Abstract
Halophytes can survive and complete their life cycle in the presence of ≥200 mM NaCl. These remarkable plants have developed various strategies to tolerate salinity and thrive in high-salt environments. At the appropriate levels, salt has a beneficial effect on the vegetative growth of halophytes but inhibits the growth of non-halophytes. In recent years, many studies have focused on elucidating the salt-tolerance mechanisms of halophytes at the molecular, physiological, and individual level. In this review, we focus on the mechanisms, from the macroscopic to the molecular, underlying the successful growth of halophytes in saline environments to explain why salt has beneficial effects on halophytes but harmful effects on non-halophytes. These mechanisms include the specialized organs of halophytes (for example, ion compartmentalization in succulent leaves), their unique structures (salt glands and hydrophobic barriers in roots), and their salt-tolerance genes. We hope to shed light on the use of halophytes for engineering salt-tolerant crops, soil conservation, and the protection of freshwater resources in the near future.
Keywords: Hydrophobic barriers, Ion compartmentalization, Molecular, Salt gland, Salt tolerance
1. Introduction
Salt water accounts for approximately 97% of the Earth’s water supply, and humans can only use 1% of the fresh water found worldwide [1]. Saving fresh water and making good use of salt water pose new challenges, especially in arid and semi-arid countries. Halophytes naturally grow in saline environments [2,3], and some species, such as mangroves, can even grow in seawater [4]. Halophyte plants can be used as forage grasses, in medicines, as vegetables, and as papermaking materials [1]. Therefore, investigating the mechanisms by which halophytes tolerate saline environments is crucial for sustainable development.
Salt can damage plants through its osmotic effect (physiological drought under high-salinity conditions), ion toxicity (especially Na+ and Cl-), and secondary stresses such as oxidative stress [5,6]. Halophytes and non-halophytes show distinct differences in maximum salt tolerance [7, 8, 9]. Plants that can survive and complete their life cycle in a salt concentration of ≥200 mM NaCl are usually defined as halophytes [5,10,11, 12, 13]. Halophytes actively control the uptake, storage, exclusion, and secretion of ions under saline conditions [14, 15, 16, 17]. The most salt-tolerant halophytes such as Suaeda salsa can complete their life cycle in soils containing 200 to 500 mM NaCl [18, 19, 20], whereas non-halophytes show limited salt tolerance and can be damaged in soils with NaCl concentrations <50 mM [21].
Halophytes do not simply tolerate high-salt conditions. True halophytes thrive at the appropriate salt concentrations and show optimal growth in the presence of significant amounts of NaCl, e.g, 200 mM for S. salsa [22,23], 150 mM for Chenopodium quinoa [24], and 100 mM for Cakile maritima [25]. The halophyte Plantago crassifolia exhibits highly efficient responses to salt stress during early seedling development [26,27]. Appropriate salt concentrations can promote the vegetative growth of halophytes and are conducive to the completion of their life cycle, as described by Flowers & Colmer [10]. Salt has a beneficial effect on halophytes, as they grow larger and more rapidly in the presence of the appropriate salt concentration, compared to conditions with little or no salt [20]. More specifically, the majority of halophytes benefit from the presence of high concentrations of salt during processes ranging from seed germination to seedling growth. For example, many halophytes such as Cakile maritima [25] and Chloris virgata [28] have been shown to have higher germination percentages at slightly elevated salinity levels (0.5% NaCl, or around 50–90 mM) vs. distilled water [28,29]. In addition to seed germination, appropriate NaCl concentrations also enhance the seedling growth of halophytes compared to non-salt conditions. This is evidenced by higher seedling biomass, larger leaf area [30], and enhanced photosynthetic efficiency [17,31] and yield, thus leading to increased seed production for the next generation [18,19].
By contrast, non-halophytes are salt sensitive and suffer from salt-induced damage. These plants are classified as salt-sensitive and salt-tolerant non-halophytes based on their level of salt tolerance. Plants in both categories show inhibited growth under saline conditions, but salt-sensitive non-halophytes, such as soybean and rice, may suffer irreparable damage in response to low concentrations of NaCl (less than 50 mM) [32,33], whereas salt-tolerant non-halophytes such as cotton, beets, and barley can tolerate higher salt concentrations (200–300 mM NaCl) [34, 35, 36]. However, all non-halophytes show decreased biomass when grown in the presence of salt with one exception: Eutrema salsugineum (formerly misclassified as Thellungiella salsuginea, Brassicaceae [37,38]). This plant is widely considered to be a model halophyte [39] because it has a certain degree of salt tolerance and was reported to survive under 250 mM NaCl conditions [40,41], although its growth sharply declines with increasing NaCl level [42,43]. Studies of E. salsugineum performed over the past 15 years have contributed to our understanding of salt tolerance mechanisms in halophytes.
Why does the appropriate salinity level enhance the vegetative growth of halophytes and inhibit the growth of non-halophytes? Do halophytes have special characteristics that allow them to adapt to saline environments? In the past decade, many studies have investigated possible underlying mechanism. In the current review, we focus on the vegetative growth of halophytes to illustrate the mechanisms underlying the robust growth of halophytes in saline environments, from the morphological to the cellular and molecular levels.
2. Morphological, cellular, and sub-cellular adaptations
All plants, including non-halophytes, compartmentalize excess ions into their vacuoles, which is considered the physiological foundation of salt tolerance in all plants [44]. Halophytes have evolved several specific structures or mechanisms to adapt to saline environments (Fig. 1). However, non-halophytes have not evolved the unique morphological features needed to cope with salt stress, and if forced to live in saline soil, their biomass is reduced and they cannot complete their life cycles. By contrast, halophytes can survive high-salt conditions due to leaf succulence and the functions of specialized organs (e.g, salt glands, as described below). There are three types of halophytes: euhalophytes, recretohalophytes, and pseudohalophytes [45]. Euhalophytes such as Kalidium foliatum and S. salsa are salt accumulators that can take up large amounts of ions and compartmentalize them in vacuoles to maintain cell turgor. These plants also develop leaf or stem succulence when the soil water potential is low [20].
Figure 1.
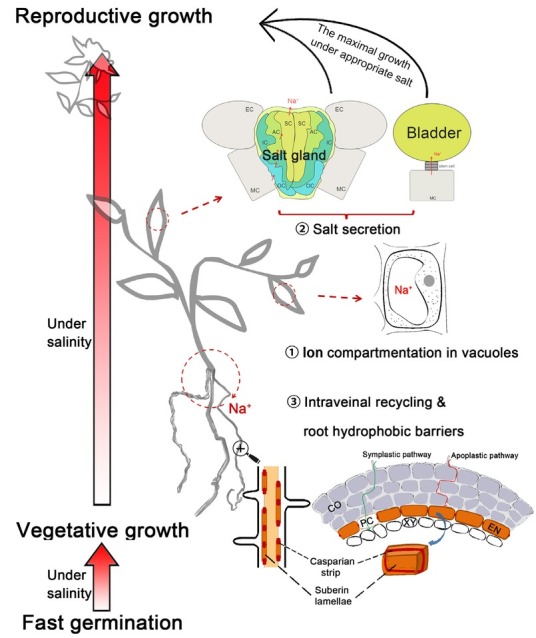
Salt-tolerance mechanisms in halophytes. Seeds that rapidly germinate under saline conditions benefit from dimorphism or mobilization. Vegetative growth is maximal and reproductive growth is stimulated under appropriate salt concentrations because of the following salt-tolerance mechanisms: 1) ion compartmentalization; 2) salt secretion; and 3) ion intraveinal recycling and the root apoplastic barrier. In the first mechanism- ion compartmentalization- Na+ actively accumulates in the vacuoles, thus preventing protoplast damage. The second mechanism- salt secretion- is described in Yuan et al [3], which showed the typical multi-cellular salt gland and salt bladder. SC, secretory cell; AC, accessory cell; IC, inner cup cell; OC, outer cup cell; MC, mesophyll cell; EC, epidermal cell. The third mechanism- the root apoplastic barrier- includes the Casparian strip and suberin lamellae, which can effectively block the apoplastic pathway. Ions can only enter endothelial cells via passage cells (PC), i.e, the symplastic pathway. XY, xylem; CO, cortex; EN, endodermis; PC: passage cell. The plant was drawn with Photoshop CS6.
Leaf succulence is a typical visible characteristic of euhalophytes such as S. salsa under high-salinity conditions (Fig. 2) [46,47], although this feature is not unique to halophytic plants as certain xerophytes, such as cacti and Kalanchoe daigremontiana also have succulent leaves under drought conditions. However, halophytes and xerophytes have evolved different strategies leading to the formation of succulence. Under saline conditions, ion accumulation in vacuoles results in succulence, which may be caused by the presence of carbon as a driving force and ion compartmentalization to relieve salt damage. For example, S. salsa actively accumulates ions and proline in its vacuoles and cytosol to reduce plant water potential [48]. A possible mechanism underlying leaf succulence in S. salsa is suggested by the finding that the presence of aquaporins in the plasma membrane is correlated with Na+ accumulation in the vacuole [23,49,50]. Under drought conditions, however, succulence is induced by the accumulation of organic compounds such as malate via a carbon gradient [51].
Figure 2.

The succulent blades of euhalophyte Suaeda salsa grown in intertidal zone (left, red leaves) and inland saline soils (right, green leaves) of the Yellow River Delta (N 37°25′; E 118°54′).
Halophytes and non-halophytes show distinct differences in ion compartmentalization. Photosynthesis and chloroplasts in non-halophytes are markedly damaged by salinity due to a weak ion compartmentalization [52]. For example, in Arabidopsis thaliana, electron transport though photosystem II is dramatically inhibited and nonphotochemical quenching of chlorophyll fluorescence increases in response to 150 mM NaCl [39]. By contrast,
the chloroplasts and mitochondria of halophytes are protected under salt-stress conditions due to a strong ion compartmentalization. The ultrastructure of thylakoids in the chloroplasts of two euhalophytic species Haloxylon ammodendron and Suaeda physophora showed no observable damage when treated with 700 mM NaCl [17]. In the halophyte Artemisia anethifolia, the thylakoids in chloroplasts were intact, and the number of cristae in the mitochondria did not decrease until the plants were treated with 200 mM NaCl [53]. The halophyte Suaeda altissima also retained normal chloroplast function under 750 mM NaCl conditions [31].
The two other types of halophytes are considered to be salt regulators. Recretohalophytes can secrete excessive ions via specific salt-secreting structures, e.g, salt bladders in Atriplex centralasiatica [54] and salt glands in L. bicolor [55,56]. These unique epidermal structures distinguish these plants from other halophytes and all non-halophytes [3,57]. Vesicle transport is the main pathway for salt secretion [2,58]. The subcellular structures of recretohalophytes also exhibit specific characteristics. Most reports have focused on salt-secretory ultrastructures such as highly developed plasmodesmata, mitochondria, vesicles, the lack of chloroplasts, cuticles, and so on (this information can be found in Yuan et al [3] and Shabala et al [54]). A review by Dassanayake [59] discusses the morphology and evolution of salt glands, suggesting that these structures emerged independently at least 12 times in recretohalophytes.
The roots of non-halophytes and halophytes behave quite differently. In root cells of the non-halophyte common bean (Phaseolus vulgaris), the addition of 80 mM NaCl leads to membrane vesiculation and increased solute leakage [60]. By contrast, the roots of pseudohalophytes such as mangroves in the Rhizophoraceae family and reeds in the Gramineae family show high salt exclusion ability, thereby protecting the shoots from salinity. The possible mechanism underlying salt exclusion in plants such as reeds is described as interveinal recycling and apoplastic barriers in the roots. During interveinal recycling, the Na+ absorbed by roots is transported into the shoots through xylem vessels and is then loaded into the phloem by HKT1 (a high-affinity K+ transporter) [61,62]. Finally, this Na+ is unloaded back into the soil by SOS1 (a plasma membrane Na+/H+ antiporter) [63,64] in roots cells [65,66] (detailed in the “Salt-tolerance genes” section). In recent years, lignin and suberin lamellae in the root endodermis have also been shown to be involved in the salt exclusion pathway. Root hydrophobic barriers play an important role in salt exclusion in Avicennia officinalis [4]. The same group reported that, although rice is a representative non-halophyte, it can also tolerate low concentrations of salt (50–100 mM NaCl), mainly due to the presence of apoplastic transport barriers in the roots [67].
As mentioned in the Introduction, E. salsugineum is a special type of halophyte that has been used as a model plant to unravel the molecular mechanisms of salt tolerance in halophytes [68,69]. Although this plant does not possess the typical characteristics of halophytes (such as salt glands or salt bladders) and shows a marked decrease in vegetative growth under high salinity, studies of E. salsugineum have shed light on the mechanisms underlying salt tolerance. Under high-salt conditions, E. salsugineum undergoes differential regulation of Na+/K+ ions and re-establishes Na+/K+ homeostasis [70], including a reduction in Na+ absorption [71] and an increase in Na+ compartmentalization [72,73]. The genes controlling Na+ absorption are described in the following section and listed in Table 1. The osmotic balance in E. salsugineum can also be maintained by proline accumulation in addition to ion accumulation [74], which helps this plant survive in saline environments.
Table 1.
Genes involved in Na+ influx and the three salt tolerance mechanisms of halophytes
| Salt-tolerance mechanism | Gene | Likely function in salt tolerance | Halophyte species | References |
|---|---|---|---|---|
| Na+ influx | HKT1 | High-affinity K+ transporter 1 | Suaeda salsa | [92] |
| Salicornia dolichostachya | [111] | |||
| Leptochloa fusca | [112] | |||
| Aeluropus lagopoides | [113] | |||
| AKT1 | Inward-rectifying K channel 1 | Suaeda maritima | [114] | |
| KUP/HAK/KT | KUP/HAK/KT type transporter | Suaeda maritime | [115] | |
| Eutrema salsugineum | [71] | |||
| KT | Potassium transporter | Reaumuria trigyna | [104] | |
| Limonium bicolor | [56] | |||
| 1) Ion compart- | NHX | Encodes a vacuolar-type Na+/H+ antiporter that | Limonium gmelinii | [116] |
| mentation in vacuoles | is located on the vacuolar membrane and pumps | Karelinia caspica | [81] | |
| excessive Na+ into the vacuole to avoid toxic Na+ | Salicornia brachiate | [114] | ||
| concentrations in the cytoplasm. | Aeluropus littoralis | [118,91] | ||
| CLC | Chloride channel on vacuolar membrane | Mesembryanthemum crystallinum | [119] | |
| AQP | Encodes aquaporin | Sesuvium portulacastrum | [76] | |
| Suaeda salsa | [50] | |||
| 2) Salt secretion | SOS1 | Encodes a Na+/H+ antiporter located on the plasma membrane that pumps excess Na+ out of the cell. | Avicennia marina | [120] |
| HA1 | PM H+-ATPase | Avicennia marina | [120] | |
| NHX | Na+/H+ antiporter on the vacuolar membrane | Avicennia marina | [120] | |
| VAMP | Vesicle-associated membrane protein | Limonium bicolor | [56] | |
| CLC | Chloride channel on the plasma membrane | Limonium bicolor | [56] | |
| PIP and TIP | Aquaporin genes | Avicennia officinalis | [121] | |
| 3) Intravein recycling and root hydrophobic barriers | SOS1 | Encodes a Na+/H+ antiporter located on the plasma membrane that plays a role in Na+ efflux from roots | Salicornia dolichostachya | [111] |
| AoCYP86B1 | Encodes cytochrome P450 that regulates suberin biosynthesis and prevents some Na+ from entering the roots | Avicennia officinalis | [4] | |
In short, halophytes have evolved several structural or ultrastructural adaptations to salt stress, whereas non-halophytes do not develop these adaptive structures, and their ultrastructure is significantly injured under low-salt conditions. Therefore, specific cellular and subcellular structures facilitate the strong growth of halophytes under the appropriate salt concentrations.
3. Salt-tolerance genes
All traits, including salt tolerance and salt sensitivity, are ultimately controlled by genes. Certain salt tolerance genes are constitutively expressed in halophytes while other genes are induced by salt [75], exhibiting increased expression under salt treatment [76,77]. Although many reports involving salt-tolerance genes have focused on non-halophytes such as Arabidopsis [64,78,79] and rice [80], we will concentrate on salt-tolerance genes in halophytes. Table 1 lists the genes involved in Na+ transport across the membrane and the three salt-tolerance mechanisms used by halophytes (also see Fig. 1). Na+ flux occurs from root to leaf in halophytes based on the genes described to date. Na+ may enter the cell by HKT1, KT, KUP/HAK/KT-type transporters, AKT1-type channels, and NSCCs (nonselective cation channels). To avoid salt damage to the cytoplasm, many genes involved in the three salt-tolerance mechanisms are upregulated, such as NHX (encoding a vacuolar-type Na+/H+ antiporter that participates in ion compartmentation in vacuoles);
SOS pathway genes such as SOS1; PIP (aquaporin involved in salt secretion); and cytochrome P450 (involved in the root hydrophobic barrier).
To date, to the best of our knowledge, only one halophyte gene has been tested in a halophyte to verify its function. Silencing KcNHX1 in the halophyte Karelinia caspica led to reduced tolerance to high concentrations of NaCl, suggesting that KcNHX1 plays an essential role in the response of K. caspica to salt stress [81]. Most of the same genes may be present in halophytes and non-halophytes but exhibit different expression patterns due to different long-term survival strategies [82]. Therefore, all salt-tolerance genes that have been cloned in halophytes to date have been tested by heterologous expression in non-halophytes to explore their functions [83, 84, 85, 86, 87, 88]. The highest concentration of NaCl that these transgenic plants could tolerate was reported as 400 mM [89,90]. For example, transgenic tobacco (Nicotiana tabacum) transformed with AlNHX (encoding a vacuolar-typed Na+/H+ antiporter) from the halophyte Aeluropus littoralis exhibited high salt tolerance (400 mM NaCl) [91]. Transgenic tobacco also compartmentalized more Na+ in its roots than wild type tobacco to maintain a relatively high K+/Na+ ratio in its leaves [91]. Overexpression of a similar gene SsNHX1 (encoding a putative vacuolar Na+/H+ antiporter) from Salsola soda allowed Medicago sativa to survive in high concentrations of NaCl (up to 400 mM) due to improved Na+ sequestration in the vacuole [90].
In addition to the role of NHX genes in ion compartmentation, studies in non-halophytes have also verified the functions of many other groups of halophyte genes controlling primary salt-tolerance traits, showing that heterologous expression of these genes significantly improved the salt tolerance of these plants. The first group of genes includes HKT1 (encoding a high-affinity K+ transporter) and SOS1 (encoding a plasma membrane Na+/ H+ antiporter). Transgenic Arabidopsis transformed with SsHKT1;1 from S. salsa showed enhanced salt tolerance and increased K+ concentrations in shoots [92]. Transgenic tobacco harboring SbSOS1 from Salicornia brachiata showed a high degree of salt tolerance, growing in 200 mM NaCl [93].
The second group of genes, including H+-pyrophosphatase and vacuolar ATPase genes, is involved in energy supply. For example, transgenic Arabidopsis transformed with SsVP (encoding a vacuolar H+ -pyrophosphatase) from S. salsa [94] or KfVP1 (encoding H+-pyrophosphatase) from Kalidium foliatum [95] showed increased salt tolerance due to enhanced V-ATPase and V-PPase activity. Transgenic rice transformed with SaVHAc1 (a vacuolar H+-ATPase subunit c1 gene) from the halophyte Spartina alterniflora performs better under salt stress than control [96].
The third group of genes is involved in the ROS scavenging system. Transgenic tobacco transformed with SbpAPX (encoding Peroxisomal Ascorbate Peroxidase) from S. brachiata showed enhanced vegetative growth compared to wild type when grown at 300 mM NaCl [97]. Transformation with Ss.sAPX (encoding a stromal ascorbate peroxidase) from S. salsa improved the growth of Arabidopsis plants under high-salt conditions [84].
The remaining groups of genes are related to plant hormones and aquaporin. Transgenic tobacco expressing high levels of SbASR-1 (encoding abscisic acid stress ripening-1) from S. brachiata showed better germination and seedling growth than wild type when grown on 400 mM NaCl [89]. Transgenic tobacco harboring SpAQP1 (aquaporin-related gene induced by salt) from Sesuvium portulacastrum showed enhanced seed germination and root growth under high-salt conditions due to increased antioxidant enzyme activity [76].
The heterologous expression of halophytic salt-tolerance genes improves salt resistance in non-halophytes to some degree, but transgenic plants often cannot finish their life cycles in naturally saline soils due to the great spatial and temporal variation of salt content. Moreover, to the best of our knowledge, no transgenic non-halophytes show typical halophyte characteristics such as improved growth under the appropriate salt concentration. In general, salt-tolerance traits are controlled by a series of genes rather than one or two genes. Therefore, it might be necessary to identify salt-tolerance gene networks and explore their effects under controlled conditions.
4. Conclusions and Perspective
The vegetative growth of halophytes can benefit from appropriate salt concentrations. Although different halophytes have evolved diverse salt-tolerance mechanisms, these can primarily be divided into three categories: the use of specialized organs (succulent leaves via ion compartmentalization), unique structures (salt glands and hydrophobic barriers in roots), and salt-tolerance genes. In this review, we focused on the mechanisms that could explain the beneficial effects of salt on vegetative growth in halophytes (i.e, better and more rapid growth than under non-salt conditions, resulting in increased seed production), including the morphological, cellular, and molecular aspects of these mechanisms. Additional reviews about various salt-tolerance mechanisms can be found in [82,98,99,100]. Many reports emphasize the important role of halophytes in improving saline soil conditions and the cultivation of salt-tolerant crops [1,3,20,44,82,100,101]. Several researchers have proposed a series of possible ways to realize these dreams, such as transforming non-halophytes with salt-tolerance genes to improve their salt resistance [82]. Indeed, salt-tolerance genes isolated from halophytes are often used to transform non-halophytes.
However, it is still difficult to apply these solutions to plants grown in the field and these solutions face many challenges. To date, no glycophytes/non-halophytes transformed with salt-tolerance genes have been successfully grown in natural saline environments. On the one hand, all known salt-tolerance genes have been heterologously overexpressed in non-halophytes to clarify their functions, which is not a very precise method. The functions of salt-tolerance genes should be verified in the halophyte itself via silencing or knockout, but this type of experiment has only been reported for the halophyte K. caspica [81]. On the other hand, salt tolerance in halophytes is a complex trait that is controlled by gene families or networks. Transforming one or several related genes into glycophytes may not cause radical changes in salt tolerance; instead, the transformed genes must function coordinately. Nevertheless, these solutions appear feasible, but additional time is needed to carry out such experiments.
For the discovery of salt-tolerance genes and networks, high-throughput RNA-seq has been used in several halophytes such as L. bicolor [13], M. crystallinum [102,103], and Reaumuria trigyna [104]. Although many salt-tolerance genes have been identified in halophytes, which genes should we focus on first? Perhaps we can focus on the genes controlling primary salt-tolerance traits as mentioned in this review (such as succulent leaves, salt glands, and root hydrophobic barriers), followed by regulatory genes (such as transcription factor genes) that control these traits (e.g., Table 1) by transforming the halophyte itself. Using this procedure, we can target the key traits directly involved in salt tolerance and the corresponding phenotypes, allowing a single trait to be improved in non-halophytes via the transformation of these genes. Good transformation systems are clearly needed for this strategy and, therefore, there is an urgent need to establish such systems for use in various halophytes, such as Leymus chinensis and L. bicolor [105,106]. Based on this system, CRISPR/Cas9-mediated genome editing will likely prove to be a useful tool for verifying target gene function [107]. In addition, many recent studies have found that long non-coding RNAs play an important role in salt tolerance in plants [108, 109, 110]. Therefore, more attention should be paid to non-coding RNAs that participate in the unique salt-tolerance strategies of halophytes via high-throughput RNA sequencing.
Overall, given that the expanding saline lands threaten human existence, there are two ways to make good use of halophytes to preserve soils and fresh water: 1) increasing the planting areas of halophytes in arid and semi-arid areas to help prevent water loss and 2) transforming non-halophytes with salt-tolerance genes to enable them to tolerate irrigation with full-strength or diluted seawater in the near future.
Acknowledgments
This work was supported by the NSFC (National Natural Science Research Foundation of China, project No. 31600200; 31570251; 31770288), Shandong Province key research and development plan (2015ZDJS03002; 2017CXGC0313), the Natural Science Research Foundation of Shandong Province (ZR2014CZ002), and the Higher Educational Science and Technology Program of Shandong Province (J15LE08).
Footnotes
Conflict of interest: Authors state no conflict of interest.
References
- [1].Abd El-Hack ME, Samak DH, Noreldin A E. Towards saving freshwater: halophytes as unconventional feedstuffs in livestock feed: a review. Environmental Science and Pollution Research. 2018. pp. 1–10. [DOI] [PubMed]
- [2].Feng Z, Sun Q, Deng Y. Study on pathway and characteristics of ion secretion of salt glands of Limonium bicolor. Acta Physiologiae Plantarum. 2014;36(10):2729–2741. [Google Scholar]
- [3].Yuan F, Leng BY, Wang B S. Progress in Studying Salt Secretion from the Salt Glands in Recretohalophytes: How Do Plants Secrete Salt? Frontiers in Plant Science. 2016;7(977):977. doi: 10.3389/fpls.2016.00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Krishnamurthy P, Jyothi-Prakash P A, Qin L. Role of root hydrophobic barriers in salt exclusion of a mangrove plant Avicennia officinalis. Plant Cell & Environment. 2014;37(7):1656–1671. doi: 10.1111/pce.12272. [DOI] [PubMed] [Google Scholar]
- [5].Flowers T J, Munns R, Colmer T D. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Annals of Botany. 2015;115(3) doi: 10.1093/aob/mcu217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yang Y, Guo Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytologist. 2018;217(2):523–539. doi: 10.1111/nph.14920. [DOI] [PubMed] [Google Scholar]
- [7].Leng BY, Yuan F, Dong XX. Distribution pattern and salt excretion rate of salt glands in two recretohalophyte species of Limonium (Plumbaginaceae) South African Journal of Botany. 2018;115:74–80. [Google Scholar]
- [8].Wang F, Xu Y G, Wang S. Salinity affects production and salt tolerance of dimorphic seeds of Suaeda salsa. Plant Physiology & Biochemistry Ppb. 2015;95:41. doi: 10.1016/j.plaphy.2015.07.005. [DOI] [PubMed] [Google Scholar]
- [9].Zhou J, Zhao W, Yin C H. The role of cotyledons in the establishment of Suaeda physophora seedlings. 2014. pp. 1–7. Plant Biosystems-An International Journal Dealing with all Aspects of Plant Biology. ahead-of-print.
- [10].Flowers T J, Colmer T D. Salinity tolerance in halophytes. New Phytologist. 2008;179(4):945–963. doi: 10.1111/j.1469-8137.2008.02531.x. [DOI] [PubMed] [Google Scholar]
- [11].Santos J, Al-Azzawi M, Aronson J. eHALOPH a database of salt-tolerant plants: helping put halophytes to work. Plant and Cell Physiology. 2015;pcv155 doi: 10.1093/pcp/pcv155. [DOI] [PubMed] [Google Scholar]
- [12].Yuan F, Chen M, Yang J C. The optimal dosage of 60Co gamma irradiation for obtaining salt gland mutants of Exo-recretohalophyte Limonium bicolor (Bunge) O. Kuntze. Pak. J. Bot. 2015;47(1):71–76. [Google Scholar]
- [13].Yuan F, Lyv M J, Leng BY. Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant, Cell & Environment. 2015;38:1637–1657. doi: 10.1111/pce.12514. [DOI] [PubMed] [Google Scholar]
- [14].Ma Q, Yue L-J, Zhang J-L. Sodium chloride improves photosynthesis and water status in the succulent xerophyte Zygophyllum xanthoxylum. Tree Physiology. 2011;32(1):4–13. doi: 10.1093/treephys/tpr098. [DOI] [PubMed] [Google Scholar]
- [15].Song J, Shi W, Liu R. The role of the seed coat in adaptation of dimorphic seeds of the euhalophyte Suaeda salsa to salinity. Plant Species Biology. 2017;32(2) [Google Scholar]
- [16].Song J, Zhou J, Zhao W. Effects of salinity and nitrate on production and germination of dimorphic seeds applied both through the mother plant and exogenously during germination in Suaeda salsa. Plant Species Biology. 2016;31(1):19–28. [Google Scholar]
- [17].Zhang S, Song J, Wang H. Effect of salinity on photosynthesis and chloroplasts ultrastructure in cotyledons of desiccated seeds of halophytes or xerophyte growing in central Asia. Journal of Plant Ecology. 2010;3:259–267. [Google Scholar]
- [18].Guo J, Li Y, Han G. NaCl markedly improved the reproductive capacity of the euhalophyte Suaeda salsa. Functional Plant Biology. 2018;44(3):350–361. doi: 10.1071/FP17181. [DOI] [PubMed] [Google Scholar]
- [19].Guo J, Suo S, Wang B S. Sodium chloride improves seed vigour of the euhalophyte Suaeda salsa. Seed Science Research. 2015;25(3):335–344. [Google Scholar]
- [20].Song J, Wang B. Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Annals of Botany. 2015;mcu194 doi: 10.1093/aob/mcu194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Galvan-Ampudia C, Christa T. Salt stress signals shape the plant root. Current Opinion in Plant Biology. 2011;14:296–302. doi: 10.1016/j.pbi.2011.03.019. [DOI] [PubMed] [Google Scholar]
- [22].Liu Q Q, Liu RR, Ma Y C. Physiological and molecular evidence for Na+ and Cl- exclusion in the roots of two Suaeda salsa populations. Aquatic Botany. 2018;146:1–7. [Google Scholar]
- [23].Yang M F, Song J, Wang B S. Organ-Specific Responses of Vacuolar H+-ATPase in the Shoots and Roots of C3 Halophyte Suaeda salsa to NaCl. Journal of Integrative Plant Biology. 2010;52(3):308–314. doi: 10.1111/j.1744-7909.2010.00895.x. [DOI] [PubMed] [Google Scholar]
- [24].Shabala L, Mackay A, Tian Y. Oxidative stress protection and stomatal patterning as components of salinity tolerance mechanism in quinoa Chenopodium quinoa. Physiologia Plantarum. 2012;146(1):26–38. doi: 10.1111/j.1399-3054.2012.01599.x. [DOI] [PubMed] [Google Scholar]
- [25].Debez A, Hamed KB, Grignon C. Salinity effects on germination, growth, and seed production of the halophyte Cakile maritima. Plant & Soil. 2004;262(1/2):179–189. [Google Scholar]
- [26].Alhassan M, Pacurar A, Gaspar A. Growth and reproductive success under saline conditions of three Plantago species with different levels of stress tolerance. Notulae Botanicae Horti Agrobotanici Cluj-Napoca. 2014;42(1):180–186. [Google Scholar]
- [27].Boscaiu M, Soriano E E, Vicente O. Effects of salt stress on the reproductive biology of the halophyte Plantago crassifolia. Biologia Plantarum. 2005;49(1):141–143. [Google Scholar]
- [28].Zhang H, Irving L J, Tian Y. Influence of salinity and temperature on seed germination rate and the hydrotime model parameters for the halophyte, Chloris virgata and the glycophyte, Digitaria sanguinalis. South African Journal of Botany. 2012;78(1):203–210. [Google Scholar]
- [29].Qu XX, Huang Z Y, Baskin J M. Effect of temperature, light and salinity on seed germination and radicle growth of the geographically widespread halophyte shrub Halocnemum strobilaceum. Annals of Botany. 2007;101(2):293–299. doi: 10.1093/aob/mcm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yuan F, Liang X, Li Y. Methyl Jasmonate Improves Salinity Tolerance in Limonium bicolor by Enhancing Photosynthesis and Abaxial Salt Gland Density. Functional Plant Biology. 2018. [DOI] [PubMed]
- [31].Balnokin Y V, Kurkova EB, Myasoedov N A. Structural and Functional State of Thylakoids in a Halophyte Suaeda altissima before and after Disturbance of Salt–Water Balance by Extremely High Concentrations of NaCl. Russian Journal of Plant Physiology. 2004;51(6):815–821. [Google Scholar]
- [32].Kaneda Y, Tabei Y, Nishimura S. Combination of thidiazuron and basal media with low salt concentrations increases the frequency of shoot organogenesis in soybeans [Glycine max (L.) Merr.] Plant Cell Reports. 1997;17(1):8–12. doi: 10.1007/s002990050342. [DOI] [PubMed] [Google Scholar]
- [33].Kawasaki S, Borchert C, Deyholos M. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell. 2001;13(4):889. doi: 10.1105/tpc.13.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Farooq M, Hussain M, Wakeel A. Salt stress in maize: effects, resistance mechanisms, and management. A review. Agronomy for Sustainable Development. 2015;35(2):461–481. [Google Scholar]
- [35].Golan-Goldhirsh A, Hankamer B, Lips S H. Hydroxyproline and proline content of cell walls of sunflower, peanut and cotton grown under salt stress. Plant Science. 1990;69(1):27–32. [Google Scholar]
- [36].Wang B. Ho T H D; Wu R. Expression of a Late Embryogenesis Abundant Protein Gene, HVA1, from Barley Confers Tolerance to Water Deficit and Salt Stress in Transgenic Rice. Plant Physiology. 1996;110(1):249. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Koch M A, German D A. Taxonomy and systematics are key to biological information: Arabidopsis Eutrema Thellungiella Noccaea and Schrenkiella (Brassicaceae) as examples. Frontiers in Plant Science. 2013;4(267):267. doi: 10.3389/fpls.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang X J, Shi DC, Wang X Y. Evolutionary Migration of the Disjunct Salt Cress Eutrema salsugineum (= Thellungiella salsuginea Brassicaceae) between Asia and North America. Plos One. 2015;10(5):e0124010. doi: 10.1371/journal.pone.0124010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Stepien P, Johnson G N. Contrasting Responses of Photosynthesis to Salt Stress in the Glycophyte Arabidopsis and the Halophyte Thellungiella: Role of the Plastid Terminal Oxidase as an Alternative Electron Sink. Plant Physiology. 2009;149(2):1154. doi: 10.1104/pp.108.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gong Q, Li P, Ma S. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant Journal. 2005;44(5):826–839. doi: 10.1111/j.1365-313X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- [41].Sui N, Han G L. Salt-induced photoinhibition of PSII is alleviated in halophyte Thellungiella halophila by increases of unsaturated fatty acids in membrane lipids. Acta Physiologiae Plantarum. 2014;36(4):983–992. [Google Scholar]
- [42].Guo Y H, Jia W J, Song J. Thellungilla halophila is more adaptive to salinity than Arabidopsis thaliana at stages of seed germination and seedling establishment. Acta Physiologiae Plantarum. 2012;34(4):1287–1294. [Google Scholar]
- [43].Guo Y H, Wang D, Jia W J. Effects of seed vernalisation and photoperiod on flowering induction in the halophyte Thellungiella halophila. Australian Journal of Botany. 2012;60(8):743. [Google Scholar]
- [44].Flowers T J, Galal H K, Bromham L. Evolution of halophytes: multiple origins of salt tolerance in land plants. Functional Plant Biology. 2010;37(7):604–612. [Google Scholar]
- [45].Breckle S. How do halophytes overcome salinity. Biology of Salt Tolerant Plants. 1995;23:199–203. [Google Scholar]
- [46].Li X, Liu Y, Chen M. Relationships between ion and chlorophyll accumulation in seeds and adaptation to saline environments in Suaeda salsa populations. Plant Biosystems-An International Journal Dealing with all Aspects of Plant Biology. 2012;146(sup1):142–149. [Google Scholar]
- [47].Song J, Shi G, Gao B. Waterlogging and salinity effects on two Suaeda salsa populations. Physiologia Plantarum. 2011;141(4):343–351. doi: 10.1111/j.1399-3054.2011.01445.x. [DOI] [PubMed] [Google Scholar]
- [48].Song J, Chen M, Feng G. Effect of salinity on growth, ion accumulation and the roles of ions in osmotic adjustment of two populations of Suaeda salsa. Plant and Soil. 2009;314(1-2):133–141. [Google Scholar]
- [49].Chen M, Song J, Wang B S. NaCl increases the activity of the plasma membrane H+-ATPase in C3 halophyte Suaeda salsa callus. Acta Physiologiae Plantarum. 2010;32(1):27–36. [Google Scholar]
- [50].Qi C H, Chen M, Song J. Increase in aquaporin activity is involved in leaf succulence of the euhalophyte Suaeda salsa under salinity. Plant Science. 2009;176(2):200–205. [Google Scholar]
- [51].North GB, Nobel P S. Water uptake and structural plasticity along roots of a desert succulent during prolonged drought. Plant Cell & Environment. 1998;21(7):705–713. [Google Scholar]
- [52].Schröppelmeier G, Kaiser W M. Ion Homeostasis in Chloroplasts under Salinity and Mineral Deficiency: II. Solute Distribution between Chloroplasts and Extrachloroplastic Space under Excess or Deficiency of Sulfate, Phosphate, or Magnesium. Plant Physiology. 1988;87(4):828–832. doi: 10.1104/pp.87.4.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Fan H, Dong K H, Hou Y P. Effect of NaCl Stress on Ultrastructure of Halophytes Artemisia anethifolia. Acta Agrestia Sinica. 2011;19(3):482–486. [Google Scholar]
- [54].Shabala S, Bose J, Hedrich R. Salt bladders: do they matter? Trends in Plant Science. 2014;19(11):687–691. doi: 10.1016/j.tplants.2014.09.001. [DOI] [PubMed] [Google Scholar]
- [55].Yuan F, Chen M, Leng BY. An efficient autofluorescence method for screening Limonium bicolor mutants for abnormal salt gland density and salt secretion. South African Journal of Botany. 2013;88:110–117. [Google Scholar]
- [56].Yuan F, Lyu M J A, Leng BY. The transcriptome of NaCl-treated Limonium bicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Molecular Biology. 2016;91(3):241–256. doi: 10.1007/s11103-016-0460-0. [DOI] [PubMed] [Google Scholar]
- [57].Deng Y, Feng Z, Yuan F. Identification and Functional Analysis of the Autofluorescent Substance in Limonium bicolor Salt Glands. Plant Physiology and Biochemistry. 2015;97:20–27. doi: 10.1016/j.plaphy.2015.09.007. [DOI] [PubMed] [Google Scholar]
- [58].Feng Z, Deng Y, Zhang S. K+ accumulation in the cytoplasm and nucleus of the salt gland cells of Limonium bicolor accompanies increased rates of salt secretion under NaCl treatment using NanoSIMS. Plant Science. 2015;238:286–296. doi: 10.1016/j.plantsci.2015.06.021. [DOI] [PubMed] [Google Scholar]
- [59].Dassanayake M, Larkin J C. Making Plants Break a Sweat: the Structure, Function, and Evolution of Plant Salt Glands. Frontiers in Plant Science. 2017;8(715) doi: 10.3389/fpls.2017.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cachorro P, Olmos E, Ortiz A. Salinity-induced changes in the structure and ultrastructure of bean root cells. Biologia Plantarum. 1995;37(2):273–283. [Google Scholar]
- [61].Horie T, Hauser F, Schroeder J I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends in Plant Science. 2009;14(12):660–668. doi: 10.1016/j.tplants.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lan W Z, Wang W, Wang SM. A rice high-affinity potassium transporter (HKT) conceals a calcium-permeable cation channel. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(15):7089–7094. doi: 10.1073/pnas.1000698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Shi HZ, Ishitani M, Kim C. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proceedings of the national academy of sciences. 2000;97(12):6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zhu J K. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Tester M, Davenport R. Na+ Tolerance and Na+ Transport in Higher Plants. Annals of Botany. 2003;91(5):503. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhang H, Wang S. Advances in study of Na+ uptake and transport in higher plants and Na+ homeostasis in the cell. Chinese Bulletin of Botany. 2007;24(5):561–571. [Google Scholar]
- [67].Krishnamurthy P, Ranathunge K, Franke R. The role of root apoplastic transport barriers in salt tolerance of rice Oryza sativa L.) Planta. 2009;230(1):119. doi: 10.1007/s00425-009-0930-6. [DOI] [PubMed] [Google Scholar]
- [68].Mucha S, Walther D, Müller TM. Substantial reprogramming of the Eutrema salsugineum Thellungiella salsuginea transcriptome in response to UV and silver nitrate challenge. BMC Plant Biology,15,1(2015-06-12) 2015;15(1):137. doi: 10.1186/s12870-015-0506-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Yang P L, Funk C, Erban A. Salt stress responses in a geographically diverse collection of Eutrema/Thellungiella spp. accessions. Functional Plant Biology. 2016;43(7) doi: 10.1071/FP15285. [DOI] [PubMed] [Google Scholar]
- [70].Kant S, Kant P, Raveh E. Evidence that differential gene expression between the halophyte, Thellungiella halophila and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila Plant Cell & Environment. 2010;29(7):1220–1234. doi: 10.1111/j.1365-3040.2006.01502.x. [DOI] [PubMed] [Google Scholar]
- [71].Alemán F, Nieves-Cordones M, Martínez V. Differential regulation of the HAK5 genes encoding the high-affinity K+ transporters of Thellungiella halophila and Arabidopsis thaliana. Environmental & Experimental Botany. 2009;65(2):263–269. [Google Scholar]
- [72].Ulanov A, Bressan R A, Bohnert H J. Sodium Stress in the Halophyte Thellungiella halophila and Transcriptional Changes in a thsos1-RNA Interference Line. Journal of Integrative Plant Biology. 2007;49(10):1484–1496. [Google Scholar]
- [73].Wu C, Gao X, Kong X. Molecular Cloning and Functional Analysis of a Na+/H+ Antiporter Gene ThNHX1 from a Halophytic Plant Thellungiella halophila. Plant Molecular Biology Reporter. 2009;27(1):1. [Google Scholar]
- [74].Ghars M A, Parre E, Debez A. Comparative salt tolerance analysis between Arabidopsis thaliana and Thellungiella halophila , with special emphasis on K+ /Na+ selectivity and proline accumulation. Journal of Plant Physiology. 2008;165(6):588–599. doi: 10.1016/j.jplph.2007.05.014. [DOI] [PubMed] [Google Scholar]
- [75].Han N, Shao Q, Bao HY. Cloning and Characterization of a Ca2+/H+ Antiporter from Halophyte Suaeda salsa L. Plant Molecular Biology Reporter. 2011;29(2):449–457. [Google Scholar]
- [76].Chang W, Liu X, Zhu J. An aquaporin gene from halophyte Sesuvium portulacastrum SpAQP1, increases salt tolerance in transgenic tobacco. Plant Cell Reports. 2016;35(2):385–395. doi: 10.1007/s00299-015-1891-9. [DOI] [PubMed] [Google Scholar]
- [77].Chen Y, Li L, Zong J. Heterologous expression of the halophyte Zoysia matrella H⁺-pyrophosphatase gene improved salt tolerance in Arabidopsis thaliana. Plant Physiology & Biochemistry Ppb. 2015;91:49. doi: 10.1016/j.plaphy.2015.04.004. [DOI] [PubMed] [Google Scholar]
- [78].Han G, Wang M, Yuan F. The CCCH zinc finger protein gene AtZFP1 improves salt resistance in Arabidopsis thaliana. Plant Molecular Biology. 2014;86(3):237–253. doi: 10.1007/s11103-014-0226-5. [DOI] [PubMed] [Google Scholar]
- [79].Zhu J K. Regulation of ion homeostasis under salt stress. Current Opinion in Plant Biology. 2003;6(5):441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- [80].Kong X-Q, Gao X-H, Sun W. Cloning and functional characterization of a cation–chloride cotransporter gene OsCCC1. Plant Molecular Biology. 2011;75(6):567–578. doi: 10.1007/s11103-011-9744-6. [DOI] [PubMed] [Google Scholar]
- [81].Liu L, Zeng Y, Pan X. Isolation, molecular characterization, and functional analysis of the vacuolar Na+/H+ antiporter genes from the halophyte Karelinia caspica. Molecular Biology Reports. 2012;39(6):7193–7202. doi: 10.1007/s11033-012-1551-x. [DOI] [PubMed] [Google Scholar]
- [82].Himabindu Y, Chakradhar T, Reddy MC. Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environmental & Experimental Botany. 2016;124:39–63. [Google Scholar]
- [83].Han N, Lan W, He X. Expression of a Suaeda salsa vacuolar H+/Ca2+ transporter gene in Arabidopsis contributes to physiological changes in salinity. Plant Molecular Biology Reporter. 2012;30(2):470–477. [Google Scholar]
- [84].Li K, Pang C, Ding F. Overexpression of Suaeda salsa stroma ascorbate peroxidase in Arabidopsis chloroplasts enhances salt tolerance of plants. South African Journal of Botany. 2012;78:235–245. [Google Scholar]
- [85].Meng X, Zhou J, Sui N. Mechanisms of salt tolerance in halophytes: current understanding and recent advances. Open Life Sciences. 2018;13(1):149–154. doi: 10.1515/biol-2018-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Pang C H, Li K, Wang B S. Overexpression of SsCHLAPXs confers protection against oxidative stress induced by high light in transgenic Arabidopsis thaliana. Physiologia Plantarum. 2011;143(4):355–366. doi: 10.1111/j.1399-3054.2011.01515.x. [DOI] [PubMed] [Google Scholar]
- [87].Sui N, Tian S, Wang W. Overexpression of glycerol-3-phosphate acyltransferase from Suaeda salsa improves salt tolerance in Arabidopsis. Frontiers in Plant Science. 2017;8:1337. doi: 10.3389/fpls.2017.01337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zhang J X, Wang C, Yang CY. The role of Arabidopsis AtFes1A in cytosolic Hsp70 stability and abiotic stress tolerance. The Plant Journal. 2010;62(4):539–548. doi: 10.1111/j.1365-313X.2010.04173.x. [DOI] [PubMed] [Google Scholar]
- [89].Jha B, Lal S, Tiwari V. The SbASR -1 Gene Cloned from an Extreme Halophyte Salicornia brachiata Enhances Salt Tolerance in Transgenic Tobacco. Marine Biotechnology. 2012;14(6):782–792. doi: 10.1007/s10126-012-9442-7. [DOI] [PubMed] [Google Scholar]
- [90].Li W, Wang D, Jin T. The Vacuolar Na+/H+ Antiporter Gene SsNHX1 from the Halophyte Salsola soda Confers Salt Tolerance in Transgenic Alfalfa Medicago sativa L.) Plant Molecular Biology Reporter. 2011;29(2):278–290. [Google Scholar]
- [91].Zhang G H, Su Q, An L J. Characterization and expression of a vacuolar Na+/H+ antiporter gene from the monocot halophyte Aeluropus littoralis. Plant Physiology & Biochemistry. 2008;46(2):117–126. doi: 10.1016/j.plaphy.2007.10.022. [DOI] [PubMed] [Google Scholar]
- [92].Shao Q, Han N, Ding T. SsHKT1;1 is a potassium transporter of a C3 halophyte Suaeda salsa involving in salt tolerance. Functional Plant Biology. 2014;41:790–802. doi: 10.1071/FP13265. [DOI] [PubMed] [Google Scholar]
- [93].Yadav NS, Shukla PS, Jha A. The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na+ loading in xylem and confers salt tolerance in transgenic tobacco. Bmc Plant Biology. 2012;12(1):188. doi: 10.1186/1471-2229-12-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Guo S, Yin H, Zhang X. Molecular cloning and characterization of a vacuolar H+-pyrophosphatase gene, SsVP, from the halophyte Suaeda salsa and its overexpression increases salt and drought tolerance of Arabidopsis. Plant Molecular Biology. 2006;60(1):41–50. doi: 10.1007/s11103-005-2417-6. [DOI] [PubMed] [Google Scholar]
- [95].Yao M, Zeng Y, Liu L. Overexpression of the halophyte Kalidium foliatum H+-pyrophosphatase gene confers salt and drought tolerance in Arabidopsis thaliana. Molecular Biology Reports. 2012;39(8):7989–7996. doi: 10.1007/s11033-012-1645-5. [DOI] [PubMed] [Google Scholar]
- [96].Baisakh N, RamanaRao M, Rajasekaran K. Enhanced salt stress tolerance of rice plants expressing a vacuolar H+ -ATPase subunit c1 SaVHAc1 gene from the halophyte grass Spartina alterniflora Löisel. Plant Biotechnol J. 2012;10(4):453–464. doi: 10.1111/j.1467-7652.2012.00678.x. [DOI] [PubMed] [Google Scholar]
- [97].Singh N, Mishra A, Jha B. Over-expression of the Peroxisomal Ascorbate Peroxidase ( SbpAPX ) Gene Cloned from Halophyte Salicornia brachiata Confers Salt and Drought Stress Tolerance in Transgenic Tobacco. Marine Biotechnology. 2014;16(3):321–332. doi: 10.1007/s10126-013-9548-6. [DOI] [PubMed] [Google Scholar]
- [98].Flowers T, Troke P, Yeo A. The mechanism of salt tolerance in halophytes. Annual review of plant physiology. 1977;28(1):89–121. [Google Scholar]
- [99].Flowers T J, Munns R, Colmer T D. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Annals of Botany. 2014;115(3) doi: 10.1093/aob/mcu217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ozfidankonakci C, Uzilday B, Ozgur R. Halophytes as a source of salt tolerance genes and mechanisms: a case study for the Salt Lake area, Turkey. Functional Plant Biology. 2016;43(7) doi: 10.1071/FP15288. [DOI] [PubMed] [Google Scholar]
- [101].Wang F X, Yin C H, Song Y P. Reproductive allocation and fruit-set pattern in the euhalophyte Suaeda salsa in controlled and field conditions. Plant Biosystems. 2017:1–10. [Google Scholar]
- [102].Barkla B J, Vera-Estrella R. Single cell-type comparative metabolomics of epidermal bladder cells from the halophyte Mesembryanthemum crystallinum. Frontiers in Plant Science. 2015;6:435. doi: 10.3389/fpls.2015.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Tsukagoshi H, Suzuki T, Nishikawa K. RNA-seq analysis of the response of the halophyte, Mesembryanthemum crystallinum (ice plant) to high salinity. Plos One. 2015;10(2) doi: 10.1371/journal.pone.0118339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Dang Z H, Zheng LL, Wang J. Transcriptomic profiling of the salt-stress response in the wild recretohalophyte Reaumuria trigyna. Bmc Genomics. 2013;14(1):29. doi: 10.1186/1471-2164-14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Sun Y L, Hong S K. Agrobacterium tumefaciens -Mediated Transformation of the Halophyte Leymus chinensis (Trin.) Plant Molecular Biology Reporter. 2012;30(5):1253–1263. [Google Scholar]
- [106].Yuan F, Chen M, Yang J C. A system for the transformation and regeneration of the recretohalophyte Limonium bicolor. In Vitro Cellular & Developmental Biology-Plant. 2014;50:610–617. [Google Scholar]
- [107].Duan YB, Li J, Qin RY. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Molecular Biology. 2016;90(1-2):49–62. doi: 10.1007/s11103-015-0393-z. [DOI] [PubMed] [Google Scholar]
- [108].Huanca-Mamani W, Arias-Carrasco R, Cã r-N S. Long Non-Coding RNAs Responsive to Salt and Boron Stress in the Hyper-Arid Lluteño Maize from Atacama Desert. Genes. 2018;9(3):170. doi: 10.3390/genes9030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Qin T, Zhao H, Cui P. A Nucleus-localized Long Non-Coding RNA Enhances Drought and Salt Stress Tolerance. Plant Physiology. 2017;175(3):1321. doi: 10.1104/pp.17.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Song Y, Zhang D. The Role of Long Noncoding RNAs in Plant Stress Tolerance. Methods Mol Biol. 2017;1631:41–68. doi: 10.1007/978-1-4939-7136-7_3. [DOI] [PubMed] [Google Scholar]
- [111].Katschnig D, Bliek T, Rozema J. Constitutive high-level SOS1 expression and absence of HKT1;1 expression in the salt-accumulating halophyte Salicornia dolichostachya. Plant Science. 2015;234:144–154. doi: 10.1016/j.plantsci.2015.02.011. [DOI] [PubMed] [Google Scholar]
- [112].Rauf M, Shahzad K, Ali R. Cloning and characterization of Na+/H+ antiporter (LfNHX1) gene from a halophyte grass Leptochloa fusca for drought and salt tolerance. Molecular Biology Reports. 2014;41(3):1669–1682. doi: 10.1007/s11033-013-3015-3. [DOI] [PubMed] [Google Scholar]
- [113].Sanadhya P, Agarwal P, Khedia J. A Low-Affinity K+ Transporter AlHKT2;1 from Recretohalophyte Aeluropus lagopoides Confers Salt Tolerance in Yeast. Molecular Biotechnology. 2015;57(6):489–498. doi: 10.1007/s12033-015-9842-9. [DOI] [PubMed] [Google Scholar]
- [114].Wang SM, Zhang J L, Flowers T J. Low-Affinity Na⁺ Uptake in the Halophyte Suaeda maritima. Plant Physiology. 2007;145(2):559–571. doi: 10.1104/pp.107.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Zhang J L, Flowers T J, Wang S M. Differentiation of low-affinity Na+ uptake pathways and kinetics of the effects of K+ on Na+ uptake in the halophyte Suaeda maritima. Plant & Soil. 2013;368(1-2):629–640. [Google Scholar]
- [116].Zhou LL, Miao J K, Zhu J B. Cloning and sequence analysis of a Na+/H+ antiporter gene in the halophyte Limonium gmelinii. Acta Prataculturae Sinica. 2009 [Google Scholar]
- [117].Jha A, Joshi M, Yadav NS. Cloning and characterization of the Salicornia brachiata Na+/H+ antiporter gene SbNHX1 and its expression by abiotic stress. Molecular Biology Reports. 2011;38(3):1965–1973. doi: 10.1007/s11033-010-0318-5. [DOI] [PubMed] [Google Scholar]
- [118].Khan M S. Role of sodium and hydrogen Na+/H+ antiporters in salt tolerance of plants: Present and future challenges. African Journal of Biotechnology. 2011;10(63):13693–13704. [Google Scholar]
- [119].Wissing F. Vacuolar chloride transport in the extreme halophyte Mesembryanthemum crystallinum. 1999. of University of Oxford. [DOI] [PubMed]
- [120].Chen J, Xiao Q, Wu F. Nitric oxide enhances salt secretion and Na+ sequestration in a mangrove plant, Avicennia marina through increasing the expression of H+-ATPase and Na+/H+ antiporter under high salinity. Tree Physiology. 2010;30(12):1570–1585. doi: 10.1093/treephys/tpq086. [DOI] [PubMed] [Google Scholar]
- [121].Tan WK, Lin Q, Lim TM. Dynamic secretion changes in the salt glands of the mangrove tree species Avicennia officinalis in response to a changing saline environment. Plant, Cell & Environment. 2013;36:1410–1422. doi: 10.1111/pce.12068. [DOI] [PubMed] [Google Scholar]


