Abstract
Objective
This study aimed to evaluate the expression of progestin and adipoQ receptor family member VI (PAQR6, mPRδ) in prostate cancer and to explore its role in prostate cancer progression.
Methods
PAQR6 mRNA expression was evaluated based on the data obtained from the TCGA database and the GEO database. The prognostic value of PAQR6 was explored by Kaplan-Meier analysis. To investigate the role of PAQR6, it was depleted by siRNA in DU145 cells. The effects of depleting PAQR6 on DU145 cell viability and migration were determined by CCK8 assay, colony formation assay, and wound healing assay, respectively. The activation of MEK and ERK were analyzed by western blot.
Results
PAQR6 mRNA expression was significantly up-regulated in prostate cancer tissues and correlated with lower survival rates (p=0.014). Furthermore, qPCR revealed that PAQR6 expression was elevated in DU145 and LNCaP cells compared with RWPE-2 cells. Depleting PAQR6 obviously suppressed DU145 cell proliferation and migration (p<0.01). In addition, the ratio of p-MEK/MEK and p-ERK/ERK was significantly reduced after silencing PAQR6 (p<0.01).
Conclusion
PAQR6 might play a facilitating role in prostate cancer development by regulating the MAPK signaling pathway. Moreover, it might serve as a potential predictor and therapeutic target in prostate cancer.
Keywords: PAQR6, Kaplan-Meier analysis, proliferation, migration, MAPK signaling pathway
1. Introduction
Prostate cancer is one of the most common malignancies in the male genitourinary system as well as a leading cause of cancer-related death in developed countries [1, 2]. The high incidence of prostate cancer is related to many factors such as age, ethnicity, family history, and genetic susceptibility [3]. Although some progress has been made in the pathogenesis, detection, and therapy of prostate cancer during the past years, metastasis of the lesion and the risk of recurrence after surgery remain major problems [4]. Hence, finding effective prognostic and diagnostic biomarkers and therapeutic targets is of great significance for the treatment of prostate cancer.
Progesterone, one of the sex steroid hormones, possibly acts through multiple receptors and has been reported to be involved in many cell biology process (proliferation, differentiation, etc.) as well as in the progression of various cancers [5, 6] . It has been found that high progesterone receptor (PGR) density is an independent poor prognostic factor in prostate cancer [5]. Membrane progesterone receptors (mPRs) that belong to the progestin and adipoQ receptor (PAQR) family are putative non-classical progesterone signaling molecules [7, 8, 9]. To date, five of these molecules have been identified in humans: PAQR5 (mPRγ), PAQR6 (mPRδ), PAQR7 (mPRα), PAQR8 (mPRβ), and PAQR9 (mPRε) [10]. Differential expression of PAQR5 and PAQR8 was found in ovarian cystadenomas, borderline tumors, and carcinomas, suggesting that they have important roles in epithelial ovarian tumors [11]. PAQR5 and PAQR8 were also supposed to be potential prognostic biomarkers of endometrial cancer [10]. In human glioblastoma cells, the activation of PAQR7 has been found to exhibit a promoting effect on cell proliferation and motility, implying its involvement in glioblastoma development [12]. In addition, LI ZHOU et al. found that progesterone inhibits triple-negative breast cancer growth and metastasis via PAQR7 [13]. This evidence suggests that mPRs play an important role in cancer development and have the potential to be therapy targets and biomarkers in cancer.
Here, we first analyzed the expression of PAQR6 in prostate cancer tissues based on data obtained from the public database. Then the role of PAQR6 in prostate cancer cell growth and migration was investigated by depleting PAQR6 using siRNA. Our observation demonstrated for the first time that PAQR6 expression is elevated in prostate cancer and plays a facilitating role in prostate cancer progression.
2. Methods
2.1. Data collection
The expression data of 13 malignant prostate tissue samples and 8 benign prostate tissue samples were obtained from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database with the access number of GSE55945. The RNA-Seq expression data of 499 prostate cancer samples and 52 normal samples were downloaded from the TCGA database (https://cancergenome.nih.gov/) Moreover, the clinic-data of these 499 prostate cancer patients were downloaded together.
2.2. Cell culture
The human normal prostate epithelial cell line RWPE-2 was obtained from American Type Culture Collection (ATCC, USA) and cultured in Keratinocyte Serum Free Medium (K-SFM) supplemented with 0.05 mg/ml bovine pituitary extract (BPE) and 5 ng/ml epidermal growth factor (EGF). The prostate cancer cell lines DU145 and LNCaP were purchased from the Chinese Academy of Sciences Shanghai Academy of Sciences Cell Resource Center (Shanghai, China). DU145 cells were cultured in MEM medium supplemented with 10% fetal bovine serum (FBS; HyClone: Logan, UT). LNCaP cells were grown in RPMI-1640 medium (Corning: Manassas, VA) supplemented with 10% FBS. All the cells were cultivated at 37°C with 5% CO2
2.3. Transfection
Transfections with PAQR6 and scrambled small interfering RNA (siRNA) were executed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) in line with the description of the specification. The PAQR6 siRNAs (PAQR6 siRNA1: 5’-GCUGCCCCAACUUCUUCAA-3’ and siRNA2: 5’-CAUCUGGACUCACUU CCUG-3’) and scrambled siRNA (5’-CUUCUUCAACCGUCGCCAA-3’) were all purchased from GENEWIZ, Inc. (Beijing, China). After 6 h of culture, the transfection medium was replaced with complete medium and incubated for 48 h. Following that, the cells were subjected to the following experiments.
2.4. RNA extraction and real-time quantitative PCR
Total RNA of prostate cancer cell lines was isolated using RNAiso Plus (TaKaRa Biotechnology, Dalian, China) following the manufacturer’s protocol. Synthesis of cDNA using RNA as a template was conducted with the HiFiScript cDNA Synthesis Kit (CwBio, Beijing, China). The mRNA expression level of PAQR6 was determined by reverse transcription polymerase chain reaction (RT-PCR) using the Applied Biosystems 7300 Sequence Detection System (Applied Biosystems). The following primers for PAQR6 were used: forward: 5’-GGTGTTCTGGGAAGATGGCA-3’ and reverse: 5’-TCTGGAAGGAGCTGAGGACA-3’. GAPDH (forward: 5’-GGAGCGAGATCCCTCCAAAAT-3’; reverse: 5’-GGCTGTTGTCATACTTCTCATGG-3’) was used as the endogenous reference gene. The PAQR6 mRNA expression was calculated using a 2-ΔΔCt method.
2.5. Cell proliferation assay
Cell Counting Kit-8 (CCK8, Dojindo, Kumamoto, Japan) was used to examine the effect of silencing of PAQR6 by siRNA on prostate cancer cell proliferation. Cells after 48 h of transfection were seeded into 96-well plates (1000 cells/well) and cultured at 37°C with 5% CO2. Cell viability was measured with CCK8 according to the manufacturer’s protocol at the time points of 24 h, 48 h, 72 h, and 96 h of incubation. The absorbance at 450 nm was determined using a microplate reader. All experiments were performed in triplicate and done at least 3 independent times.
2.6. Colony formation assay
Single cell suspensions were prepared and plated onto fresh 60 mm dishes (700 cells/dish). The cells were cultured in media with 10% FBS for 2 weeks. During these 2 weeks, the medium was replaced with fresh media every 4 days. The colonies were fixed by methanol and stained with 0.1% crystal violet. The visible colonies were counted, and the relative ratio of colonies was calculated by considering the colony number of the si-con group as 100%.
2.7. Wound healing assay
For the purpose of investigating the effect of PAQR6 knockdown on DU145 cell migration, we implemented a wound healing assay. A micropipette tip was used to make a horizontal wound in the cell monolayer. The width of the wounds was photographed and measured 0 h and 24 h post scratching. The relative migration distance of DU145 cells with silenced PAQR6 was calculated by considering the migration distance of the control DU145 cells (si-con) as 100%.
2.8. Western blot analysis
Total protein of prostate cancer cells was extracted using RIPA lysis buffer (Beyotime, Jiangsu, China) and measured using the BCA method. Then the proteins were fractionated by 10% SDS-PAGE and electrotransferred to a PVDF membrane (Millipore, Bedford, MA) followed by blocking with 5% skim milk for 1 h at room temperature. Subsequently, the membranes were incubated with rabbit anti-human primary antibodies at 4°C for 16 h. Then the membranes were incubated with secondary antibodies (1/1000, Cell Signaling Technology) at room temperature for 1 h. Lastly, the signals were detected using an Enhanced chemi-luminescence (ECL) plus detection kit (Thermo Fisher Scientifc, Inc.). The expression of GAPDH was used as an internal control.
2.9. Statistical analysis
Kaplan-Meier analysis and log-rank tests were performed to draw the survival curve and assess the difference between groups. The samples were divided into high and low expression groups according to the median of PAQR6 expression. Statistical analysis was performed using SPSS 15.0 software. One-way ANOVA analysis was used to compare the mean of multiple samples. It was considered as statistically significant when p<0.05.
Ethical approval: The conducted research is not related to either human or animals use.
3. Results
3.1. PAQR6 mRNA expression was enhanced in prostate cancer tissues and associated with poor prognosis
We investigated the relative PAQR6 mRNA expression level in prostate cancer tissues (n=499) compared with normal prostate tissues (n=52) based on the data downloaded from the TCGA database. Our results showed that PAQR6 mRNA expression was obviously up-regulated in prostate cancer tissues compared with normal prostate tissues (p=1.29E-22, Figure 1A). Based on the data obtained from the GEO database, the up-regulation of PAQR6 was also observed in malignant prostate tissue (n=13) compared with benign prostate tissue (n=8, p=0.001687, Figure 1B).
Figure 1.
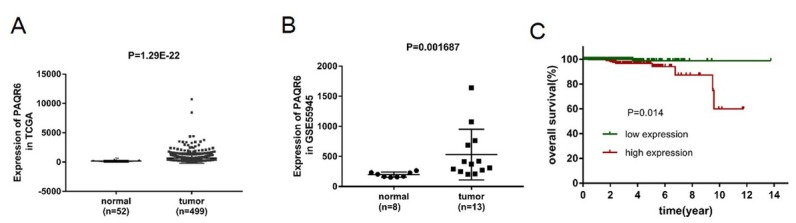
The mRNA expression level of PAQR6 was up-regulated in prostate cancer and positively related to poor prognosis in prostate cancer patients. (A) Analysis of PAQR6 mRNA expression in prostate cancer samples (N=499) and normal prostate samples (N=52) based on the expression profile downloaded from the TCGA database. (B) Analysis of PAQR6 mRNA expression in malignant prostate tissues (N=13) and benign prostate tissues (N=8) based on the expression profile obtained from the GEO database with access number GSE55945. (C) The correlation between PAQR6 expression and overall survival time was analyzed using Kaplan-Meier method based on the TCGA database. The samples were divided into high PAQR6 expression (N=250) and low PAQR6 expression (N=249) according the median of PAQR6 mRNA expression.
The Kaplan-Meier analysis demonstrated that PAQR6 expression was a potential predictor of survival. Patients with higher PAQR6 mRNA expression presented a shorter survival time compared to those with lower expression (p=0.014, Figure 1C).
3.2. Silencing PAQR6 inhibits DU145 cell proliferation
To further explore the role of PAQR6 in prostate cancer progression, we explored the mRNA expression of PAQR6 in prostate cancer cell lines by qPCR. Our results showed that PAQR6 mRNA expression levels in DU145 and LNCaP cells were remarkably higher than in RWPE-2 cells (p<0.01, Figure 2A). Since the DU145 cell line showed the highest PAQR6 levels, it was used for subsequent experiments. Si- PAQR6 (1) and si- PAQR6 (2) were synthesized and transfected into DU145 cells to deplete PAQR6 expression. As shown in Figure 2B, PAQR6 mRNA expression was down-regulated to 36.7% and 15.2% by using si- PAQR6 (1) and si- PAQR6 (2), respectively. Western blot analysis showed similar down regulation of PAQR6 protein levels (p<0.01, Figure 2C-D). Subsequent CCK8 assays showed that the OD value of the si- PAQR6 group was significantly lower at 48 h, 72 h, and 96 h compared to that of the si-con group (p<0.01, Figure 3A). The results of the colony formation assay were consistent with the outcomes observed in the CCK8 assay. We found that the number of colonies formed in the si- PAQR6 group was significantly smaller than that of the si-con group (p<0.01, Figure 3 B-C). These data suggest that down-regulating PAQR6 significantly inhibits cell proliferation.
Figure 2.
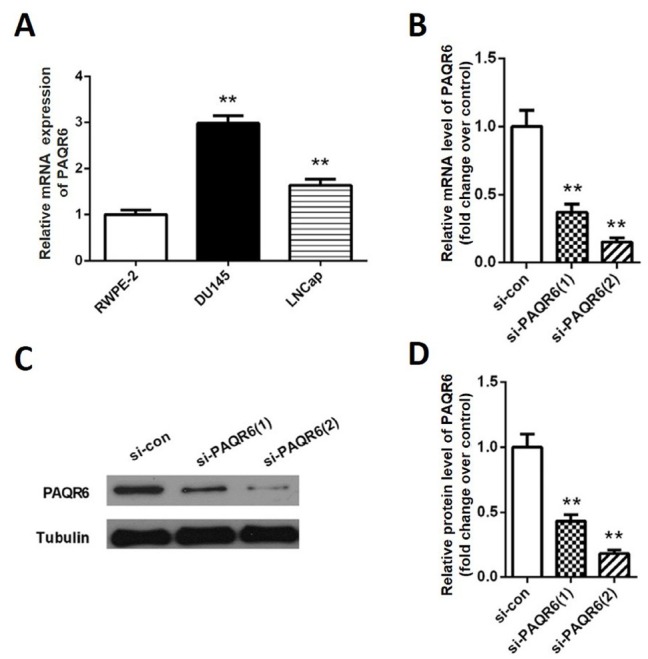
Knockdown of PAQR6 by PAQR6 siRNA in prostate cancer cells. (A) The mRNA expression levels of PAQR6 in prostate cancer cell lines DU145 and LNCaP as well as normal RWPE-2 cells line were determined by qPCR. (B) The mRNA expression levels of PAQR6 in DU145 cells were analyzed by qPCR 48 h after transfection. (C) PAQR6 protein expression in DU145 cells was determined by western blot 48 h after transfection. (D) Quantification of PAQR6 protein expression. Columns, mean (n= 6); bars, SD. **P<0.01, vs. PAQR6 expression in DU145 cells transfected with scramble siRNA (si-con).
Figure 3.
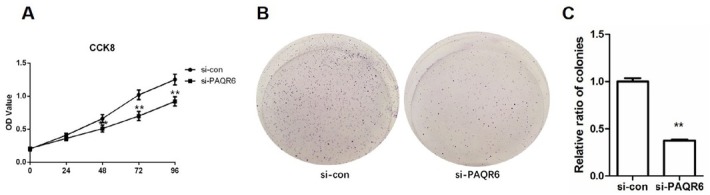
Depletion of PAQR6 inhibits DU145 cell proliferation. DU145 cell proliferation ability was determined by CCK8 assay 48 h after transfection. (B) DU145 cell proliferation ability was determined by colony formation assay. (C) The relative ratio of colonies in Figure (B). N=6. **P<0.01, vs. DU145 cells transfected with scramble siRNA (si-con).
3.3. Silencing PAQR6 inhibits DU145 cell migration
Wound healing assays were performed to determine the migration ability of DU145 cells. We found that knockdown of PAQR6 significantly reduced the migration distance to 47% of that observed in the cells transfected with scrambled siRNA (Figure 4 A-B, p<0.01). These results demonstrated that DU145 cell migration ability was markedly suppressed after depleting PAQR6.
Figure 4.
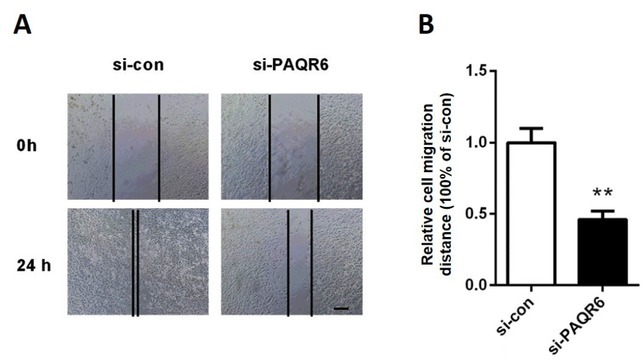
Knockdown of PAQR6 suppressed DU145 cell migration. (A) The effects of silencing PAQR6 on DU145 cell migration were determined by wound healing assays 0 h and 24 h after scratching. The bar represents 200 μm. (B) The relative migration distance of DU145. **p<0.01, vs. DU145 cells transfected with scramble control siRNA (si-con).
3.4. Silencing PAQR6 inhibits the MAPK signaling pathway
To study the potential mechanistic role of PAQR6 in cell proliferation and migration, we examined the expression changes of MAPK-related proteins in DU145 cells 8 h after transfection by western blot. Our results showed that the ratio of p-MEK/MEK and p-ERK/ERK was significantly reduced in PAQR6-depleted DU145 cells compared with cells transfected with scrambled siRNA (Figure 5 A-B, p<0.01). These results suggested that silencing PAQR6 remarkably suppresses activation of the MAPK signaling pathway.
Figure 5.
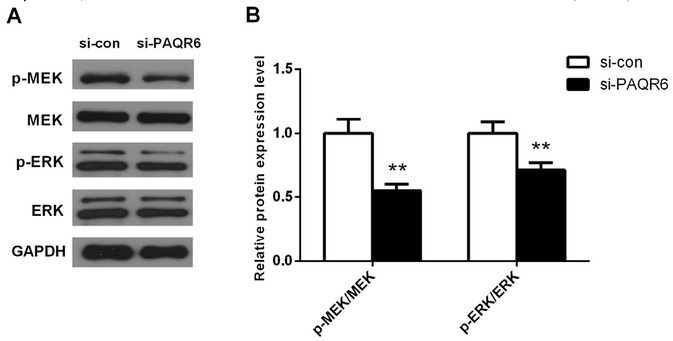
Knockdown of PAQR6 significantly suppressed the activation of the MAPK signaling pathway. (A) The protein expression of p-MEK, MEK, p-ERK, and ERK in DU145 cells was determined by western blot. (B) Quantification of the ratio of p-MEK/MEK, p-ERK/ERK obtained by western blot. Columns, mean (n= 6); bars, SD. **P<0.01, vs. expression of these proteins in DU145 cells transfected with scramble siRNA (si-con).
4. Discussion
In the present study, we observed that PAQR6 is up-regulated in prostate cancer tissues and that high expression of PAQR6 is correlated with lower overall survival rate. Through a series of in vitro experiments, we demonstrated that PAQR6 might play a facilitating role in prostate cancer cell proliferation and migration by regulating the MAPK signaling pathway.
PAQR6, a member of the PAQR family, has been characterized to be capable of responding to progesterone and suggested to possess progestin-binding characteristics [14, 15]. It is considered to be a unique type of G protein-coupled receptor that acts via cAMP in the nervous system [16]. Previous studies showed that PAQR6 is highly expressed in the human brain [17] and also in the small intestine and back fat of swine [15]. However, the expression and the function of this gene, particularly in prostate cancer, remains unclear.
In this study, we analyzed the PAQR6 mRNA expression in prostate cancer tissues based on the TCGA database and the GEO database. Our results indicated that PAQR6 mRNA expression was enhanced in prostate cancer tissues compared with normal tissues (Figure 1 A-B). The up-regulation of PAQR6 was also observed in prostate cancer cell lines DU145 and LNCaP compared with normal RWPE-2 cells (Figure 2A). Further prognostic analysis revealed that higher PAQR6 expression was closely associated with poor prognosis in prostate cancer patients (p=0.014, Figure 1C). These data suggest that PAQR6 might promote prostate cancer development and can be used as a potential independent prognostic marker.
In order to further investigate the role of PAQR6 in prostate cancer, we performed a knockdown of this gene in DU145 cells using si-PAQR6 (Figure 2B-D). Our results showed that DU145 expression was significantly down-regulated after transfection with si- PAQR6. It is well known that excessive proliferation is one of the main features of cancer cells. Therefore, we then examined the proliferation ability of DU145 cells following the depletion of PAQR6 using CCK8 assays and colony formation assays. Our results demonstrated that the viability of DU145 cells decreased significantly after silencing PAQR6 (p<0.01 Figure 3), suggesting that PAQR6 might promote the proliferation of prostate cancer cells. Another characteristic of cancer cells is that invasion and migration are often activated [18]. Hence, we performed a wound healing assay and found that the migration distance of DU145 cells with silenced PAQR6 was shorter compared with the si-con group (p<0.01, Figure 4). This phenomenon indicated that PAQR6 might promote cell migration in vitro. In summary, our results revealed that PAQR6 possibly acts as a pro-tumor factor in prostate cancer occurrence and progression.
MAPK signaling pathway is known as an important downstream signaling cascade in several biological events as well as various types of cancers [19, 20]. It is essential for the transcription of genes participating in cell growth and survival [20, 21]. Hence, we next studied the effect of silencing PAQR6 on the expression of MAPK signaling pathway related proteins by western blot. Our results demonstrated that the ratio of p-MEK/MEK and p-ERK/ERK decreased remarkably after silencing PAQR6 (p<0.01, Figure 5). We conjectured that PAQR6 may inhibit cell proliferation and migration by modulating the MAPK signaling pathway.
Taken together, our results demonstrate for the first time that PAQR6 expression is enhanced in prostate cancer tissues and positively associated with poor prognosis in prostate cancer patients. In addition, PAQR6 might act as a tumor promoter by regulating the MAPK signaling pathway and might be a new therapeutic target for prostate cancer. However, the promoting role of this gene in the cancer development process needs to be further validated in animal models as soon as possible.
Acknowledgements
The authors are grateful for the help of Yue-Jian Wang.
Footnotes
Conflict of interest: Authors state no conflict of interest.
References
- [1].Ramalho-Carvalho J, Gonçalves C, Graça I, Bidarra D, Pereira-Silva E, Salta S. A multiplatform approach identifies miR-152-3p as a common epigenetically regulated onco-suppressor in prostate cancer targeting. Clin Epigenetics. 2018;10:40. doi: 10.1186/s13148-018-0475-2. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016, CA: a cancer journal for clinicians. 2016;66:7. doi: 10.3322/caac.21332. –. [DOI] [PubMed] [Google Scholar]
- [3].Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011. doi: 10.1016/j.cell.2015.10.025. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zoni E, Van dHG, Af VDM, Chen L, Rane JK, Pelger RC. miR-25 Modulates Invasiveness and Dissemination of Human Prostate Cancer Cells via Regulation of αv- and α6-Integrin Expression. Cancer Research. 2015;75:2326. doi: 10.1158/0008-5472.CAN-14-2155. et al. –. [DOI] [PubMed] [Google Scholar]
- [5].Grindstad T, Andersen S, Al-Saad S, Donnem T, Kiselev Y, Nordahl M-JC. High progesterone receptor expression in prostate cancer is associated with clinical failure. PloS one. 2015;10:e0116691. doi: 10.1371/journal.pone.0116691. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Migliaccio A, Castoria G, Auricchio F. Src-dependent signalling pathway regulation by sex-steroid hormones: therapeutic implications. International Journal of Biochemistry & Cell Biology. 2007;39:1343. doi: 10.1016/j.biocel.2006.12.009. –. [DOI] [PubMed] [Google Scholar]
- [7].Petersen SL, Intlekofer KA, Mouraconlon PJ, Brewer DN, Sans JDP, Lopez JA. Novel progesterone receptors: neural localization and possible functions. Frontiers in Neuroscience. 2013;7:164. doi: 10.3389/fnins.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jia CY, Li HH, Zhu XC, Dong YW, Fu D, Zhao QL. MiR-223 suppresses cell proliferation by targeting IGF-1R. PLoS One. 2011;6:e27008. doi: 10.1371/journal.pone.0027008. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Valadezcosmes P, Vázquezmartínez ER, Cerbón M, Camachoarroyo I. Membrane progesterone receptors in reproduction and cancer. Mol. Cell. Endocrinol. 2016;434:166. doi: 10.1016/j.mce.2016.06.027. –. [DOI] [PubMed] [Google Scholar]
- [10].Sinreih M, Knific T, Thomas P, Grazio SF, Rižner TL. Membrane progesterone receptors β and γ have potential as prognostic biomarkers of endometrial cancer, J. Steroid Biochem. Mol. Biol. 2018:178. doi: 10.1016/j.jsbmb.2018.01.011. [DOI] [PubMed] [Google Scholar]
- [11].Romero-Sánchez M, Peiper S, Evans B, Wang Z, Catasús L, Ribe A. Expression profile of heptahelical putative membrane progesterone receptors in epithelial ovarian tumors. Hum. Pathol. 2008;39:1026. doi: 10.1016/j.humpath.2007.11.007. et al. –. [DOI] [PubMed] [Google Scholar]
- [12].González-Orozco JC, Hansberg-Pastor V, Valadez-Cosmes P, Nicolas-Ortega W, Bastida-Beristain Y, Fuente-Granada MDL. Activation of membrane progesterone receptor-alpha increases proliferation, migration, and invasion of human glioblastoma cells, Mol. Cell. Endocrinol. 2018 doi: 10.1016/j.mce.2018.06.004. et al. [DOI] [PubMed] [Google Scholar]
- [13].Li Z, Wei Z, Zhang H, Yan H, Lei Y, Zhang Y. Progesterone suppresses triple-negative breast cancer growth and metastasis to the brain via membrane progesterone receptor α. Int J. Mol. Med. 2017;40:755. doi: 10.3892/ijmm.2017.3060. et al. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Smith JL, Kupchak BR, Garitaonandia I, Hoang LK, Maina AS, Regalla LM. Heterologous expression of human mPRα. mPRβ and mPRγ in yeast confirms their ability to function as membrane progesterone receptors, Steroids. 2008;73:1160. doi: 10.1016/j.steroids.2008.05.003. et al. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhao X, Mo D, Li A, Gong W, Zhang Y, Qian W. Characterization and transcriptional regulation analysis of the porcine PAQR6 gene. DNA Cell Biol. 2011;30:947. doi: 10.1089/dna.2011.1262. et al. [DOI] [PubMed] [Google Scholar]
- [16].Thomas P, Pang Y. Membrane progesterone receptors: evidence for neuroprotective. neurosteroid signaling and neuroendocrine functions in neuronal cells, Neuroendocrinology. 2012;96:162. doi: 10.1159/000339822. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tang YT, Hu T, Arterburn M, Boyle B, Bright JM, Emtage PC. PAQR proteins: a novel membrane receptor family defined by an ancient 7-transmembrane pass motif. J. Mol. Evol. 2005;61:372. doi: 10.1007/s00239-004-0375-2. et al. –. [DOI] [PubMed] [Google Scholar]
- [18].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646. doi: 10.1016/j.cell.2011.02.013. –. [DOI] [PubMed] [Google Scholar]
- [19].Li P, Xue W, Feng Y, Mao Q. Long non-coding RNA CASC2 suppresses the proliferation of gastric cancer cells by regulating the MAPK signaling pathway. Am J Transl Res. 2016;8:3522. –. [PMC free article] [PubMed] [Google Scholar]
- [20].Chakraborty C, Sharma AR, Patra BC, Bhattacharya M, Sharma G, Lee SS. MicroRNAs mediated regulation of MAPK signaling pathways in chronic myeloid leukemia. Oncotarget. 2016;7:42683. doi: 10.18632/oncotarget.7977. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta. 2010;1802:396. –. [Google Scholar]


