Abstract
Herbal medicinal products (HMPs) have grown increasingly popular in the United States, many of them with imported raw materials and sold online. Yet due to the lack of regulation from the US Food and Drug Administration (FDA), manufacturers of the products can substitute or add in other herbs that are not advertised on the label. In this study, as part of the Urban Barcode Research Program (UBRP), an education initiative to engage New York City high school students in science, we aimed to taxonomically authenticate single-ingredient online-sold HMPs containing non-native plants through DNA barcoding of the internal transcribed spacer 2 region (ITS2) and matK. We were able to successfully barcode 20 HMPs, but four of these did not match the expected species. It was concluded that the four HMPs advertising astragalus, epazote, ginseng, and chanca piedra were contaminated/ substituted because their ITS2 and matK DNA sequences did not match the expected taxonomy in GenBank, a government database. Our study highlights the importance of herbal pharmacovigilance in the absence of strict government regulation of herbal supplements and motivates crowd-sourced DNA barcoding to enable American consumers make informed choices and be more empowered to safeguard their health.
Keywords: drug safety, herbal medicinal products, taxonomic authentication
1. Introduction
Herbal medicine, the use of plants as medicine based on traditional knowledge, has been a growing industry even in developed nations where conventional medicine is well established, with sales reaching over $US 5B in 2011 in the US alone. Four out of 10 adult Americans have used some form of alternative medicine, including herbal medicine (National Center for Complementary and Integrative Health https://nccih.nih.gov/). Due to the different waves of immigration that the United States has experienced over the years, non-native plants have made their way into the US herbal medicine market. Preference for traditional medicine among immigrant populations over conventional medicine may be due to cultural beliefs, lack of insurance, and/or language barriers [1,2,3].
Many herbal medicinal products (HMPs) are sold online, thus easily accessible. Unfortunately, the US Food and Drug Administration (FDA) does not regulate HMPs as strictly as pharmaceutical drugs, with HMPs regarded as dietary supplements that can be produced, sold, and marketed without first demonstrating safety and efficacy [4]. Thus, many HMPs have been found to not contain the herbs advertised, often containing substitutions, contaminations or fillers [5, 6, https://ag.ny.gov/press-release/ag-schneiderman-asks-major-retailers-halt-sales-certain-herbal-supplements-dna-tests]. These HMPs will not have the expected therapeutic effect, or worse, the undeclared plant/s may trigger allergic reactions and/or potentially interfere with medications.
DNA barcoding has been demonstrated to be an effective way to authenticate HMPs [5,6,7,8]. DNA barcoding is a biotechnological tool that uses short genetic markers from a standard part of a genome to identify an organism [9], and can be proposed as a routine procedure to promote herbal pharmacovigilance, or herbal drug safety, which ensures correct product labels and adherence to good manufacturing practices [10]. Labels must include all source ingredients including the common name of the botanical species, fillers, excipients, and binders used (Code of Federal Regulations, 2015 https://www.ecfr.gov/). However, traditional DNA barcoding, which uses Sanger sequencing, has been shown to be problematic when applied to finished/processed HMPs because of degraded DNA [11], as well as to mixed herbs because it cannot resolve mixed signals. Next-generation sequencing techniques [12] combined with chemical analyses [11] may be more appropriate for these types of samples.
However, unprocessed single-ingredient (i.e. one plant species) HMPs abound in the US market, and Sanger-based DNA barcoding would be capable of authenticating such samples. As part of the Urban Barcode Research Program (UBRP), a science education initiative to promote biodiversity research among New York City high schools, we aimed to taxonomically validate single-ingredient herbal supplements containing imported/non-native plants sold by a major online retailer through DNA barcoding. Among the authors in our study are high school students (CS, JN, TS) and children of immigrants who use HMPs, and this barcoding project was an auspicious opportunity to promote and crowd-source pharmacovigilance. In this study we used the nuclear internal transcribed spacer 2 (ITS2), which has been shown to be a practical DNA barcode for herbal supplements [6, 13]. In cases where ITS2 could not be amplified or resulted in a taxonomic identification different from the expected species, the plastid marker matK was also amplified [6]. The DNA barcodes allowed us to conclude on the authenticity and reliability of the labels on imported medicinal herbal products.
2. Methods
Thirty-two (32) single-ingredient HMPs (i.e. single plant species) from different manufacturers were purchased from a major online retailer. Constituent plant species are not native to the US. Eleven of the sampled HMPs declared foreign origins. Nineteen were American brands but it was unclear if the non-native plant was cultivated in the US or was imported and packaged in the US. Two HMPs did not disclose the origin. HMP brand names and the name of the online retailer were withheld to prevent legal consequences. Only HMPs that contained ground herbs, not extracts/tinctures, were bought to be sure that plant cells are still intact in the product. Product labels were carefully read prior purchase to rule out plant ingredients that were extracted/processed in a solvent. Some examples of ingredient labels on purchased HMPs included “400 mg devil’s claw root powder”, “300 mg noni fruit powder”, “400 mg black cumin seed”, “1000 mg moringa leaf”, etc. We sampled non-native herbal species used by various cultural groups from Asia, Latin America, and Africa. Since many of these herbs are not easily accessible, there is a risk that constituent plants have been supplanted by easily accessible species. Table 1 lists the herbal medicines used in this study. Scientific names were obtained from the label, or searched for based on the common name provided.
Table 1.
List of single-ingredient ground HMPs purchased from a major online retailer. The common name, expected scientific names (corresponds to accepted names from The Plant List http://www.theplantlist.org/, with alternative scientific name if on the label), botanical family, and indigenous culture/native distribution for constituent plant species are provided. Taxonomic identifications (ID) of HMPs from web BLAST (part of JM workflow) and from DNA Subway BLAST (database as of 4/10/17) are indicated for ITS2 DNA barcodes, and in some case for matK. “spp.” refers to multiple species within the genus. “*” refers to an HMP that has no corresponding sequence for the expected species in GenBank, though a closely related species is available. Twelve HMPs could not be barcoded in spite of multiple attempts. When BLAST results were ambiguous, phylogenetic analysis was performed to confirm taxonomic identity and is indicated with “P”, with the corresponding phylogeny in Supplement (Fig. S). Potentially contaminated/substituted HMPs (i.e. HMP not in the same family as expected species) are indicated with !!!
| Botanical family | Expected Species (scientific name) | Indigenous culture/ native distribution | Taxonomic ID based on web BLAST | Taxonomic ID based on DNA Subway |
|---|---|---|---|---|
| Acanthaceae | fah talai jone (Andrographis paniculata) | Southeast Asia | ITS2: not barcoded; matK: Andrographis paniculata | ITS2: not barcoded; matK: Andrographis paniculata |
| Amaranthaceae | wormseed/ epazote (Dysphania ambrosioides= Chenopodium ambrosioides) !!! | Latin America (Central America, South America) | ITS2: Artemisia absinthium; matK: Artemisia absinthium (Asteraceae) | ITS2: Artemisia vulgaris; matK: Athanasia flexuosa (Asteraceae) |
| Annonaceae | graviola (Annona muricata*) | Latin America (Caribbean and Central America) | ITS2: Annonaceae sp. | ITS2: Goniothalamus cheliensis |
| Apiaceae | dong quai (Angelica sinensis) | Asia (China) | not barcoded | not barcoded |
| Apiaceae | gotu kola (Centella asiatica) | Tropical Asia | not barcoded | not barcoded |
| Aquifoliaceae | yerba mate (Ilex paraguariensis) | Latin America (South America) | ITS2: Ilex paraguariensis | ITS2: Ilex sp. |
| Araliaceae | eleuthero (Eleutherococcus senticosus) | Northeastern Asia | not barcoded | not barcoded |
| Araliaceae | Korean ginseng (Panax ginseng)!!! | East Asia | ITS2: Astragalus membranaceus=A. propinquus; matK: Astragalus sp. (Fabaceae) | ITS2: Astragalus membranaceus=A. propinquus; matK: Mimosa pudica (Fabaceae) |
| Asparagaceae | shatavari (Asparagus racemosus) | Asia (Nepal, India, Sri Lanka) | matK (P, Fig S): Asparagus spp. | matK: Asparagus rubicundus |
| Asteraceae | yacon (Smallanthus sonchifolius*) | Latin America (northern and central Andes) | ITS2: not barcoded; matK (P, Fig S): Smallanthus sp. | ITS2: not barcoded; matK: Rudbeckia hirta |
| Berberidaceae | horny goat weed (Epimedium spp.) | Asia (China) | ITS2: Epimedium spp. | ITS2: Epimedium wushanense |
| Bignoniaceae | pau d’ arco (Handroanthus heptaphyllus= Tabebuia heptaphylla) | Latin America (Central America, South America) | not barcoded | not barcoded |
| Brassicaceae | maca (Lepidium meyenii*) | Latin America (Peruvian Andes) | ITS2 (P, Fig S): Lepidium spp. | ITS2: Lepidium virginicum |
| Convolvulaceae | dodder (Cuscuta chinensis) | Asia (China) | ITS2 (P, Fig S): Cuscuta spp. | ITS2: Cuscuta australis |
| Fabaceae | Astragalus (Astragalus propinquus)!!! | Asia (China) | ITS2: Plantago spp.; matK: Plantago ovata (Plantaginaceae) | ITS2: Plantago spp. matK: Plantago patagonica (Plantaginaceae) |
| Fabaceae | Butea superba | Asia (Thailand, Vietnam, India) | not barcoded | not barcoded |
| Lamiaceae | tulsi powder (Ocimum tenuiflorum=O. sanctum) | Asia (India) | ITS2 (P, Fig S): Ocimum basilicum/tenuiflorum | ITS2: Hyptidendron sp. |
| Lauraceae | Ceylon cinnamon (Cinnamomum verum) | Asia (Sri Lanka) | not barcoded | not barcoded |
| Meliaceae | neem leaf (Azadirachta indica) | Asia (India) | ITS2: Azadirachta indica | ITS2: Azadirachta indica |
| Moringaceae | moringa (Moringa oleifera) | Asia (India) | ITS2 (P, Fig S): Moringa oleifera/peregrina | ITS2: Moringa oleifera |
| Oleaceae | muira puama (Ptychopetalum olacoides) | Latin America (South America) | not barcoded | not barcoded |
| Pedaliaceae | devil’s claw (Harpagophytum procumbens) | South Africa | ITS2; Harpagophytum spp. | ITS2: Harpagophytum zeyheri |
| Phyllanthaceae | chanca piedra (Phyllanthus niruri)!!! | Originally Asia (India) but now pantropical | ITS2: Desmodium canadense; matK: not barcoded (Fabaceae) | ITS2: Desmodium spp.; matK: not barcoded (Fabaceae) |
| Plantaginaceae | brahmi (Bacopa monnieri) | Asian wetlands | not barcoded | not barcoded |
| Ranunculaceae | black cumin (Nigella sativa) | South and southwest Asia | ITS2: Nigella sativa | ITS2: Nigella damascena |
| Rubiaceae | cat’s claw (Uncaria tomentosa) | Latin America (South and Central America) | ITS2: Uncaria tomentosa | ITS2: Uncaria lancifola |
| Rubiaceae | noni fruit (Morinda citrifolia) | Southeast Asia | not barcoded | not barcoded |
| Rubiaceae | yohimbe (Pausinystalia johimbe) | Western and central Africa | not barcoded | not barcoded |
| Sapindaceae | guarana (Paullinia cupana*) | Latin America (Amazon basin) | ITS2: Paullinia sp. | ITS2: Paullinia pachycarpa |
| Simaroubaceae | tongkat ali (Eurycoma longifolia) | Southeast Asia | not barcoded | not barcoded |
| Solanaceae | ashwagandha (Withania somnifera) | Asia (India) | not barcoded | not barcoded |
| Zygophyllaceae | goat’s head (Tribulus terrestris) | Asian and African tropics | ITS2: Tribulus terrestris | ITS2: Tribulus terrestris |
We extracted DNA using the Qiagen DNeasy Plant Mini Kit (Qiagen cat. no. 69104). In some cases, when PCR amplification of Qiagen extracts was not successful, we also used the Phire Plant Direct PCR kit (Thermofisher cat no. F130WH), which simply involves agitating a speck of the ground HMP in 20 uL of the manufacturer’s dilution buffer and using 1 uL of the supernatant as PCR template. PCR amplification of the ITS2 marker (and in some cases, matK), visualization of PCR products, and purification were performed using primers and protocols in [6] and [13]. In one experiment, the portable and inexpensive MiniPCR (Amplyus, Cambridge, MA) was also used for PCR amplification to test its utility in crowd-sourced DNA barcoding. Purified PCR products were submitted for sequencing to Genewiz Inc. (South Plainfield, NJ).
Sequences were assembled, edited, compared to sequences from GenBank’s NCBI using the “blue line” workflow (=determine sequence relationships) in the open-access DNA Subway interface (https://dnasubway.cyverse.org/, developed by DNA Learning Center at Cold Spring Harbor Laboratory, NY under CyVerse). UBRP high school students were trained in DNA Subway. The same raw sequences (forward, reverse sequences from Genewiz, Inc.) were analyzed using the DNA barcoding workflow described in [6] and [13], hereafter referred to as the JM workflow: raw sequences were edited and assembled manually in Geneious 7.1.9 (Biomatters, New Zealand) with the consensus sequences then compared against the GenBank NCBI nucleotide database using BLASTn [14]. Species with highest max score/bit score were considered the taxonomic identity of the HMP, unless results were ambiguous (i.e. different species with similarly high pairwise identity to the query). In this case, HMP specific identity was verified by phylogenetic analyses.
3. Results
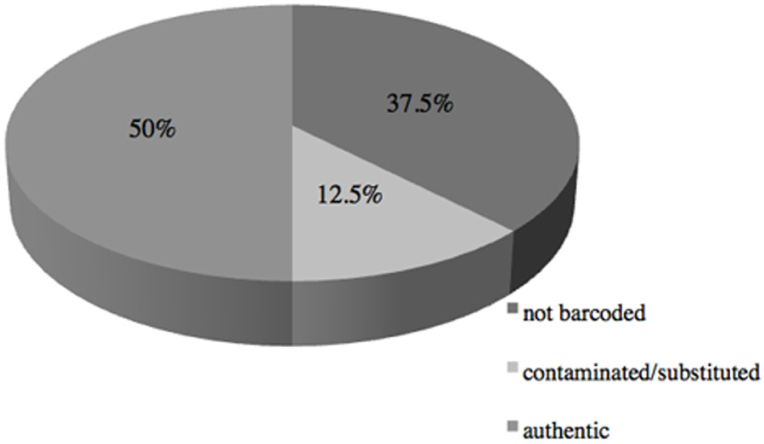
In an effort to authenticate single-ingredient HMPs containing ground parts of non-native plant species, we performed Sanger-based DNA barcoding, with sequences analyzed using DNA Subway and results compared to those produced using the JM workflow. Of the 32 HMPs, 20 HMPs were successfully barcoded, with 17 ITS2 barcodes (271-514 bp) and 6 matK (633-875 bp) sequences generated (Table 1, GenBank accession numbers MG548803-MG548815). We find that of 20 successfully barcoded samples, 16 were taxonomically authentic (Fig. 1, 16/32= 50%) and matched expected species or at least genus, if the sequence for the expected species is not represented in GenBank. However, 4 did not match the expected species, nor the family (4/32= 12.5%, Fig. 1), even after barcoding ITS2 and matK from each of these HMPs, although the sequences for their confamilial species are represented in GenBank and would be expected to come up as the top hits if they had been taxonomically accurate. These were HMPs labeled astragalus (supposedly Astragalus propinquus, but came up as Plantago ovata), epazote (=Dysphania ambrosioides but was Artemisia absinthium), ginseng (=Panax ginseng but was Astragalus propinquus), and chanca piedra (=Phyllanthus niruri but was Desmodium canadense) [Table 1]. Only the latter declared that the plant material originated in Peru and packaged in the US; the rest did not disclose origin, but were American brands containing non-native plants.
Figure 1.
Pie chart showing proportion of taxonomically authentic and contaminated HMPs. Twenty of 32 HMPs were successfully barcoded, with 16/32 (=50%) as taxonomically authentic, and 4/32 (=12.5%) substituted/contaminated. Twelve of 32 (37.5%) samples could not be barcoded in spite of multiple attempts.
4. Discussion
Of the 20 successfully barcoded samples, 16 were taxonomically authentic, yet 4 did not match the expected species, nor the family. We know that this is not a contamination on our part because we extracted, amplified and sequenced DNA from each HMP twice, barcodings both ITS2 and matK, so these HMPs have more than one barcode (except for chanca piedra; Table 1), and in all cases the BLAST hits were not the expected taxonomic affiliation. Thus, these HMPs may have been substituted, deliberately or inadvertently, or possible, but unlikely— accidentally contaminated with a different species during processing, and the two primer sets used (for ITS2 and matK) coincidentally amplified the same contaminant. However, the fact that they contained species that were not declared on the label is still disconcerting, even if they were only present in trace amounts, as consumers may be allergic to these components. For epazote (D. ambrosiodes), it is possible that there was a taxonomic misidentification as A. absinthium superficially looks similar to epazote. For astragalus (A. propinquus), Plantago ovata has been naturalized in the US and easier to access and may have been used as substitute. In chanca piedra, the raw material, Phyllanthus niruri, is claimed to have been imported from Peru, but during packaging in the US may have been substituted/contaminated with the American native Desmodium canadense. In the fourth HMP, where A. propinquus was substituted for P. ginseng, they could have been switched accidentally as both look similar superficially, or A. propinquus may have been used as an intentional filler or substitute, since ginseng is expensive.
Table 1 shows that in some cases, top species hits following the JM workflow differed from those of DNA Subway, though these species belonged to the same family. This may be because DNA Subway references a local database created from NCBI sequences < 20 kb and longer than 400kb matching only certain terms, and has to be constantly updated (latest update used here as of 4/10/17 when project concluded), which is in contrast to the web-based NCBI nucleotide database that is updated in real time. This was brought to UBRP’s attention, and DNA Subway’s sequence database is now updated more frequently. Prior to this, DNA Subway could at least identify HMPs at the generic/family level, which is adequate to determine substitutions/adulterations at these taxonomic levels. For pedagogical purposes, DNA Subway has an intuitive interface that simplifies comparative genomic analyses, guiding users on “different subway lines”, with the blue line geared towards determining sequence relationships, from sequence editing to BLAST and phylogenetic analyses, and would be sufficient for crowd-sourced DNA barcoding projects. For a more rigorous analysis, DNA Subway needs to be supplemented by web-based BLAST sequence comparison and/or phylogenetic analyses that outputs node/clade support such as in MEGA [15].
Supplementary Figure.
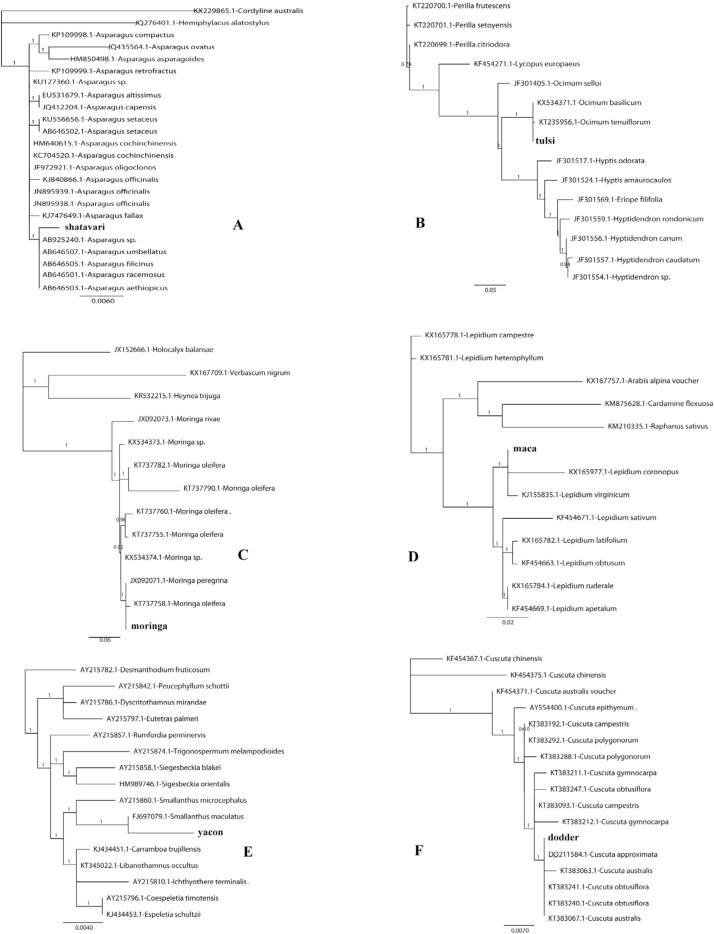
Phylogenies for shatavari (A. Asparagus racemosus) ITS2; B. tulsi (Ocimum tenuiflorum=O.sanctum) ITS2; C.moringa (Moringa oleifera) ITS2; D. maca (Lepidium meyenii) ITS2; E. yacon (Smallanthus sonchifolius) matK; F. dodder (Cuscuta chinensis) ITS2. Barcoded HMP is represented by its common name in bold and compared with GenBank-obtained sequences (top BLAST hits) with accession numbers preceding the scientific name. Numbers on branches indicate clade support with >0.9 as strong support. Maca and yacon cluster with other congeneric species and not the expected species as sequences for latter are not yet in GenBank.
Twelve HMPs (12/32=37.5%, Fig. 1) could not be barcoded even after 3-4 attempts were tried for each sample using different extraction techniques (Qiagen, Phire) and/or barcodes/primers (ITS2, matK). Apart from the absence of plant material, other reasons for barcoding failure may be the high amounts of secondary metabolites in plants that often result in PCR inhibition [16]. Dilution of the DNA extract for use as PCR template may help [17]. Additionally, primer sets used were not suited for amplifications of all species and new primers may be warranted [18]. Most of the samples that were not barcoded originated from roots and barks. Michel et al. [6] reported that leaves are the easiest to barcode, and new extraction techniques are needed for other plant parts.
Six of the 20 barcoded samples had to be phylogenetically analyzed (Supplement Figure) because the BLAST hits were ambiguous (congeneric species, in addition to the expected species, were top hits). For shatavari (Asparagus racemosus), tulsi (Ocimum tenuiflorum=O. sanctum), and moringa (Moringa oleifera), the sequence of the HMP still clustered with other congeneric species including the expected species, which could indicate that ITS2 does not have enough genetic variation to differentiate among these closely-related species. HMPs maca (Lepidium meyenii) and yacon (Smallanthus sonchifolius) do not have their expected species represented in GenBank but they did cluster with other congeneric species, which may suggest they may be taxonomically accurate, at least at the genus level. The HMP dodder is expected to be Cuscuta chinensis but clustered with other Cuscuta spp., and not C. chinensis even if this was represented in the phylogeny, and may represent a substitution, but not an egregious one considering the HMP is still Cuscuta, unlike the 4 mentioned above (astragalus, epazote, ginseng, and chanca piedra), where completely unrelated species to the declared species were contained in the HMP. However, as described before, the GenBank NCBI database does have some problems, including submitted sequences may have been misidentified or sequences for the query are not yet available, as in the case for maca and yacon. However, it is still adequately informative [6].
Synergistic approaches that combine next-generation sequencing and chemical analytical methods [11, 12] may be preferable and more appropriate in authenticating different types of HMPs especially processed products, but these can only be afforded by big labs, such as government or well-funded academic labs. Though they may have done some DNA testing on commercially available products, these results are not readily accessible to the general public because of the technical jargon, with the actual brand names also not disclosed to avoid legal consequences. Thus, these studies will not be able to help consumers. However, for single-ingredient unprocessed HMPs consumers themselves take, they may test for the taxonomic accuracy of these products given the accessibility and low cost of Sanger-based DNA barcoding.
Though Sanger-based DNA barcoding may be limited in the HMPs it can handle, with the right samples, it is still a pragmatic and effective way to integrate molecular biology and herbal pharmacovigilance in the high school curriculum, as the workflow is simple and materials are relatively inexpensive. The Phire DNA extraction kit costs only $256 for 500 reactions (Thermofisher cat no. F130WH), while PCR amplification can be performed on the portable $650 MiniPCR (Amplyus, Cambridge, MA), which worked well for us (5/8 amplification reactions successful) and has been used in many crowd-sourced biotechnological applications (http://www.minipcr.com/educational-resources/). The barcoding workflow presented here makes crowdsourcing pharmacovigilance in high school or even in home labs feasible and empowers consumers to choose quality products among the wide array of HMPs available, none of which have been evaluated by the FDA. More informed, discriminating consumers could eventually drive US herbal manufacturers to improve their quality control and strictly guarantee the authenticity of their products. Our study highlights the importance of consumer awareness and pharmacovigilance when buying herbal medicines in the US, and motivates more crowd-sourced DNA barcoding projects to enable consumers like us, and our families, make informed choices and be more empowered to safeguard our own health.
Acknowledgments
We are indebted to the Urban Barcode Research Program of the DNA Learning Center at Cold Spring Harbor Laboratory, without which, this would not have been possible. The Urban Barcode Research Program is partially funded by the Pinkerton Foundation. Christine Marizzi has been an enormous help throughout. We also thank Joseph Morin and Allan Howell of LIU-Brooklyn. Finally, we are grateful to the families of CS, JN, and TS for being supportive and allowing them to stay in JM’s lab after school.
Footnotes
Conflict of interest: Authors state no conflict of interest.
References
- [1].Kraut A.M.. Healers and strangers. Immigrant attitudes toward the physician in America--a relationship in historical perspective, JAMA. 1990;263:1807–1811. doi: 10.1001/jama.263.13.1807. [DOI] [PubMed] [Google Scholar]
- [2].Kronenberg F., Cushman L.F., Wade C.M., Kalmuss D., Chao M.T.. Race/ethnicity and women’s use of complementary and alternative medicine in the United States: results of a national survey. Am. J. Public Health. 2006;96:1236–1242. doi: 10.2105/AJPH.2004.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ransford H.E., Carrillo F. R., Rivera Y.. Health care-seeking among latino immigrants: blocked access, use of traditional medicine, and the role of religion. J. Health Care Poor Underserved. 2010;21:862–878. doi: 10.1353/hpu.0.0348. [DOI] [PubMed] [Google Scholar]
- [4].Bent S.. Herbal medicine in the United States: review of efficacy, safety, and regulation: grand rounds at University of California, San Francisco Medical Center, Int. J. Gen. Med. 2008;23:854–859. doi: 10.1007/s11606-008-0632-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Newmaster S., Grguric M., Shanmughanandhan D., Ramalingam S., Ragupathy S.. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013;11:222. doi: 10.1186/1741-7015-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [6].Michel C.I., Meyer R.S., Taveras Y., Molina J.. The nuclear internal transcribed spacer (ITS2) as a practical plant DNA barcode for herbal medicines. J. Appl. Res. Med. Aromat. Plants. 2016;3:94–100. [Google Scholar]
- [7].Li M., Cao H., But P.P., Shaw P.. Identification of herbal medicinal materials using DNA barcodes. J. Syst. Evol. 2011;49:271–283. [Google Scholar]
- [8].Mishra P., Kumar A., Nagireddy A., Mani D.N., Shukla A.K., Tiwari R.. et al. DNA barcoding: an efficient tool to overcome authentication challenges in the herbal market. Plant Biotech, J. 2015;14:8–21. doi: 10.1111/pbi.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kress W.J., Erickson D.L. DNA Barcodes: Methods and Protocols. Humana Press; New York, NY: 2012. [DOI] [PubMed] [Google Scholar]
- [10].De Boer H.J., Ichim M.C., Newmaster S.G.. DNA barcoding and pharmacovigilance of herbal medicines. Drug Saf. 2015;38:611–620. doi: 10.1007/s40264-015-0306-8. [DOI] [PubMed] [Google Scholar]
- [11].Pawar R.S., Handy S.M., Cheng R., Shyong N., Grundel E.. Assessment of the authenticity of herbal dietary supplements: comparison of chemical and DNA barcoding methods. Planta Med. 2017;83:921–936. doi: 10.1055/s-0043-107881. [DOI] [PubMed] [Google Scholar]
- [12].Ivanova N.V., Kuzmina M.L., Braukmann T.W., Borisenko A.V., Zakharov E.V.. Authentication of herbal supplements using next-generation sequencing. PLoS One. 2016;26(11)(5):e0156426. doi: 10.1371/journal.pone.0156426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pedales R.D., Damatac A, Marquez C.M., Limbo C.A., Navarro A.I., Molina J.. DNA barcoding of Philippine herbal medicinal products. J. AOAC Int. 2016;99:1479–1489. doi: 10.5740/jaoacint.16-0185. [DOI] [PubMed] [Google Scholar]
- [14].Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W.. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kumar S., Stecher G., Tamura K.. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Moyo M., Amoo S.O., Bairu M.W., Finnie J.F., Van Staden J.. Optimising DNA isolation for medicinal plants. S. Afr. J. Bot. 2008;74:771–775. [Google Scholar]
- [17].Li F.W., Kuo L.Y., Huang Y.M., Chiou W.L., Wang C.N.. Tissue-direct PCR, a rapid and extraction-free method for barcoding of ferns. Mol. Ecol. Resour. 2010;10:92–95. doi: 10.1111/j.1755-0998.2009.02745.x. [DOI] [PubMed] [Google Scholar]
- [18].Cheng T., Xu C., Lei L., Li C., Zhang Y., Zhou S.. Barcoding the kingdom Plantae: new PCR primers for ITS regions of plants with improved universality and specificity. Mol. Ecol. Resour. 2016;16:138–149. doi: 10.1111/1755-0998.12438. [DOI] [PubMed] [Google Scholar]