Abstract
The purpose of the present investigation is to examine the function of the C2H2-type zinc finger transcription factor of Arabidopsis thaliana 6 (ZAT6) in salt stress tolerance in cells of rice (Oryza sativa L.), cotton (Gossypium hirsutum L.) and slash pine (Pinus elliottii Engelm.). Cells of O. sativa, G. hirsutum, and P. elliottii overexpressing ZAT6 were generated using Agrobacterium-mediated genetic transformation. Molecular and functional analysis of transgenic cell lines demonstrate that overexpression of ZAT6 increased tolerance to salt stress by decreasing lipid peroxidation and increasing the content of abscisic acid (ABA) and GA8, as well as enhancing the activities of antioxidant enzymes such as ascorbate peroxidise (APOX), catalase (CAT), glutathione reductase (GR), and superoxide dismutase (SOD). In rice cells, ZAT6 also increased expression of Ca2+-dependent protein kinase genes OsCPK9 and OsCPK25 by 5–7 fold under NaCl stress. Altogether, our results suggest that overexpression of ZAT6 enhanced salt stress tolerance by increasing antioxidant enzyme activity, hormone content and expression of Ca2+-dependent protein kinase in transgenic cell lines of different plant species.
Keywords: NaCl stress tolerance, Agrobacterium-mediated genetic transformation, Ca2+-dependent protein kinase, Transcription factor, ZAT6 gene
1. Introduction
Salt stress causes decreased growth, development, and reduced productivity in many plant species [1, 2, 3, 4, 5]. Plants respond to salt stress by increasing expression of stress-inducible genes and activating expression of transcription factors [6, 7, 8, 9, 10, 11]. Understanding the molecular mechanisms of salt stress tolerance in plants is essential for molecular biologists to improve plant growth and production of crop plant [12, 13, 14, 15, 16, 17, 18]. Although different mechanisms can be adapted by plant in response to salt stress, transcription factors have been reported to play a crucial roles in salt tolerance in many plant species [19, 20, 21, 22, 23, 24, 25, 26].
Among different transcription factors (TFs) investigated in plant salt tolerance, basic region/leucine zipper (bZIP) TFs [27, 28, 29], AP2/ERF TFs [30, 31, 32], WRKY TFs [33, 34, 35], and MYB TFs [36, 37, 38] have been reported to enhance salt tolerance in a large number of plant species. The bZIP TFs are very important in regulating salt stress signaling, as well as modulating plant growth and development [29, 39, 40, 41]. AP2/ERF TFs interact with a cis-acting DRE (dehydration-responsive element)/ CRT (C-repeat) DNA sequence and activate the expression of downstream genes that are involved in salt stress tolerance in plants [24, 31, 42, 43]. WRKY TFs could bind to the W-box (TTGAC) and function on the stress-induced genes to increased sensitivity and confer salt and drought tolerance [35, 44, 45, 46]. MYB TFs improve salt and osmotic stress tolerance by affecting the expression of genes such as SOD, POD and P5CS to increase reactive oxygen species scavenging level and reduce water loss [15, 16, 36, 37, 38].
The C2H2-type zinc finger proteins (ZFPs) represent a large family of eukaryotic TFs [48, 49, 50, 51, 52]. A total of 176 proteins that contain one or more zinc finger domains have been reported in Arabidopsis [50]. It has been reported that the ZFP transcription factors play important roles in stress responses [49, 53, 54]. Expression of Zinc finger of Arabidopsis thaliana 6 (ZAT6) is regulated by phytohormones and low concentration of phosphate in Arabidopsis [49, 55]. Recently, ZAT6 has been reported to be involved in regulating responses of plants to abiotic stress including drought and freezing stresses [5, 48, 56]. It has been reported that overexpression of ZAT6 improves seed germination of Arabidopsis under salt and osmotic stress and that phosphorylation of ZAT6 by MPK6 is required for the enhanced salt and osmotic stress tolerance [57]. However, the possible mechanisms of ZAT6 in salt stress tolerance is not reported in slash pine (Pinus elliottii Engelm.), rice (Oryza sativa L.), and cotton (Gossypium hirsutum L.)”.
In this study, we have investigated the function of ZAT6 in plant responses to salt stress in transgenic cells overexpressing the ZAT6 gene. Cells of three different plant species slash pine (P. elliottii Engelm.), rice (O. sativa L.), and cotton (G. hirsutum L.), were genetically engineered via Agrobacterium tumefaciens LBA4404 harboring pBI-ZAT6. Increased cell growth was observed in transgenic cells of all three species that overexpress the ZAT6 transcription factor under the condition of salt stress. In cells overexpressing the ZAT6 gene, lipid peroxidation was decreased and the content of abscisic acid (ABA) and GA8, as well as the activities of antioxidant enzymes ascorbate peroxidase (APOX), glutathione reductase (GR), superoxide dismutase (SOD), and catalase (CAT) were increased. In rice cells overexpressing the ZAT6 gene, expression of Ca2+-dependent protein kinase genes OsCPK9 and OsCPK25 was increased under treatment of NaCl. These results showed that overexpression of the Arabidopsis ZAT6 transcription factor in plant cells of angiosperm and gymnosperm improved salt stress tolerance through multiple mechanisms.
2. Materials and methods
2.1. Plasmid constructs
The cDNA of ZAT6 and the pBI121 binary vector were used to generate the expression vector. After the pBI121 vector and the ZAT6 DNA were digested by Kpn I and Xba I (Promega, Madison, WI, USA) at 37°C, the digested DNA was purified using QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA, USA). The 717-bp protein encoding fragment of the ZAT6 gene was inserted into the vector pBI121 [58] to produce the expression vector pBI-ZAT6. Vector pBI-ZAT6 was introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation.
2.2. Agrobacterium-mediated transformation
Transgenic cell lines of O. sativa, G. hirsutum, and P. elliottii were generated as described before [59], using Agrobacterium tumefaciens strain LBA4404 harboring the pBI-ZAT6 expression vector to transform cultured cells. To generate large amount of transformed cell cultures for molecular and enzyme measurements, cells from different species were cultured on a liquid proliferation medium. Six weeks after culture, the cell cultures of different species were growing 50–70 mg of cells/L each week. These cell cultures were used for PCR, Southern, and Northern blot analysis.
2.3. Polymerase chain reaction and Southern blot analyses of transgenic cells of rice, cotton, and pine
Polymerase chain reaction (PCR) and Southern blot analysis of transgenic cells were conducted as previously described [58]. Five hundred mg cells of control and putative transgenic cell lines of rice, cotton, and pine were used to extract DNA using a Genomic DNA Isolation Kit (Sigma), by following the manufacturer’s protocol. PCR was performed with a PTC-100TM Programmable Thermal Controller (MJ Research, San Francisco, CA, USA). The primers used are the transcription factor ZAT6 forward primer (zf) 5’-GTCGACATGGCGGAGGAATTTGGAAGCATAG-3’ the reverse primer (zr) 5’-CCATGGTAGACTCCTGCTTCGACATCATGG-3’, nrp and nfp for NPTII gene [58]. The DNA template was 300 ng and the reaction mix 50 ml. The PCR mixture, the PCR conditions, and Southern blot analysis was carried out as described previously [58]. Five grams of control cells and transgenic cells of rice, cotton, and pine were used to isolate genomic DNA, using a Genomic DNA Isolation Kit (Sigma). Twenty-five micrograms of DNA were digested for 16 hours with the enzymes Xba I (Boehringer Mannheim) at 37°C. The molecular probes (717 bp fragment of ZAT6) were labeled by Digoxigenin (DIG) (Roche Diagnostics, Indianapolis, IN, USA).
2.4. RNA isolation and Northern blot analysis
Five grams of fresh cultures of transgenic and control cells were used to isolate total RNA, using a RNeasy Mini Plant Kit (Germantown, MD, USA) by following the manual. Six micrograms of total RNA were used for northern blotting as described before [58]. The hybridization probe is the Digoxigenin (DIG)-labelling ZAT6 DNA (717 bp) (Roche Diagnostics). The tobacco 25SrRNA was used as the loading control of RNA samples. After Southern and Northern blotting analyses, 6 cell lines of rice (Os1, Os2, Os3, Os4, Os5, and Os6), 6 cell lines of cotton (Gh1, Gh2, Gh3, Gh4, Gh5, andGh6), and 6 cell lines of slash pine (Pe1, Pe2, Pe3, Pe4, Pe5, and Pe6), each carrying only one copy of the pBI-ZAT6 T-DNA, were used for salt-induced oxidative damage experiments.
2.5. Salt treatment of transgenic cell lines
Salt treatment was applied by adding different concentrations of NaCl (50, 100, 150, 200, 250, and 300 mM) to the media used for transgenic cells, which consisted of TE medium [58] supplemented with 0.5 mM indole-3-butyric acid, 8.9 mM BA. The influence of NaCl on cell growth of rice (Os1, Os2, Os3, Os4, Os5, and Os6), cotton (Gh1, Gh2, Gh3, Gh4, Gh5, andGh6), and slash pine (Pe1, Pe2, Pe3, Pe4, Pe5, and Pe6) were examined by culture of cell on medium supplemented with different concentrations of NaCl, as previously described [59]. The average growth was expressed as mg/ g FW/day. The control cells (non-transgenic cultures) were proliferated for 3 weeks and then transferred to media containing different concentrations of NaCl (0, 50, 100, 150, 200, 250, and 300 mM). The growth rate of cells was determined 7 days after salt stress treatment. The results demonstrated that 50–100 mM NaCl did not significantly decrease the growth rate. The 150 mM NaCl significantly reduced the growth rate. The 200–300 mM NaCl leads to no cell growth (Fig. 3). On medium without NaCl, rate of cell growth does not change significantly.
Fig. 3.
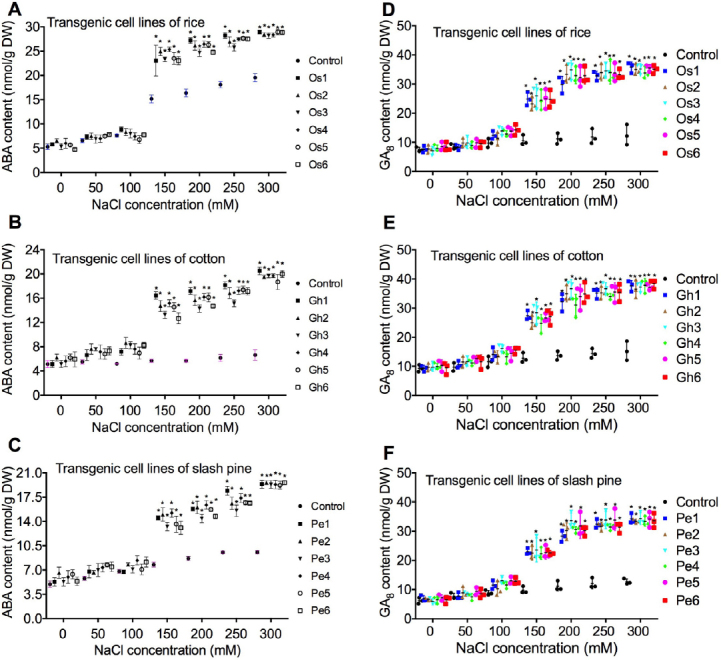
Effect of ZAT6 overexpression on ABA and GA8 content. ABA content (A-C) and GA8 (D-F) content changes in transgenic cell lines of rice (Os1, Os2, Os3, Os4, Os5, and Os6), cotton (Gh1, Gh2, Gh3, Gh4, Gh5, and Gh6), and slash pine (Pe1, Pe2, Pe3, Pe4, Pe5, and Pe6) were measured 21 days after cell cultures were transferred into media containing different concentrations of NaCl (50, 100, 150, 200, 250, and 300 mM). Each experiment was replicated three times, and each replicate consisted of five to ten 250-ml flasks of transgenic cell cultures. Values represent the means ± S.D.
2.6. Thiobarbituric acid reactive substances (TBARS) determination
Lipid peroxidation was determined as the amount of thiobarbituric acid reactive substances (TBARS) measured by the thiobarbituric acid (TBA) reaction as described previously (Tang and Page 2013). Cell cultures (1 g) of rice (Os1, Os2, Os3, Os4, Os5, and Os6), cotton (Gh1, Gh2, Gh3, Gh4, Gh5, andGh6), and slash pine (Pe1, Pe2, Pe3, Pe4, Pe5, and Pe6) were homogenized in 3 ml of 20 % (w/v) trichloroacetic acid (TCA). The homogenate was centrifuged at 5,000 rpm for 20 min and mixed with 20% TCA containing 0.5% (w/v) TBA and100 ml 4% butylated hydroxytoluene (BHT) in ethanol at 1:1. After the extracts of cell cultures were heated at 95°C for 30 min, they were cooled on ice for 5 minutes, centrifuged at 10,000 x g for 15 min. The absorbance of extracts from different cell lines was measured at 532 nm. The control of non-specific absorption at 600 nm was subtracted from the samples. The value of TBARS was calculated using the method described previously [59].
2.7. Determination of the antioxidant enzymes glutathione reductase (GR), ascorbate peroxidase (APOX), superoxide dismutase (SOD), and CAT activity
The activities of APOX, GR, SOD, and CAT were determined as described previously [59]. Two grams of control and transgenic cells of rice (Os1, Os2, Os3, Os4, Os5, and Os6), cotton (Gh1, Gh2, Gh3, Gh4, Gh5, and Gh6), and slash pine (Pe1, Pe2, Pe3, Pe4, Pe5, and Pe6) were homogenized under ice-cold conditions in 3 ml of extraction buffer, consisting of 50 mM phosphate buffer (pH 7.4), 1 mM EDTA, 1 g PVP, and 0.5% (v/v) Triton X-100 at 4°C. The extracts were centrifuged at 10,000 × g for 20 min. The supernatant was used to determine the enzyme activity. APOX activity was measured immediately in fresh extracts and was assayed as described [59]. GR activity was determined by following the decrease in absorbance at 340 nm due to NADPH oxidation [58, 60]. SOD activity was measured by the inhibition of the photochemical reduction of nitroblue tetrazolium (NBT), as described previously [58, 60]. CAT activity was determined in a 3 ml 50 mM potassium phosphate buffer (pH 7.8) containing 3 mM H2O2, as described previously [60].
2.8. Determination of the endogenous ABA and GA8 levels
The ABA content was determined using the radioimmunoassay method as described previously [58, 59, 61]. The GA8 content was determined as previously described by Okamoto et al. [62]. The homogenates of 100 g transgenic cells of rice (Os1, Os2, Os3, Os4, Os5, and Os6), cotton (Gh1, Gh2, Gh3, Gh4, Gh5, andGh6), and slash pine (Pe1, Pe2, Pe3, Pe4, Pe5, and Pe6) were centrifuged at 12,000 x g for 10 min at 4°C. The supernatant of different cell lines was used to determine ABA concentration. Fifty microliters of supernatant derived from different cell lines were mixed with 200 μl phosphate-buffered saline (pH 6.0), 100 μl [3H] ABA solution, and 100 μl diluted antibody solution. After the mixture was incubated at 4°C for 45 min, the bound radioactivity in each sample of different plant species was determined with a liquid scintillation counter.
2.9. Measurement of the OsCPK gene expression
Expression of OsCPK9 and OsCPK25 in different cell lines was examined using Northern blotting by the method of Tang et al. [59, 60]. Twenty micrograms of total RNA of rice (Os1, Os2, Os3, Os4, Os5, and Os6) was applied. The PCR-amplified fragments of OsCPK9 (amplified by forward primer 5’-AAGTCGACACCGACAAGGAT-3’ and reverse primer 5’-TCTCAAAGCCTGAATCGACT-3’) and OsCPK25 (amplified by forward primer 5’-ACGTACTCCATCGGCAAAGT-3’ and reverse primer 5’-GATGATGCGGTCGAAGAGTT-3’) were labeled by Digoxigenin (DIG) (Roche Diagnostics Corporation, Roche Applied Science, Indianapolis, IN, USA). The labeled fragments were used as a hybridization probe for Northern blotting analysis.
2.10. Statistical analyses
Statistical analysis was performed using the General Linear Model procedure of SAS (Cary, NC, USA), employing ANOVA models. The significant differences between mean values of different cell lines of O. sativa, G. hirsutum, and P. elliottii were made at 5% level of probability. Each value of different cell lines of O. sativa, G. hirsutum, and P. elliottii is presented as mean ± standard deviations of the mean.
3. Results
3.1. Generation of transgenic cell lines
A. tumefaciens (Strain LBA4404) mediated genetic transformation was used to generate transgenic O. sativa, G. hirsutum, and P. elliottii lines (Fig. 1a), as described previously by Tang et al. [58]. Thirty-six O. sativa cell lines, thirty-eight G. hirsutum cell lines, and thirty-nine P. elliottii cell lines were infected with the A. tumefaciens strain LBA4404 containing pBI-ZAT6 (Fig. 1a). Transgenic cell lines were selected by kanamycin. Integration of the T -DNA into the genome was confirmed by PCR analysis (Fig. 1b). Seven, nine, and eight cell lines each with one copy of the ZAT6 gene were generated from G. hirsutum, O. sativa, and P. elliottii, respectively. After molecular analysis by PCR (Fig. 1b and c), Southern (Fig. 1d), and Northern blotting analysis (Fig. 1e), eighteen stable transgenic cell lines each containing only one copy of the pBI-ZAT6 T-DNA were obtained from rice, cotton, and slash pine and used for salt stress experiments.
Fig. 1.
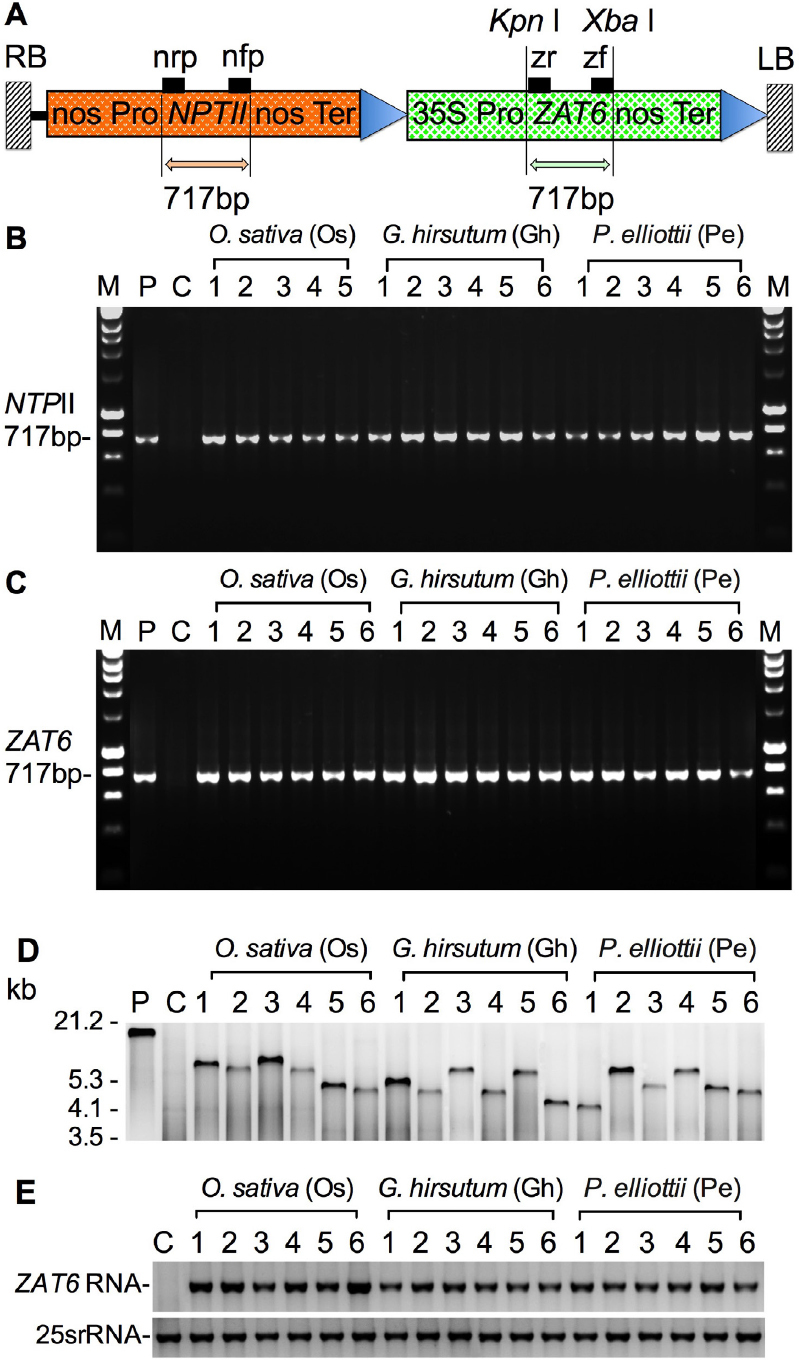
Plasmid map and molecular analyses of transgenic cell lines. (A) A linear plasmid map of pBI-ZAT6. (B) PCR analysis of NPTII gene in transgenic cell lines. (C) PCR analysis of ZAT6 gene in transgenic cell lines. (D) Southern blot analysis of transgenic cell lines. (E) Northern blot analysis of transgenic cell lines.s
3.2. Influence of ZAT6 overexpression on NaCl tolerance
Among different concentrations of NaCl tested, NaCl at 50–100 mM did not significantly decrease the rate of cell growth of transgenic lines in O. sativa, G. hirsutum, and P. elliottii, compared to the control (Fig. 2). NaCl at 150-300 mM significantly decrease the rate (Fig. 2). On medium supplemented with 150 mM NaCl, the rate of O. sativa cell growth reduced 49% (Fig. 2a), the rate of G. hirsutum cell growth decreased 49% (Fig. 2b), and the rate of P. elliottii cell growth decreased 82% (Fig. 2c). On medium supplemented with 300 mM NaCl, the rate of cell growth has the highest reduction in transgenic cell lines (Fig. 2a-c) of O. sativa, G. hirsutum, and P. elliottii.
Fig. 2.
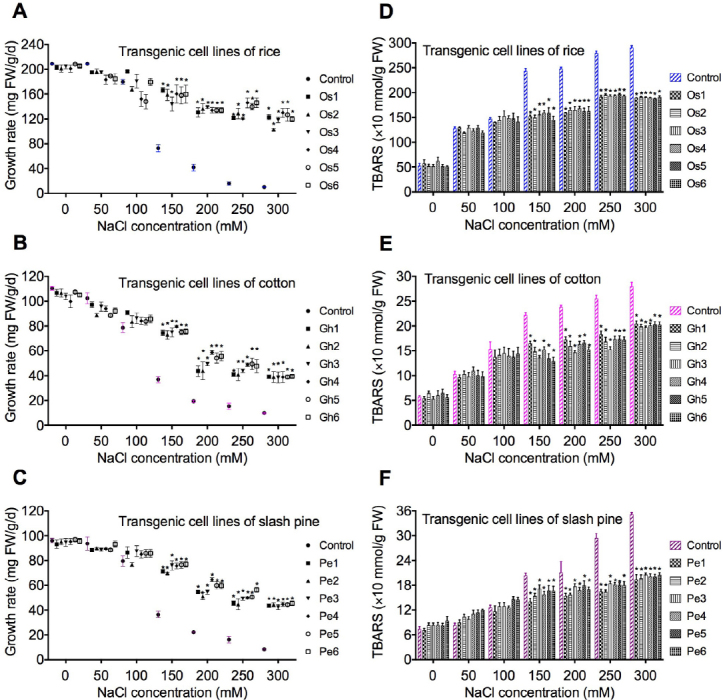
Growth of cell lines under different concentrations of NaCl and thiobarbituric acid reactive substance changes (TBARS). (A-C) Effect of different concentrations of NaCl (50, 100, 150, 200, 250, and 300 mM) on the growth of transgenic cell lines of rice (Os1, Os2, Os3, Os4, Os5, and Os6), cotton (Gh1, Gh2, Gh3, Gh4, Gh5, and Gh6), and slash pine (Pe1, Pe2, Pe3, Pe4, Pe5, and Pe6) were measured 21 days after callus transgenic cell cultures were transferred into media containing NaCl. (D-F) TBARS changes in transgenic cell lines of rice (Os1, Os2, Os3, Os4, Os5, and Os6), cotton (Gh1, Gh2, Gh3, Gh4, Gh5, and Gh6), and slash pine (Pe1, Pe2, Pe3, Pe4, Pe5, and Pe6). The growth and TBARS were measured 21 days after cell cultures were transferred into media containing different concentrations of NaCl (50, 100, 150, 200, 250, and 300 mM). Each experiment was replicated three times, and each replicate consisted of five to ten 250-ml flasks of transgenic cell cultures. Values represent the means ± S.D.
3.3. Influence of ZAT6 overexpression on lipid peroxidation
To examine salt stress-induced oxidative damage in transgenic O. sativa, G. hirsutum, and P. elliottii cells, TBARS content that acts as marker of lipid peroxidation was measured in salt stressed cells and in medium. Compared to the control, the total amounts of TBARS (cell cultures + incubation medium) were significantly reduced in transgenic cells overexpressing ZAT6 in O. sativa, G. hirsutum, and P. elliottii on medium supplemented with 150–300 mM NaCl (Fig. 2d-f). TBARS were not significantly reduced in transgenic O. sativa, G. hirsutum, and P. elliottii cells under 50–100 mM NaCl (Fig. 2d-f). Significant reduction in the products of lipid peroxidation in transgenic O. sativa, G. hirsutum, and P. elliottii cells demonstrated that the protection on membranes in transgenic O. sativa, G. hirsutum, and P. elliottii cells with overexpression of ZAT6 was demonstrated by the decrease of TBARS (Fig. 2d-f).
3.4. Effect of ZAT6 overexpression on ABA and GA8 content
Among different concentrations of NaCl applied, NaCl at 50–100 mM did not significantly decrease ABA content of transgenic cell lines in cotton, rice, and slash pine, compared to the control (Fig. 3). Compared to the control, NaCl at concentration of 150-300 mM significantly reduce ABA content in O. sativa, G. hirsutum, and P. elliottii (Fig. 3), on medium supplemented with 150 mM NaCl, ABA content of O. sativa cells decreased 49% (Fig. 3a), ABA content of G. hirsutum cell cultures decreased 49% (Fig. 3b), and ABA content of P. elliottii cell cultures decreased 82% (Fig. 3c). Under stress of 300 mM NaCl, ABA content has the highest increase (Fig. 3a-c) in O. sativa, G. hirsutum, and P. elliottii cells. A similar change of GA8 was obtained (Fig. 3a-c) in O. sativa, G. hirsutum, and P. elliottii cells.
3.5. Antioxidant enzymes APOX, GR, SOD, and CAT activities
Antioxidant enzymes APOX (Fig. 4a-c), GR (Fig. 4d-f), SOD (Fig. 5a-c), and CAT (Fig. 5d-f) were selected to evaluate the oxidative damage caused by NaCl stress in transgenic cell lines of rice, cotton, and slash pine. APOX, GR, SOD, and CAT activities were significantly reduced from the concentration point 150mM to 300 mM of NaCl in non-transgenic control (Figs. 4 and 5), however, the enzyme activity was stable in transgenic cells of O. sativa, G. hirsutum, and P. elliottii. Compared to controls, the levels of APOX, GR, SOD, and CAT activities were not significantly decreased from 50 to 150 mM NaCl in rice, cotton, and slash pine transgenic cell lines (Figs. 4 and 5).
Fig. 4.
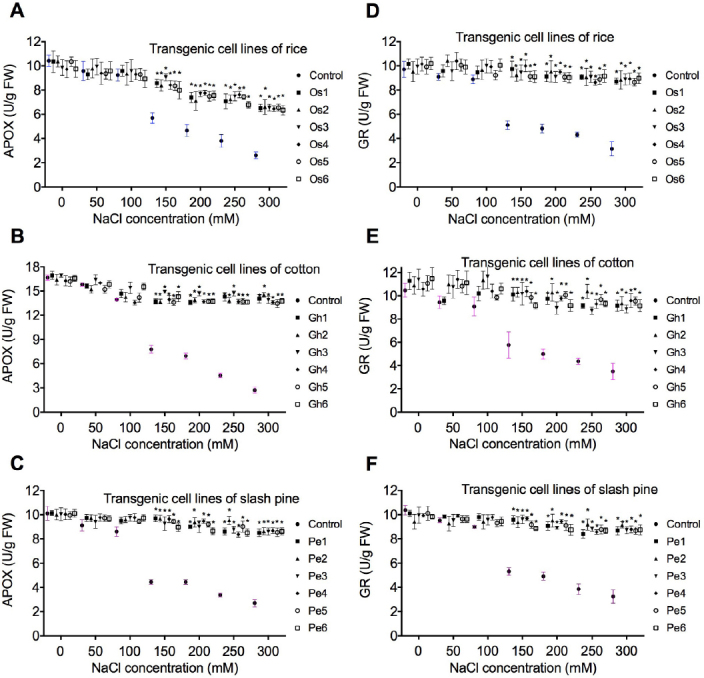
Effect of ZAT overexpression on APOX and GR activity. APOX (A-C) and GR (D-F) changes in transgenic cell lines of rice (Os1, Os2, Os3, Os4, Os5, and Os6), cotton (Gh1, Gh2, Gh3, Gh4, Gh5, and Gh6), and slash pine (Pe1, Pe2, Pe3, Pe4, Pe5, and Pe6) were measured 21 days after cell cultures were transferred into media containing different concentrations of NaCl (50, 100, 150, 200, 250, and 300 mM). Each experiment was replicated three times, and each replicate consisted of five to ten 250-ml flasks of transgenic cell cultures. Values represent the means ± S.D.
Fig. 5.
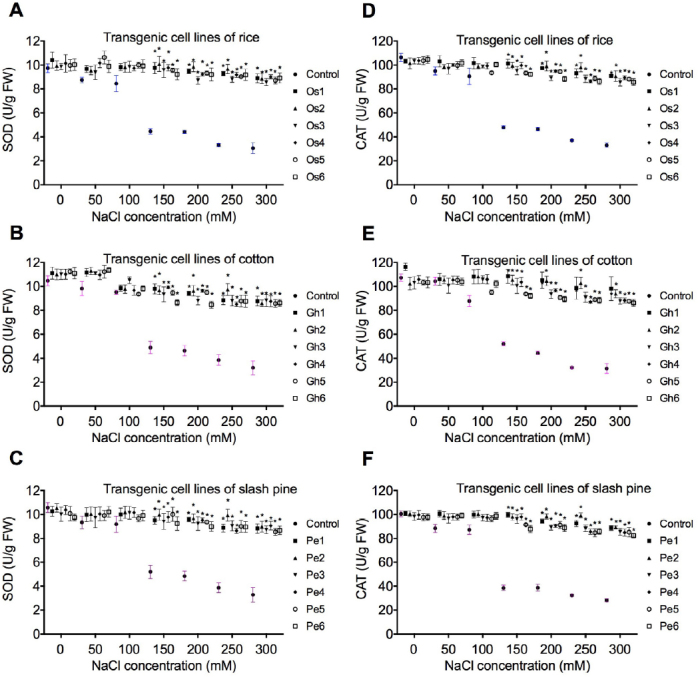
Effect of ZAT6 overexpression on SOD (A-C) and CAT (D-F) activity. SOD and CAT changes in transgenic cell lines of rice (Os1, Os2, Os3, Os4, Os5, and Os6), cotton (Gh1, Gh2, Gh3, Gh4, Gh5, and Gh6), and slash pine (Pe1, Pe2, Pe3, Pe4, Pe5, and Pe6) were measured 21 days after cell cultures were transferred into media containing different concentrations of NaCl (50, 100, 150, 200, 250, and 300 mM). Each experiment was replicated three times, and each replicate consisted of five to ten 250-ml flasks of transgenic cell cultures. Values represent the means ± S.D
3.6. Effect of ZAT6 on expression of Ca2+-dependent protein kinase genes
The Ca2+-dependent protein kinase genes OsCPK9 and OsCPK25 have been reported to be associated with stress tolerance in rice (Wan et al. 2007). To examine the effect of the ZAT6 on expression of OsCPK9 and OsCPK25, we have analyzed the expression of OsCPK9 and OsCPK25 in ZAT6 transgenic cells of O. sativa (Os1, Os2, Os3, Os4, and Os5) on medium supplemented with 150 mM NaCl (Fig. 6). These results showed that the expression level of OsCPK9 (Fig. 6 a-f) and OsCPK25 (Fig. 6g-l) increased 5 to 7-fold in transgenic cells of O. sativa, G. hirsutum, and P. elliottii, compared to the control.
Fig. 6.
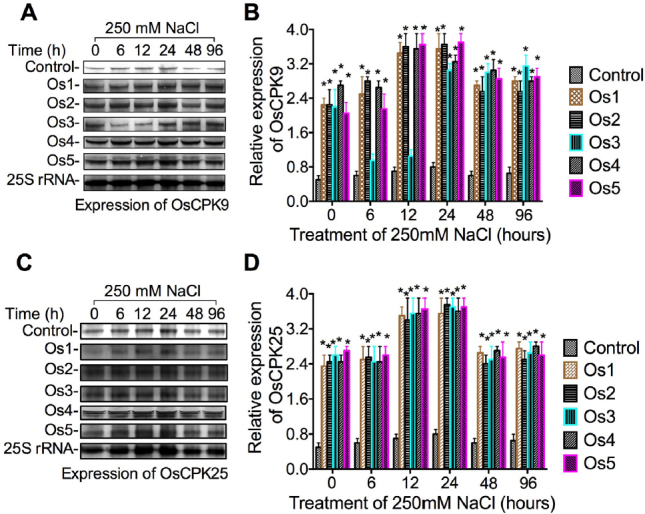
Influence of NaCl on expression of rice Ca2+-dependent protein kinase gene OsCPK9 and OsCPK25 in ZAT6 overexpression cells. (A) Northern blotting analysis of expression of OsCPK9 in ZAT6 transgenic cell lines Os1, Os2, Os3, Os4, and Os5 under 200 mM NaCl treatment at 6, 12, 24, 49, and 96 h. (B) Quantitative analysis of expression of OsCPK9. (c) Northern blotting analysis of expression of OsCPK25 in ZAT6 transgenic cell lines Os1, Os2, Os3, Os4, and Os5 under 200 mM NaCl treatment at 6, 12, 24, 49, and 96 h. (D) Quantitative analysis of expression of OsCPK25. Each experiment was replicated three times. Values represent the mean ± SD.
4. Discussion
TFs regulate NaCl stress response through different molecular mechanisms in a large number of plant species. It has been reported that overexpression of bZIP, AP2/ERF, MYB, WRKY TFs and other stress related genes in plant cells could increase NaCl stress tolerance [24, 25, 27, 34, 36, 38, 45, 63]. TF-regulated signaling transduction pathways and metabolic changes are essential for plant cells to counteract damages caused by NaCl stress [24, 27, 36, 38, 64, 65]. Overexpression of transcription factors in cells lead to improvement of NaCl stress tolerance in many plant species including Arabidopsis, wheat, rice, and Brachypodium distachyon [47, 66, 67, 68]. In potato, soybean, poplar, and pine, transcription factors have been reported to protect cells from NaCl stress through the decrease of oxidative damage and the increase of auxin signaling [43, 69, 70, 71]. Overexpression of some TFs lead to enhanced expression of antioxidant enzyme genes, induced metabolic reprogramming, and changes of stomatal closure in different plant species including Arabidopsis, tomato, tobacco, and pine, which resulted in increased NaCl tolerance [27, 43, 72, 73]. However, molecular mechanisms associated with TF ZAT6 increased NaCl stress tolerance is elusive.
Overexpression of stress-related transcription factor genes regulates salt stress response through regulation of ABA and GA metabolism and signaling [1, 74, 75]. It has been reported that, RAS1 (Response to ABA and Salt 1) enhance salt stress tolerance and ABA sensitivity as a negative regulator during early seedling growth [76], AtSAT32 [salt tolerance32 (SAT32)] functions on both salinity tolerance and ABA signaling as a positive regulator in Arabidopsis [1, 74, 77]. Endogenous ABA and GAs influence salt stress and endophytic fungal association in cucumber [78, 79]. GAs in fungal culture filtrate are involved in salinity induced oxidative stress in soybean plants through reduction of lipid peroxidation and regulation of the activities of the antioxidant enzymes [75, 80]. To examine if the enhanced salt stress via overexpression of ZAT6 is related to ABA and GA metabolism, we examined the content of ABA and GA8 in ZAT6 transgenic cell lines of O. sativa, G. hirsutum, and P. elliottii under treatment of different concentrations (50, 100, 150, 200, 250, and 300 mM) of NaCl (Fig. 3). Our experimental results showed that the content of ABA and GA8 was increased in transgenic O. sativa, G. hirsutum and P. elliottii cells, compared to the controls (Fig. 3). Our experimental results demonstrated that the increased ABA and GA8 could be contributed to the increased NaCl stress tolerance in transgenic G. hirsutum, O. sativa, and P. elliottii cells.
NaCl stress affects the activity of antioxidative enzymes and lipid peroxidation in plants. In maze, NaCl stress increase the activitis of superoxide dismutase (SOD), ascorbate peroxidase (APOX), and glutathione reductase (GR) to protect cells from oxidative damage, compared to the controls [81, 82]. In Broussonetia papyrifera, NaCl stress increases the activities of superoxide dismutase (SOD), peroxidase (POD) and catalase (CAT) in the leaves, stems and roots. The changed activities of antioxidant defense enzymes were associated with distinct SOD and POD isoenzymes [60, 61]. It has been reported that overexpression of ZAT6 in Arabidopsis enhances seed germination under salt and osmotic stress and that phosphorylation of ZAT6 by MPK6 is required for the increased salt and osmotic stress tolerance [60, 61]. To determine if the ZAT6-overexpression increased salt stress tolerance is related to the reduced oxidative damage, we have examined the activities of antioxidant enzymes APOX, GR, SOD, and CAT in ZAT6 transgenic O. sativa, G. hirsutum, and P. elliottii cells under treatment of NaCl (Figs. 4 and 5). Our results demonstrated that the activities of antioxidant enzymes APOX, GR, SOD, and CAT was increased in transgenic cells of rice, cotton, and pine, compared to the controls (Figs. 4 and 5), indicating that the increased activities of antioxidant enzymes could be related to the increased tolerance to NaCl stress in examined transgenic O. sativa, G. hirsutum and P. elliottii cell lines. Overexpression of ZAT6 may decrease oxidative damage, at least in part, through the increase of the activity of antioxidant enzymes.
Ca2+ dependent protein kinases (CPKs) are involved in NaCl stress tolerance through regulating expression of transcriptional activators or transducing stress signals to other signaling molecular. Cross talk between CPKs-mediated signal transduction and ABA signaling may regulate cellular homeostasis for NaCl stress tolerance. In rice, OsCPK9 improves NaCl stress tolerance by increasing the osmotic adjustment ability, expression of stress-responsive genes, and stomatal closure in plants [83]. In Arabidopsis, CPKs can phosphorylate the biotinylated peptide at the threonine residue for stress-response signaling pathway, regulate expression of several ABA responsive genes, be involved in oxidative stress and lipid metabolism [83]. To examine if the increasing salt stress tolerance through overexpression of ZAT6 is related to certain Ca2+ dependent protein kinases, we have investigated the expression of OsCPK9 and OsCPK25 in ZAT6 overexpressing transgenic cells of Os1, Os2, Os3, Os4, and Os5 under treatment of 250 mM NaCl (Fig. 6). Our experimental results showed that the expression of OsCPK9 and OsCPK25 was increased 5 to 7 fold in transgenic cells, compared to the non-transgenic control cells (Fig. 6), indicating that ZAT6 overexpression-enhanced expression of OsCPK9 and OsCPK25 could be associated with the increased NaCl stress tolerance in transgenic O. sativa, G. hirsutum and P. elliottii cell lines and that overexpression of ZAT6 may contribute to CPK-mediated signaling through cross talk between CPKs and ZAT6 under NaCl stress condition.
Although enhanced salt tolerance via overexpression of TFs has been documented in many different plant species [66, 67, 71, 84], molecular mechanisms of transcription factor-enhanced salt stress tolerance are not fully understood [70, 72, 73]. Overexpression of ZAT6 enhanced salt stress tolerance is not reported in cells of P. elliottii, O. sativa, and G. hirsutum. Different mechanisms could be contributed to salt tolerance in different plant species (Fig. 7). Overexpression of stress-responsive transcription factors plays important roles in the response to salt stress by coordinating the phytohormone signaling networks [15, 16, 85]. Overexpression of the TabHLH1 gene plays critical roles in plant tolerance to osmotic stresses through an ABA-dependent pathway [67]. Although cells use multiple strategies to improve salt tolerance, NaCl induces adverse effects by producing oxidative damage [5, 29, 34, 37, 86, 87]. In this study, we showed that increased tolerance to NaCl stress in cells of different plant species expressing transcription factor ZAT6 was related to the increased content of ABA and GA8, the increased activities of antioxidant enzymes, the increased expression of Ca2+- dependent protein kinase genes, and the decreased lipid peroxidation.
Fig. 7.
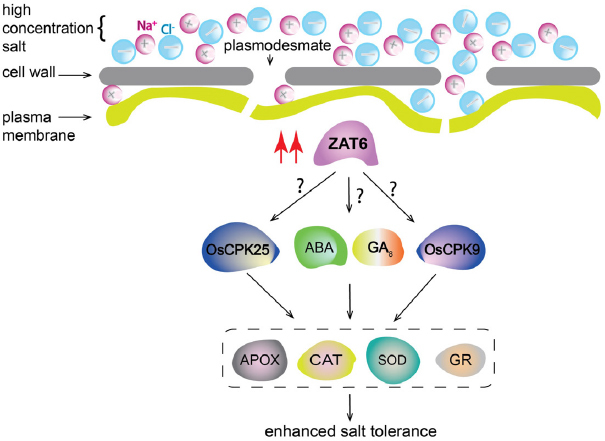
Molecular mechanism of ZAT6 overexpression enhanced salt tolerance. ZAT6 overexpression enhances expression of OsCPK25 and OsCPK25, as well as contents of ABA and GA8 in cells of angiosperm and gymnosperm, which may contribute to increased activity of Antioxidant enzymes APOX, CAT, SOD, and GR. Increased activity of APOX, CAT, SOD, and GR enhance salt tolerance in cells of angiosperm and gymnosperm.
Compared to use whole plants, it is easy to get consistent phenotypes of transformed cells. This is one reason that we use transgenic cell lines to examine the function of ZAT6 in P. elliottii, O. sativa, and G. hirsutum in the present investigation. Although we demonstrated that overexpression of ZAT6 enhanced salt stress tolerance and that the salt stress-associated changes of phytohormones, antioxidant enzymes, and CPKs in ZAT6 overexpression transgenic cell lines of P. elliottii, O. sativa, and G. hirsutum extended the discovery of ZAT6-improved seed germination under salt and osmotic stress in Arabidopsis, further investigation is needed to examine the function of ZAT6 in transgenic plants of P. elliottii, O. sativa, and G. hirsutum for practical application of transcription factor ZAT6 in agriculture and forestry
Although it has been reported that overexpression of ZAT6 improves seed germination under salt and osmotic stress and that phosphorylation of ZAT6 by MPK6 is required for the enhanced salt and osmotic stress tolerance in Arabidopsis [54, 57], overexpression of ZAT6 enhanced salt stress tolerance is not reported in crop plants and conifers. To explore the possible molecular mechanisms of ZAT6 overexpression-enhanced salt stress tolerance in crop plants and conifers, we overexpressed ZAT6 in cell lines of P. elliottii, O. sativa, and G. hirsutum and analyzed the changes of phytohormones, antioxidant enzymes, and CPKs in transgenic cell lines under salt stress. Overexpression of ZAT6 enhanced salt stress and the salt stress-associated changes of phytohormones, antioxidant enzymes, and CPKs in ZAT6 overexpression transgenic cell lines of P. elliottii, O. sativa, and G. hirsutum extended the discovery of ZAT6 improved seed germination under salt and osmotic stress in Arabidopsis. Overexpression of stress-related transcription factor genes regulates salt stress response [74, 77, 88]. It has been reported that, RAS1 (Response to ABA and Salt 1) enhance salt stress tolerance and ABA sensitivity as a negative regulator during early seedling growth [74, 77, 88], AtSAT32 [salt tolerance32 (SAT32)] functions on both salinity tolerance and ABA signaling as a positive regulator in Arabidopsis [1, 74]. Endogenous ABA and GAs influence salt stress and endophytic fungal association in cucumber (Khan et al. 2012). GAs in fungal culture filtrate are involved in salinity induced oxidative stress in soybean plants through reduction of lipid peroxidation and regulation of the activities of the antioxidant enzymes [89]. Cross talk among phytohormones signaling, activity of antioxidant enzymes, and expression of CPKs genes may be associated with ZAT6 overexpression-enhanced salt stress tolerance.
In conclusion, we have investigated the rate changes of cell growth, the content of ABA and GA8, the activities of antioxidant enzymes APOX, GR, SOD, and CAT, and lipid peroxidation in ZAT6 transgenic G. hirsutum, O. sativa, and P. elliottii cells, as well as expression of OsCPK9 and OsCPK25 in O. sativa. Our experimental results showed that overexpression of ZAT6 decreased NaCl-induced oxidative damage by elevating the expression level of Ca2+-dependent protein kinases and by elevating the activities of enzyme APOX, GR, and SOD and reducing lipid peroxidation (Fig. 7). The protection of transcription factor ZAT6 overexpression against salt stress-induced oxidative damage was related to the increasing expression of Ca2+-dependent protein kinase genes OsCPK9 and OsCPK25 in transgenic O. sativa cells. Overexpression of the ZAT6 transcription factor could be valuable approach for engineering plant abiotic stress tolerance.
Acknowledgments
The authors appreciate Nicki Karungo, Whitley Harikrishnan, Nicole Moskvin, Kuribayashi Bloom, and Ambrosia Nishimura for their help in maintaining transgenic cell cultures. The authors are grateful to Dr. Page and Dr. Thompson for their critical reading of the manuscript. This work was supported by a grant from the Education Committee of Hubei Providence of China (Grant No. D20101306).
Footnotes
Conflict of interest: Authors state no conflict of interest.
References
- [1].Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH. Overexpression of the tobacco Tsi1 gene encoding an EREBP/ AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell. 2001;13:1035–1046. doi: 10.1105/tpc.13.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Qin Y, Tian Y, Liu X. A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem Biophys Res Commun. 2015;464:428–433. doi: 10.1016/j.bbrc.2015.06.128. [DOI] [PubMed] [Google Scholar]
- [3].Sun P, Zhu X, Huang X, Liu JH. Overexpression of a stress-responsive MYB transcription factor of Poncirus trifoliata confers enhanced dehydration tolerance and increases polyamine biosynthesis. Plant Physiol Biochem. 2014;78:71–79. doi: 10.1016/j.plaphy.2014.02.022. [DOI] [PubMed] [Google Scholar]
- [4].Tsuzuki M, Moskvin OV, Kuribayashi M, Sato K, Retamal S, Abo M. Salt stress-induced changes in the transcriptome, compatible solutes, and membrane lipids in the facultatively phototrophic bacterium Rhodobacter sphaeroides. Appl Environ Microbiol. 2011;77:7551–7559. doi: 10.1128/AEM.05463-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vanacloig-Pedros E, Bets-Plasencia C, Pascual-Ahuir A, Proft M. Coordinated gene regulation in the initial phase of salt stress adaptation. J Biol Chem. 2015;290:10163–10175. doi: 10.1074/jbc.M115.637264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Allu AD, Soja AM, Wu A, Szymanski J, Balazadeh S. Salt stress and senescence: identification of crossߝtalk regulatory components. J Exp Bot. 2014;65:3993–4008. doi: 10.1093/jxb/eru173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dong L, Cheng Y, Wu J, Cheng Q, Li W, Fan S. Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J Exp Bot. 2015;66:2635–2647. doi: 10.1093/jxb/erv078. [DOI] [PubMed] [Google Scholar]
- [8].Fang L, Su L, Sun X, Li X, Sun M, Karungo SK. Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J Exp Bot. 2016;67:2829–2845. doi: 10.1093/jxb/erw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ganesan G, Sankararamasubramanian HM, Harikrishnan M, Ganpudi A, Parida A. A MYB transcription factor from the grey mangrove is induced by stress and confers NaCl tolerance in tobacco. J Exp Bot. 2012;63:4549–4561. doi: 10.1093/jxb/ers135. [DOI] [PubMed] [Google Scholar]
- [10].He Y, Li W, Lv J, Jia Y, Wang M, Xia G. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J Exp Bot. 2012;63:1511–1522. doi: 10.1093/jxb/err389. [DOI] [PubMed] [Google Scholar]
- [11].Meng LS, Wang YB, Yao SQ, Liu A. Arabidopsis AINTEGUMENTA mediates salt tolerance by trans-repressing SCABP8. J Cell Sci. 2015;128:2919–2927. doi: 10.1242/jcs.172072. [DOI] [PubMed] [Google Scholar]
- [12].Le Henanff G, Profizi C, Courteaux B, Rabenoelina F, Gerard C, Clement C. Grapevine NAC1 transcription factor as a convergent node in developmental processes, abiotic stresses, and necrotrophic/biotrophic pathogen tolerance. J Exp Bot. 2013;64:4877–4893. doi: 10.1093/jxb/ert277. [DOI] [PubMed] [Google Scholar]
- [13].Liu D, Chen X, Liu J, Ye J, Guo Z. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J Exp Bot. 2012;63:3899–3911. doi: 10.1093/jxb/ers079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nagaoka S, Takano T. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J Exp Bot. 2003;54:2231–2237. doi: 10.1093/jxb/erg241. [DOI] [PubMed] [Google Scholar]
- [15].Xu R, Wang Y, Zheng H, Lu W, Wu C, Huang J. Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J Exp Bot. 2015;66:5997–6008. doi: 10.1093/jxb/erv312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xu Z, Gongbuzhaxi Wang C, Xue F, Zhang H, Ji W. Wheat NAC transcription factor TaNAC29 is involved in response to salt stress. Plant Physiol Biochem. 2015;96:356–363. doi: 10.1016/j.plaphy.2015.08.013. [DOI] [PubMed] [Google Scholar]
- [17].Yang A, Dai X, Zhang WH. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot. 2012;63:2541–2556. doi: 10.1093/jxb/err431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot. 2009;60:3781–3796. doi: 10.1093/jxb/erp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen Y, Li F, Ma Y, Chong K, Xu Y. Overexpression of OrbHLH001, a putative helix-loop-helix transcription factor, causes increased expression of AKT1 and maintains ionic balance under salt stress in rice. J Plant Physiol. 2013;170:93–100. doi: 10.1016/j.jplph.2012.08.019. [DOI] [PubMed] [Google Scholar]
- [20].Cheng L, Li X, Huang X, Ma T, Liang Y, Ma X. Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol Biochem. 2013;70:252–260. doi: 10.1016/j.plaphy.2013.05.025. [DOI] [PubMed] [Google Scholar]
- [21].Diedhiou CJ, Popova OV, Golldack D. Transcript profiling of the salt-tolerant Festuca rubra ssp. litoralis reveals a regulatory network controlling salt acclimatization. J Plant Physiol. 2009;166:697–711. doi: 10.1016/j.jplph.2008.09.015. [DOI] [PubMed] [Google Scholar]
- [22].Kitashiba H, Ishizaka T, Isuzugawa K, Nishimura K, Suzuki T. Expression of a sweet cherry DREB1/CBF ortholog in Arabidopsis confers salt and freezing tolerance. J Plant Physiol. 2004;161:1171–1176. doi: 10.1016/j.jplph.2004.04.008. [DOI] [PubMed] [Google Scholar]
- [23].Li XW, Wang Y, Yan F, Li JW, Zhao Y, Zhao X. Overexpression of soybean R2R3-MYB transcription factor, GmMYB12B2, and tolerance to UV radiation and salt stress in transgenic Arabidopsis. Genet Mol Res. 2016;15:1–15. doi: 10.4238/gmr.15026573. [DOI] [PubMed] [Google Scholar]
- [24].Yao W, Wang L, Zhou B, Wang S, Li R, Jiang T. Over–expression of poplar transcription factor ERF76 gene confers salt tolerance in transgenic tobacco. J Plant Physiol. 2016;198:23–31. doi: 10.1016/j.jplph.2016.03.015. [DOI] [PubMed] [Google Scholar]
- [25].Yao W, Wang S, Zhou B, Jiang T. Transgenic poplar overexpressing the endogenous transcription factor ERF76 gene improves salinity tolerance. Tree Physiol. 2016;36:896–908. doi: 10.1093/treephys/tpw004. [DOI] [PubMed] [Google Scholar]
- [26].Zhao J, Zhi D, Xue Z, Liu H, Xia G. Enhanced salt tolerance of transgenic progeny of tall fescue (Festuca arundinacea) expressing a vacuolar Na+H+ antiporter gene from Arabidopsis. J Plant Physiol. 2007;164:1377–1383. doi: 10.1016/j.jplph.2007.04.001. [DOI] [PubMed] [Google Scholar]
- [27].Hartmann L, Pedrotti L, Weiste C, Fekete A, Schierstaedt J, Gottler J. Crosstalk between Two bZIP Signaling Pathways Orchestrates Salt-Induced Metabolic Reprogramming in Arabidopsis Roots. Plant Cell. 2015;27:2244–2260. doi: 10.1105/tpc.15.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liu C, Mao B, Ou S, Wang W, Liu L, Wu Y. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol. 2014;84:19–36. doi: 10.1007/s11103-013-0115-3. [DOI] [PubMed] [Google Scholar]
- [29].Moon SJ, Han SY, Kim DY, Yoon IS, Shin D, Byun MO. Ectopic expression of a hot pepper bZIP-like transcription factor in potato enhances drought tolerance without decreasing tuber yield. Plant Mol Biol. 2015;89:421–431. doi: 10.1007/s11103-015-0378-y. [DOI] [PubMed] [Google Scholar]
- [30].Pan Y, Seymour GB, Lu C, Hu Z, Chen X, Chen G. An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 2012;31:349–360. doi: 10.1007/s00299-011-1170-3. [DOI] [PubMed] [Google Scholar]
- [31].Tian Z, He Q, Wang H, Liu Y, Zhang Y, Shao F. The Potato ERF Transcription Factor StERF3 Negatively Regulates Resistance to Phytophthora infestans and Salt Tolerance in Potato. Plant Cell Physiol. 2015;56:992–1005. doi: 10.1093/pcp/pcv025. [DOI] [PubMed] [Google Scholar]
- [32].Wu D, Ji J, Wang G, Guan C, Jin C. LchERF, a novel ethylene-responsive transcription factor from Lycium chinense, confers salt tolerance in transgenic tobacco. Plant Cell Rep. 2014;33:2033–2045. doi: 10.1007/s00299-014-1678-4. [DOI] [PubMed] [Google Scholar]
- [33].Shen Z, Yao J, Sun J, Chang L, Wang S, Ding M. Populus euphratica HSF binds the promoter of WRKY1 to enhance salt tolerance. Plant Sci. 2015;235:89–100. doi: 10.1016/j.plantsci.2015.03.006. [DOI] [PubMed] [Google Scholar]
- [34].Wang L, Wang C, Qin L, Liu W, Wang Y. ThERF1 regulates its target genes via binding to a novel cis-acting element in response to salt stress. J Integr Plant Biol. 2015;57:838–847. doi: 10.1111/jipb.12335. [DOI] [PubMed] [Google Scholar]
- [35].Yan H, Jia H, Chen X, Hao L, An H, Guo X. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 2014;55:2060–2076. doi: 10.1093/pcp/pcu133. [DOI] [PubMed] [Google Scholar]
- [36].Guo H, Wang Y, Wang L, Hu P, Wang Y, Jia Y. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol J. 2017;15:107–121. doi: 10.1111/pbi.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang R, Jing W, Xiao L, Jin Y, Shen L, Zhang W. The Rice High-Affinity Potassium Transporter1;1 Is Involved in Salt Tolerance and Regulated by an MYB-Type Transcription Factor. Plant Physiol. 2015;168:1076–1090. doi: 10.1104/pp.15.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhu N, Cheng S, Liu X, Du H, Dai M, Zhou DX. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 2015;236:146–156. doi: 10.1016/j.plantsci.2015.03.023. [DOI] [PubMed] [Google Scholar]
- [39].Orellana S, Yanez M, Espinoza A, Verdugo I, Gonzalez E, Ruiz-Lara S. The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 2010;33:2191–2208. doi: 10.1111/j.1365-3040.2010.02220.x. [DOI] [PubMed] [Google Scholar]
- [40].Tak H, Mhatre M. Cloning and molecular characterization of a putative bZIP transcription factor VvbZIP23 from Vitis vinifera. Protoplasma. 2013;250:333–345. doi: 10.1007/s00709-012-0417-3. [DOI] [PubMed] [Google Scholar]
- [41].Xiang Y, Tang N, Du H, Ye H, Xiong L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008;148:1938–1952. doi: 10.1104/pp.108.128199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Matsukura S, Mizoi J, Yoshida T, Todaka D, Ito Y, Maruyama K. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol Genet Genomics. 2010;283:185–196. doi: 10.1007/s00438-009-0506-y. [DOI] [PubMed] [Google Scholar]
- [43].Tang W, Newton RJ, Lin J, Charles TM. Expression of a transcription factor from Capsicum annuum in pine calli counteracts the inhibitory effects of salt stress on adventitious shoot formation. Mol Genet Genomics. 2006;276:242–253. doi: 10.1007/s00438-006-0137-5. [DOI] [PubMed] [Google Scholar]
- [44].Shi W, Hao L, Li J, Liu D, Guo X, Li H. The Gossypium hirsutum WRKY gene GhWRKY39-1 promotes pathogen infection defense responses and mediates salt stress tolerance in transgenic Nicotiana benthamiana. Plant Cell Rep. 2014;33:483–498. doi: 10.1007/s00299-013-1548-5. [DOI] [PubMed] [Google Scholar]
- [45].Wang F, Chen HW, Li QT, Wei W, Li W, Zhang WK. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 2015;83:224–236. doi: 10.1111/tpj.12879. [DOI] [PubMed] [Google Scholar]
- [46].Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J. 2008;6:486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- [47].Yuan Y, Fang L, Karungo SK, Zhang L, Gao Y, Li S. Overexpression of VaPAT1, a GRAS transcription factor from Vitis amurensis, confers abiotic stress tolerance in Arabidopsis. Plant Cell Rep. 2016;35:655–666. doi: 10.1007/s00299-015-1910-x. [DOI] [PubMed] [Google Scholar]
- [48].Chen J, Yang L, Yan X, Liu Y, Wang R, Fan T. Zinc-Finger Transcription Factor ZAT6 Positively Regulates Cadmium Tolerance through the Glutathione-Dependent Pathway in Arabidopsis. Plant Physiol. 2016;171:707–719. doi: 10.1104/pp.15.01882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Devaiah BN, Nagarajan VK, Raghothama KG. Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol. 2007;145:147–159. doi: 10.1104/pp.107.101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Englbrecht CC, Schoof H, Bohm S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics. 2004;5:39. doi: 10.1186/1471-2164-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Li X, Zhang D, Li H, Wang Y, Zhang Y, Woody AJ. EsDREB2B, a novel truncated DREB2-type transcription factor in the desert legume Eremosparton songoricum, enhances tolerance to multiple abiotic stresses in yeast and transgenic tobacco. BMC Plant Biol. 2014;14:44. doi: 10.1186/1471-2229-14-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Li XL, Yang X, Hu YX, Yu XD, Li QL. A novel NAC transcription factor from Suaeda liaotungensis K. enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance. Plant Cell Rep. 2014;33:767–778. doi: 10.1007/s00299-014-1602-y. [DOI] [PubMed] [Google Scholar]
- [53].Liu HC, Charng YY. Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol. 2013;163:276–290. doi: 10.1104/pp.113.221168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Liu QL, Zhong M, Li S, Pan YZ, Jiang BB, Zhang HQ. Overexpression of a chrysanthemum transcription factor gene, DgWRKY3, in tobacco enhances tolerance to salt stress. Plant Physiol Biochem. 2013;69:27–33. doi: 10.1016/j.plaphy.2013.04.016. [DOI] [PubMed] [Google Scholar]
- [55].Mito T, Seki M, Shinozaki K, Ohme-Takagi M, Matsui K. Generation of chimeric repressors that confer salt tolerance in Arabidopsis and rice. Plant Biotechnol J. 2011;9:736–746. doi: 10.1111/j.1467-7652.2010.00578.x. [DOI] [PubMed] [Google Scholar]
- [56].Shi H, Chan Z. The cysteine2/histidine2-type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA 6-activated C-REPEAT-BINDING FACTOR pathway is essential for melatonin-mediated freezing stress resistance in Arabidopsis. J Pineal Res. 2014;57:185–191. doi: 10.1111/jpi.12155. [DOI] [PubMed] [Google Scholar]
- [57].Liu XM, Nguyen XC, Kim KE, Han HJ, Yoo J, Lee K. Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochem Biophys Res Commun. 2013;430:1054–1059. doi: 10.1016/j.bbrc.2012.12.039. [DOI] [PubMed] [Google Scholar]
- [58].Tang W, Newton RJ, Li C, Charles TM. Enhanced stress tolerance in transgenic pine expressing the pepper CaPF1 gene is associated with the polyamine biosynthesis. Plant Cell Rep. 2007;26:115–124. doi: 10.1007/s00299-006-0228-0. [DOI] [PubMed] [Google Scholar]
- [59].Tang W, Page M. Transcription factor AtbZIP60 regulates expression of Ca2+ -dependent protein kinase genes in transgenic cells. Mol Biol Rep. 2013;40:2723–2732. doi: 10.1007/s11033-012-2362-9. [DOI] [PubMed] [Google Scholar]
- [60].Tang W, Newton RJ. Peroxidase and catalase activities are involved in direct adventitious shoot formation induced by thidiazuron in eastern white pine (Pinus strobus L.) zygotic embryos. Plant Physiol Biochem. 2005;43:760–769. doi: 10.1016/j.plaphy.2005.05.008. [DOI] [PubMed] [Google Scholar]
- [61].Tang W, Charles TM, Newton RJ. Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus Virginiana Mill.) confers multiple stress tolerance and enhances organ growth. Plant Mol Biol. 2005;59:603–617. doi: 10.1007/s11103-005-0451-z. [DOI] [PubMed] [Google Scholar]
- [62].Okamoto M, Hanada A, Kamiya Y, Yamaguchi S, Nambara E. Measurement of abscisic acid and gibberellins by gas chromatography/mass spectrometry. Methods Mol Biol. 2009;495:53–60. doi: 10.1007/978-1-59745-477-3_5. [DOI] [PubMed] [Google Scholar]
- [63].Wang T, Tohge T, Ivakov A, Mueller-Roeber B, Fernie AR, Mutwil M. Salt-Related MYB1 Coordinates Abscisic Acid Biosynthesis and Signaling during Salt Stress in Arabidopsis. Plant Physiol. 2015;169:1027–1041. doi: 10.1104/pp.15.00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wang X, Han H, Yan J, Chen F, Wei W. A New AP2/ERF Transcription Factor from the Oil Plant Jatropha curcas Confers Salt and Drought Tolerance to Transgenic Tobacco. Appl Biochem Biotechnol. 2015;176:582–597. doi: 10.1007/s12010-015-1597-z. [DOI] [PubMed] [Google Scholar]
- [65].Wang Z, Zhang N, Zhou X, Fan Q, Si H, Wang D. Isolation and characterization of StERF transcription factor genes from potato Solanum tuberosum L.) C R Biol. 2015;338:219–226. doi: 10.1016/j.crvi.2015.01.006. [DOI] [PubMed] [Google Scholar]
- [66].Kang H, Zhang M, Zhou S, Guo Q, Chen F, Wu J. Overexpression of wheat ubiquitin gene, Ta-Ub2, improves abiotic stress tolerance of Brachypodium distachyon. Plant Sci. 2016;248:102–115. doi: 10.1016/j.plantsci.2016.04.015. [DOI] [PubMed] [Google Scholar]
- [67].Yang T, Yao S, Hao L, Zhao Y, Lu W, Xiao K. Wheat bHLH-type transcription factor gene TabHLH1 is crucial in mediating osmotic stresses tolerance through modulating largely the ABA-associated pathway. Plant Cell Rep. 2016;35:2309–2323. doi: 10.1007/s00299-016-2036-5. [DOI] [PubMed] [Google Scholar]
- [68].Zhou Y, Yang P, Cui F, Zhang F, Luo X, Xie J. Transcriptome Analysis of Salt Stress Responsiveness in the Seedlings of Dongxiang Wild Rice (Oryza rufipogon Griff.) PLoS One. 2016;11:e0146242. doi: 10.1371/journal.pone.0146242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Bouaziz D, Pirrello J, Charfeddine M, Hammami A, Jbir R, Dhieb A. Overexpression of StDREB1 transcription factor increases tolerance to salt in transgenic potato plants. Mol Biotechnol. 2013;54:803–817. doi: 10.1007/s12033-012-9628-2. [DOI] [PubMed] [Google Scholar]
- [70].Li Y, Xu B, Du Q, Zhang D. Association genetics and expression patterns of a CBF4 homolog in Populus under abiotic stress. Mol Genet Genomics. 2015;290:913–928. doi: 10.1007/s00438-014-0967-5. [DOI] [PubMed] [Google Scholar]
- [71].Pi E, Qu L, Hu J, Huang Y, Qiu L, Lu H. Mechanisms of Soybean Roots’ Tolerances to Salinity Revealed by Proteomic and Phosphoproteomic Comparisons Between Two Cultivars. Mol Cell Proteomics. 2016;15:266–288. doi: 10.1074/mcp.M115.051961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Campos JF, Cara B, Perez-Martin F, Pineda B, Egea I, Flores FB. The tomato mutant ars1 (altered response to salt stress 1) identifies an R1-type MYB transcription factor involved in stomatal closure under salt acclimation. Plant Biotechnol J. 2016;14:1345–1356. doi: 10.1111/pbi.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Chen H, He H, Yu D. Overexpression of a novel soybean gene modulating Na+ and K+ transport enhances salt tolerance in transgenic tobacco plants. Physiol Plant. 2011;141:11–18. doi: 10.1111/j.1399-3054.2010.01412.x. [DOI] [PubMed] [Google Scholar]
- [74].Park HY, Seok HY, Woo DH, Lee SY, Tarte VN, Lee EH. AtERF71/HRE2 transcription factor mediates osmotic stress response as well as hypoxia response in Arabidopsis. Biochem Biophys Res Commun. 2011;414:135–141. doi: 10.1016/j.bbrc.2011.09.039. [DOI] [PubMed] [Google Scholar]
- [75].Rahaie M, Xue GP, Naghavi MR, Alizadeh H, Schenk PM. A MYB gene from wheat (Triticum aestivum L.) is up-regulated during salt and drought stresses and differentially regulated between salt-tolerant and sensitive genotypes. Plant Cell Rep. 2010;29:835–844. doi: 10.1007/s00299-010-0868-y. [DOI] [PubMed] [Google Scholar]
- [76].Reis RR, da Cunha BA, Martins PK, Martins MT, Alekcevetch JC, Chalfun A. Induced over–expression of AtDREB2A CA improves drought tolerance in sugarcane. Plant Sci. 2014;221-222:59–68. doi: 10.1016/j.plantsci.2014.02.003. [DOI] [PubMed] [Google Scholar]
- [77].Park J, Yoon YS, Han HS, Kim YH, Ogawa Y, Park KG. Diabetes. 2014;63:SIK2 is critical in the regulation of lipid homeostasis and adipogenesis in vivo. 3659–3673. doi: 10.2337/db13-1423. [DOI] [PubMed] [Google Scholar]
- [78].Kang HG, Kim J, Kim B, Jeong H, Choi SH, Kim EK. Overexpression of FTL1/DDF1, an AP2 transcription factor, enhances tolerance to cold, drought, and heat stresses in Arabidopsis thaliana. Plant Sci. 2011;180:634–641. doi: 10.1016/j.plantsci.2011.01.002. [DOI] [PubMed] [Google Scholar]
- [79].Kant P, Kant S, Gordon M, Shaked R, Barak S. STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol. 2007;145:814–830. doi: 10.1104/pp.107.099895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ramegowda V, Senthil-Kumar M, Nataraja KN, Reddy MK, Mysore KS. Expression of a finger millet transcription factor, EcNAC1, in tobacco confers abiotic stress-tolerance. PLoS One. 2012;7:e40397. doi: 10.1371/journal.pone.0040397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhang H, Zhang J, Quan R, Pan X, Wan L, Huang R. EAR motif mutation of rice OsERF3 alters the regulation of ethylene biosynthesis and drought tolerance. Planta. 2013;237:1443–1451. doi: 10.1007/s00425-013-1852-x. [DOI] [PubMed] [Google Scholar]
- [82].Zhang XX, Tang YJ, Ma QB, Yang CY, Mu YH, Suo Hc. OsDREB2A, a rice transcription factor, significantly affects salt tolerance in transgenic soybean. PLoS One. 2013;8:e83011. doi: 10.1371/journal.pone.0083011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wei W, Jiang J, Li X, Wang L, Yang SS. Isolation of salt-sensitive mutants from Sinorhizobium meliloti and characterization of genes involved in salt tolerance. Lett Appl Microbiol. 2004;39:278–283. doi: 10.1111/j.1472-765X.2004.01577.x. [DOI] [PubMed] [Google Scholar]
- [84].Tang W, Tang AY. MicroRNAs associated with molecular mechanisms for plant root formation and growth. Journal of Forestry Research. 2016;27:1–12. [Google Scholar]
- [85].Tang W, Tang AY. Transcriptional mechanisms regulating gene expression and determining cell fates in plant development. Journal of Forestry Research. 2017;28:863–880. [Google Scholar]
- [86].Lehti-Shiu MD, Uygun S, Moghe GD, Panchy N, Fang L, Hufnagel DE. Molecular Evidence for Functional Divergence and Decay of a Transcription Factor Derived from Whole-Genome Duplication in Arabidopsis thaliana. Plant Physiol. 2015;168:1717–1734. doi: 10.1104/pp.15.00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sakuraba Y, Piao W, Lim JH, Han SH, Kim YS, An G. Rice ONAC106 Inhibits Leaf Senescence and Increases Salt Tolerance and Tiller Angle. Plant Cell Physiol. 2015;56:2325–2339. doi: 10.1093/pcp/pcv144. [DOI] [PubMed] [Google Scholar]
- [88].Rao SS, El-Habbak MH, Havens WM, Singh A, Zheng D, Vaughn L. Overexpression of GmCaM4 in soybean enhances resistance to pathogens and tolerance to salt stress. Mol Plant Pathol. 2014;15:145–160. doi: 10.1111/mpp.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Roychoudhury A, Paul S, Basu S. Cross–talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 2013;32:985–1006. doi: 10.1007/s00299-013-1414-5. [DOI] [PubMed] [Google Scholar]


