Abstract
Agriculture, food industry, and manufacturing are just some of the areas where anaerobic technology can be used. Currently, anaerobic technologies are mainly used for wastewater treatment, solid waste treatment, or for the production of electrical and thermal energy from energy crops processing. However, a clear trend is towards more intensive use of this technology in biomass and biodegradable waste processing and hydrogen or biomethane production. An enormous number of anaerobic digesters are operating worldwide but there is very little information about the effect of different substrate combinations on the methanogens community. This is due to the fact that each of the anaerobic digesters has its own unique microbial community. For the most effective management of anaerobic processes it would be important to know the composition of a consortium of anaerobic microorganisms present in anaerobic digesters processing different input combinations of raw material. This paper characterizes the effect of the input raw materials on the diversity of the methanogen community. Two predominant microorganisms in anaerobic digesters were found to be 99% identity by the sequences of the 16S rRNA gene to the Methanoculleus and Thermogymnomonas genera deposited in GenBank.
Keywords: methanogenic microorganisms, Archaea, anaerobic digesters, biogas, methane production
1. Introduction
Anaerobic digestion is a natural process in which microorganisms decompose organic materials, the major final product is biogas and other fermentation products. This process can occur in swamps, wetlands and in digestive tract of ruminants [1]. Anaerobic microorganisms are also active in landfill sites where they may degrade landfilled bio wastes. Biogas can be collected and used as a potential source of renewable energy [2]. Basically the process occurs in an anaerobic environment through the activities of wide groups of microorganisms that decompose organic material and produce methane (CH4) and carbon dioxide (CO2) in a gaseous form known as biogas [3, 4, 5, 6]. Anaerobes play an important role in establishing a stable environment at various stages of methane digestion [7, 8]. Methane digestion offers an effective means of pollution reduction, which is superior to that achieved via conventional aerobic processes. Anaerobic digesters have been used for decades at municipal wastewater treatment plants, and more recently have been used to process industrial and agricultural wastes [9, 10, 11, 12, 13]. Typically, using organic materials as the major input, the systems produce biogas that contains from 55% to 70% CH4 and from 30% to 45% CO2 [14, 15, 16].
Recent progress in biogas technology in development of the molecular biology of methanogens and in operation of improved anaerobic digesters is discussed. However, the prevalence of the methanogenic populations of microorganisms as influenced by the type of substrate in methane anaerobic digesters has not been reported widely.
The aim of this research was to compare the diversity of methanogenic populations in methane anaerobic digesters with unusual input ratio of initial amount of substrate using the amplification of gene fragments and Illumina sequencing. The chosen biogas plants have never been studied before.
2. Materials and methods
The anaerobic digesters are located in Modřice, Bratčice, Pánov, Úvalno, Horní Benešov, Rusín, and Loděnice in the Czech Republic (Fig.1).
Fig. 1.
The map of localization of biogas plants (Czech Republic)
2.1. The sampling and characteristic of substrates in the anaerobic digesters
The samples were collected from the different biogas plant reactors. The samples were taken directly from the reactors into sterile sampling vessels. After sampling, the samples were stored in thermocontainers and transported to the laboratory for further analysis. Each of the reactors processed a different type of substrate which is described in Table 1.
Table 1.
The type of substrate in anaerobic digester
| Number of the sample | Location of the fermenter | Main substrate | Input Substrate Combination (%) |
|---|---|---|---|
| 1 | Modřice | primary sludge, biological sludge | 50 : 50 |
| 2 | Bratčice | maize silage, whole crop silage, poultry litter | 63:31 : 6 |
| 3 | Pánov | maize silage, poultry litter | 92 : 8 |
| 4 | Úvalno | maize silage, sugar beet pulp, whole crop silage, cattle manure | 44 : 44 : 6 : 6 |
| 5 | Horní Benešov | maize silage, sugar beet pulp, whole crop silage, cattle manure, grass silage | 29 : 39 : 12 : 15 : 5 |
| 6 | Rusín | maize silage, sugar beet pulp | 70 : 30 |
| 7 | Loděnice | maize silage, sugar beet pulp | 75 : 25 |
2.2. Analytical methods
The pH, redox potential, temperature, total solids content, volatile solids content, and biogas composition at each anaerobic digester was determined (Table 2). Total solids (TS) content was determined by drying at 105±5°C followed by cooling in a desiccator and weighed when a constant weight was reached, EcoCELL 111 (BMT Medical Technology Ltd., Brno, the Czech Republic) used according to Czech Standard Method (CSN EN 14346, 2007) [17]. Volatile solids content (VS) content was determined by the combustion of the samples in a muffle furnace at 550°C ±5°C according to Czech Standard Method (CSN EN 15169, 2007) [18]by using a furnace LMH 11/12 (LAC, Ltd., Rajhrad, the Czech Republic). The pH and redox potential were determined by using pH/Cond meter 3320 (WTW GmbH, Dinslaken, Germany) in accordance with standard procedures (CSN EN 12176, 1999) [19].
Table 2.
Physical and chemical characteristics of fermentation in the bioreactors
| Number of the bioreactor | Temperature (°C) | pH | Redox (mV) | Total solids (%) | Volatile solids (%) | Biogas composition | |||
|---|---|---|---|---|---|---|---|---|---|
| CH4(%vol) | CO2(%vol) | H2(%vol) | Other(%vol) | ||||||
| 1 | 34 | 7 | -3.1 | 5.09 | 59.13 | 47 | 48 | 0.0055 | 4.99 |
| 2 | 43 | 8.3 | -75 | 10.16 | 75.23 | 51.5 | 47 | 0.0045 | 1.49 |
| 3 | 49 | 8 | -58 | 10.33 | 79.46 | 48 | 47 | 0.0050 | 4.99 |
| 4 | 48 | 7.69 | -38.5 | 8.84 | 78.85 | 49 | 48 | 0.0035 | 2.99 |
| 5 | 49 | 7.85 | -47.4 | 7.87 | 77.52 | 52 | 46 | 0.0060 | 1.99 |
| 6 | 48 | 7.63 | -34.7 | 8.52 | 79.15 | 48 | 48 | 0.0040 | 3.99 |
| 7 | 44 | 7.65 | -36 | 7.9 | 78.51 | 50.5 | 47 | 0.0035 | 2.49 |
The temperature of samples was determined by using high accuracy PT100 RTD thermometer HH804U (OMEGA Engineering, Stamford, USA). Biogas composition was determined by using the gas analyser Dräger X-am 7000 (Dräger Safety AG&Co. KGaA, Lübeck, Germany).
2.3. Isolation of DNA from collected samples
The QIAamp Fast DNA Stool Mini Kit (QIAGEN GmbH, Hilden, Germany) provides fast and easy purification of total DNA from fresh or frozen stool samples and was used for DNA extraction from anaerobic digesters samples. DNA extractions were carried out according to the handbook of the manufacturer with minor adjustments as described below. Briefly, 100 mg of each sample was mixed with 1.4 ml of ASL buffer (QIAGEN GmbH, Hilden, Germany) incubated at 95°C for 10 minutes. After centrifugation, an InhibitEX tablet was added to the supernatant to remove impurities and PCR inhibitors. After centrifugation, 200 μl of the supernatant was added to 15 μl of proteinase K solution, and 200 μl of buffer AL (QIAGEN GmbH, Hilden, Germany) was also added. The mixture was incubated at 70°C for 10 minutes, cooled and 200 μl of ethanol (96-100%) was added. The supernatant was then centrifuged through the QIAamp kit column followed by two washes with buffers AW1 and AW2 (QIAGEN GmbH, Hilden, Germany). For DNA elution, 200 μl of elution buffer was used.
2.4. Amplification and sequencing
For amplification of the V3 and V4 variable regions of the 16S rRNA gene fragments universal primers were used [20]. The primers were marked by molecular barcodes for sample identification. Maxima™ Probe qPCR Master Mix (Thermo Fisher Scientific, Waltham, USA), was used for PCR reaction. Cycling conditions, 95°C for 10 min, followed by 30 cycles of incubation at 94°C for 30 s, 60°C for 30 s and 72°C for 120 s, and a final extension step at 72°C for 2 minutes. PCR products were visualized using electrophoresis on 1.5% agarose gels using DNA purified from the gel using the QIAquick Gel Extraction Kit (Qiagen GmbH, Hilden, Germany). DNA was quantified using the Quant-iTPicoGreen dsDNA Assay (Thermo Fisher Scientific, Waltham, USA) and equimolar amounts of the PCR products were pooled together.
Purified amplicons were paired-end sequenced on an Illumina Mi-Seq platform. QIIME data analysis package was used for 16S rRNA data analysis [21]. Quality filtering on raw sequences was performed according to base quality score distributions, average base content per read and GC distribution in the reads. Chimeras and reads that did not cluster with other sequences were removed. The obtained sequences with qual scores higher than 20 were shortened to the same length of 350 bp and classified with RDP Seqmatch with an operational taxonomic unit (OTU) discrimination level set to 97%. The relative abundance of the taxonomic groups was calculated to the microorganisms detected in this study. Sequences were compared using the BLAST feature of the National Center for Biotechnology Information (NCBI) [22].
The sequences were uploaded to Geneious 7.1.9 for comparative genomic analyses [23]. Alignments of sequences were performed in Geneious 7.1.9 using Clustal W with the BLOSUM cost matrix, and clustering was performed by the neighbor-joining method [24].
2.5. Statistical analysis
The results were processed by methods of variation statistics and analyzed using software Statistica 12 (www.statistica.software.informer.com) and Origin 7.0 (www.origin-lab.com). The value of the statistical reliability of the parameters was tested using Fisher test (F-test). The assessment of the reliability of the difference between the statistical characteristics of alternative sets of data was tested using t-test. Obtaining equations of functions approximation of the experimental data were performed by least squares [25]. The share of impact (η2, %) of the temperature, pH and redox on the microbial diversity, Fisher coefficient and the reliability of the effect was calculated [26].
3. Results
As our research results have shown, the production of biogas composition was depended on the ratio of main substrate in specific anaerobic digesters. The lowest level of the produced methane (47%) was detected in the anaerobic digesters located in Modřice. This can be attributed to the fact that this anaerobic digester is a wastewater treatment plant which is not a typical biogas reactor unlike anaerobic digesters 2–7. The highest methane production (52%) among all typical anaerobic digesters was found in the reactor with a mixture of the substrates including maize silage, sugar beet pulp, whole crop silage, cattle manure, and grass silage in ratio of 29:39:12:15:5, respectively.
The main genera were investigated in different fermenters by amplification of 16S rRNA gene and using Illumina sequencing. The greatest diversity of methanogenic microorganisms was detected in the anaerobic digesters from Modřice where two types of substrate were used (primary sludge and biological sludge, 50:50). The most abundant genera identified included: Methanoculleus, Thermogymnomonas, Methano-bacterium, Methanolinea, Methanosaeta, Methanobrevibacter, Methanospirillum, Thermoplasmata and Thermoprotei (Table 3).
Table 3.
The most widespread of Archaea genera in anaerobic digesters
| Number of the fermenter | Number of Archaea genera (OUT·ml–1) | Total number | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Methanoculleus | Thermogymnomonas | Methanobacterium | Methanimicrococcus | Methanolinea | Methanosaeta | Methanobrevibacter | Methanospirillum | Thermoplasmata | Thermoprotei | ||
| 1 | 195 | 135 | 65 | nd | 95 | 25 | 10 | 25 | 10 | 5 | 565 |
| 2 | 1630 | 70 | 5 | 10 | nd | nd | nd | Nd | nd | nd | 1715 |
| 3 | 270 | 10 | nd | nd | nd | nd | nd | Nd | nd | nd | 280 |
| 4 | 865 | 300 | nd | nd | nd | nd | nd | Nd | nd | nd | 1165 |
| 5 | 390 | 50 | 5 | nd | nd | nd | nd | Nd | nd | nd | 445 |
| 6 | 1695 | 90 | nd | nd | nd | nd | nd | Nd | nd | nd | 1785 |
| 7 | 485 | 75 | 5 | nd | nd | nd | nd | Nd | nd | nd | 565 |
Comment: “nd” is not detected
However, not all of these genera among this wide range were detected in other typical biogas reactors. The Methanimicrococcus genus was not detected in the fermenter from the wastewater treatment plant but it was present (0.6%) only in the anaerobic digesters (2) which included the following substrates: maize silage, whole crop silage, and poultry litter in proportion of 63:31:6, respectively. Methanobacterium was present in the same anaerobic digesters (0.29%) as well as in anaerobic digesters 5 (1.12%) and 7 (0.88%). Two genera, Methanoculleus and Thermogymnomonas, were dominant methanogenic microorganisms in all the anaerobic digesters. The total number of Archaea genera was different in each anaerobic digester; however, there was no statistically significant difference in methane production. The greatest number of methanogenic genera (Methanoculleus and Thermogymnomonas) was detected in anaerobic digester 6 (maize silage and sugar beet pulp were of 70:30), where the methane production reached up to 48%. Methane production was highest in anaerobic digester 5 (52%), despite the fact that these genera were not found in significantly high numbers (445 OUT·ml4). Obviously, methane production was not dependent on the number of methanogens but on the type and ratio of specific consumed substrate in the anaerobic digesters.
Based on our data, the percentage ratio of Methanoculleus and Thermogymnomonas in the anaerobic digesters (in %) was of 35:24 at Modřice, 95:4 at Bratčice, 96:4 at Pánov, 74:26 at Úvalno, 88:11 at Horní Benešov, 95:5 at Rusín, and 86:13 at Loděnice (Fig. 2A). In view of the fact that greatest diversity of Archaea populations was observed in anaerobic digesters of the wastewater treatment plant located in Modřice, the abundance of the following genera was determined as follows Methanoculleus (34.51%), Thermogymnomonas (23.89%), Methanobacterium (11.50%), Methanolinea (16.81%), Methanosaeta (4.42%), Methanobrevibacter (1.77%), Methanospirillum (4.42%), Thermo-plasmata (1.77%), and Thermoprotei (0.88%) (Fig. 2B).
Fig. 2.
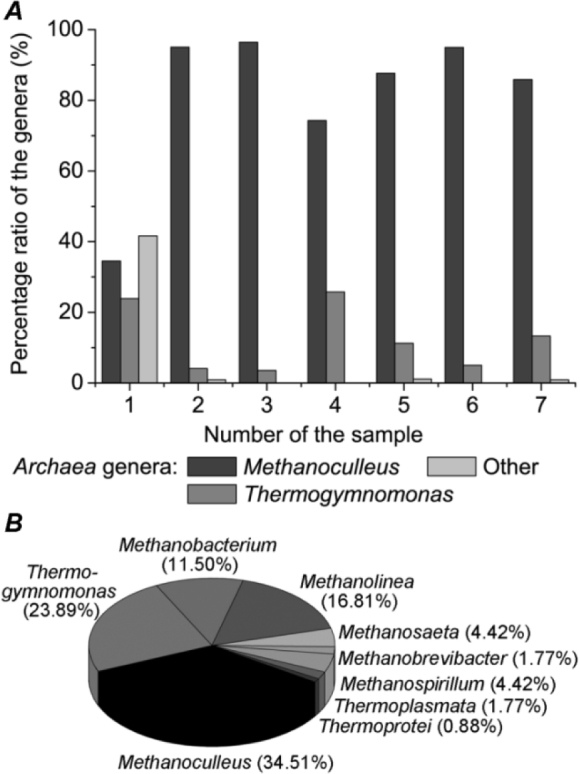
Ratio of Archaea genera in all anaerobic digesters analyzed (A), diversity of Archaea populations observed in anaerobic digester of wastewater treatment plant (B)
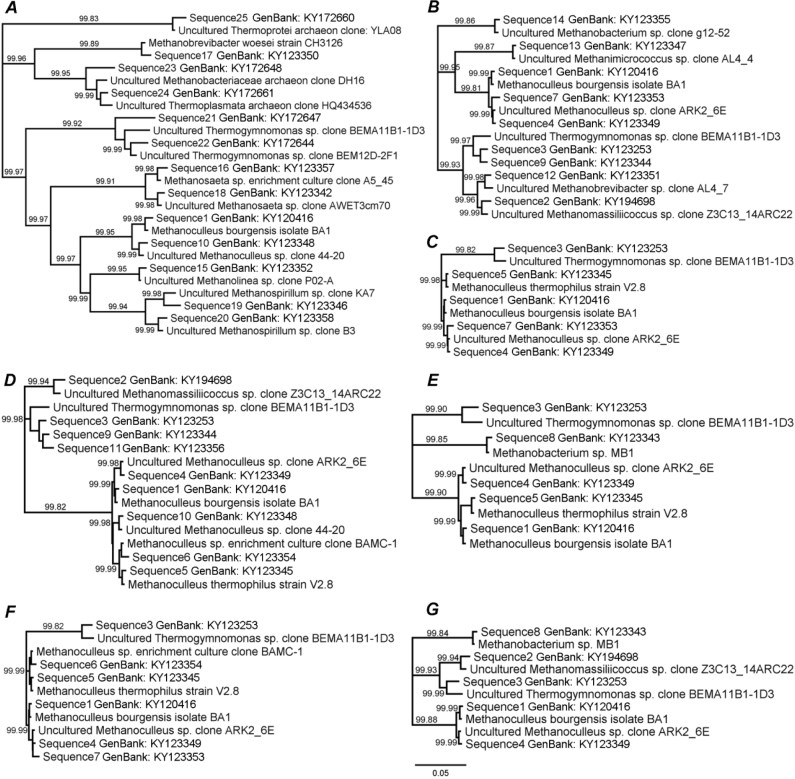
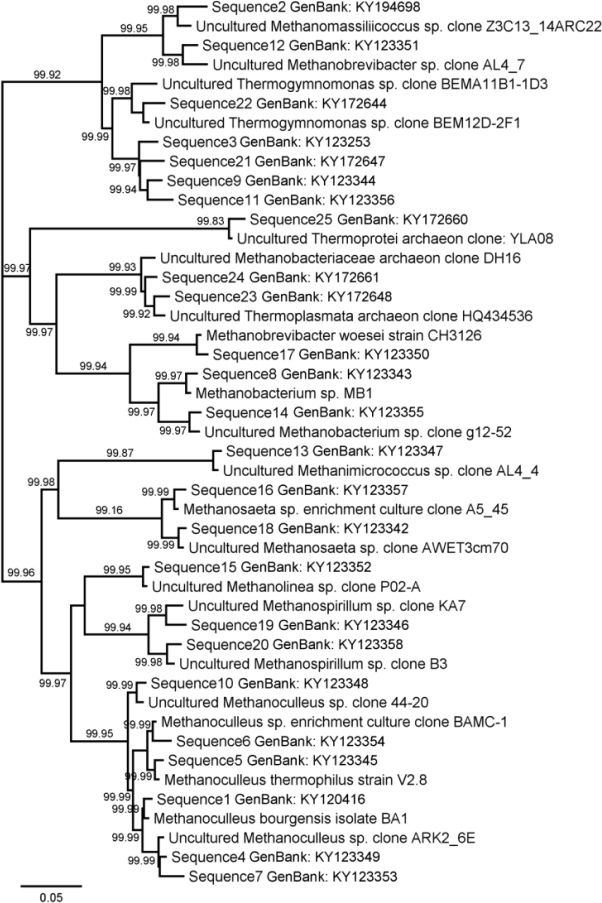
To estimate the genetic relations of the methanogenic Archaea populations in each anaerobic digester, the sequences of their 16S rRNA gene and sequences of the strains from GenBank were compared. The genomic sequences of methanogenic Archaea are stored in GenBank under accession no. KY120416, KY194698, KY123253, KY123349, KY123345, KY123354, KY123353, KY123343, KY123344, KY123348, KY123356, KY123351, KY123347, KY123355, KY123352, KY123357, KY123350, KY123342, KY123346, KY123358, KY172647, KY172644, KY172648, KY172661, KY172660. The phylogenetic tree of these relationships in each fermenter was constructed (Fig. 3). The identity of sequences of 16S rRNA gene Archaea from different anaerobic digesters with the strains from GenBank is demonstrated in Fig. 3, 4.
Fig. 3.
Phylogenetic tree of Archaea relationships separately in each anaerobic digester: Modřice (A), Bratčice (B), Pánov (C), Úvalno (D), Horní Benešov (E), Rusín (F), and Loděnice (G)
Fig. 4.
Phylogenetic tree showing of Archaea relationships together in all anaerobic digesters
Based on all sequences of 16S rRNA, gene Archaea from different anaerobic digesters, a phylogenetic tree demonstrating genetic relationship in each fermenter together was constructed. The identity of sequences of 16S rRNA gene with different strains from GenBank is shown on Fig. 4. The detected Archaea genera were identical to Methanoculleus bourgensis isolate BA1, Methanomassiliicoccus sp. clone Z3C13_14ARC22, Thermogymnomonas sp. clone BEMA11B1-1D3, Methanoculleus sp. clone ARK275, Methanoculleus thermophilus strain V2.8, Methanoculleus sp. clone BAMC-1, Methanoculleus sp. clone ARK2_6E, Methanobacterium sp. MB1, Thermogymnomonas sp. clone BEM12D-2F1, Methanoculleus sp. clone 44-20, Thermogymnomonas sp. clone BEM12D-2F1, Methanobrevibacter sp. clone AL4_7, Methanimicrococcus sp. clone AL4_4, Methanobacterium sp. clone g12-52, Methanolinea sp.clone P02-A, Methanosaeta sp. clone A5_45, Methanobrevibacter woesei strain CH3126, Methanosaeta sp. clone AWET3cm70, Methanospirillum sp. clone KA7, Methanospirillum sp. (LN717042.1), Thermogymnomonas sp. clone BEMA11B1-1D3, Thermogymnomonas sp. clone BEM12D-2F1, Thermoplasmata archaeon (LN796137.1), Methanobacteriaceae archaeon clone DH16, Thermoprotei archaeon clone: YLA08.
To investigate the influence of temperature, pH, redox, TS and VS on biogas composition (CH4, CO2, H2 and other gas), multiple regression analysis was conducted. Using regression analysis, the function demonstrating the nature of the influence of the model parameters (temperature, pH, redox etc.) on the dependent variables (CH4, CO2, H2, and rest) was determined. In general, the regression equation is as follows:
where X1 is temperature, X2 is pH, X3 is redox, X4 is total solids, X5 is volatile solids, and a1, a2, a3, a4, a5, ads coefficient for eachX1,2,3,4,5, Y is the dependent variable.
By the results of this analysis, the dependence on the studied factors was apportioned based on the experimental data for temperature (a1), pH (a2), redox (a3), total solids (a4), and volatile solids (a5) (Table 4).
Table 4.
The coefficients of the investigated factors effect on the biogas composition
| The dependent variable (Y) | R | R2 | The coefficients of the factor | F | ||||
|---|---|---|---|---|---|---|---|---|
| a1 | a2 | a3 | a4 | a5 | ||||
| CH4 | 0.999± | 0.999 | -0.106± | 6.948± | -0.069± | -2.46± | 0.241± | 9105.31 |
| 0.61 | 0.13 | 0.49** | 0.03 | 0.45* | 0.11 | |||
| co2 | 0.999± | 0.999 | -0.079± | 7.019± | 0.196± | 1.029± | -0.051± | 11212.99 |
| 0.53 | 0.11 | 0.42** | 0.02* | 0.38 | 0.10 | |||
| H2 | 0.997± | 0.995 | 0.0003± | 0.002± | 0.001± | 0.001± | 0.001± | 73.896 |
| 0.001 | 0.0001 | 0.001* | 0.001 | 0.001 | 0.001 | |||
| Rest | 0.987± | 0.975 | 0.194± | 0.985± | 0.141± | 1.695± | -0.284± | 16.12 |
| 1.02 | 0.22 | 0.82 | 0.05 | 0.75 | 0.19 | |||
Comment: R is a determination coefficient, R2 is a correlation coefficient, F is Fisher coefficient, ***P > 0.999 were statistical significantly.
P > 0.99
P > 0.95
Analysis of regression coefficients allowed the ascertaining of the extent and the influence of the factor for each effective variable. The most important parameters affecting methane production were detected pH (a2) and total solids (a4), which reached up to 6.948±0.49 (P>0.99) and -2.46±0.45 (P > 0.95), respectively. The negative sign of the coefficient (a4) in the multiple regression models indicated that the increased total solids (a4) led to a reduction of CH4 level. In turn, increase in pH (a2) caused the accumulation of CH4 in the fermenter. Levels of other gases was dependent (1.695 ± 0.75) on total solids (a4). There is no significant effect of the pH on H2 level. Multiple correlation coefficients and coefficients of determination of CH4, CO2, H2, and other gases in this study indicated to a close relationship of variables with the effective factors. The coefficient of determination indicated that the share of influence of each parameter on level of CH4 and H2 was calculated in the range from 0.975 to 0.999. Regression analysis showed that the temperature (a1), pH (a2), redox (a3), total solids (a4), and volatile solids (a5) for the variability parameters of CH4 and H2 production in the anaerobic digesters were important from 97.5% to 99.9%.
4. Discussion
Multiple correlation coefficients and determination coefficients indicated a close relationship with the percentage of biogas production and investigated factors.
The coefficient of the correlation and determination was up to 0.99. The coefficient of determination indicated the share of influence of selected parameters on effective sign. These results show that the model is adequate to the experimental data and tested by using Fisher criterion [26].
Biogas, composed from methane and carbon dioxide, is the end product of the anaerobic digestion process. In this process a wide range of microorganisms is involved, including hydrolytic, acidogenic and acetogenic bacteria and finally methanogenic Archaea [27]. Methane producing microorganisms are very stable and flexible, but they are also very often unclassified. The results of our work are in agreement with other research articles [1, 9, 28, 29, 30]. The species of Methanoculleus genus seems to play the key role in different biogas fermenter systems [30, 31, 32, 33].
The dominant methanogen detected in all evaluated anaerobic digesters was Methanoculleus bourgensis.Other papers state that Methanoculleus bourgensis was isolated from fermenters performing syntrophic acetate oxidation under high ammonium concentrations [34, 35, 36, 37]. Unfortunately, Methanosaeta, Methanobacterium, and Methanocorpusculum negatively correlated with high ammonium concentrations, which can be suggest that the application of methanogenic Archaea adapted to specific feedstock [38]. Fotidis et al, (2013) and Schnürer et al., (1996) stated that syntrophic association can be observed in anaerobic digesters between Methanoculleus bourgensis and Clostridium ultunense, acetate-oxidizing bacterium. Bioaugmentation involving Methanoculleus spp. in culture together with syntrophic acetate oxidation bacteria seems to be a feasible approach to decrease the period of adaptation in anaerobic digesters processing substrates with high ammonium/ammonia concentrations [35, 39].
The results of 16S rRNA gene sequence analysis classified the isolate as a member of the species Methanoculleus bourgensis, sequence identity 99% to the 16S rRNA gene of strain MS2T [40, 41]. Genomic DNA of strain BA1 was isolated using sequenced applying the paired-end protocol on an Illumina MiSeq system. In our work, using the same approach, the sequences of Methanoculleus bourgensis was prevailing in all anaerobic digesters [30, 41].
Another genus which was detected in the digesters was Thermogymnomonas. This strain, thermoacidophilic, cell wall-less archaeon was isolated from a solfataric field in Ohwaku-dani, Hakone, Japan. The cells were irregular cocci, sometimes lobed, cup-shaped or squares forming, and were variable in size. The diameter of cells varied from 0.8 to 8.0 μm [29]. This strain was identified by Itoh et al. (2007) as Thermogymnomonas acidicola. The strain grew at temperatures from 38 °C to 68 °C (optimum 60 °C) and at pH range 1.8–4.0 (optimum pH 3.0).
To the order of Thermoplasmatales also belongs the strain IC-189T despite its different metabolism. It is an obligate aerobe microorganism with the heterotrophic metabolism what diverges it from species of the genera Thermoplasma, Picrophilus and Ferroplasma (order Thermoplasmatales). Strain IC-189 T requires yeast extract for its growth, in combination with glucose and mannose as energy and carbon source [29].
The methanogenic populations of the microorganisms depend on the different type and initial amount of substrate ratioin the anaerobic digesters as was described in the paper [42]. Two dominant morphotypes of these microorganisms in the anaerobic digesters were 99% identical to the sequences of 16S rRNA gene to the Methanoculleus and Thermogymnomonas genera deposited in GenBank. The greatest variety of morphotypes, genetically similar to genera of Methanoculleus, Thermogymnomonas, Methanobacterium, Methanolinea, Methanosaeta, Methanobrevibacter, Methanospirillum, Thermoplasmata and Thermoprotei, was detected in the wastewater treatment plant.
On the other hand, it should be noted that the diversity of methanogenic microorganisms and their biogas production can depend on other bacteria in the bioreactor including sulfate-reducing bacterial population [5]. These bacteria can use also organic compounds and produce toxic hydrogen sulfide, and can compete for substrate or electron donor (hydrogen) [43, 44, 45, 46]. This competition and production of high concentrations of hydrogen sulfide can inhibit methanogenic Archaea. However, one of the solutions can be usage of different compounds [47, 48, 49, 50, 51], which can inhibit this bacterial group and their sulfate reduction.
The sequences of 16S rRNA gene of these microorganisms from methane anaerobic digesters compared to the sequences of the strains from GenBank and the phylogenetic trees can show their genetic relationship. The number and diversities of the genera as well as the production of methane depends on the ratio of the principle substrates in each anaerobic digester. The highest methane production (52%) was found in the reactor with a mixture of substrates which included maize silage, sugar beet pulp, whole crop silage, cattle manure, and grass silage in a ratio of 29:39:12:15:5, respectively.
5. Conclusions
Methanogenic Archaea have been isolated from various anaerobic digesters and their diversity under the effect of different substrates has been observed. New isolates of these microorganisms and their study are important for the optimization of biogas production and its quality. Also, this research provides better understanding on interaction between methanogenic Archaea and other physiological bacterial groups during the process of methanogenesis in the anaerobic digesters and how different substrates have impact to this interaction.
Acknowledgements
This study was financed by Masaryk University (project TAČR GAMA – internal project CTT MU “Technology for qualitative biogas treatment” Registration Project ID: 51047). The authors wish to acknowledge the institutional support of the Faculty of AgriSciences, Mendel University in Brno funded by the Ministry of Education, Youth and Sports of the Czech Republic.
Footnotes
Conflict of interest: Authors state no conflict of interest.
Reference
- [1].Krich K., Augenstein D., Batmale J.P., Benemann J., Rutledge B., Salour D.. Biomethane from Dairy Waste. A Sourcebook for the Production and Use of Renewable Natural Gas in California. USDA Rural Development. 2005 [Google Scholar]
- [2].Wilkie A. Harwood C., Demain A. Biowaste and Biofuels. ASM Press; Washington.: 2008. Biomethane from Biomass; pp. 195–205. [Google Scholar]
- [3].Ahring B., Ibrahim A.A., Mladenovska Z.. Effect of temperature increase from 55 to 65°C on performance and microbial population dynamics of an anaerobic reactor treating cattle manure. Water Resour. 2001;35:2446–2452. doi: 10.1016/s0043-1354(00)00526-1. [DOI] [PubMed] [Google Scholar]
- [4].Ziemiński K., Frąc M.. Methane fermentation process as anaerobic digestion of biomass: Transformations, stages and microorganisms. African. J. Biotech. 2012;11:4127–4139. [Google Scholar]
- [5].Kushkevych I., Vítězová M., Vítěz T., Bartoš M.. Production of biogas: relationship between methanogenic and sulfate-reducing microorganisms. Open Life Sciences. 2017;12:82–91. [Google Scholar]
- [6].Bouallagui H., Torrijos M., Godon J., Moletta R., Cheikh R., Touhami Y.. et al. Microbial monitoring by molecular tools of a two-phase anaerobic fermenter treating fruit and vegetable wastes. Biotechnol. Lett. 26:857–862. doi: 10.1023/b:bile.0000025892.19733.18. [DOI] [PubMed] [Google Scholar]
- [7].Conrad R.. Contribution of hydrogen to methane production and control of hydrogen concentration in methanogenic soils and sediments. FEMS Microbiol. Ecol. 1999;28:193–202. [Google Scholar]
- [8].Demirel B., Scherer P.. The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: a review. Rev. Environ. Sci. Biotechnol. 2008;7:173–190. [Google Scholar]
- [9].Griffin M.E., McMahon K.D., Mackie R.I., Raskin L.. Methanogenic population dynamics during start-up of anaerobic digesters treating municipal solid waste and biosolids. Biotechnol. Eng. 2000;57:342–355. doi: 10.1002/(sici)1097-0290(19980205)57:3<342::aid-bit11>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- [10].Grothenhuis J.T., Smith M., Plugge C.M., Yuansheng X., Lammeren A.A., Stams A.J.. Bacteriological composition and structure of granular sludge adapted to different substrates. Appl. Environ. Microbiol. 1991;57:1942–1949. doi: 10.1128/aem.57.7.1942-1949.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ilyin V.K., Korniushenkova I.N., Starkova L.V., Lauriniavichius K.S.. Study of methanogenesis during bioutilization of plant residuals. Acta Astronautica. 2005;56:465–470. doi: 10.1016/j.actaastro.2004.05.077. [DOI] [PubMed] [Google Scholar]
- [12].Jäckel U., Thummes K., Kämpfer P.. Thermophilic methane production and oxidation in compost. FEMS Microbiol. Ecol. 2005;52:175–184. doi: 10.1016/j.femsec.2004.11.003. [DOI] [PubMed] [Google Scholar]
- [13].Yadvika Santosh, Sreekrishnan T.R., Kohli S., Rana V.. Enhancement of biogas production from solid substrates using different techniques. Bioresour Technol. 2004;95:1–10. doi: 10.1016/j.biortech.2004.02.010. [DOI] [PubMed] [Google Scholar]
- [14].Scherer P.A., Vollmer G.R., Fakhouri T., Martensen S.. Development of methanogenic process to degrade exhaustively the organic fraction of municipal grey waste under thermophilic and hyperthermophilic conditions. Water Sci. Technol. 2000;41:83–91. [PubMed] [Google Scholar]
- [15].Schink B.. Energetics of syntrophic cooperation in methanogenic degradation. Microb. Mol. Biol. Rev. 1997;61:262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Weiland P.. Biogas production: current state and perspectives. Appl. Microbiol. Biotechnol. 2010;85:849–860. doi: 10.1007/s00253-009-2246-7. [DOI] [PubMed] [Google Scholar]
- [17].CSN EN 14346. Characterization of waste – Calculation of dry matter by determination of dry residue or water content. Czech Standards Institute 2007 [Google Scholar]
- [18].CSN EN 15169. Characterization of waste – Determination of loss on ignition in waste, sludge and sediments. Czech Standards Institute 2007 [Google Scholar]
- [19].CSN EN 12176. Characterization of sludge – Determination of pH-value. Czech Standards Institute 1999 [Google Scholar]
- [20].Nossa C.W., Oberdorf W.E., Yang L., Aas J.A., Paster B.J., Desantis T.Z.. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J. Gastroenterol. 2010;16:4135–4144. doi: 10.3748/wjg.v16.i33.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K.. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Altschul S.F., Gish W., Mille W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- [23].Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S.. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Clustal W.. and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- [25].Chen P.Y., Popovich P.M.. Correlation: Parametric and Nonparametric Measures. Sage University Papers Series on Quantitative Applications in the Social Sciences. 2002 [Google Scholar]
- [26].Bailey N.T.J. Statistical Methods in Biology. third. Cambridge University Press; Cambridge: 1995. [Google Scholar]
- [27].Zeikus J.G.. The biology of methanogenic bacteria. Bact. Rev. 1977;41:514–541. doi: 10.1128/br.41.2.514-541.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Amon T., Amon B., Kryvoruchko V., Zollitsch W., Mayer K., Gruber L.. Biogas production from maize and dairy cattle manure – influence of biomass composition on the methane yield. Agric. Ecosys. Environ. 2007;118:173–182. [Google Scholar]
- [29].Itoh T., Yoshikawa N., Takashina T.. Thermogymnomonas acidicola gen. nov., sp. nov., a novel thermoacidophilic, cell wall-less archaeon in the order Thermoplasmatales, isolated from a solfataric soil in Hakone. Japan. Int. J. Syst. Evol. Microbiol. 2007;57:2557–2561. doi: 10.1099/ijs.0.65203-0. [DOI] [PubMed] [Google Scholar]
- [30].Maus I., Wibberg D., Winkler A., Pühler A., Schnürer A., Schlütera A.. Complete Genome Sequence of the Methanogen Methanoculleus bourgensis BA1 Isolated from a Biogas Reactor Genome. Announcements. 2016;4:e00568–16. doi: 10.1128/genomeA.00568-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chynoweth D.P., Turick C.E., Owens J.M., Jerger D.E., Peck M.W.. Biochemical methane potential of biomass and waste feedstocks. Biomass Bioen. 1993;5:95–111. [Google Scholar]
- [32].Jaenicke S., Ander C., Bekel T., Bisdorf R., Dröge M., Gartemann K.H.. Comparative and joint analysis of two metagenomic datasets from a biogas fermenter obtained by 454-pyrosequencing. PLoS One. 2011;6:e14519. doi: 10.1371/journal.pone.0014519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Stolze Y., Zakrzewski M., Maus I., Eikmeyer F., Jaenicke S., Rottmann N.. Comparative metagenomics of biogas-producing microbial communities from production-scale biogas plants operating under wet or dry fermentation conditions. Biotechnol Biofuels. 2015;8:14. doi: 10.1186/s13068-014-0193-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Moestedt J., Müller B., Westerholm M., Schnürer A.. Ammonia threshold for inhibition of anaerobic digestion of thin stillage and the importance of organic loading rate. Microb. Biotechnol. 2016;9:180–194. doi: 10.1111/1751-7915.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Westerholm M., Levén L., Schnürer A.. Bioaugmentation of syntrophic acetate-oxidizing culture in biogas reactors exposed to increasing levels of ammonia. Appl. Environ. Microbiol. 2012;78:7619–7625. doi: 10.1128/AEM.01637-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Westerholm M., Müller B., Isaksson S., Schnürer A.. Trace element and temperature effects on microbial communities and links to biogas digester performance at high ammonia levels. Biotechnol. Biofuels. 2015;8:154. doi: 10.1186/s13068-015-0328-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ziganshina E.E., Belostotskiy D.E., Shushlyaev R.V., Miluykov V.A., Vankov P.Y., Ziganshin A.M.. Microbial Community Diversity in Anaerobic Reactors Digesting Turkey, Chicken, and Swine Wastes. J. Microbiol. Biotechnol. 2014;24:1464–772. doi: 10.4014/jmb.1404.04043. [DOI] [PubMed] [Google Scholar]
- [38].Ziganshin A.M., Ziganshina E.E., Kleinsteuber S., Nikolausz M.. Comparative Analysis of Methanogenic Communities in Different Laboratory-Scale Anaerobic Digesters. Archaea. 2016:12. doi: 10.1155/2016/3401272. Article ID 3401272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fotidis I.A., Wang H., Fiedel N.R., Luo G., Karakashev D.B., Angelidaki I.. Bioaugmentation as a solution to increase methane production from an ammonia-rich substrate. Environ. Sci. Technol. 2014;48:7669–7676. doi: 10.1021/es5017075. [DOI] [PubMed] [Google Scholar]
- [40].Maus I., Wibberg D., Stantscheff R., Stolze Y., Blom J., Eikmeyer F.G.. Insights into the annotated genome sequence of Methanoculleus bourgensis MS2(T), related to dominant methanogens in biogas-producing plants. J. Biotechnol. 2014;201:43–53. doi: 10.1016/j.jbiotec.2014.11.020. [DOI] [PubMed] [Google Scholar]
- [41].Maus I., Wibberg D., Stantscheff R., Eikmeyer F.G., Seffner A., Boelter J.. Complete genome sequence of the hydrogenotrophic, methanogenic archaeon Methanoculleus bourgensis strain MS2(T), isolated from a sewage sludge digester. J. Bacteriol. 2012;194:5487–5488. doi: 10.1128/JB.01292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sundberg C., Al-Soud W.A., Larsson M., Alm E., Yekta S.S., Svensson B.H., Sørensen S.J., Karlsson A.. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol. 2013;85:612–626. doi: 10.1111/1574-6941.12148. [DOI] [PubMed] [Google Scholar]
- [43].Kushkevych I.V.. Kinetic Properties of Pyruvate Ferredoxin Oxidoreductase of Intestinal Sulfate-Reducing Bacteria Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9. Polish J Microbiol. 2015;64:107–114. [PubMed] [Google Scholar]
- [44].Kushkevych I., Fafula R., Parak T., Bartos M.. Activity of Na+ / K+-activated Mg2+-dependent ATP hydrolase in the cell-free extracts of the sulfate-reducing bacteria Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9. Acta Vet Brno. 2015;84:3–12. [Google Scholar]
- [45].Kushkevych I.V.. Activity and kinetic properties of phosphotransacetylase from intestinal sulfate-reducing bacteria. Acta Biochimica Polonica. 2015;62:1037–108. doi: 10.18388/abp.2014_845. [DOI] [PubMed] [Google Scholar]
- [46].Kushkevych I., Vítězová M., Fedrová M., Vochyanová Z., Paráková L., Hošek J.. Kinetic properties of growth of intestinal sulphate-reducing bacteria isolated from healthy mice and mice with ulcerative colitis. Acta Vet Brno. 2017;86:405–411. [Google Scholar]
- [47].Kushkevych I., Kollar P., Suchy P., Parak K., Pauk K., Imramovsky A.. Activity of selected salicylamides against intestinal sulfate-reducing bacteria. Neuroendocrinol Lett. 2015;36:106–113. [PubMed] [Google Scholar]
- [48].Kushkevych I., Kollar P., Ferreira A.L., Palma D.. Antimicrobial effect of salicylamide derivatives against intestinal sulfate-reducing bacteria. J Appl Biome. 2016;14:125–130. [Google Scholar]
- [49].Kushkevych I., Vítězová M., Kos J., Kollár P., Jampílek J.. Effect of selected 8-hydroxyquinoline-2-carboxanilides on viability and sulfate metabolism of Desulfovibrio piger. J. App.Biomed. 2018;16:1–6. [Google Scholar]
- [50].Kushkevych I., Kováč J., Vítězová M., Vítěz T., Bartoš M.. The diversity of sulfate-reducing bacteria in the seven bioreactors. Arch. Microbiol. 2018;200:1–6. doi: 10.1007/s00203-018-1510-6. [DOI] [PubMed] [Google Scholar]
- [51].Kováč J., Kushkevych I.. New modification of cultivation medium for isolation and growth of intestinal sulfate-reducing bacteria. Proceed. Intern. PhD Stud. Conf. MendelNet. 2017:702–707. [Google Scholar]