Abstract
The present study was performed to investigate the effects of dietary supplementation with Lactobacillus plantarum (CGMCC1.557) on egg production and fecal microbiota composition in laying hens. Sixty Hy-Line Brown laying hens (18 weeks old) were randomly divided into two groups. The control group was fed a basal diet only, and the test group was fed basal diet supplemented with a final concentration of 1.0 × 109 CFU/mL during the 10-week experimental period. Egg production and fecal microbiota composition were both assessed in 28-week-old hens using high-throughput sequencing technology. The results showed that, compared with the control group, the test group exhibited increased laying and feed intake rates (p < 0.05). At the genus level, Lactobacillus was more abundant in the test group compared with the control group (p < 0.05). Conversely, Romboutsia was more abundant in the control group compared with the test group (p < 0.05). This study provides us with an insight into the potential use of L. plantarum as a food supplement in the laying hen industry. the study also provides us with a better understanding of the interplay between L. plantarum and the fecal microbiota of laying hens.
Keywords: additive, fecal microbiota, Lactobacillus plantarum, laying hens, performance
1. Introduction
During the past few decades, antibiotics have been widely used at subtherapeutic doses to improve growth rates and performance in the poultry industry. However, antibiotics can result in side effects including bacterial resistance; these side effects often have detrimental consequences for human health [1]. In response to this apparent threat, the European Commission (EC) decided to ban the use of antibiotics as growth promoters in livestock feed (EC Regulation NO. 1831/2003) [2]. Consequently, the poultry sector needs to address the consequences of this ban by seeking alternative strategies to promote animal performance [3]. One environmentally friendly approach to growth promotion involves the use of probiotics, which have been used in the poultry industry for decades [4, 5].
Probiotics are defined as living microorganisms that are beneficial to the host [6]. Probiotics have been shown to improve poultry performance, modulate intestinal microbiota and reduce disease risk in the poultry industry [7, 8]. Lactic acid bacteria (LAB) are the most widely used bacterial order in relation to probiotic administration. LAB include the genera Lactobacillus, Lactococcus, Enterococcus, and Streptococcus [9]. Substantial evidence indicates that LAB are beneficial to both humans and animals [10, 11]. These beneficial effects are underpinned by the capacity of LAB to prevent colonization by pathogenic organisms, enhance the natural flora, and optimize nutrient absorption from feed [12]. However, LAB species elicit differential effects due to variations pertaining to dose, administration route, age of the animals, the appetites of the animals, the quality of farm management, farm hygiene and biosecurity [13, 14].
Lactobacillus plantarum is generally regarded as a safe species and has been used for a long time in both animal and human food [15, 16]. Over recent decades, attention has been focused on the deployment of L. plantarum in commercial poultry activities. L. plantarum P-8 has the potential to improve metabolic activity and nutrient utilization, while also modulating the intestinal microbiota of broiler chickens [17]. L. plantarum BS22 has been shown to promote immune function, production performance and intestinal microbiota homeostasis in broilers [18]. Moreover, metabolite combinations generated from L. plantarum RI11, RG14, and RG11 strains (COM456) have been shown to improve egg production, fecal LAB population abundance, and small intestinal villus height, while also reducing fecal pH, Enterobacteriaceae abundance, and plasma and yolk cholesterol concentrations [19, 20].
In recent years, next-generation sequencing technology has been used to determine the composition, establishment and function of gut microbiota in chickens [21, 22]. However, only a limited number of studies pertaining to the effects of L. plantarum on the microbiota of laying hens have been published. In the current study, we hypothesized that supplementing feed with L. plantarum would improve production performance, while also modulating the host fecal microbiome of laying hens. The study also provides us with some interesting insights into the use of L. plantarum in laying hens.
2. Materials and methods
2.1. Experimental animals
Hy-Line Brown layers obtained from Chunya Poultry Company Limited (Xuchang, China) were allocated into cages. A total of 60 Hy-Line layers (aged 18 weeks; mean body weight 1.50 kg) were randomly assigned into 2 dietary treatment groups. Each dietary group was composed of 5 identical sub-groups containing 6 birds each. The 2 dietary groups were fed either the control diet (control group) or an L. plantarum (CGMCC 1.557)-supplemented diet (test group). The strain of L. plantarum used in this current study was isolated from silage and deposited in the China General Microbiological Culture Collection Center (CGMCC, Beijing, China). This strain was characterized by acid and bile tolerance, adhesion capacity, antibacterial activity and immunomodulatory activity [23-24]. The L. plantarum strains were cultured using de Man, Rogosa, and Sharpe (MRS) medium at 37°C for 18 h under anaerobic conditions. The cultured L. plantarum cells were subsequently centrifuged at 2000 g for 15 min at 4°C and washed three times with 0.5% NaCl solution. The L. plantarum was resuspended to 1.0 × 109 colony-forming units (CFU)/mL using 0.5% NaCl solution. Finally, the L. plantarum re-suspension was mixed at a level of 1 g/kg (0.1%, m/m) each day during the 10-week experimental period to ensure that the bacterial cells in the feed were viable. This level of supplementation has been shown to have positive effects on production performance, excreta microflora, ammonia emission and nutrient utilization [25]. The hens were provided with feed and water ad libitum during the experimental period. No antibiotics were used throughout the trial period. The temperature was maintained at 25–27°C, with 16 h of light per day. The basal diet (Table 1) used in the present study was formulated according to the NRC (1994) recommendations, which have been shown to be suitable for laying hens. All animal experiments were conducted according to the Guidelines for the Care and Use of Experimental Animals established and approved by the Laboratory Animal Management Committee of Henan University of Animal Husbandry and Economy (HNMY1618).
Table 1.
Composition of the basic diet
| Ingredient | Amount (percentage of the dried weight) |
|---|---|
| Corn | 60.51 |
| Soybean meal | 24.00 |
| Wheat | 2.80 |
| Crude palm oil | 1.25 |
| L-lysine | 0.06 |
| DL-methionine | 0.15 |
| MDCP | 2.40 |
| Limestone | 7.30 |
| Common salt | 0.50 |
| Vitamin premix† | 0.07 |
| Mineral premix‡ | 0.06 |
| Choline chloride | 0.90 |
| Total Calculated values | 100 |
| ME (cal/kg) | 2805.32 |
| Crude protein,% | 16.00 |
| Calcium,% | 3.70 |
| Aval. Phophprus,% | 0.45 |
Vitamin premix provided (per kilogram of diet): retinol 3.00 mg; cholecalciferol 0.06 mg; ɑ-tocopherol 15.00 mg; thiamine 1.20 mg; riboflavin 4.00 mg; pantothenic acid 8.00 mg; pyridoxine 2.00 mg; niacin 30.00 mg; cobalamin 0.02 mg; folic acid 0.50 mg; biotin 0.03 mg; menadione 3.00 mg.
‡Mineral premix provided (per kilogram of diet): manganese 100.0 mg; copper 8.0 mg; iodine 0.8 mg; cobalt 0.25 mg; selenium 0.3 mg; zinc 80.0 mg; iron 40.0 mg.
Ethical approval
The research related to animals use has been compl ied with all the relevant national regulations and institutional policies for the care and use of animal.
2.2. Egg production performance
Egg production, egg weight and the number of cracked eggs were recorded daily. Feed consumption was recorded weekly per replicate sub-group. Laying and feed (feed conversion ratio; FCR) efficiencies were also calculated.
2.3. Fecal collection and DNA extraction
Thirty fresh fecal samples (15 each from the control group and test group, 3 samples per sub-group) were randomly collected at 28 wk. The fecal samples were stored immediately at −20°C prior to DNA extraction. Two hundred milligrams of feces from each bird were utilized for DNA isolation using a DNA isolation kit (Tiangen Biotech Corporation, Beijing, China) following the manufacturer’s instructions. The quality of the extracted DNA was assessed by 0.8% agarose gel electrophoresis and spectrophotometry (optical density at 260/280 nm). All extracted DNA samples were stored at −20°C prior to further analysis.
2.4. Library preparation and Illumina MiSeq sequencing
NGS library preparations and Illumina MiSeq sequencing were conducted at GENEWIZ, Inc. (Suzhou, China). For the library preparation, the V3 and V4 regions of the 16S rRNA gene were amplified using a 10-ng DNA aliquot isolated from each fecal sample. The V3 and V4 regions were amplified by polymerase chain reaction (PCR) using a forward primer (5′–CCTACGGRRBGCASCAGKVRVGAAT–3′) and a reverse primer (5′–GGACTACNYVGGGTWTCTAATCC–3′). The first-round PCR products were used as templates for a second round of amplicon enrichment by PCR (94°C for 3 min, followed by 24 cycles at 94°C for 5 s, 57°C for 90 s, 72°C for 10 s and a final extension at 72°C for 5 min). At the same time, indexed adapters were added to the ends of the 16S rDNA amplicons to generate indexed libraries that were ready for downstream NGS on the MiSeq platform (Illumina, San Diego, CA, USA). DNA libraries were validated by Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and quantified using a Qubit 2.0 Fluorometer. Next, the DNA libraries were multiplexed and loaded on an Illumina MiSeq instrument according to the manufacturer’s instructions (Illunmina, San Diego, CA, USA). Sequencing was performed using a 2 × 300/250 paired-end (PE) configuration; image analysis and base calling were performed using MiSeq Control Software (MCS) embedded in the MiSeq instrument. The sequences generated in this study have been deposited in the National Center for Biotechnology Information sequence read archive (https://www.ncbi.nlm.nih.gov/biosample) under the accession number SRA: SRS2480324.
2.5. Statistical analysis
The QIIME data analysis package was used for the 16S rRNA data analysis. The forward and reverse reads were joined, assigned to samples based on barcodes and truncated by cutting off the barcode and primer sequences. Quality filtering of the joined sequences was performed, and sequences that did not fulfill the following criteria were discarded: sequence length < 200 bp, no ambiguous bases, and a mean quality score ≥ 20. Next, the sequences were compared with sequences from a reference database (the Ribosomal Database Project [RDP] Gold database) using the UCHIME algorithm to detect chimeric sequences; the chimeric sequences were subsequently removed.
The quality filtered-sequences were utilized in the final analysis. Sequences were grouped into Operational Taxonomic Units (OTUs) using the clustering program VSEARCH (1.9.6) against the SILVA 119 database that was pre-clustered at a 97% sequence identity. The RDP classifier was used to assign a taxonomic category to all OTUs at a confidence threshold of 0.8. The RDP classifier uses the SILVA 119 database, which predicts taxonomic categories at the species level. Sequences were rarefied prior to calculating alpha and beta diversity indices. Alpha diversity indices were calculated in QIIME from rarefied samples, using the Shannon index for diversity and the Chao1 index for richness. Beta diversity was calculated using a PCoA analysis. A heatmap was clustered using R (2.15.3) analysis. Differences between the 2 groups were compared using STAMP (2.1.3) analysis. All results were analyzed by AVOVA using the general linear models (GLM) procedure (SAS Inst. Inc., Cary, NC) and expressed as mean ± standard errors. p < 0.05 was considered statistically significant.
3. Results
3.1. Egg production performance
The egg production performance results are presented in Table 2. There were significant differences in the laying rate and feed intake between control and test groups (p < 0.05). However, egg weight, feed conversion ratios and the number of cracked eggs between the groups were not different between groups (p > 0.05).
Table 2.
Effect of L. plantarum on the performance of laying hens at 28 weeks of age*
| Performance variable | Test group | Control group | SEM | P-value |
|---|---|---|---|---|
| Laying rate (%) | 93.51a | 90.97b | 0.31 | 0.04 |
| Egg weight (g) | 55.27 | 55.12 | 0.89 | 0.91 |
| Egg mass(g/hen/d) | 48.89 | 48.69 | 0.72 | 0.86 |
| Feed intake (g/hen/d) | 108.27a | 106.30b | 1.54 | 0.03 |
| Feed conversion ratio (g/g egg) | 2.09 | 2.12 | 0.37 | 0.59 |
| Cracked-egg rate (%) | 0.09 | 0.12 | 0.02 | 0.54 |
Means in the same row not sharing a common superscript differ significantly at P < 0.05.
SEM = Standard error of means.
3.2. Data acquisition and analysis of the groups
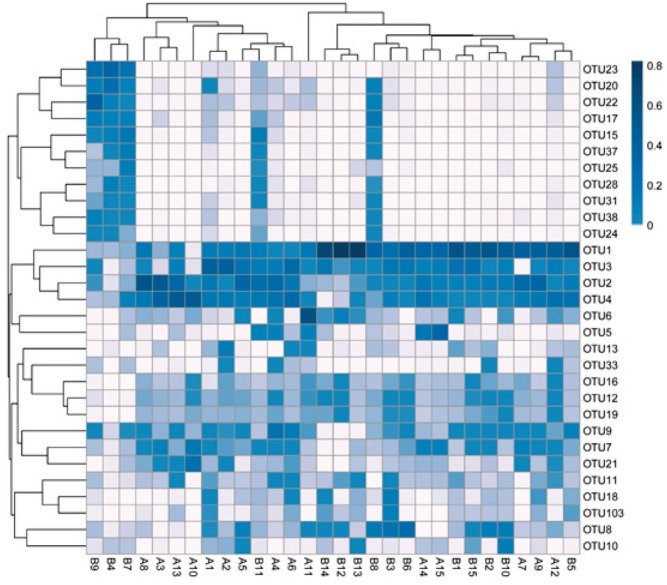
In this study, a 16S rRNA gene sequence analysis of 30 fecal samples generated 3,505,668 high-quality sequences, with an average of 116,855 sequences per sample. The sequences were clustered into 205 OTUs using a 97% similarity cut-off. A clustering analysis of 30 OTUs with the highest abundances revealed both similarities and differences between the samples (Fig. 1). For example, similar abundances of OTU1, OTU2, OTU3 and OTU4 were
Figure 1.
Cluster analysis of the 30 most abundant OTUs in the two groups. A: test group; B: control group.
observed across nearly 30 samples, and these OTUs belong to one cluster. There were some similarities in the OTUs (OTU31, OTU28, OTU25, OTU37, OTU15) among samples B8, B4, B7, B9 and B11, while other samples showed similarities in abundances as well. Other OTUs had some dissimilarity among the rest of the samples. Using an abundance-based coverage estimator (ACE) along with Chao1, Simpson and Shannon indices, we observed the species richness between the two groups (Table 3). The Shannon index for the control group was significantly higher (p < 0.05) than for the test group, while there were no significant differences in the ACE, Chao1 index and Simpson index between the two groups. These results indicate that species diversity was more abundant in the test group compared with the control group.
Table 3.
Diversity estimations for the 16S rRNA gene libraries of the 30 samples from the 16S rRNA sequences
| Group | Chao1 | ACE | Simpson | Shannon |
|---|---|---|---|---|
| Control (n = 15) | 113.66 ± 33.75 | 117.01 ± 34.74 | 0.72 ± 0.08 | 2.47 ± 0.41a |
| Test (n = 15) | 119.37 ± 38.13 | 121.91 ± 36.03 | 0.72 ± 0.20 | 3.10 ± 1.43b |
Different superscript letters in the same column indicate significant differences.
3.3. Microbial beta diversity analysis
In a beta-diversity analysis, variations in the composition of the microbial communities for the 30 samples were presented using principal coordinates analysis (PCoA) plots, with PC1 accounting for 28.28% of the total variation and PC2 accounting for 25.55% (Fig. 2). Thus, there were 2 main clusters between the test group and the control group; however, 3 laying hens in the control group were classified as outliers. This result suggested that the microbial population structures between the two groups were slightly different.
Figure 2.
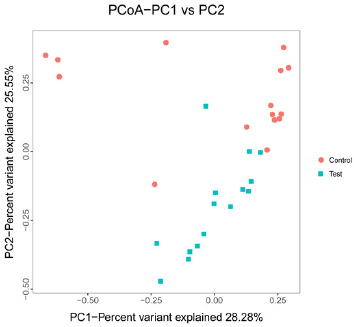
PCoA plot. Three clusters were observed for the 30 samples from the 2 groups.
3.4. Bacterial community composition
At the phylum level, a total of 6 main phyla were identified in the 2 groups as shown in Fig. 3. Most of the sequences in the test group belonged to Firmicutes (92.24%), Bacteroidetes (0.43%), and Proteobacteria (6.69%), while Firmicutes (74.11%), Bacteroidetes (19.04%), and Proteobacteria (5.44%) were also abundant in the control group. The phylum-level microbiota analysis demonstrated that Firmicutes were the predominant bacteria in the fecal samples from laying hens. There was a significant difference between the 2 groups at the phylum level, mainly in the Bacteroidetes (p < 0.05).
Figure 3.
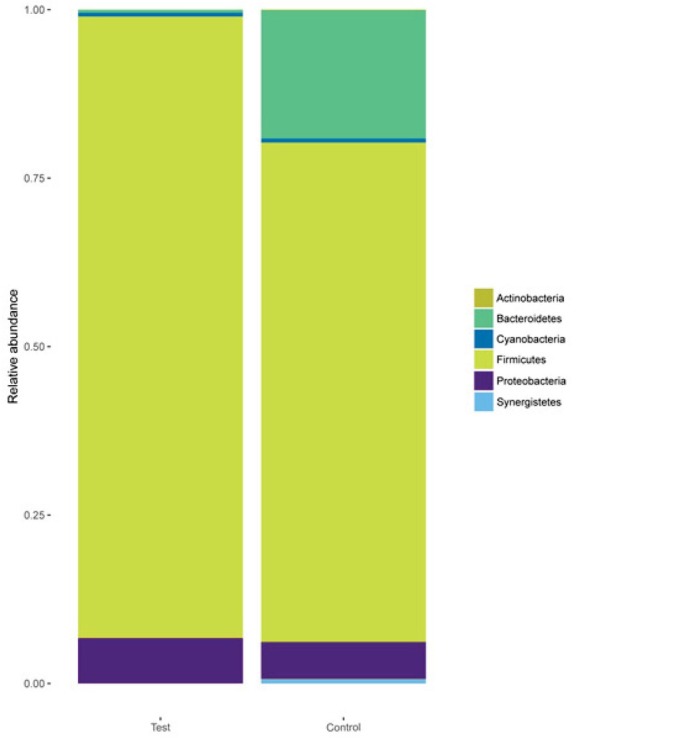
Phylum-level analyses of the 30 samples. Comparison of the relative abundances of the main bacterial phyla in the 2 groups.
As shown in Fig. 4, at the genus level, an analysis of the most abundant taxa revealed that Lactobacillus (58.55%), Romboutsia (18.29%), and Enterococcus (12.09%) were abundant in the test group, while Lactobacillus (13.39%), Romboutsia (39.02%), and Enterococcus (8.26%) were also abundant in the control group. The differences in the abundances of Romboutsia and Lactobacillus between the two groups were significant (p < 0.05). In addition, the test group contained higher Escherichia-Shigella, while in the control group, Turicibater and Unclassed were higher.
Figure 4.
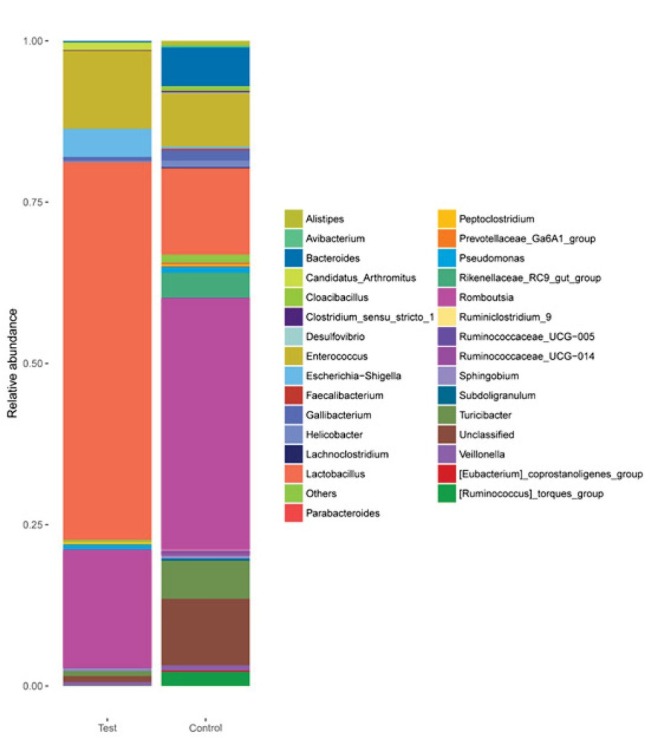
Genus-level analyses of the 30 samples. Comparison of the relative abundances of the main bacterial genera in the 2 groups.
3.5. Community composition heat-map combined with a cluster analysis
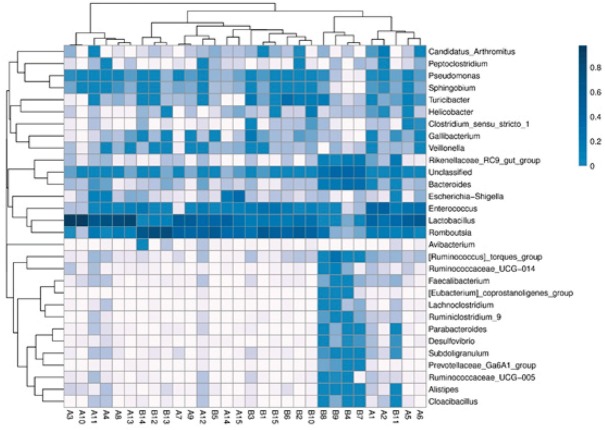
As shown in Fig. 5, the top 30 genera in terms of abundance were clustered and plotted using R software. The heat-map reveals that the abundances of Lactobacillus, Romboutsia, and Enterococcus were the most similar among the 30 samples. However, samples B4, B7, B8, B9 and B11 displayed the greatest diversity in relation to genus composition when compared with the other samples analyzed.
Figure 5.
Heat-map analysis of the 30 samples. A: test group; B: control group Heat-map showing the abundances of the top 30 genera was clustered and plotted using R software. Blue represents genera with higher abundances in the corresponding sample, and white represents genera with lower abundances.
Except for the three samples B4, B7 and B9, the abundance of Pseudomonas, Sphingobium and Turicibacter genera was relatively high. Compared to other samples, Escherichia-Shigella was higher in abundance among five samples of B11, A15, A14, A4 and A11.
3.6. STAMP analysis
At the genus level, we compared the differences between the 2 groups using STAMP software and Welch’s t-test. As shown in Fig. 6, the abundance of Lactobacillus was significantly (p < 0.05) higher in the test group compared with the control group. However, the abundance of Romboutsia was significantly (p < 0.05) higher in the control group compared with the test group.
Figure 6.
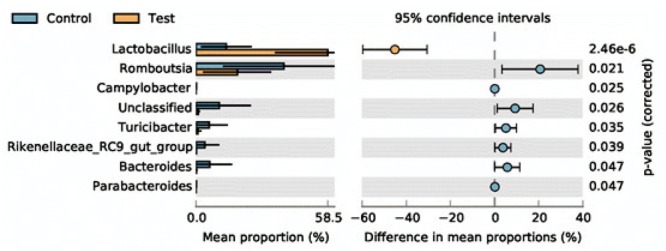
STAMP analysis of the test and control groups. Lactobacillus was more abundant in the test group compared with the control group.
4. Discussion
Many studies have investigated the effects of dietary supplementation with L. plantarum on the performance of laying hens; however, only a limited amount of research
has been performed using 16S rRNA sequencing technology to investigate fecal microbiota composition in laying hens. In this study, we observed that the fecal microbiota composition of laying hens fed a diet supplemented with L. plantarum was different from the control group. The results of this study support our hypothesis that feed with L. plantarum would improve production performance, while also modulating the host fecal microbiome of laying hens.
In this study, we observed that diets supplemented with L. plantarum improved feed intake and laying rate. Previous studies reported that supplementation with Lactobacillus improved feed efficiency in hens [26]. Additional studies reported that supplementation with octacosanol or E. faecium also improved egg production [27, 25], while diets supplemented with Lactobacillus johnsonii BS15 promoted growth performance in broilers [28]. In contrast, Mahdavi et al. reported that egg production was not affected by the inclusion of LAB in the diet [29]. The associated differences resulting from supplementation with probiotics were dependent upon a number of factors including the different bacterial strains present, the ages of the animals, the effectiveness of associated farm-management practices, the method of administration used, the appetites of the animals, and the hygiene conditions of the farm [30].
At the phylum level, Firmicutes, Bacteroidetes, and Proteobacteria were the most common phyla in the fecal samples from the laying hens (Fig. 3). In the control group, the microbiota of laying hens was dominated by Firmicutes
(>50%); This result is consistent with previously published findings [31,22]. However, our results differ from results published following a study on broiler chickens published by Singh [32], where it was shown that Proteobacteria was the most dominant phylum. In this current study, we also observed that, upon supplementation with L. plantarum, the abundance of Firmicutes was higher in the test group compared with the control group. Moreover, the addition of L. plantarum increased the abundance of Firmicutes, but reduced the abundance of Bacteroidetes, which was conducive to nutrient digestion. In a separate study performed by our group, we observed that Firmicutes (71.39%) were abundant in young laying hens [33]. Combining the results from the two studies, we found that the abundance of Firmicutes was the same in young laying hens and older laying hens. However, in older hens, the abundance of Firmicutes decreased gradually to 58.8% at 66 weeks [31]. These results revealed that L. plantarum can affect fecal microbiota composition in laying hens. The reason for the latter result might be explained by the fact that L. plantarum can improve intestinal development and digestive ability.
As shown in Fig. 4, an analysis of the most abundant taxa revealed that at the genus level, Lactobacillus (58.55%), Romboutsia (18.29%), and Enterococcus (12.09%) were abundant in the test group, while Lactobacillus (13.39%), Romboutsia (39.02%), and Enterococcus (8.26%) were also abundant in the control group. The differences in the abundances of Lactobacillus and Romboutsia were significant (p < 0.05). In this study, we detected genus Romboutsia in fecal samples from both older and younger laying hens[33]. Analysis of the two experiments with the laying hens from the same hen farm indicated that Romboutsia genus might be related to the environment of the hen farm. Romboutsia genus has been previously observed only in human gut [34], lake sediment [35] and rat gastro-intestinal tract [36]. In our experiment, feed supplemented with L. plantarum promoted Lactobacillus abundance while inhibiting Rombousia. The difference in abundance of the two genera is most likely underpinned
by competitive exclusion. We speculate that the reason for this competitive exclusion may due to the reduction of pH at the intestinal (duodenum, ileum, cecum) level or the colonization of the border or intestinal villi with L. plantarum. Another reason may be the effect of a quorum-sensing mechanism of some strains of L. plantarum spp. against other bacteria, producing protein bacteriocins [37].
The heat-map and STAMP analyses revealed that Lactobacillus was more abundant in the test group compared with the control group. Lactobacillus has long been known to be beneficial in both humans and animals, and a previous study demonstrated that this genus promotes a healthy gut in animals by producing several short-chain fatty acids [38]. In this current study, Lactobacillus was the dominant genus, and we speculate that when used as a feed supplement, L. plantarum can modulate fecal microbial composition and promote nutrient digestion and absorption. In addition, Escherichia-Shigella, which can be associated with diarrhea, was found in five samples (B11, A15, A14, A4 and A11). However, one of the limitations of this study is the use of fecal samples instead of intestinal contents. The composition of the intestinal microbiota may better reflect the animal’s digestive function. However, taking fecal samples is more conducive to animal welfare protection. Moreover, it is well known that fecal microbial diversity and composition affect the development of the immune system in poultry [39]. This suggests that we can promote the health of livestock, and more specifically poultry, by modulating the fecal microbiome. Moreover, supplementation of diets with probiotics might prove to be an effective alternative to antibiotics [18].
5. Conclusion
Our data suggest that administration of an L. plantarum strain can increase the laying rate and feed intake of laying hens. The current study also suggests that administration of L. plantarum plays a vital role in the modulation of fecal microbiota. The study also provides us with a better understanding of the interplay between L. plantarum and the fecal microbiota of laying hens. Future investigations should be performed to help us understand the mechanism in hens fed with L. plantarum.
Acknowledgments
This work was supported by Henan Province Natural Science Planning Fund Projects [grant no. 162300410128], by the Henan Province Science and Technology Open Cooperation Project [grant no. 182106000042],by the Key Discipline of Preventive Veterinary Medicine of Henan University of Animal Husbandry and Economy [grant no. mxk2016102], and the Research and Innovation Team of Henan University of Animal Husbandry and Economy.
Footnotes
Conflict of interest
Authors state no conflict of interest
References
- [1].Ghasemian M, Jahanian R. Dietary mannan-oligosaccharides supplementation could affect performance, immunocompetence, serum lipid metabolites, intestinal bacterial populations, and ileal nutrient digestibility in aged laying hens. Anim Feed Sci Tech. 2016;213:81–9. [Google Scholar]
- [2].Dahiya R, Berwai RS, Sihag S, Patil CS. Lalit. The effect of dietary supplementation of salts of organic acid on production performance of laying hens. Vet World. 2016;12:1478–84. doi: 10.14202/vetworld.2016.1478-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhao L, Zhang X, Cao F, Sun D, Wang T, Wang G. Effect of dietary supplementation with fermented Ginkgo-leaves on performance, egg quality, lipid metabolism and egg-yolk fatty acids composition in laying hens. Livest Sci. 2013;155:77–85. [Google Scholar]
- [4].Smith JM. A review of avian probiotics. J Avian Med Surg. 2014;28:87–94. doi: 10.1647/2012-031. [DOI] [PubMed] [Google Scholar]
- [5].Murshed MA, Abudabos AM. Effects of dietary inclusion of probiotic, prebiotic and symbiotic on growth performance of broiler chickens. Braz J Poultry Sci.2015; 117(5):99–104. [Google Scholar]
- [6].Upadhaya S, Devi SM, Lee BI, Kim I. Poteintials of probiotics RX7 and C14 strains as an alternative to antibiotics in challenged weaning pigs. J Anim Sci. 2016;94(Suppl 2):81. [Google Scholar]
- [7].Perić I, Milošević N, Žikić D, Bjedov S, Cvetković D, Markov S. Effects of probiotics and phytogenic products on performance, gut morphology and cecal microflora of broiler chickens. Archiv Tierzucht. 2010;53(3):350–59. et al. [Google Scholar]
- [8].Abudabos AM, Al-Batshan HA, Murshed MA. Effects of Prebiotics and Probiotics on the Performance and Bacterial Colonization of Broiler Chickens. South African Journal of Animal Science. 2015;45(4):419. [Google Scholar]
- [9].Rinkinen M, Jalava K, Westermarck E, Salminen S, Ouwehand AC. Interaction between probiotic lactic acid bacteria and canine enteric pathogens: a risk factor for intestinal Enterococcus faecium colonization? Vet microbiolgy. 2003;92(1-2):111–19. doi: 10.1016/s0378-1135(02)00356-5. [DOI] [PubMed] [Google Scholar]
- [10].Fuller R. Probiotcs in man and animals-A review. J Appl Microbiol. 1989;66:365–78. [PubMed] [Google Scholar]
- [11].Smoragiewicz W, Bielecka M, Babuchowski A, Boutard A, Dubeau H. Les probiotiqes. Can J Microbiol. 1993;39:1089–95. [Google Scholar]
- [12].Vanbelle M, Teller E, Focant M. Probiotcis in animal nutrition: A review. Berlin Arc Anim Nutr. 1990;7:543–67. doi: 10.1080/17450399009428406. [DOI] [PubMed] [Google Scholar]
- [13].Chen YJ, Min BJ, Cho JH, Kwon OS, Son KS, Kim HJ. Effects of dietary Bacillus-based probiotic on growth performance, nutrients digestibility, blood characteristics and focal noxious gas content in finishing pigs. Asian Australas J Anim Sci. 2006;19:587–92. et al. [Google Scholar]
- [14].Zhang JL, Xie QM, Ji J, Yang WH, Wu YB, Li C. Different combinations of probiotics improve the production performance, egg quality, and immune response of layer hens. Poult Sci. 2012;91:2755–60. doi: 10.3382/ps.2012-02339. et al. [DOI] [PubMed] [Google Scholar]
- [15].De Vries MC, Vaughan EE, Kleerebezem M, Willem M de Vosab. Lactobacillus plantarum-survival, functional and potential probiotic properties in the human intestinal tract. International Dairy J. 2006;16:1016–28. [Google Scholar]
- [16].Kanmani P, Staish kumar R, Yuvaraj N, Paari KA, Pattukumar V, Arul V. Probiotics and its functionally valuable products-a review. Crit Rev Food Sci. 2003;53:641–58. doi: 10.1080/10408398.2011.553752. [DOI] [PubMed] [Google Scholar]
- [17].Gao PF, Hou QC, Kwok LY, Huo DX, Feng SZ, Zhang HP. Effect of feeding Lactobacillus plantarum P-8 on the faecal microbiota of the broiler chickens exposed to lincomycin. Sci Bull. 2017;62:105–13. doi: 10.1016/j.scib.2017.01.001. [DOI] [PubMed] [Google Scholar]
- [18].Shen X, Yi D, Ni X, Zeng D, Jing B, Lei M. Effects of Lactobacillus plantarum on production performance, immune characteristics, antioxidant status, and intestinal microflora of bursin-immunized broilers. Can J Microbiol. 2013;60:193–202. doi: 10.1139/cjm-2013-0680. et al. [DOI] [PubMed] [Google Scholar]
- [19].Choe DW, Loh TC, Foo HL, Hair-Bejo M, Awis QS. Egg production, faecal pH and microbial population, small intestine morphology, and plasma and yolk cholesterol in laying hens given liquid metabolites produced by Lactobacillus plantarum-strains. Br Poul Sci. 2012;1:106–15. doi: 10.1080/00071668.2012.659653. [DOI] [PubMed] [Google Scholar]
- [20].Loh TC, Choe DW, Foo HL, Sazili AQ, Bejo MH. Effects of feeding different postbiotic metabolite combinations produced by Lactobacillus plantarum strains on egg quality and production performance, faecal parameters and plasma cholesterol in laying hens. BMC Vet Res. 2014;10:149. doi: 10.1186/1746-6148-10-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ding JM, Dai RH, Yang LY, He C, Xu K, Liu SY. Inheritance and establishment of gut microbiota in chickens. Front microbiol. 2017;8:1967. doi: 10.3389/fmicb.2017.01967. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Xu YH, Yang HX, Zhang LL, Su YH, Shi DH, Xiao HD. High-throughput sequencing technology to reveal the composition and function of cecal microbiota in Dagu chicken. BMC Microbiol. 2016;16:259–68. doi: 10.1186/s12866-016-0877-2. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ren DY, Li C, Qin YQ, Yin RL, Du SW, Ye F. Lactobacilli reduce chemokine IL-8 production in response to TNF-α and Salmonella challenge of Caco-2 clles. BioMed Res Int. 2013:925219. doi: 10.1155/2013/925219. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ren DY, Li C, Qin YQ, Yin RL, Du SW, Liu HF. Evaluation of immunomodulatetory activity of two potential probiotic Lactobacillus strains by in vivo tests. Anaerobe. 2015;35(10):22–7. doi: 10.1016/j.anaerobe.2015.06.008. et al. [DOI] [PubMed] [Google Scholar]
- [25].Park JW, Jeong JS, Lee SI, Kim IH. Effect of dietary supplementation with a probiotic Enterococcus faecium on production performance, excreta microflora, ammonia emission, and mutrient utilization in ISA brown laying hens. Poult Sci. 2016;12:2829–35. doi: 10.3382/ps/pew241. [DOI] [PubMed] [Google Scholar]
- [26].Yan W, Sun CJ, Yuan JW, Yang N. Gut metagenomic analysis reveals prominent roles of Lactobacillus and cecal microbiota in chicken feed efficiency. Sci Rep. 2017;7:45308. doi: 10.1038/srep45308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Long L, Wu SG, Yuan F, Zhang HJ, Wang J, Qi GH. Effects of dietary octacosanol supplementation on laying performance, egg quality, serum hormone levels, and expression of genes related to the reproductive axis in laying hens. Poult Sci. 2017;4:894–903. doi: 10.3382/ps/pew316. [DOI] [PubMed] [Google Scholar]
- [28].Wang HS, Ni XQ, Qing XD, Zeng D, Luo M, Liu L. Live probiotic Lactobacillus johnsonil BS15 promotes growth performance and lowers fat deposition by improving lipid metabolism, intestinal development, and gut microflora in broilers. Front Microbiol. 2017;8:1073–87. doi: 10.3389/fmicb.2017.01073. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mahdavi AH, Rahman AR, Pourreza J. Effect of probiotic supplements on egg quality and laying hen’s performance. Inter J Poult Sci. 2005;4:488–92. [Google Scholar]
- [30].Kers JG, Velkers FC, Fischer EA, Hermes GDA, Stegeman JA, Smidt H. Host and environmental factors affecting the intestinal microbiota in chickens. Front Microbiol. 2018;9:235. doi: 10.3389/fmicb.2018.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Videnska P, Rahman MM, Faldynova M, Babak V, Matulova ME, Prukner-Radovcic E. Charaterization of egg laying hen and broiler fecal microbiota in poultry farms in Croatia, Czech Republic, Hungary and Slovenia. Plos One. 2014;10:e110076. doi: 10.1371/journal.pone.0110076. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Singh KM, Shah T, Deshpande S, Jakhesara SJ, Koringa PG, Rank DN. High through put 16S rRNA gene-based pyrosequencing analysis of the fecal microbiota of high FCR and low FCR broiler growers. Mol Biol Rep. 39(201):10595–602. doi: 10.1007/s11033-012-1947-7. et al. [DOI] [PubMed] [Google Scholar]
- [33].Qiao HX, Zhang LH, Shi HT, Song YZ, Bian CZ. Astragalus affects fecal microbial composition of young hens as determined by 16S rRNA sequencing. AMB Express. 2018;8:70. doi: 10.1186/s13568-018-0600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ricaboni D, Mailhe M, Khelaifia S, Raoult D, Million M. Romboutsia timonensis, a new species isolated from human gut. New Microbes New Infect. 2016;12:6–7. doi: 10.1016/j.nmni.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang YW, Song JL, Zhai Y, Zhang C, Gerritsen J, Wang HM. Romboutsia sedimentorum sp. nov., isolated from an alkalinesaline lake sediment and emended description of the genus Romboutsia Int Journal Sys Evol Microbiol. 2015;65:1193–8. doi: 10.1099/ijs.0.000079. et al. [DOI] [PubMed] [Google Scholar]
- [36].Gerritsen J, Fuentes S, Grievink W, Niftrik LV, Tindall BJ, Timmerman HM. Characterization of Romboutsia ilealis gen. nov., sp. nov., isolated from the gastro-intestinal tract of a rat, and proposal for the reclassification of five closely related members of the genus Clostridiuminto the genera Romboutsia gen. nov., Intestinibacter gen. nov.,Terrisporobacter gen. nov. and Asaccharospora gen nov. Int J Sys Evol Microbiol. 2014;64:1600–16. doi: 10.1099/ijs.0.059543-0. et al. [DOI] [PubMed] [Google Scholar]
- [37].Piewngam P, Zheng Y, Nguyen TH, Dickey SW, Joo HS, Villaruz AE, Glose KA, Fisher EL, Hunt RL, Li B, Chiou J, Pharkjaksu S, Khongthong S, Cheung GYC, Kiratisin P, Otto M. Pathogen elimination by probiotic Bacillus via signalling interference. Nature. 2018;562:532–537. doi: 10.1038/s41586-018-0616-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sidira M, Kandylis P, Kanellaki M, Kourkoutas Y. Effect of immobilized lactobacillus casei on the evolution of flavor compounds in probiotic dry-fermented sausages during ripening. Meat Sci. 2015;100:41–51. doi: 10.1016/j.meatsci.2014.09.011. [DOI] [PubMed] [Google Scholar]
- [39].Sugiharto S, Yudiarti T, Isroli I, Widiastuti E, Kusumanti E. Dietary supplementation of probiotics in poultry exposed to heat stress-a review. Ann Anim Sci. 2016;3:591–604. [Google Scholar]