Abstract
Skewed sex development is prevalent in fish hybrids. However, the histological observation and molecular mechanisms remain elusive. In this study, we showed that the interspecific hybrids of the two fish species, Oreochromis niloticus and Oreochromis aureus, had a male ratio of 98.02%. Microscopic examination revealed that the gonads of both male and female hybrids were developmentally retarded. Compared with the ovaries, the testes of both O. niloticus and hybrids showed higher DNA methylation level in two selected regions in the promoter of cyp19a, the gonadal aromatase gene that converts androgens into estrogens, cyp19a showed higher level gene expression in the ovary than in the testis in both O. niloticus and hybrid tilapia. Methylation and gene expression level of cyp19a were negative correlation between the testis and ovary. Gene transcription was suppressed by the methylation of the cyp19a promoter in vitro. While there is no obvious difference of the methylation level in testis or ovary between O. niloticus and hybrids. Thus, the DNA methylation of the promoter of cyp19a may be an essential component of the sex maintenance, but not a determinant of high male ratio and developmental retardation of gonads in tilapia hybrids.
Keywords: Oreochromis niloticus, Oreochromis aureus, hybridization, DNA Methylation, gonadal aromatase
1. Introduction
Tilapias are the second most-farmed fish group in world fisheries. Nile tilapia (Oreochromis niloticus) is an excellent animal model for teleost sex control [1], fish threpsology [2], or environmental adaption [3]. Many studies have explored the mechanisms of sex determination in Nile tilapia that confer commercial value, such as the high growth rates of all-male progenies [4, 5]. Hormone treatment, temperature control and interspecific hybridization are the main approaches to produce all male fish. Of the three methods, interspecific hybridization is the environmentally most sustainable one. Hybridization between some species of tilapias, such as Nile tilapia and blue tilapia (Oreochromis aureus), results in predominantly male offspring [6]. Nevertheless, the nature and complexity of hybrid incompatibility remain poorly understood.
DNA methylation is essential for normal development and is associated with various key processes, such as gene expression regulation, genomic imprinting, X-chromosome inactivation, transposable element repression, and aging. Cytosine methylation is widespread in both eukaryotes and prokaryotes [7, 8]. In our previous study, we utilized methylated DNA immunoprecipitation (Me-DIP) to generate the methylome of the Nile tilapia testis and ovary. We produced a candidate gene repository that provides options for exploring the relationship between DNA methylation and sex maintenance, the gene repository includes the cyp19a, estrogen receptor (esr) and other sex-associated genes [9]. DNA methylation is a potential regulatory mechanism for temperature induced sex change in Nile tilapia [10]. While the involvement of DNA methylation and abnormal expression of cyp19a in the sex development of tilapia hybrids remains unclear. We explored the possibility that the methylation state of cyp19a could lead to high male ratio and developmental retardation of gonads in tilapia hybrids.
2. Materials and Methods
2.1. Fish and histology examination
Nile tilapias and Blue tilapias were maintained at the Xinchang Aquafarm of Shanghai Ocean University. The natural fertilization of these two fish species (O. niloticus ♀× O. aureus ♂) was accomplished. Fertilized hybrid eggs were hatched in parallel to fertilized Nile tilapia eggs. Fish larvae were bred in an automatic recycled water cement pool. Water temperature was maintained at 26 ± 1°C. All experimental fishes were fed with formula feed three times a day.
The implemented procedures were approved by the Ethics and Animal Care Committee of China. All experimental procedures were conducted in conformity with institutional guidelines for the care and use of laboratory animals. On 60 days after hatching (DAH), male and female Nile tilapia or hybrids were sacrificed and gonadal samples were collected (n > 5 fish per group). A portion of the right gonad was processed for the histological identification of sex. The remaining portions were snap-frozen in liquid nitrogen and stored at −80°C until further analysis to determine methylation levels and gene expression. Mature gonads (from 120 DAH fish) were collected for histological observation.
For histological examination, the gonads of Nile tilapia and hybrid fish were fixed with Bouin solution, dehydrated in ethanol gradient series, and then embedded in paraffin. All samples were cut into 50 serial sections, followed by staining with hematoxylin and eosin for anatomical visualization. Immunohistochemical studies were conducted as our previous study [11]. Microscopic observations and photographs were taken using a Nikon Eclipse Ni-E microscope (Nikon, Japan).
Ethical approval: The research related to animals use has been complied with all the relevant national regulations and institutional policies for the care and use of animals.
2.2. cyp19a sequence analysis and qRT-PCR
Genomic DNA and Total RNA were extracted using a QIAamp DNA Mini Kit (Qiagen, Germany) and RNeasy Mini kit (Qiagen, Germany) in accordance with the manufacturer’s instructions. Three pairs of conserved primers (cyp19a1s/cyp19a1s, cyp19a2s/cyp19a2a and cyp19a3s/cyp19a3a) were designed based on the sequence of the Nile tilapia cyp19a. The genomic DNA of hybrid fish was used as a template to amplify and obtain the whole length of the hybrid tilapia cyp19a gene. Synteny comparisons across different vertebrate species were performed in silico via BLAST searches using the ENSEMBL database.
The cyp19a gene was amplified via qRT-PCR using the specific primers cyp19aqs and cyp19aqa (Table S1). qRT-PCR was performed on a Bio-Rad PCR system using iTaq Universal SYBR Green Supermix (BioRad, USA) in accordance with the manufacturer’s instructions. Relative cyp19a mRNA expression was evaluated using the 2−ΔΔCT method with an initial normalization of cyp19a against β actin. Each PCR trial was conducted with three samples and repeated at least three times in all experiments.
Table S1.
Primers used in this study.
| Primer | Sequences | Application |
|---|---|---|
| cyp19a1s | CACTCTCTGTTGCACTAGCTTG | cyp19a cloning |
| cyp19a1a | AATGCAAAAGCCACAACACTGC | |
| cyp19a2s | GCAGTGTTGTGGCTTTTGCATT | cyp19a cloning |
| cyp19a2a | GATCACCATCTCCAGCACGCAC | |
| cyp19a3s | GTGCGTGCTGGAGATGGTGATC | cyp19a cloning |
| cyp19a3a | CATTTGTGACACAATAACTTAC | |
| M40s | TGTATTAGTTTGTAATGTGTAGTGG | P1 methylation analysis |
| M465a | TCTAAATCAATCTCTCTAAAAAAAA | |
| M692s | GGTTGATTATAATTTAGAGTTTAGAGA | P2 methylation analysis |
| M913a | ACATAAAAAACCCTACAAACTCACC | |
| cyp19apros | CAGTAAAGGCTACACTCTCTG | Promoter TA cloning |
| cyp19aproa | TCTGCCACCACGGCGTCTAA | |
| cyp19alucLs | GGCTCGAGTGCACTAGCTTGTAATG | Luciferase assay vector construction |
| cyp19alucLa | GGTTCGAATGAGAAGGGTGATGATGTA | |
| cyp19alucSs | GGCTCGAGCATGTACTAGTGATAAAG | Luciferase assay vector construction |
| cyp19alucSa | GGTTCGAATGAGAAGGGTGATGATGTA | |
| cyp19aqs | GGCAGATACGCTGGACAACA | qRT-PCR of cyp19a |
| cyp19aqa | TGGCAGGCGGAAAAGAAAT | |
| NTactins | CAGCAGATGTGGATCAGCAAGC | qRT-PCR reference gene |
| NTactina | TGAAGTTGTTGGGCGTTTGG |
2.3. Bisulfite specific PCR
DNA bisulfite treatment of tilapia DNA were conducted with the EpiTect Bisulfite Kit (Qiagen, USA) in accordance with the manufacturer’s instructions. The product was diluted with 20 μL of elution buffer. Bisulfate-treated DNA was amplified by PCR using specific primers (M40s/M465a and M692s/M913a) (Table 1S). The PCR product was cloned and sequenced. The total number of methylated cytosine was calculated as the average of the total number of methylated cytosines across the sequenced clones. Five fish were tested in every group, the results were expressed as mean ± standard deviation. P<0.05 indicated statistically significant difference.
2.4. Plasmid construction, cell culture and luciferase assay
The fragment of the cyp19a promoter region was amplified by PCR using the primers cyp19apros/cyp19aproa. The cloned products were subcloned into a pMD 19-T vector (Takara, Japan). Then, the 2172- and 1357-bp fragments were amplified with a second PCR run using the primer pairs cyp19alucLs/cyp19alucLa and cyp19alucSs/cyp19alucSa (Table S1), and were then inserted into the Xhol and HindIII restriction enzyme cutting sites of the pGL3-Basic plasmid, respectively.
Recombinant pGL3-Basic plasmids were cytosine-methylated using SssI methylase (New England BioLabs, USA) in accordance with the manufacturer’s instructions. SssI methylates all cytosine residues within the double-stranded dinucleotide recognition sequence (5′-CpG-3′), by using 10 mM Tris, pH 7.9, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, and 160 μM S-adenosylmethionine at 37°C for 1 h. After methylation, reaction plasmids were purified by phenol extraction. Successful vector methylation was confirmed by gel electrophoresis after digesting with the McrBC enzyme (New England BioLabs), which digests only methylated DNA. Fully methylated plasmids were utilized for transient transfection assays.
Human embryonic kidney (HEK) 293 cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, USA) that was supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 incubator. The luciferase activity assay was conducted as as mentioned by Gu et al. [12] HEK293 cells were seeded in 24-well plates overnight and co-transfected with 100 ng of the luciferase reporter plasmids lpCYP19a, lmCYP19a, spCYP19a, and smpCYP19a, and 10 ng of pRL-TK (Promega, USA). At 48 hours after transfection, the transfected cells were harvested and lysed in accordance with the Dual-Luciferase Reporter Assay System (Promega, USA). Luciferase activities were measured with a Junior LB9509 Luminometer (Berthold Detection System, Germany) and normalized to Renilla luciferase activities in accordance with standard protocol. The experiment was repeated three times. Each independent experiment was performed in triplicate.
Statistical analysis was dong by SPSS 17.0 (SPSS, Inc., Chicago, IL). Values were expressed as the mean±standard deviation. Student’s t-test was used to compare the difference in means between the two groups. P<0.05 was considered to be statistically significant.
3. Results
3.1. Culture of tilapia hybrids and histological observation
The male Nile tilapia and hybrid tilapia weighed 10.96±1.01 g and 8.91±1.57 g respectively, the female Nile tilapia and hybrid tilapia reached 7.99±1.64 g and 6.76±1.27 g at the end of the 2-month culture period. Therefore, the males were heavier than the females. Moreover, male Nile tilapia were heavier than male hybrid tilapia. Body length was also significantly different. More importantly, the male ratio of hybrids was 98.02% (Figure 1A).
Figure 1.
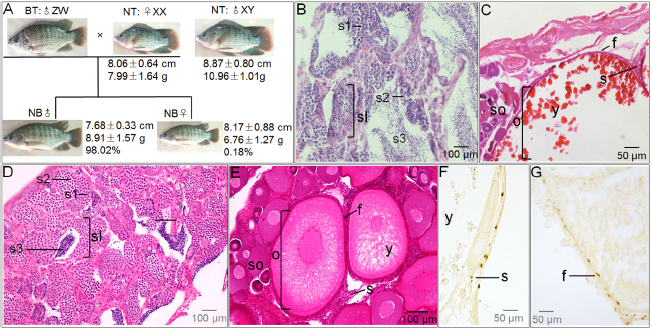
Fish morphology and gonad histology of NT and NB. (A) Average body length and body weight of 2-month-old female NT, male NT, and NB. (B) and (C) indicate transverse sections of the testis and ovary of adult NT (4 months old). (D) and (E) indicate transverse sections of the testis and ovary of adult NB (4 months old). (F) and (G) indicate the immunological analysis of Cyp19a in the ovary of NT and NB. sl: seminiferous lobula; s1: spermatogonia; s2: spermatocytes; s3: spermatides; o: oocytes containing numerous vacuoles and deeply staining nucleus; so: small oocytes surrounded by still small oocytes; f: follicle cell; y: yolk; s: stroma of connective tissue; NT: Nile tilapia; BT: Blue tilapia; NB: tilapia hybrid.
Sections of testes from 4-month-old Nile tilapia exhibited normal tissue architecture and sperm cell distribution. Tissue architecture and cell development in male hybrid fish were not different from those in Nile tilapia. But fewer spermatozoids were observed in the seminiferous tubules of male hybrid fish. Spermatogonia, spermatocytes, and spermatids occupied the majority of the testes of male hybrid fish (Fig. 1B, D). Female Nile tilapia exhibited normal ovarian histology that was characterized by oocytes in advanced vitellogenesis (Fig. 1C). By contrast, the gonadal development of female hybrid fish was characterized by oocytes in initial, mid, and immature vitellogenesis (Fig. 1E). Immunohistochemical analysis showed that Cyp19a was distributed in the follicle and stroma cells in ovaries of both Nile tilapia and hybrid fish (Fig. 1F, G).
3.2. Structure analysis of the tilapia cyp19a gene
Cross-species comparison of the chromosomal location indicates the cyp19a gene is flanked by several genes, including gliomedin (gldn), tnf alpha induced protein 8 like 3 (tnfaip813), and secretogranin 3 (scg3) etc. They are located on different chromosomes of four organisms, which is indicated that the cyp19a-containing region in Nile tilapia chromosome LG1 is syntenic to medaka chromosome 3, zebrafish chromosome 18, and human chromosome 15. The tilapia shares complete similarity with medaka and minor differences with zebrafish and human (Fig. 2A). The full-length cyp19a gene, including promoter regions, from Nile and hybrid tilapia was cloned using three pairs (cyp19a1s/cyp19a1a, cyp19a2s/cyp19a2a, and cyp19a3s/cyp19a3a) of primers. Cloning and sequencing results confirmed that Nile tilapia and hybrid fish shared 100% identical. Similar to the cyp19a of zebrafish, Chinese softshell turtle, and chicken, the cyp19a gene of tilapia has nine exons. The cyp19a in medaka and humans has 10 exons. These homologous genes have significantly different intron lengths (Fig. 2B).
Figure 2.
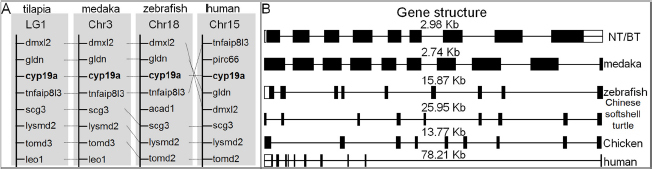
Gene structure analysis of tilapia cyp19a. (A) Chromosome synteny of tilapia cyp19a. (B) Comparison of cyp19a gene structures from different species. Exons are numbered with their sizes indicated in base pairs. Chr: chromosome; LG: linkage group.
3.3. DNA methylation and gene expression of tilapia cyp19a
The 5191-bp DNA sequence that encompassed the promoter and transcription regions with the 103 CpG sites of cyp19a gene was analyzed (Fig. S1). The CpG sites methylation ratio of male to female from positions 1–35 and positions 60–80 were all greater than 0.3. The methylation ratio of the promoter was higher than that of the transcript region in total (Figure 3A). cyp19a expression was higher in the ovary than in the testis in both O. niloticus and hybrids (Figure 3B). Two DNA regions, P1 and P2, in the promoter were verified by BSP analysis. The methylation level of males was all higher than those of females in both Nile tilapia and hybrid fish (P<0.05). Methylation and gene expression level of cyp19a were negatively correlated in the testis and ovary in both species (Figure 3C–D). All the methylated CpG sites in P1 and P2 showed significant difference between the ovary and testis. In particular, two or six CpGs were differentially methylated in the testis or ovary between Nile tilapia and hybrid tilapia respectively (Figure 3E).
Figure 3.
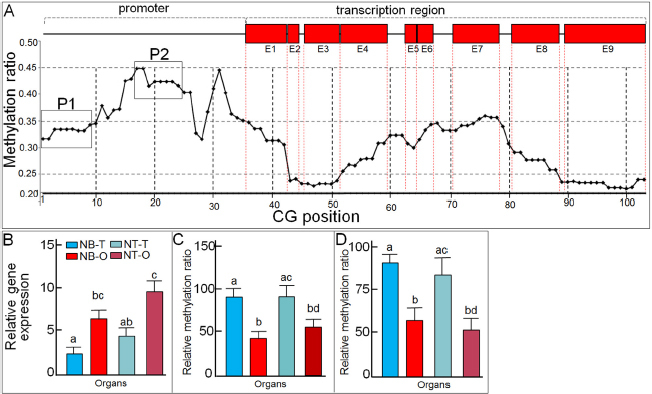
Analysis of DNA methylation and mRNA expression of cyp19a. (A) Methylation ratio of all CpG sites of NT cyp19a. The red box in E1-9 indicates nine exons, and the line indicates intron and promoter regions. The methylation ratio level of testis to ovary of 103 CpG sites is shown in the coordinate axis. (B) The relative mRNA expression of cyp19a in the testis and ovary of NT and BT. (C) and (D) shows the methylation level of P1 and P2 regions on the promoter of cyp19a. Columns with different colors indicate different organs of NT and BT as shown in figure (B). Values with different superscripts on the columns in (B), (C), and (D) are significantly different (P< 0.05). O: ovary; T: testis; NT: Nile tilapia; NB: tilapia hybrid.
3.4. Methylation of tilapia cyp19a promoter blocks its expression in vitro
The luciferase reporter assay was used to determine the activation of the cyp19a promoter under methylated and unmethylated conditions. Promoter length was related to the reported gene transcription, the longer promoter showed higher transcriptional activity than the shorter one. Remarkably, the induced hypermethylation of the cyp19a promoter suppressed promoter transcription in both the experimental groups (Figure 4).
Figure 4.
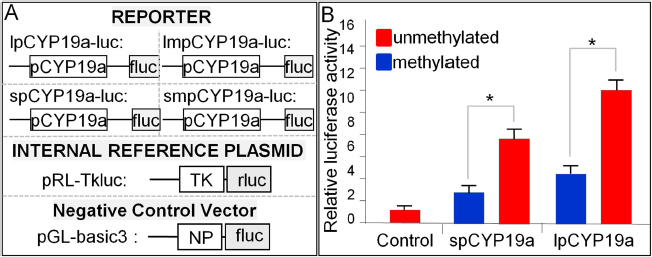
Luciferase reporter assay. (A) Plasmid construction. TK, human thymidine kinase promoter; fluc, firefly luciferase; rluc, renilla luciferase; lpCYP19a: long promoter of tilapia cyp19a; lmpCYP19a: long methylated promoter of tilapia cyp19a; spCYP19a: short promoter of tilapia cyp19a; smpCYP19a: short methylated of tilapia cyp19a; NP: no promoter. (B) Luciferase assay. Different groups of reporter plasmid of cyp19a promoter plus pRL-TK (internal control reporter) were co-transfected into HEK293 cells. At 24 h post-transfection, the cells were lysed for the luciferase assay. The asterisks indicates P<0.05 between two groups.
4. Discussion
In aquaculture, the preferred sex of tilapia is often produced via exogenous steroid treatment or temperature manipulations to artificially induce sex reversal. Hybrids of O. niloticus♀ ×O. aureus♂, which are commonly bred in aquaculture have growth rates that are significantly higher than those of their parent species [6, 13, 14]. The results of the current study are consistent with above reports. O. niloticus ♀×O. aureus♂ hybrids have significantly higher growth performance and male ratio than their parents. The male hybrids grow slower than male Nile tilapia but faster than female Nile tilapia. The growth improvement of hybrid fish results from the combination of the heterosis effect. These traits make the almost all-male hybrids more suitable for culture than either parental species. The available data on the growth traits of hybrids reported have minor difference [14, 15] because of different experimental fish strains and aquaculture environments, including temperature, density, and daily feeding dose. Here, we studied the “new GIFT” strain of Nile tilapia, which has been artificially selected for more than 20 years based on growth rate. This specific strain has excellent and stable disease resistance. Thus, the O. niloticus and O. aureus in this study are both perfect fundamental breeding materials.
Although tilapia hybrids were fertile in our aquafarm, we confirmed that the gonads of hybrids acquired developmental retardation compared with those of Nile tilapia. The ovaries of the hybrids remained in immature vitellogenesis. The nature and complexity of hybrid incompatibility remains poorly understood. Misexpression has been reported in many animal hybrids, including fruit flies [16, 17], mice [18, 19], African clawed frog [20], and whitefish [21]. Several known hybrid incompatibility genes affect the transcription of other genes, including a regulatory homeodomain protein [16] and the mouse sterility gene histone-lysine N-methyltransferase (ehmt1) [22]. Gene expression mapping showed that the X chromosome has a massive effect on testis gene expression in hybrid mice. The majority of the X chromosome is significantly enriched for quantitative trait loci that affect the expression of autosomal genes [23].
DNA methylation is essential for normal development including sex development. Numerous approaches exist to decipher whole-genome DNA methylation profile, such as methylated DNA immunoprecipitation (MeDIP-seq), which is more sensitive to highly methylated and high-CpG densities. BS-seq involving treating DNA with sodium bisulfite combined with subsequent high-throughput sequencing is the gold standard for whole genome DNA methylation analysis[24]. It has been reported that the methylation of various chromosome regions in both female and male tilapia increase after high-temperature induction [10]. We utilized MeDIP-seq to generate the methylome of the Nile tilapia testis and ovary. It produced a candidate gene repository to explore the relationship between DNA methylation and sex maintenance, we also performed a thorough analyses of whole genome DNA methylome data of testis and ovary from Nile tilapia [9]. And on this basis, cyp19a, an enzyme with clear function of the biosynthesis of estrogens, was selected to analyze in different fish individuals and species in this study. Comprehensive analysis of all CpG and transcription factor binding sites is very important in future research. Generally speaking, the majority of gene promoter regions are poorly methylated. Promoter DNA methylation results in a compact chromatin structure and acts as a repressive signal for gene transcription [25]. Maintenance of very low cyp19a mRNA levels is a prerequisite for testicular differentiation, whereas increased cyp19a mRNA levels are required for ovarian differentiation [26,27,28]. We compared all CpG methylation sites and cyp19a gene expression in Nile tilapia. The results indicated that the average methylation ratio level of testis to ovary of the promoter region is higher than that of the transcription region. DNA methylation in the promoter regions of cyp19a are widely observed in fish [27,28,29]. In protogynous rice-field eels (Monopterus albus), DNA methylation levels at cyp19a promoter regions are inversely correlated with cyp19a expression during natural sex change [30]. Abdominal implants that contain DNA methylation inhibitors could impede or even reverse gonadal transition towards males in this species. Similarly, in European sea bass (Dicentrarchus labrax), a gonochoristic species in which sex is determined by both genotype and temperature, high temperatures increase cyp19a promoter methylation and cause the masculinization of genetic females during early development [27]. In this study, cyp19a was expressed at low levels in the testis and at high levels in the ovary of both Nile tilapia and hybrid tilapia, but there are no obvious difference of the testis or ovary between Nile tilapia and hybrids. Therefore, the expression of cyp19a are not likely related to the ovary developmental retardation of hybrid fish. Our results confirmed the inverse correlation between gene expression level and DNA methylation in sex maintenance. However, the control mechanisms of hybrid sterility or inviability remain largely unknown. The epigenetic regulation of sex change will be a very active topic of research with crucial and promising applications.
Acknowledgments
China Agriculture Research System (CARS-49); Shanghai Collaborate Innovation Center for Aquatic Animal Genetics and Breeding (ZF1206).
Footnotes
Conflict of interest: Authors state no conflict of interest.
References
- [1].Li M, Sun Y, Zhao J, Shi H, Zeng S, Ye K. A Tandem Duplicate of Anti-Mullerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia, Oreochromis niloticus. PLoS Genet. 2015;11(11):e1005678. doi: 10.1371/journal.pgen.1005678. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ng WK, Romano N. A review of the nutrition and feeding management of farmed tilapia throughout the culture cycle. Reviews in Aquaculture. 2013;5(4):220. –. [Google Scholar]
- [3].Xu Z, Gan L, Li T, Xu C, Chen K, Wang X. Transcriptome Profiling and Molecular Pathway Analysis of Genes in Association with Salinity Adaptation in Nile Tilapia Oreochromis niloticus. PLoS One. 2015;10(8):e0136506. doi: 10.1371/journal.pone.0136506. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mair GC, Abucay JS, Abella TA, Beardmore JA, Skibinski D. Genetic manipulation of sex ratio for the large-scale production of all-male tilapia Oreochromis niloticus. Canadian Journal of Fisheries & Aquatic Sciences. 1997;54(2):396. –. [Google Scholar]
- [5].Kwon JY, Haghpanah V, Kogson-Hurtado LM, McAndrew BJ, Penman DJ. Masculinization of genetic female nile tilapia (Oreochromis niloticus) by dietary administration of an aromatase inhibitor during sexual differentiation. J Exp Zool. 2000;287(1):46. –. [PubMed] [Google Scholar]
- [6].Hulata G. Genetic manipulations in aquaculture: a review of stock improvement by classical and modern technologies. Genetica. 2001;111(1-3):155. doi: 10.1023/a:1013776931796. –. [DOI] [PubMed] [Google Scholar]
- [7].Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916. doi: 10.1126/science.1186366. –. [DOI] [PubMed] [Google Scholar]
- [8].Capuano F, Mülleder M, Kok R, Blom HJ, Ralser M. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Analytical chemistry. 2014;86(8):3697. doi: 10.1021/ac500447w. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen X, Wang Z, Tang S, Zhao Y, Zhao J. Genome-wide mapping of DNA methylation in Nile Tilapia. Hydrobiologia. 2016. pp. 1–11. –.
- [10].Sun LX, Wang YY, Zhao Y, Wang H, Li N, Ji XS. Global DNA Methylation Changes in Nile Tilapia Gonads during High Temperature-Induced Masculinization. PLoS One. 2016;11(8):e0158483. doi: 10.1371/journal.pone.0158483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen XW, Jiang S, Gu YF, Shi ZY. Molecular characterization and expression of cyp19a gene in Carassius auratus. J Fish Biol. 2014;85(2):516. doi: 10.1111/jfb.12418. –. [DOI] [PubMed] [Google Scholar]
- [12].Gu YF, Wei Q, Tang SJ, Chen XW, Zhao JL. Molecular characterization and functional analysis of IRF3 in tilapia (Oreochromis niloticus) Dev Comp Immunol. 2016;55:130. doi: 10.1016/j.dci.2015.10.011. –. [DOI] [PubMed] [Google Scholar]
- [13].Mcandrew BJ, Majumdar KC. Growth studies on juvenile tilapia using pure species, hormone-treated and nine interspecific hybrids. Aquaculture Research. 2010;20(20):35. –. [Google Scholar]
- [14].El-Hawarry WN. Growth Performance, Proximate Muscle Composition and Dress-Out Percentage of Nile Tilapia (Oreochromis niloticus), Blue Tilapia (Oreochromis aureus) and their Interspecific Hybrid ( ⍰ O. aureus X ⍰ O. niloticus) Cultured in Semi-Intensive Culture System. World’s Veterinary Journal. 2012;2(2):17. –. [Google Scholar]
- [15].Gomez-Ponce MA, Granados-Flores K, Padilla C, Lopez-Hernandez M, Nunez-Nogueira G. [Age and growth of the hybrid tilapia Oreochromis niloticus x Oreochromis aureus (Perciformes: Cichlidae) in the dam “Zimapan” Mexico] Rev Biol Trop. 2011;59(2):761. –. [PubMed] [Google Scholar]
- [16].Michalak P, Noor MA. Association of misexpression with sterility in hybrids of Drosophila simulansand D. mauritiana. J Mol Evol. 2004;59(2):277. doi: 10.1007/s00239-004-2622-y. –. [DOI] [PubMed] [Google Scholar]
- [17].Mishra PK, Singh BN. Genetic interactions underlying hybrid male sterility in the Drosophila bipectinata species complex. Genes Genet Syst. 2006;81(3):193. doi: 10.1266/ggs.81.193. –. [DOI] [PubMed] [Google Scholar]
- [18].Good JM, Giger T, Dean MD, Nachman MW. Widespread over-expression of the X chromosome in sterile F(1)hybrid mice. PLoS Genet. 2010;6(9):e1001148. doi: 10.1371/journal.pgen.1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Voolstra C, Tautz D, Farbrother P, Eichinger L, Harr B. Contrasting evolution of expression differences in the testis between species and subspecies of the house mouse. Genome Res. 2007;17(1):42. doi: 10.1101/gr.5683806. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Malone JH, Michalak P. Gene expression analysis of the ovary of hybrid females of Xenopus laevis and X. muelleri. BMC Evol Biol. 2008;8:82. doi: 10.1186/1471-2148-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Renaut S, Nolte AW, Bernatchez L. Gene expression divergence and hybrid misexpression between lake whitefish species pairs (Coregonus spp. Salmonidae). Mol Biol Evol. 2009;26(4):925. doi: 10.1093/molbev/msp017. –. [DOI] [PubMed] [Google Scholar]
- [22].Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323(5912):373. doi: 10.1126/science.1163601. –. [DOI] [PubMed] [Google Scholar]
- [23].Turner LM, White MA, Tautz D, Payseur BA. Genomic networks of hybrid sterility. PLoS Genet. 2014;10(2):e1004162. doi: 10.1371/journal.pgen.1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li N, Ye M, Li Y, Yan Z, Butcher LM, Sun J. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods. 2010;52(3):203. doi: 10.1016/j.ymeth.2010.04.009. et al. –. [DOI] [PubMed] [Google Scholar]
- [25].Azaza MS, Legendre M, Kraiem MM, Baras E. Size-dependent effects of daily thermal fluctuations on the growth and size heterogeneity of Nile tilapia Oreochromis niloticus. J Fish Biol. 2010;76(3):669. doi: 10.1111/j.1095-8649.2009.02524.x. –. [DOI] [PubMed] [Google Scholar]
- [26].Vanselow J, Pohland R, Furbass R. Promoter-2-derived Cyp19 expression in bovine granulosa cells coincides with gene-specific DNA hypo-methylation. Mol Cell Endocrinol. 2005;233(1-2):57. doi: 10.1016/j.mce.2005.01.007. –. [DOI] [PubMed] [Google Scholar]
- [27].Navarro-Martin L, Vinas J, Ribas L, Diaz N, Gutierrez A, Di Croce L. DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet. 2011;7(12):e1002447. doi: 10.1371/journal.pgen.1002447. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wen AY, You F, Sun P, Li J, Xu DD, Wu ZH. CpG methylation of dmrt1 and cyp19a promoters in relation to their sexual dimorphic expression in the Japanese flounder Paralichthys olivaceus. J Fish Biol. 2014;84(1):193. doi: 10.1111/jfb.12277. et al. –. [DOI] [PubMed] [Google Scholar]
- [29].Chen S, Zhang G, Shao C, Huang Q, Liu G, Zhang P. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet. 2014;46(3):253. doi: 10.1038/ng.2890. et al. –. [DOI] [PubMed] [Google Scholar]
- [30].Zhang Y, Zhang S, Liu Z, Zhang L, Zhang W. Epigenetic modifications during sex change repress gonadotropin stimulation of cyp19a1a in a teleost ricefield eel (Monopterus albus) Endocrinology. 2013;154(8):2881. doi: 10.1210/en.2012-2220. –. [DOI] [PubMed] [Google Scholar]


