few structural variants were detected within a short 500-bp sequence immediately upstream of the transcriptional start site, the vast majority of these rearrangements (which have primarily been reported in neuroblastomas) occur over a 50-kb region proximal to the TERT gene.7, 8, 9 This assay is therefore not designed to detect these alterations when they occur further upstream of the transcription start site. Finally, hypermethylation at specific CpG islands upstream of the transcriptional start site has been correlated with increased TERT gene expression, and this assay does not document methylation status at these sites.6,10,11 Methylation status was determined for a smaller subset of cases using an alternate assay.
Relatively few large-scale pan-cancer studies have studied TERT alterations across tumor types.2,12,13 Herein, we present pan-cancer data pertaining to both promoter mutations and amplification events across 30,773 specimens that were profiled as part of an institutional clinical sequencing cohort. This data set is unique as TERT promoter mutations are often not evaluated in whole exome sequencing assays, whereas TERT amplification events are rarely documented in large pan-cancer data sets.14 In addition, a limited number of cases in this study have been profiled for rarer alterations, such as structural variants and hypermethylation events upstream of the TERT transcription start site, which have been shown to up-regulate TERT expression, as well as relative gene expression status.
Materials and Methods
Case Selection
This study was approved by the institutional review board and involved analysis of molecular profiling data of multiple solid tumors profiled by a next-generation sequencing–based assay (MSK-IMPACT), as part of an institutional clinical cancer genomics initiative.14, 15, 16 DNA sequencing results for 30,773 tumor samples obtained from formalin-fixed, paraffin-embedded tissue, performed in a Clinical Laboratory Improvement Amendments–approved setting, were analyzed for TERT alterations.
Next-Generation Sequencing: MSK-IMPACT
Details of the MSK-IMPACT assay have been previously reported.14, 15, 16 In brief, this assay involves paired analysis of tumor and normal specimens to filter out germ-line variants. Specifically, hybridization capture-based library preparation is followed by deep sequencing of select noncoding regions and 6614 protein-coding exons of 468 genes. Noncoding sequences include the TERT promoter (extending to 500 bp upstream of the transcriptional start site), intronic sequences of commonly rearranged genes, microsatellite sites, and several single-nucleotide polymorphisms. In all, the capture probes target approximately 1.5 megabases of the human genome. Accurate genome-wide copy number assessment is facilitated by homogeneous distribution of single-nucleotide polymorphism tiling probes across the genome. On the basis of previously reported criteria, amplifications were defined as a fold change ≥2.0.17, 18, 19, 20 Rearrangements involving the TERT promoter (500-bp sequence immediately upstream of the transcriptional start site) were identified on the basis of previously published criteria, which included the following: five paired or split reads, a mapping quality of 20, and a length >500-bp sequence.15 This assay is currently approved by the US Food and Drug Administration as a class II in vitro diagnostic test.
TERT Gene Expression Analysis
Details pertaining to RNA extraction and the MSK-Fusion assay have been previously reported.21,22 Eight gene-specific primers were designed by ArcherDx (Boulder, CO) and added to the MSK-Fusion panel to specifically detect TERT expression levels. The total number of unique RNA reads for TERT, obtained using all eight gene-specific primers, were normalized to the total number of unique RNA reads for five separate housekeeping genes interrogated as part of the assay. The relative TERT gene expression status was evaluated for 155 cases that were profiled using MSK-IMPACT from available archived formalin-fixed, paraffin-embedded tissue. Broad categories of tumor types evaluated included colorectal adenocarcinoma (n = 35), lung adenocarcinoma (n = 25), sarcoma (n = 20), gliomas (n = 19), breast carcinoma (n = 14), melanoma (n = 12), pancreaticobiliary cancer (n = 10), salivary cancer (n = 4), thyroid carcinoma (n = 4), carcinoma of unknown primary (n = 4), and miscellaneous tumor types (n = 8).
TERT Promoter Methylation Analysis
TERT promoter hypermethylation status was detected via bisulfite conversion followed by methylation array for 57 cases profiled by MSK-IMPACT, as previously described.23 Herein, methylation status of three CpG sites upstream of the TERT transcription start site was evaluated. This included the cg11625005 CpG site, where hypermethylation has been previously reported to be associated with high-grade tumors and increased TERT gene expression.6,10,11
Statistical Analysis
All statistical tests were two sided, and P < 0.05 was considered statistically significant.
Results
TERT Promoter Mutation and Amplification
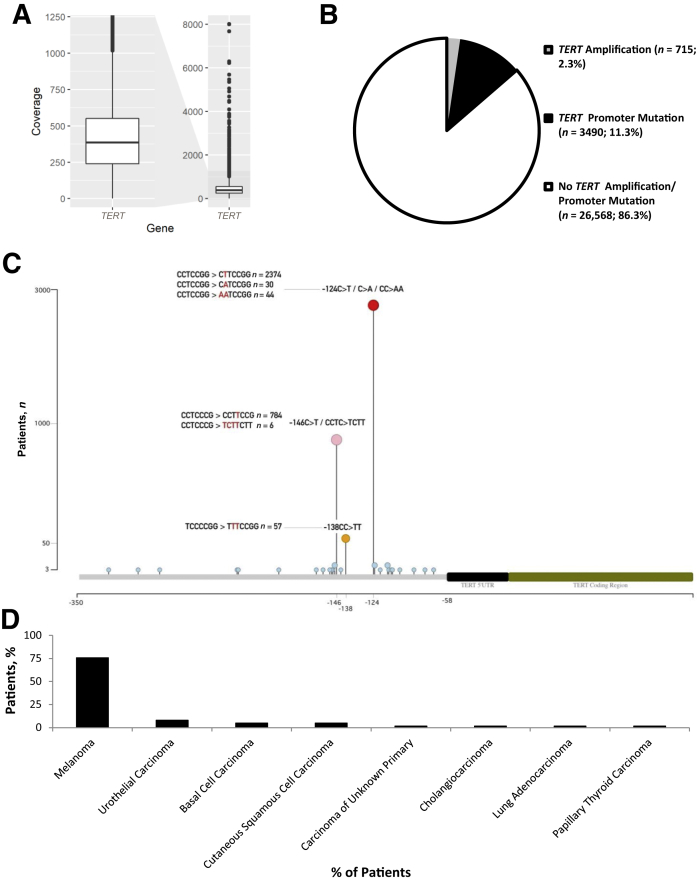
A hybrid capture library preparation strategy was used to sequence the TERT promoter region, including 500 bases upstream of the transcriptional start site. The mean depth of coverage for the promoter region across all 30,773 specimens that were profiled was 425×, with an interquartile range of 240× to 551× (Figure 1A). In all, 4205 of these specimens (13.6%) harbored either a TERT promoter mutation or an amplification. Of note, TERT promoter mutations (11.3%) and amplifications (2.3%) were mutually exclusive, with the promoter mutations occurring 4.9 times as frequently compared with amplification events (Figure 1B).
Figure 1.
Next-generation sequencing (MSK-IMPACT) for TERT alterations. A: A box plot depicting the mean depth of coverage for the TERT promoter region (500 bp upstream of the transcription start site) for 30,773 specimens is depicted. The y axis demonstrates the depth of coverage. The range of coverage across 30,773 specimens is depicted on the right side of the figure, whereas the left side of the figure shows a magnified area of the box plot that highlights the interquartile range. B: The frequency of TERT genomic amplifications at the 5p15.33 locus and promoter mutations, both mutually exclusive events, for all cases that were profiled is shown. C: The spatial distribution of TERT promoter mutations relative to the transcription start site is depicted. Specific nucleotide changes that contribute to presumptive ETS transcription factor binding sites are highlighted for three mutational hot spots (−124, −146, and −138 bases upstream of the transcriptional start site). Variant nucleotide sequences are highlighted in red. D: Furthermore, tumor types that exhibit TERT promoter mutations at the −138 position are shown.
Although TERT promoter mutations showed a wide spatial distribution, consistent with our prior studies, three recurring hot spots that were mutually exclusive were identified.14 Relative to the transcription start site, hot spot promoter alterations at position −124 were the most common (n = 2448; 74%), followed by those at positions −146 (n = 790; 24%) and −138 (n = 57; <2%) (Figure 1C). Although TERT promoter mutations at the −138 position were seen in varied tumor types, they predominantly occurred in melanoma (75.8% of cases) (Figure 1D). Consistent with prior reports, G-to-A substitutions at these sites led to the formation of presumptive ETS transcription factor binding sites, including at position −138.14
Relative Frequency of TERT Alterations in Multiple Tumor Types
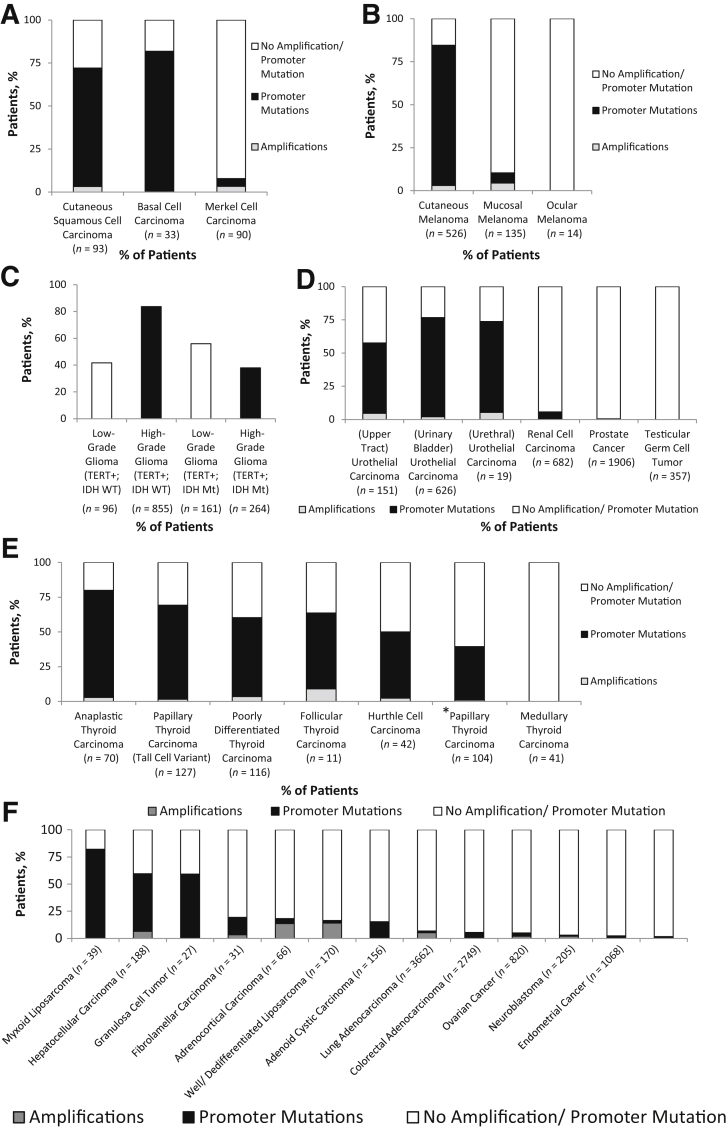
Solid tumor types with high rates of TERT alterations (in combined primary and metastatic tumors) have been highlighted (Figure 2). Cutaneous neoplasia, such as squamous cell carcinoma, basal cell carcinoma, and cutaneous melanoma, primarily harbored TERT hot spot promoter mutations in association with a UV-induced mutational signature (ie, predominant G-to-A and C-to-T mutations), whereas amplifications were relatively rare (Figure 2, A and B). In contrast, similar alterations were infrequently identified in Merkel cell carcinoma and mucosal/ocular melanoma. Consistent with prior reports, TERT hot spot mutations in central nervous system glial neoplasia predominantly occurred in an IDH1 wild-type setting in high-grade gliomas (84% of IDH1 wild-type cases) compared with low-grade gliomas, where they tended to occur in association with IDH1 mutations (56% of IDH1 mutant cases) (Figure 2C).24, 25, 26 Among genitourinary malignancies, the highest incidence was seen for urothelial carcinoma, with a relatively lower frequency observed for upper tract disease (53% versus 74%) (Figure 2D). As previously reported, TERT promoter mutations showed a strong association with the underlying subtype in thyroid carcinoma (Figure 2E).13,27, 28, 29, 30, 31, 32, 33 No alterations were identified in medullary thyroid carcinoma, and up to 77% of anaplastic thyroid carcinomas harbored these alterations. Interestingly, although a lower frequency was seen for papillary thyroid carcinoma (including classic and follicular variants; 38%), the aggressive tall cell variant showed a much higher frequency (68%). As expected, other tumor types with a high frequency of TERT promoter mutations included myxoid liposarcoma, hepatocellular carcinoma, and granulosa cell tumors (Figure 2F).34, 35, 36 In contrast, tumor types where TERT amplification was the prevalent alteration included well-differentiated/dedifferentiated liposarcoma (14.1%) and adrenocortical carcinoma (13.6%) (Figure 2F).37,38
Figure 2.
Relative frequency of TERT alterations in multiple tumor types. A–F: The relative frequency of TERT promoter mutations and amplifications in nonmelanoma skin cancer (A), melanoma (B), gliomas (C), genitourinary neoplasia, including urothelial carcinoma (D), thyroid carcinoma (E), and miscellaneous tumor types (F) is depicted. C:TERT alterations in high- and low-grade gliomas are further stratified on the basis of IDH1 mutation status. The papillary thyroid carcinoma category includes all well-differentiated subtypes and excludes the tall cell variant. The asterisk indicates papillary thyroid carcinoma excluding tall cell variant. IDH Mt, IDH1 mutant; IDH WT, IDH1 wild type; TERT+, TERT alteration present.
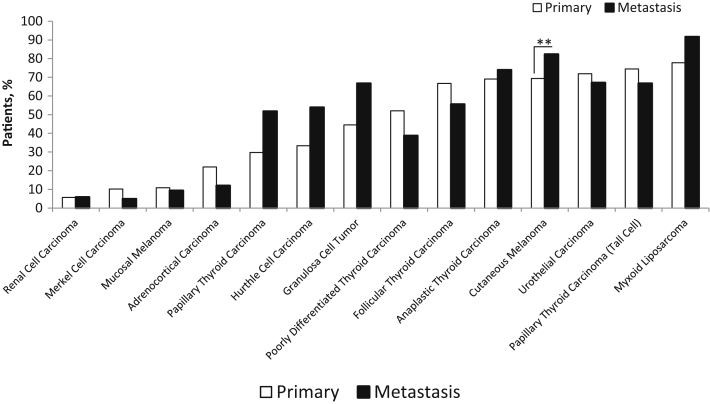
Of note, high rates of TERT alterations were documented in the primary tumors themselves for aggressive variants of thyroid cancer, such as poorly differentiated (n = 73; 52%), anaplastic (n = 55; 69%), and tall cell variant of papillary thyroid cancer (n = 43; 74%), as is depicted in Figure 3. In contrast, these alterations were predominantly documented in metastatic/recurrent tumors for classic well-differentiated papillary thyroid cancer (n = 58; 52%) and Hurthle cell carcinoma (n = 26; 54%).
Figure 3.
Relative distribution of TERT alterations in primary compared with metastatic/recurrent tumors. TERT promoter mutations and amplifications combined, for specific tumor types, are depicted for primary compared with metastatic/recurrent tumors. ∗∗P < 5 × 10−3.
TERT Promoter Rearrangements
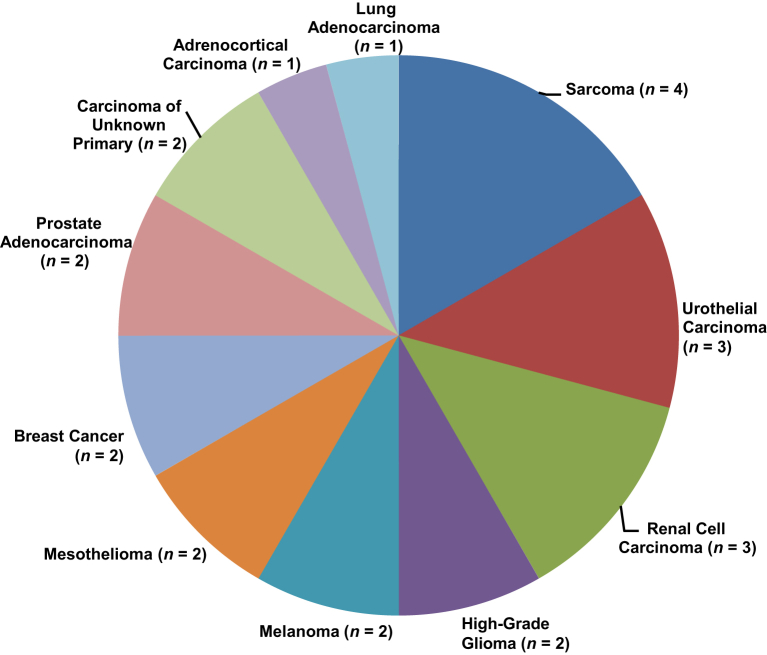
Prior studies have identified recurrent genomic rearrangements at the TERT locus, upstream of the TERT coding sequence, primarily in high-risk neuroblastoma.7,8 These cases showed substantial induction of TERT gene expression and downstream increases in telomerase activity, likely secondary to the juxtaposition of strong enhancer elements adjacent to the TERT coding sequence.7,8 Although subsequent studies have confirmed the presence of TERT promoter rearrangements, these remain challenging to identify as these rearrangements often occur over a 50-kb region proximal of the TERT gene.9,11,39,40 Although the MSK-IMPACT assay only tiles for 500 bases of this 50-kb region, 24 cases occurring in multiple tumor types were found to harbor similar rearrangements upstream of the TERT coding sequence (Figure 4). No recurrent break point or partner gene was identified for these cases. In addition, these events were mutually exclusive with TERT hot spot promoter mutations/genomic amplifications.
Figure 4.
TERT promoter rearrangements. Diverse tumor types for which TERT promoter rearrangements were identified within 500 bp immediately upstream of the transcriptional start site are shown.
Relative TERT Gene Expression
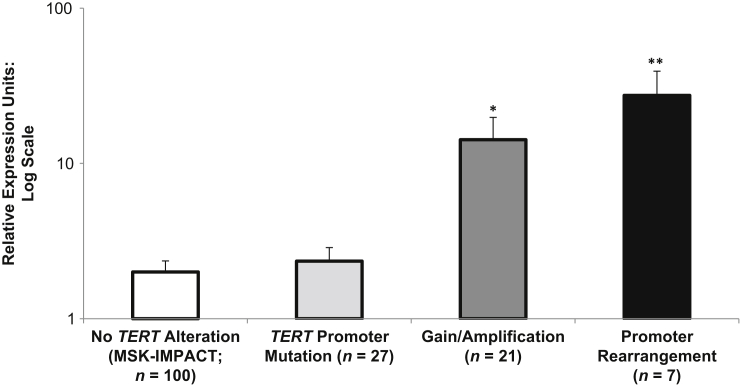
Herein, 155 specimens of archived formalin-fixed, paraffin-embedded specimens that were initially evaluated using MSK-IMPACT were retrieved and further interrogated for TERT mRNA expression using the MSK-Fusion assay. Unique TERT-specific RNA reads were normalized to the expression of multiple housekeeping genes and quantified. This included cases that lacked TERT alterations or harbored amplifications, hot spot promoter mutations, or rearrangements. Interestingly, cases with genomic amplifications and promoter rearrangements showed statistically significant increases in TERT gene expression, in contrast to cases with hot spot promoter mutations (Figure 5).
Figure 5.
Relative TERT gene expression. Relative TERT gene expression from archived formalin-fixed, paraffin-embedded material for 155 cases is depicted. n = 100 for no TERT alteration on the basis of Memorial Sloan Kettering Cancer Center Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT); n = 27 for TERT promoter mutation; n = 21 for TERT genomic amplification; n = 7 for TERT promoter rearrangement. ∗P < 5 × 10−5, ∗∗P < 5 × 10−9.
Although multiple studies have attempted to demonstrate TERT protein expression using immunohistochemistry, this has historically been controversial. Such efforts have been hampered by poor reproducibility, unexpected patterns of subcellular localization, as well as documented cross-reactivity with other proteins.41,42 Validation of such an immunohistochemical assay would require establishing a baseline status for all cases that was inclusive of not only promoter mutations, but also amplifications, upstream rearrangements, and methylation events to correlate protein expression with underlying tumor biology. Although multiple antibodies were to be validated, the validation failed because of a lack of consistency with the underlying prediction of TERT expression status and unexpected patterns of protein localization (data not shown).
TERT Promoter Hypermethylation Status
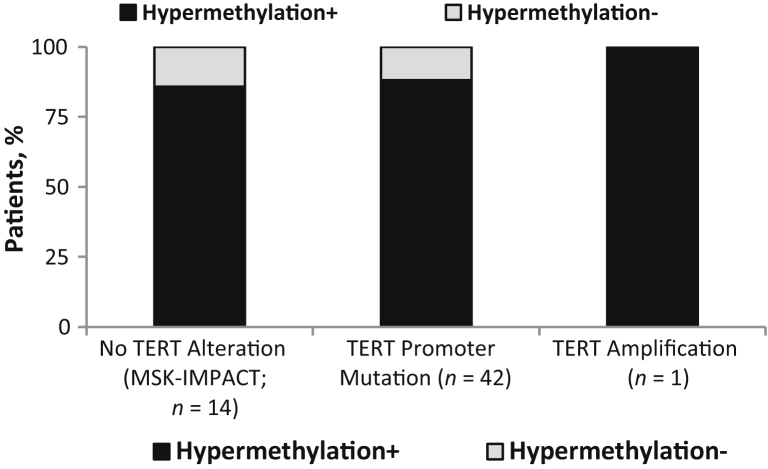
Recent studies have suggested that hypermethylation upstream of the TERT transcriptional start site is required for TERT gene expression and corresponding telomerase activity in neoplastic cells compared with nonneoplastic cells.6,9, 10, 11 At least one study has suggested that these hypermethylation events are not mutually exclusive with either TERT promoter mutations or rearrangements.11 Although cases with hypermethylation were not evaluated for relative gene expression, 57 cases of high-grade glioma profiled by MSK-IMPACT were further evaluated for methylation status upstream of the TERT gene. This was performed to determine if hypermethylation events were mutually exclusive with activating alterations, such as hot spot promoter mutations and amplifications. Our results suggest that these are not mutually exclusive events as 37 of 42 (88%) of cases with promoter mutations also exhibited hypermethylation upstream of the transcriptional start site (Figure 6).
Figure 6.
TERT promoter hypermethylation status. TERT promoter hypermethylation upstream of the transcriptional start site, including at the cg11625005 CpG site, was evaluated for 57 cases profiled by Memorial Sloan Kettering Cancer Center Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) to assess for mutual exclusivity of hypermethylation with cases harboring promoter mutations and amplification. n = 42 cases harboring promoter mutations; n = 1 case harboring amplification.
Discussion
Telomerase reactivation in cancer is often driven by TERT expression, and the predominant underlying alteration involves promoter mutations that generate a de novo binding site for activating ETS family transcription factors that lead to transcriptional up-regulation.1, 2, 3, 4, 5 Other alterations that can up-regulate TERT expression include genomic amplification and rearrangements or hypermethylation events upstream of the TERT transcriptional start site.6, 7, 8, 9, 10
The mean coverage for the TERT promoter region across all 30,773 specimens evaluated by MSK-IMPACT was 425× and revealed the presence of TERT alterations in 13.7% of all cases. Promoter mutations were found to be 4.9 times more prevalent compared with amplification events. Although promoter mutations were distributed along the length of the entire promoter region that was sequenced, consistent with our prior report, three recurrent and mutually exclusive hot spots were identified.14 Among the hot spot mutations, the canonical alterations at positions −124 and −146 bases relative to the transcription start site accounted for 74% and 24% of cases, respectively. An additional noncanonical hot spot at position −138 accounted for <2% of cases, and all these alterations are predicted to generate de novo ETS transcription factor binding sites.14 Therefore, this latter category of promoter mutations (−138) might be missed in a small subset of cases in targeted assays that interrogate/report only the canonical hot spot alterations (−124 and −146). Similarly, amplification events can be missed in a much larger subset of cases. This is particularly relevant for tumor types where amplification events significantly outnumber promoter mutations, such as in well-differentiated/dedifferentiated liposarcoma (14.1% versus 2.4%) and adrenocortical carcinoma (13.6% versus 4.5%).37,38,43
TERT alterations were found to be enriched in specific tumor types, and a subset of these has been highlighted with the caveat that the institutional (tertiary referral center) clinical sequencing cohort presented herein has a bias toward patients who present with high stage and aggressive disease.
Some relevant associations include cutaneous melanomas, which showed a high rate of TERT alterations (85%) in a background of a UV-induced mutational signature, as opposed to both mucosal (10%) and ocular melanoma (0%), and these trends and frequencies are consistent with what has been previously reported in the literature.2,13,44 A similar high incidence of TERT alterations was seen in sun-exposed cutaneous squamous cell (72%) and basal cell carcinomas (82%).3
Prior studies have shown that TERT promoter mutations in high-grade gliomas occur infrequently in the background of IDH1 mutations (<3%) as opposed to low-grade gliomas (34%).26 Our results support this trend as the vast majority of TERT alterations in high-grade gliomas occurred in the IDH1 wild-type setting (n = 856; 84%) and, similarly, a large number of TERT alterations in low-grade gliomas occurred in the IDH1 mutant setting (n = 161; 56%).
Among genitourinary cancers, TERT alterations had an exceedingly low prevalence in prostatic adenocarcinoma and testicular germ cell tumors (<0.5%), followed by a low incidence in renal cell carcinoma (5.7%), as has been previously reported.2,14,45 The highest incidence was seen in urothelial carcinoma (overall: 73%). The overall frequency is consistent with published reports that suggest that on the basis of a high incidence, and the observation that TERT alterations may be an early event in urothelial carcinoma, the detection of these alterations may be exploited as a potential urothelial cancer screening tool, particularly using urine/cytology specimens.2,12,13,46, 47, 48 On the basis of our results, some variables that may confound the results of urothelial cancer screening assays that rely on the detection of TERT promoter mutations include the presence of these alterations in a subset of renal cell carcinoma (false positives), whereas the absence of TERT alterations in a subset of cases or the presence of amplification events (2.6% of cases) may contribute to false negatives.45,46,48
With regard to thyroid cancer, prior studies have shown absence of TERT promoter mutations in medullary thyroid carcinoma, a relatively low incidence in papillary carcinoma (7% to 25.5%), Hurthle cell carcinoma (13% to 33%), and follicular thyroid carcinoma (14% to 36%), and the highest incidence in poorly differentiated carcinoma (21% to 60%) and anaplastic thyroid carcinoma (13% to 73%); these studies suggest that these alterations are enriched in thyroid carcinomas that exhibit more aggressive behavior.2,13,27, 28, 29, 30, 31, 32, 33 In our series of 511 thyroid carcinomas, with many patients presenting with advanced disease, an absence of TERT promoter mutations in medullary thyroid carcinoma was confirmed, followed by progressively increasing incidence in papillary, Hurthle cell, and follicular thyroid carcinomas (39%, 48%, and 55%, respectively) compared with 57% in poorly differentiated and 77% in anaplastic thyroid carcinomas. TERT amplifications were detected in 2% of cases, which is close to the previously reported incidence of 4.9%.27 Furthermore, consistent with prior reports, our results underline the higher incidence of TERT alterations in the tall cell variant compared with classic well-differentiated papillary thyroid carcinoma (69% versus 39%).2 Among poorly differentiated, anaplastic and tall cell variants of papillary thyroid carcinoma, there was no significant difference in the frequency of TERT promoter mutations when comparisons were made between primary and metastatic tumors. This finding is consistent with their (more) aggressive biology.30,33 However, among classic well-differentiated papillary thyroid carcinoma and Hurthle cell carcinoma, TERT was more commonly mutated in metastatic/recurrent disease than in primary tumors (classic well-differentiated papillary thyroid carcinoma: 52% versus 30%; and Hurthle cell carcinoma: 54% versus 33%), suggesting that these genetic alterations may identify a relatively more aggressive subset of well-differentiated thyroid carcinomas with a tendency to metastasize/recur. However, these results must be interpreted with caution because of sampling bias (ie, the fact that the vast majority of thyroid cases subjected for testing were from patients with relatively more aggressive disease).
Other tumor types that have high reported incidences of TERT alterations include hepatocellular carcinoma (amplifications: 10%; and promoter mutations: 44%) and ovarian granulosa cell tumors (22%), whereas myxoid liposarcomas (74%) have the highest reported incidence among soft tissue tumors.34, 35, 36 Our results revealed a similar trend, revealing an overall frequency of promoter mutations/amplifications of 59%, 60%, and 82% for hepatocellular carcinoma, granulosa cell tumors, and myxoid liposarcoma, respectively. Of note, the frequency of these alterations in ovarian granulosa cell tumors has been reported to be significantly higher among recurrences, and furthermore, their presence has been correlated with poor outcomes.34 In hepatocellular carcinoma, up to 6.4% of cases harbored amplifications of TERT and are likely to be missed if they are evaluated using assays that do not evaluate copy number changes.
With regard to TERT gene expression, although a large number of studies have demonstrated the impact of TERT hot spot promoter mutations on transcriptional up-regulation in vitro, similar studies that have directly evaluated the effect of TERT promoter mutations on gene expression in tumor-derived biospecimens are limited.3 Furthermore, several studies have documented only modest increases in TERT gene expression in both in vitro studies and in tumor specimens.11,46,49 Our results confirm that cases with genomic amplifications and promoter rearrangements showed statistically significant increases in TERT gene expression and highlight the importance of identifying such cases.7, 8, 9 In contrast, relative gene expression in cases with promoter mutations did not show a statistically significant increase. This could be secondary to the age and type (archived formalin fixed and paraffin embedded) of specimen evaluated, potentially modest increases in gene expression, and/or potentially low sensitivity of the assay used. A limitation of our study involves a lack of correlation of TERT expression with downstream gene expression signatures, particularly related to ETS transcription factors.50,51 This approach was not utilized in the current study as gene expression–specific primers to perform such an analysis were not included in the MSK-Fusion assay. However, future studies may utilize such a strategy when interrogating TERT gene expression status.
Perhaps the most intriguing category of TERT alterations involves structural variants that have been previously reported to be present in neuroblastomas.7, 8, 9 Specifically, these have been reported in 23% to 31% of high-risk neuroblastomas, have exhibited a large structural diversity of TERT promoter rearrangement events, and were frequently clustered approximately 50 kb upstream of the TERT transcriptional start site.7,8 These events were reported to lead to a juxtapositioning of superenhancer elements and were associated with a massive transcriptional up-regulation of the TERT gene.7,8 Although reports of such alterations are predominantly restricted to neuroblastomas that show aggressive clinical behavior, 24 such events were identified across diverse tumor types and significantly higher TERT gene expression was reported in seven such cases where archived material was available for downstream analysis, to validate these findings.7, 8, 9 In summary, prior studies pertaining to neuroblastoma suggest that TERT promoter rearrangements are associated with high levels of TERT gene expression, and our results confirm these findings.7,8 However, future studies are needed to define the true prevalence of these alterations and their impact on tumor biology.7, 8, 9
Recent studies have shown that hypermethylation at specific CpG islands upstream of the TERT transcriptional start site has been associated with increased TERT gene expression, particularly in pediatric brain tumors and in melanoma.6,10,11 As there is a paucity of studies that have evaluated large data sets for such hypermethylation events in the context of other pathogenic alterations, 57 cases with known promoter mutation and copy number status on the basis of profiling by MSK-IMPACT were evaluated. Consistent with the results of a prior study that evaluated a limited number of cases in a similar manner, these events were not mutually exclusive with either amplification events or promoter mutations.11 However, it is possible that a subset of cases that are considered to be wild type for TERT may in fact harbor epigenetic alterations that have a significant impact on gene expression.11
Given the high prevalence of TERT alterations in cancer, there have been ongoing efforts to target components of the telomerase holoenzyme.14,52, 53, 54 Although such approaches are currently not in clinical use, they represent exciting future cancer therapy strategies. Herein, we provide a snapshot of the landscape of somatic TERT alterations across multiple cancer types. Although canonical hot spot promoter mutations (positions −124 and −146, relative to the transcription start site) are the prevalent pathogenic alteration, our data support the presence of an additional, mutually exclusive, hot spot alteration at position −138.14 Other alterations of note include genomic amplification events and rearrangements proximal to the TERT gene, which may have a significant impact on gene expression but are likely missed by targeted assays that interrogate only the conventional promoter hot spot mutations. In addition, these data suggest that upstream hypermethylation events, which have been reported to be associated with increased gene expression, are not mutually exclusive with known activating TERT alterations, and additional studies are needed to further define their role across varied cancer types. In summary, studies aimed at defining the prognostic impact of TERT alterations should attempt to incorporate other pathogenic alterations, including amplifications, upstream rearrangements, and hypermethylation events, as these might significantly impact telomerase function.
Footnotes
Supported in part through NIH/National Cancer Institute Cancer Center (NCI) under the MSK Cander Center Support Grant/Core Grant P30CA008748.
S.G. and C.M.V. contributed equally to this work.
Disclosures: None declared.
References
- 1.Maciejowski J., de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol. 2017;18:175–186. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell R.J., Rube H.T., Xavier-Magalhaes A., Costa B.M., Mancini A., Song J.S., Costello J.F. Understanding TERT promoter mutations: a common path to immortality. Mol Cancer Res. 2016;14:315–323. doi: 10.1158/1541-7786.MCR-16-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell R.J., Rube H.T., Kreig A., Mancini A., Fouse S.D., Nagarajan R.P., Choi S., Hong C., He D., Pekmezci M., Wiencke J.K., Wrensch M.R., Chang S.M., Walsh K.M., Myong S., Song J.S., Costello J.F., Cancer The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348:1036–1039. doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horikawa I., Chiang Y.J., Patterson T., Feigenbaum L., Leem S.H., Michishita E., Larionov V., Hodes R.J., Barrett J.C. Differential cis-regulation of human versus mouse TERT gene expression in vivo: identification of a human-specific repressive element. Proc Natl Acad Sci U S A. 2005;102:18437–18442. doi: 10.1073/pnas.0508964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Min J., Shay J.W. TERT promoter mutations enhance telomerase activation by long-range chromatin interactions. Cancer Discov. 2016;6:1212–1214. doi: 10.1158/2159-8290.CD-16-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castelo-Branco P., Choufani S., Mack S., Gallagher D., Zhang C., Lipman T., Zhukova N., Walker E.J., Martin D., Merino D., Wasserman J.D., Elizabeth C., Alon N., Zhang L., Hovestadt V., Kool M., Jones D.T., Zadeh G., Croul S., Hawkins C., Hitzler J., Wang J.C., Baruchel S., Dirks P.B., Malkin D., Pfister S., Taylor M.D., Weksberg R., Tabori U. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol. 2013;14:534–542. doi: 10.1016/S1470-2045(13)70110-4. [DOI] [PubMed] [Google Scholar]
- 7.Peifer M., Hertwig F., Roels F., Dreidax D., Gartlgruber M., Menon R. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valentijn L.J., Koster J., Zwijnenburg D.A., Hasselt N.E., van Sluis P., Volckmann R., van Noesel M.M., George R.E., Tytgat G.A., Molenaar J.J., Versteeg R. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015;47:1411–1414. doi: 10.1038/ng.3438. [DOI] [PubMed] [Google Scholar]
- 9.Yuan X., Larsson C., Xu D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene. 2019;38:6172–6183. doi: 10.1038/s41388-019-0872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan Y., Lee S., Wu G., Easton J., Yergeau D., Dummer R., Vogel P., Kirkwood J.M., Barnhill R.L., Pappo A., Bahrami A. Telomerase expression by aberrant methylation of the TERT promoter in melanoma arising in giant congenital nevi. J Invest Dermatol. 2016;136:339–342. doi: 10.1038/JID.2015.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seynnaeve B., Lee S., Borah S., Park Y., Pappo A., Kirkwood J.M., Bahrami A. Genetic and epigenetic alterations of TERT are associated with inferior outcome in adolescent and young adult patients with melanoma. Sci Rep. 2017;7:45704. doi: 10.1038/srep45704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Killela P.J., Reitman Z.J., Jiao Y., Bettegowda C., Agrawal N., Diaz L.A., Jr. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinagre J., Almeida A., Populo H., Batista R., Lyra J., Pinto V., Coelho R., Celestino R., Prazeres H., Lima L., Melo M., da Rocha A.G., Preto A., Castro P., Castro L., Pardal F., Lopes J.M., Santos L.L., Reis R.M., Cameselle-Teijeiro J., Sobrinho-Simoes M., Lima J., Maximo V., Soares P. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 14.Zehir A., Benayed R., Shah R.H., Syed A., Middha S., Kim H.R. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng D.T., Mitchell T.N., Zehir A., Shah R.H., Benayed R., Syed A., Chandramohan R., Liu Z.Y., Won H.H., Scott S.N., Brannon A.R., O'Reilly C., Sadowska J., Casanova J., Yannes A., Hechtman J.F., Yao J., Song W., Ross D.S., Oultache A., Dogan S., Borsu L., Hameed M., Nafa K., Arcila M.E., Ladanyi M., Berger M.F. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyman D.M., Solit D.B., Arcila M.E., Cheng D.T., Sabbatini P., Baselga J., Berger M.F., Ladanyi M. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next-generation sequencing enabling next-generation targeted therapy trials. Drug Discov Today. 2015;20:1422–1428. doi: 10.1016/j.drudis.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross D.S., Zehir A., Cheng D.T., Benayed R., Nafa K., Hechtman J.F., Janjigian Y.Y., Weigelt B., Razavi P., Hyman D.M., Baselga J., Berger M.F., Ladanyi M., Arcila M.E. Next-generation assessment of human epidermal growth factor receptor 2 (ERBB2) amplification status: clinical validation in the context of a hybrid capture-based, comprehensive solid tumor genomic profiling assay. J Mol Diagn. 2017;19:244–254. doi: 10.1016/j.jmoldx.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta S., Vanderbilt C.M., Cotzia P., Arias-Stella J.A., 3rd, Chang J.C., Zehir A., Benayed R., Nafa K., Razavi P., Hyman D.M., Baselga J., Berger M.F., Ladanyi M., Arcila M.E., Ross D.S. Next-generation sequencing-based assessment of JAK2, PD-L1, and PD-L2 copy number alterations at 9p24.1 in breast cancer: potential implications for clinical management. J Mol Diagn. 2019;21:307–317. doi: 10.1016/j.jmoldx.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta S., Cheville J.C., Jungbluth A.A., Zhang Y., Zhang L., Chen Y.B., Tickoo S.K., Fine S.W., Gopalan A., Al-Ahmadie H.A., Sirintrapun S.J., Blum K.A., Lohse C.M., Hakimi A.A., Thompson R.H., Leibovich B.C., Berger M.F., Arcila M.E., Ross D.S., Ladanyi M., Antonescu C.R., Reuter V.E. JAK2/PD-L1/PD-L2 (9p24.1) amplifications in renal cell carcinomas with sarcomatoid transformation: implications for clinical management. Mod Pathol. 2019;32:1344–1358. doi: 10.1038/s41379-019-0269-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta S., Vanderbilt C.M., Cotzia P., Arias Stella J.A., 3rd, Chang J.C., Chen Y., Tang L.H., DeLair D.F., Yao J., Ladanyi M., Ross D.S. JAK2, PD-L1, and PD-L2 (9p24.1) amplification in metastatic mucosal and cutaneous melanomas with durable response to immunotherapy. Hum Pathol. 2019;88:87–91. doi: 10.1016/j.humpath.2018.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu G., Benayed R., Ho C., Mullaney K., Sukhadia P., Rios K., Berry R., Rubin B.P., Nafa K., Wang L., Klimstra D.S., Ladanyi M., Hameed M.R. Diagnosis of known sarcoma fusions and novel fusion partners by targeted RNA sequencing with identification of a recurrent ACTB-FOSB fusion in pseudomyogenic hemangioendothelioma. Mod Pathol. 2019;32:609–620. doi: 10.1038/s41379-018-0175-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benayed R., Offin M., Mullaney K., Sukhadia P., Rios K., Desmeules P., Ptashkin R., Won H., Chang J., Halpenny D., Schram A.M., Rudin C.M., Hyman D.M., Arcila M.E., Berger M.F., Zehir A., Kris M.G., Drilon A., Ladanyi M. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res. 2019;25:4712–4722. doi: 10.1158/1078-0432.CCR-19-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cocco E., Benhamida J., Middha S., Zehir A., Mullaney K., Shia J., Yaeger R., Zhang L., Wong D., Villafania L., Nafa K., Scaltriti M., Drilon A., Saltz L., Schram A.M., Stadler Z.K., Hyman D.M., Benayed R., Ladanyi M., Hechtman J.F. Colorectal carcinomas containing hypermethylated MLH1 promoter and wild-type BRAF/KRAS are enriched for targetable kinase fusions. Cancer Res. 2019;79:1047–1053. doi: 10.1158/0008-5472.CAN-18-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan Y., Qi C., Maling G., Xiang W., Yanhui L., Ruofei L., Yunhe M., Jiewen L., Qing M. TERT mutation in glioma: frequency, prognosis and risk. J Clin Neurosci. 2016;26:57–62. doi: 10.1016/j.jocn.2015.05.066. [DOI] [PubMed] [Google Scholar]
- 25.Pekmezci M., Rice T., Molinaro A.M., Walsh K.M., Decker P.A., Hansen H., Sicotte H., Kollmeyer T.M., McCoy L.S., Sarkar G., Perry A., Giannini C., Tihan T., Berger M.S., Wiemels J.L., Bracci P.M., Eckel-Passow J.E., Lachance D.H., Clarke J., Taylor J.W., Luks T., Wiencke J.K., Jenkins R.B., Wrensch M.R. Adult infiltrating gliomas with WHO 2016 integrated diagnosis: additional prognostic roles of ATRX and TERT. Acta Neuropathol. 2017;133:1001–1016. doi: 10.1007/s00401-017-1690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eckel-Passow J.E., Lachance D.H., Molinaro A.M., Walsh K.M., Decker P.A., Sicotte H. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panebianco F., Nikitski A.V., Nikiforova M.N., Nikiforov Y.E. Spectrum of TERT promoter mutations and mechanisms of activation in thyroid cancer. Cancer Med. 2019;8:5831–5839. doi: 10.1002/cam4.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu R., Xing M. TERT promoter mutations in thyroid cancer. Endocr Relat Cancer. 2016;23:R143–R155. doi: 10.1530/ERC-15-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landa I., Ganly I., Chan T.A., Mitsutake N., Matsuse M., Ibrahimpasic T., Ghossein R.A., Fagin J.A. Frequent somatic TERT promoter mutations in thyroid cancer: higher prevalence in advanced forms of the disease. J Clin Endocrinol Metab. 2013;98:E1562–E1566. doi: 10.1210/jc.2013-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landa I., Ibrahimpasic T., Boucai L., Sinha R., Knauf J.A., Shah R.H., Dogan S., Ricarte-Filho J.C., Krishnamoorthy G.P., Xu B., Schultz N., Berger M.F., Sander C., Taylor B.S., Ghossein R., Ganly I., Fagin J.A. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016;126:1052–1066. doi: 10.1172/JCI85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ibrahimpasic T., Xu B., Landa I., Dogan S., Middha S., Seshan V., Deraje S., Carlson D.L., Migliacci J., Knauf J.A., Untch B., Berger M.F., Morris L., Tuttle R.M., Chan T., Fagin J.A., Ghossein R., Ganly I. Genomic alterations in fatal forms of non-anaplastic thyroid cancer: identification of MED12 and RBM10 as novel thyroid cancer genes associated with tumor virulence. Clin Cancer Res. 2017;23:5970–5980. doi: 10.1158/1078-0432.CCR-17-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganly I., Makarov V., Deraje S., Dong Y., Reznik E., Seshan V., Nanjangud G., Eng S., Bose P., Kuo F., Morris L.G.T., Landa I., Carrillo Albornoz P.B., Riaz N., Nikiforov Y.E., Patel K., Umbricht C., Zeiger M., Kebebew E., Sherman E., Ghossein R., Fagin J.A., Chan T.A. Integrated genomic analysis of Hurthle cell cancer reveals oncogenic drivers, recurrent mitochondrial mutations, and unique chromosomal landscapes. Cancer Cell. 2018;34:256–270.e255. doi: 10.1016/j.ccell.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research Network Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pilsworth J.A., Cochrane D.R., Xia Z., Aubert G., Farkkila A.E.M., Horlings H.M., Yanagida S., Yang W., Lim J.L.P., Wang Y.K., Bashashati A., Keul J., Wong A., Norris K., Brucker S.Y., Taran F.A., Kramer B., Staebler A., van Meurs H., Oliva E., Shah S.P., Kommoss S., Kommoss F., Gilks C.B., Baird D.M., Huntsman D.G. TERT promoter mutation in adult granulosa cell tumor of the ovary. Mod Pathol. 2018;31:1107–1115. doi: 10.1038/s41379-018-0007-9. [DOI] [PubMed] [Google Scholar]
- 35.Koelsche C., Renner M., Hartmann W., Brandt R., Lehner B., Waldburger N., Alldinger I., Schmitt T., Egerer G., Penzel R., Wardelmann E., Schirmacher P., von Deimling A., Mechtersheimer G. TERT promoter hotspot mutations are recurrent in myxoid liposarcomas but rare in other soft tissue sarcoma entities. J Exp Clin Cancer Res. 2014;33:33. doi: 10.1186/1756-9966-33-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandahl N., Magnusson L., Nilsson J., Viklund B., Arbajian E., von Steyern F.V., Isaksson A., Mertens F. Scattered genomic amplification in dedifferentiated liposarcoma. Mol Cytogenet. 2017;10:25. doi: 10.1186/s13039-017-0325-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng S., Cherniack A.D., Dewal N., Moffitt R.A., Danilova L., Murray B.A. Comprehensive pan-genomic characterization of adrenocortical carcinoma. Cancer Cell. 2016;29:723–736. doi: 10.1016/j.ccell.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dwight T., Flynn A., Amarasinghe K., Benn D.E., Lupat R., Li J., Cameron D.L., Hogg A., Balachander S., Candiloro I.L.M., Wong S.Q., Robinson B.G., Papenfuss A.T., Gill A.J., Dobrovic A., Hicks R.J., Clifton-Bligh R.J., Tothill R.W. TERT structural rearrangements in metastatic pheochromocytomas. Endocr Relat Cancer. 2018;25:1–9. doi: 10.1530/ERC-17-0306. [DOI] [PubMed] [Google Scholar]
- 40.Fahey C.C., Rathmell W.K. A tale of two cancers: complete genetic analysis of chromophobe renal cell carcinoma contrasts with clear cell renal cell carcinoma. Mol Cell Oncol. 2015;2:e979686. doi: 10.4161/23723556.2014.979686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kyo S., Masutomi K., Maida Y., Kanaya T., Yatabe N., Nakamura M., Tanaka M., Takarada M., Sugawara I., Murakami S., Taira T., Inoue M. Significance of immunological detection of human telomerase reverse transcriptase: re-evaluation of expression and localization of human telomerase reverse transcriptase. Am J Pathol. 2003;163:859–867. doi: 10.1016/S0002-9440(10)63446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y.L., Dudognon C., Nguyen E., Hillion J., Pendino F., Tarkanyi I., Aradi J., Lanotte M., Tong J.H., Chen G.Q., Segal-Bendirdjian E. Immunodetection of human telomerase reverse-transcriptase (hTERT) re-appraised: nucleolin and telomerase cross paths. J Cell Sci. 2006;119:2797–2806. doi: 10.1242/jcs.03001. [DOI] [PubMed] [Google Scholar]
- 43.Beird H.C., Wu C.C., Ingram D.R., Wang W.L., Alimohamed A., Gumbs C., Little L., Song X., Feig B.W., Roland C.L., Zhang J., Benjamin R.S., Hwu P., Lazar A.J., Futreal P.A., Somaiah N. Genomic profiling of dedifferentiated liposarcoma compared to matched well-differentiated liposarcoma reveals higher genomic complexity and a common origin. Cold Spring Harb Mol Case Stud. 2018;4:a002386. doi: 10.1101/mcs.a002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griewank K.G., Murali R., Puig-Butille J.A., Schilling B., Livingstone E., Potrony M., Carrera C., Schimming T., Moller I., Schwamborn M., Sucker A., Hillen U., Badenas C., Malvehy J., Zimmer L., Scherag A., Puig S., Schadendorf D. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Natl Cancer Inst. 2014;106:dju246. doi: 10.1093/jnci/dju246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Casuscelli J., Becerra M.F., Manley B.J., Zabor E.C., Reznik E., Redzematovic A., Arcila M.E., Tennenbaum D.M., Ghanaat M., Kashan M., Stief C.G., Carlo M., Voss M.H., Feldman D.R., Motzer R.J., Chen Y., Reuter V.E., Coleman J.A., Russo P., Hsieh J.J., Hakimi A.A. Characterization and impact of TERT promoter region mutations on clinical outcome in renal cell carcinoma. Eur Urol Focus. 2019;5:642–649. doi: 10.1016/j.euf.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borah S., Xi L., Zaug A.J., Powell N.M., Dancik G.M., Cohen S.B., Costello J.C., Theodorescu D., Cech T.R., Cancer TERT promoter mutations and telomerase reactivation in urothelial cancer. Science. 2015;347:1006–1010. doi: 10.1126/science.1260200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang M.T., Penson A., Desai N.B., Socci N.D., Shen R., Seshan V.E., Kundra R., Abeshouse A., Viale A., Cha E.K., Hao X., Reuter V.E., Rudin C.M., Bochner B.H., Rosenberg J.E., Bajorin D.F., Schultz N., Berger M.F., Iyer G., Solit D.B., Al-Ahmadie H.A., Taylor B.S. Small-cell carcinomas of the bladder and lung are characterized by a convergent but distinct pathogenesis. Clin Cancer Res. 2018;24:1965–1973. doi: 10.1158/1078-0432.CCR-17-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinde I., Munari E., Faraj S.F., Hruban R.H., Schoenberg M., Bivalacqua T., Allaf M., Springer S., Wang Y., Diaz L.A., Jr., Kinzler K.W., Vogelstein B., Papadopoulos N., Netto G.J. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. 2013;73:7162–7167. doi: 10.1158/0008-5472.CAN-13-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allory Y., Beukers W., Sagrera A., Flandez M., Marques M., Marquez M., van der Keur K.A., Dyrskjot L., Lurkin I., Vermeij M., Carrato A., Lloreta J., Lorente J.A., Carrillo-de Santa Pau E., Masius R.G., Kogevinas M., Steyerberg E.W., van Tilborg A.A., Abas C., Orntoft T.F., Zuiverloon T.C., Malats N., Zwarthoff E.C., Real F.X. Telomerase reverse transcriptase promoter mutations in bladder cancer: high frequency across stages, detection in urine, and lack of association with outcome. Eur Urol. 2014;65:360–366. doi: 10.1016/j.eururo.2013.08.052. [DOI] [PubMed] [Google Scholar]
- 50.Huang F.W., Hodis E., Xu M.J., Kryukov G.V., Chin L., Garraway L.A. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mehra R., Dhanasekaran S.M., Palanisamy N., Vats P., Cao X., Kim J.H., Kim D.S., Johnson T., Fullen D.R., Chinnaiyan A.M. Comprehensive analysis of ETS family members in melanoma by fluorescence in situ hybridization reveals recurrent ETV1 amplification. Transl Oncol. 2013;6:405–412. doi: 10.1593/tlo.13340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shay J.W., Wright W.E. Telomerase therapeutics for cancer: challenges and new directions. Nat Rev Drug Discov. 2006;5:577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 53.Arndt G.M., MacKenzie K.L. New prospects for targeting telomerase beyond the telomere. Nat Rev Cancer. 2016;16:508–524. doi: 10.1038/nrc.2016.55. [DOI] [PubMed] [Google Scholar]
- 54.Shay J.W., Reddel R.R., Wright W.E. Cancer: cancer and telomeres--an ALTernative to telomerase. Science. 2012;336:1388–1390. doi: 10.1126/science.1222394. [DOI] [PubMed] [Google Scholar]