Abstract
Characterization of the humoral response to SARS-CoV-2, the etiological agent of COVID-19, is essential to help control the infection. The neutralization activity of plasma from patients with COVID-19 decreases rapidly during the first weeks after recovery. However, the specific role of each immunoglobulin isotype in the overall neutralizing capacity is still not well understood. In this study, we select plasma from a cohort of convalescent patients with COVID-19 and selectively deplete immunoglobulin A, M, or G before testing the remaining neutralizing capacity of the depleted plasma. We find that depletion of immunoglobulin M is associated with the most substantial loss of virus neutralization, followed by immunoglobulin G. This observation may help design efficient antibody-based COVID-19 therapies and may also explain the increased susceptibility to SARS-CoV-2 of autoimmune patients receiving therapies that impair the production of immunoglobulin M (IgM).
Keywords: COVID-19, SARS-CoV-2, spike glycoprotein, IgM, IgA, IgG, neutralization, convalescent plasma
Graphical abstract
Gasser et al. highlight the importance of IgM in the capacity of plasma from convalescent donors to neutralize SARS-CoV-2.
Introduction
Since its discovery in Wuhan, China, in 2019, the causative agent of COVID-19, the SARS-CoV-2 virus (Zhu et al., 2020), has become a major global public health problem. A better understanding of immune responses induced by SARS-CoV-2 is urgently needed to help control the infection. Several studies have shown that the neutralization activity of plasma from patients with COVID-19 decreases rapidly during the first weeks after recovery (Beaudoin-Bussières et al., 2020; Long et al., 2020; Prévost et al., 2020; Robbiani et al., 2020; Seow et al., 2020). Although a good correlation between the presence of spike (S)-specific antibodies and the capacity of plasma from infected individuals to neutralize viral particles was reported, recent data looking at individual immunoglobulin (Ig) isotypes revealed a stronger correlation between the decrease in S-specific IgM antibodies and loss of neutralization compared with S-specific IgG and IgA antibodies, suggesting that IgM has an important role in the neutralization activity of plasma from individuals who suffered from COVID-19 (Beaudoin-Bussières et al., 2020; Prévost et al., 2020). To better understand the relative contribution of S-specific IgM, IgA, and IgG antibodies in SARS-CoV-2 neutralization, we selectively depleted each Ig isotype from plasma obtained from 25 convalescent donors and assessed the effect of depletion on the capacity of the plasma to neutralize SARS-CoV-2 pseudoviral particles and wild-type, infectious SARS-CoV-2 viral particles.
Results
Ig depletion
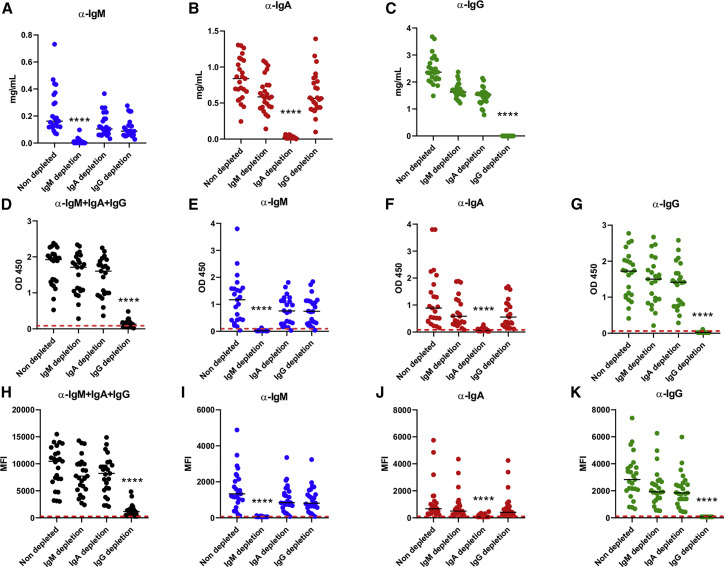
Demographic information on the 25 convalescent donors (21 males, 4 females) is presented in Table 1 . Each donor was sampled once between 25 and 69 days after the onset of symptoms, with an average time of 45 days. Selective depletion of IgM, IgA, or IgG was achieved by adsorption on isotype-specific ligands immobilized on Sepharose or agarose beads, starting with a 5-fold dilution of plasma (see details in STAR methods). The depletion protocols permitted efficient depletion of each isotype while leaving the other isotypes nearly untouched, as measured by ELISA (Figures 1 A–1C). Depletion of IgG had a much greater effect on the total level of SARS-CoV-2 receptor-binding domain (RBD) antibodies than IgM and IgA depletion (Figure 1D), although RBD-specific antibodies of each isotype were selectively removed by the depletion as shown by their respective signals close to the background level established with plasma samples collected before the outbreak of SARS-CoV-2 (Figures 1E–1G, red dashed line). The effect of IgG depletion on the level of total antibodies against the full S glycoprotein expressed on 293T cells (measured by flow cytometry) was also noticeable (Figure 1H), whereas isotype-specific detection of full S antibodies by flow cytometry confirmed the efficacy of selective depletion (Figures 1I–1K).
Table 1.
COVID convalescent plasma donor’s characteristics
| All donors | Males | Females | |
|---|---|---|---|
| Donors (n) | 25 | 21 | 4 |
| Average age, years ± SD (range) | 47 ± 16 (20–69) | 49 ± 17 (20–69) | 40 ± 14 (29–60) |
| Age, years (median) | 50 | 51 | 34.5 |
| Average time (days) between symptom onset and donation, median (range) | 45 (25–69) | 47 (25–69) | 40 (27–56) |
Figure 1.
IgM, IgA, and IgG depletion in plasma samples from convalescent donors
(A–C) Efficacy of the specific isotype depletion assessed by ELISA for total IgM, IgA, and IgG. All plasma samples were diluted 5-fold before depletion.
(A) IgM concentration in non-depleted, IgM-depleted, IgA-depleted, and IgG-depleted plasma samples, measured with an anti-human IgM (μ-chain specific) as the capture antibody.
(B) IgA concentration measured on the same plasma samples with an anti-human IgA (α-chain specific).
(C) IgG concentration measured with anti-human an IgG (γ-chain specific).
(D–G) Efficacy of SARS-CoV-2-specific antibody depletion as assessed by SARS-CoV-2 RBD ELISA.
(D) Level of total (pan-Ig) anti-SARS-CoV-2, RBD-specific antibodies in non-depleted, IgM-depleted, IgA-depleted, and IgG-depleted plasma samples.
(E) Level of IgM-specific anti-RBD.
(F) Level of IgA-specific anti-RBD. (G) Level of IgG-specific anti-RBD.
(H–K) Efficacy of full S glycoprotein-specific antibody depletion measured by flow cytometry.
(H) Level of total (pan-Ig) anti-SARS-CoV-2 S-specific antibodies in non-depleted, IgM-depleted, IgA-depleted, and IgG-depleted plasma samples.
(I) Level of IgM-specific anti-S.
(J) Level of IgA-specific anti-S.
(K) Level of IgG-specific anti-S. Red dashed lines represent the average signal given by negative controls taken from non-infected patients.
Asterisks indicate the level of statistical significance obtained by a Dunn’s test. ∗∗∗∗p < 0.0001.
Neutralizing activity of depleted plasma
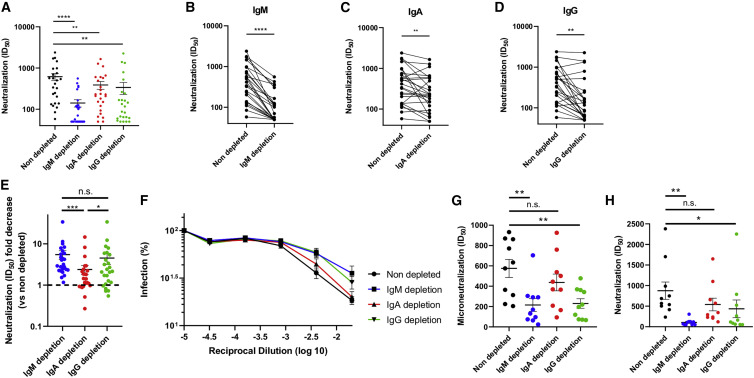
We then evaluated the capacity of non-depleted and isotype-depleted plasma samples to neutralize pseudoviral particles expressing the S glycoprotein from SARS-CoV-2 (Prévost et al., 2020) (STAR methods). Depletion of IgM, IgA, or IgG each resulted in a significant decrease of neutralization compared with non-depleted plasma (Figures 2 A–2D). However, the loss of neutralization activity was much more pronounced in IgM- and IgG-depleted plasma with a 5.5- and 4.5-fold decrease in half maximal inhibitory dilution (ID50) compared to non-depleted plasma, respectively, than in IgA-depleted plasma, in which only a 2.4-fold decrease was observed (Figure 2E). To evaluate whether the effect of isotype depletion on neutralization could be extended beyond pseudoviral particles, we tested plasma from 10 donors in microneutralization experiments using fully infectious SARS-CoV-2 viral particles, as described in the STAR methods. The neutralizing potency of plasma was greatly reduced after IgM and IgG (4.0- and 2.8-fold, respectively) but not after IgA (no decrease) depletion (Figure 2F). Despite the few samples tested with the live virus, the effect of IgM and IgG depletion on neutralization was similar to that observed with the same samples in the pseudoviral particle neutralization assay (Figures 2G and 2H). These data not only confirm the role of IgG in the neutralizing activity of convalescent plasma but also highlight the important contribution of IgM with respect to neutralization activity. To further assess the consistency of our data, we compared the ID50 obtained with pseudoviral particles and the one obtained with full SARS-CoV-2 virions, and we observed a significant correlation between the two datasets (Figure S1).
Figure 2.
Role of IgM, IgA, and IgG in neutralization
(A) Comparison of the SARS-CoV-2 pseudoviral inhibitory dilution (ID50) of all plasma samples.
(B–D) ID50 of plasma from each convalescent donor before and after IgM (B), IgA (C), and IgG (D) depletion.
(E) Fold decrease (isotype-depleted versus non-depleted plasma) in ID50 measured by SARS-CoV-2 pseudoviral particle neutralization.
(F and G) Microneutralization assay with infectious wild-type SARS-CoV-2 performed on non-depleted and isotype-depleted plasma from 10 donors. Mean percentage of infection (F) and ID50 observed from plasma from the 10 donors (G).
(H) ID50 obtained with the pseudoviral particle neutralization assay for the samples in (F)–(G).
Asterisks indicate the level of statistical significance obtained by a Wilcoxon signed rank test. n.s., not significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Discussion
Our findings detailing the important role of IgM in the neutralizing activity of convalescent plasma has several implications. First, although the therapeutic efficacy of convalescent plasma for the treatment of patients with COVID-19 remains to be established, it is likely that neutralizing antibodies will play a role. Because SARS-CoV-2-specific IgM antibodies rapidly decrease after disease onset (Beaudoin-Bussières et al., 2020; Prévost et al., 2020; Robbiani et al., 2020) and the overall neutralization capacity decreases as well (Gaebler et al., 2020; Grzelak et al., 2020; Seow et al., 2020), the collection of convalescent plasma with maximal neutralizing activity should be performed early after disease recovery.
In the present work, we did not observe trends such as a more-important contribution of IgM in convalescent plasma taken at earlier times (around day 30 after symptom onset) or of IgG at later times (around day 70) (Figure S2). Analysis of samples taken at much later times (for example around day 70) could nevertheless reveal a predominant role of IgG in the residual neutralization activity of convalescent plasma, given the reported decrease in SARS-CoV-2 IgM antibodies in most recovered patients. It should be emphasized that the present study was performed with plasma samples from 25 different convalescent plasma donors and not with sequential samples from a few donors. We felt that this strategy was important to obtain more-robust data that could be generalized to all convalescent plasma collected within a few weeks after symptom onset.
Second, our results suggest that caution should be taken when using therapeutics that impair the production of IgM. Anti-CD20 antibodies (B cell-depleting agents) are used to treat several inflammatory disorders. Their use is associated with IgM deficiency in many patients, whereas their impact on IgG and IgA levels is more limited (Kridin and Ahmed, 2020). In line with our data, recent studies reported that anti-CD20 therapy could be associated with a higher susceptibility to contract SARS-CoV-2 and develop severe COVID-19 (Guilpain et al., 2020; Hughes et al., 2020; Safavi et al., 2020; Schulze-Koops et al., 2020; Sharmeen et al., 2020; Sormani et al., 2020). Whether this is associated with the preferential depletion of IgM-producing B cells by these treatments (Looney et al., 2008) remains to be shown. Nevertheless, our results suggest that the IgM levels should be investigated as a biomarker to stratify patients on immunosuppressive therapies at higher risk for COVID-19.
One limitation of this study is that it focuses on the study of the blood compartment of recovered patients who had COVID-19 with an aim for application in convalescent plasma therapy. Therefore, our data cannot be generalized to the immune response happening during the early stages of the disease or to mucosal immunity in general. It has indeed been proposed elsewhere that secretory IgA might have a much more significant role during the early stages of the infection (Sterlin et al., 2020) and could be critical in prevention of reinfection (Wang et al., 2020).
In summary, our results extend previous observations showing a strong correlation between neutralization potency and the presence of RBD-specific IgM (Beaudoin-Bussières et al., 2020; Perera et al., 2020; Prévost et al., 2020; Seow et al., 2020). It is intriguing that IgM represents only about 5% of the total antibodies in plasma (Wang et al., 2020) yet has such an important role in SARS-CoV-2 neutralization. Whether this is due to the enhanced avidity provided by its pentameric nature remains to be formally demonstrated but is in agreement with recent work demonstrating that dimeric antibodies are more potent than their monomeric counterparts (Wang et al., 2020). The possible establishment of long-lived, IgM-producing B cells, which might contribute to long-term immunity in recovered patients, has been suggested (Brouwer et al., 2020; Newell et al., 2020). However, how plasma neutralization evolves over prolonged periods and the specific role of IgM in that activity remains to be determined.
STAR★methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SARS-CoV-2 Nucleocapsid protein monoclonal antibody, clone 1C7 | Bioss Antibodies | Cat# bs-41408P |
| Anti Human IgM (μ chain specific) | Sigma-Aldrich | Cat# I0759-1MG; RRID:AB_260109 |
| Goat anti Human serum IgA | Jackson ImmunoResearch | Cat# 109-005-011; RRID:AB_2337535 |
| AffiniPure Goat Anti-Human IgM, Fc5μfragment specific | Jackson ImmunoResearch | Cat # 109-005-129; RRID:AB_2337543 |
| AffiniPure Goat Anti-Human IgG, Fcγ fragment specific | Jackson ImmunoResearch | Cat # 109-005-098; RRID:AB_2337541 |
| Peroxidase AffiniPure Goat Anti-Human IgA + IgG + IgM (H+L) | Jackson ImmunoResearch | Cat # 109-035-064; RRID:AB_2337583 |
| Peroxidase AffiniPure Goat Anti-Human IgG (H+L) | Jackson ImmunoResearch | Cat # 109-035-003; RRID:AB_2337577 |
| Peroxidase AffiniPure Goat Anti-Human Serum IgA, α chain specific | Jackson ImmunoResearch | Cat # 109-035-011; RRID:AB_2337580 |
| Peroxidase AffiniPure Goat Anti-Human IgM, Fc5μfragment specific | Jackson ImmunoResearch | Cat # 109-035-129; RRID:AB_2337588 |
| Peroxidase AffiniPure Goat Anti-Human IgG, Fcγ fragment specific | Jackson ImmunoResearch | Cat # 109-035-098; RRID:AB_2337586 |
| Alexa Fluor® 647 anti-human IgG Fc Antibody | BioLegend | Cat # 409319; RRID:AB_2563329 |
| Alexa Fluor® 647 AffiniPure Goat Anti-Human Serum IgA, α chain specific | Jackson ImmunoResearch | Cat # 109-605-011; RRID:AB_2337883 |
| Alexa Fluor® 647 AffiniPure Goat Anti-Human IgM, Fc5μ fragment specific | Jackson ImmunoResearch | Cat # 109-605-043; RRID:AB_2337884 |
| Alexa Fluor® 647 AffiniPure Goat Anti-Human IgA + IgG + IgM (H+L) | Jackson ImmunoResearch | Cat # 109-605-064; RRID:AB_2337886 |
| Bacterial and virus strains | ||
| SARS-CoV-2 USA-WA1/2020 | GenBank | MN985325.1 |
| Biological samples | ||
| Human Plasma from SARS-CoV-2 infected or uninfected individuals | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| SIGMAFAST OPD | Millipore Sigma | Cat#P9187-50 SET |
| Dulbecco’s Modified Eagle’s medium (DMEM) | Wisent | Cat# 319-005-CL |
| Penicillin/Streptomycin | Wisent | Cat# 450-201-EL |
| Fetal Bovine Serum (FBS) | VWR | Cat# 97068-085 |
| Bovine Serum Albumin (BSA) | Sigma | Cat# A7638 |
| Phosphate Buffered Saline (PBS) | ThermoFischer Scientific | Cat# 10010023 |
| Tween 20 | Sigma | Cat# P9416-100ML |
| Puromycin Dihydrochloride | Millipore Sigma | Cat# P8833 |
| Passive Lysis Buffer | Promega | Cat# E1941 |
| Freestyle 293F expression medium | Thermo Fischer Scientific | Cat# A14525 |
| Ni-NTA Agarose | Invitrogen | Cat# R90110 |
| D-Luciferin Potassium Salt | Thermo Fischer Scientific | Cat# L2916 |
| LIVE-DEAD Fixable AquaVivid Cell Stain | Thermo Fischer Scientific | Cat# P34957 |
| Formaldehyde 37% | Thermo Fischer Scientific | Cat# F79-500 |
| Critical commercial assays | ||
| Protein G HP SpinTrap | Fisher Scientific | Cat# 11580644 |
| NHS HP SpinTrap | Fisher Scientific | Cat# 11713329 |
| Peptide M/Agarose | InvivoGen | Cat# gel-pdm-2 |
| Experimental models: cell lines | ||
| Vero | ATCC | Cat# CRL-1586; RRID: CVCL_0574 |
| 293T-ACE2 | Prévost et al., 2020 | N/A |
| 293T Spike | This paper | N/A |
| HEK293T | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| FreeStyle 293F cells | ThermoFischer Scientific | Cat# R79007; RRID: CVCL_D603 |
| Recombinant DNA | ||
| pCG1-SARS-CoV-2 Spike | Hoffmann et al., 2020 | N/A |
| pNL4.3 R-E- Luc | NIH AIDS reagent program | Cat# 3418 |
| pcDNA3.1(+)-SARS-CoV-2 RBD | Beaudoin-Bussières et al., 2020 | N/A |
| pLV-SARS-CoV-2 S C-GFPSpark tag | SinoBiological | Cat# VG40590-ACGLN |
| Software and algorithms | ||
| Microplate Reader and Image Software | BioTek | https://www.biotek.com/products/software-robotics-software/gen5-microplate-reader-and-imager-software/ |
| Flowjo v10 | Tree Star | https://www.flowjo.com/ |
| GraphPad Prism v8 | GraphPad | https://www.graphpad.com/ |
| Microsoft Excel v16 | Microsoft Office | https://www.microsoft.com/en/microsoft-365/ |
| Other | ||
| ELx405 Microplate Washer | BioTek | N/A |
| Synergy LX Multimode Reader | BioTek | N/A |
| BD LSRII Flow Cytometer | BD Biosciences | N/A |
| TriStar LB942 Microplate Reader | Berthold Technologies | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead contact (andres.finzi@umontreal.ca).
Materials availability
All unique reagents generated during this study are available from the Lead contact without restriction.
Data and code availability
This study did not generate new code.
Experimental model and subjects details
Ethics statement
All work was conducted in accordance with the Declaration of Helsinki in terms of informed consent and approval by an appropriate Ethics Review board. Convalescent plasmas were obtained from donors who consented to participate in this research project at CHUM (19.381) and at Héma-Québec (REB # 2020-004). The donors met all donor eligibility criteria: previous confirmed COVID-19 infection and complete resolution of symptoms for at least 14 days.
Human subjects
No specific criteria such as number of patients (sample size), clinical or demographic were used for inclusion, beyond PCR confirmed SARS-CoV-2 infection in adults.
Plasmids
The plasmid expressing the human coronavirus Spike of SARS-CoV-2 was kindly provided by Stefan Pöhlmann and was previously reported (Hoffmann et al., 2020). The pNL4.3 R-E- Luc was obtained from NIH AIDS Reagent Program. The codon-optimized RBD sequence (encoding residues 319-541) fused to a C-terminal hexahistidine tag was cloned into the pcDNA3.1(+) expression vector and was reported elsewhere (Beaudoin-Bussières et al., 2020). The vesicular stomatitis virus G (VSV-G)-encoding plasmid (pSVCMV-IN-VSV-G) was previously described (Lodge et al., 1997).
Cell lines
293T human embryonic kidney cells (obtained from ATCC) and Vero E6 cells (ATCC CRL-1586) were maintained at 37°C under 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) (Wisent) containing 5% fetal bovine serum (VWR), 100 UI/ml of penicillin and 100 μg/ml of streptomycin (Wisent). The 293T-ACE2 cell line has been generated by us and was previously reported (Prévost et al., 2020). For the generation of 293T cells stably expressing SARS-CoV-2 Spike the same technique than previously described has been used (Prévost et al., 2020): VSV-G pseudotyped lentivirus packaging the SARS-CoV-2 Spike gene was produced in 293T using a third-generation lentiviral vector system. Briefly, 293T cells were co-transfected with two packaging plasmids (pLP1 and pLP2), an envelope plasmid (pSVCMV-IN-VSV-G) and a lentiviral transfer plasmid coding for a GFP-tagged SARS-CoV-2 Spike (pLV-SARS-CoV-2 S C-GFPSpark tag) (SinoBiological). Supernatant containing lentiviral particles was used to infect 293T cells in presence of 5μg/mL polybrene. The 293T cells stably expressing SARS-CoV-2 Spike (GFP+) were sorted by flow cytometry. SARS-CoV-2 expression was confirmed using the CR3022 mAb and plasma from SARS-CoV-2-infected individuals (Anand et al., 2020).
Method details
Isotype depletion
Selective depletion of IgM, IgA or IgG was done by adsorption on isotype-specific ligands immobilized on Sepharose or agarose beads starting with a five-fold dilution of plasma in PBS. IgG and IgA antibodies were depleted from plasma obtained from 25 recovered COVID-19 patient using Protein G HP Spintrap (Cytiva, formerly GE Healthcare Life Sciences, Buckinghamshire, UK) and Peptide M / Agarose (InvivoGen, San Diego, CA), respectively, according to the manufacturer’s instructions with the exception that no elution step for the recovery of the targeted antibodies was done, because the elution of bound antibodies requires exposure to denaturing conditions (for example acidic pH) which, according to several reports in the literature, could alter their biological activity (Bergmann-Leitner et al., 2008; Dimitrov et al., 2007; St-Amour et al., 2009). For IgM depletion, anti-human IgM (μ-chain specific, Sigma, St.Louis, MO) was covalently coupled to NHS HP SpinTrap (Cytiva, formerly GE Healthcare) at 815 μg/mL of matrix. Depletion was performed according to the manufacturer’s instructions with the exception that no elution step for the recovery of the targeted isotype was done. All non-depleted and isotype-depleted samples were filtered on a 0.22 μm Millex GV filter (SLGV013SL, Millipore, Burlington, MA) to ensure sterility for the virus capture and neutralization assays.
Immunoglobulin isotype ELISA
To assess the extent of IgM, IgG and IgA depletion, ELISA were performed on non-depleted as well as IgM-, IgA- and IgG-depleted plasma samples. Each well of a 96-well microplate was filled with either goat anti-human IgM (μ-chain specific) at 5 μg/mL, goat anti-human serum IgA (α chain specific) at 0.3 μg/mL or goat anti-human IgG (γ-chain specific) at 5 μg/mL (all from Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Microtiter plates were sealed and stored overnight at 2- 8°C. After four (IgA) to six (IgM and IgG) washes with H2O-0.1% Tween 20 (Sigma), 200 μL of blocking solution (10 mmol/L phosphate buffer, pH 7.4, containing 0.85% NaCl, 0.25% Hammerstein casein (EMD Chemicals Inc., Gibbstown, NJ,) were added to each well to block any remaining binding sites. The blocking solution for the IgG and IgM ELISA also contained 0.05% Tween 20. After 0.5 (IgA) to 1h (IgM and IgG) incubation at 37°C and washes, samples and the standard curves (prepared with human calibrated standard serum, Cedarlane, Burlington, Canada) were added to the plates in triplicates. Plates were incubated for 1h at 37°C. After washes, 100 μL of either goat anti-human IgA+G+M (H+L) HRP conjugate (1/30 000), goat anti-human IgG (H+L) HRP conjugate (1/30 000), goat anti-human IgM (μ-chain specific) HRP conjugate (1/10 000) or goat anti-human IgA (α chain specific) HRP conjugate (1/10 000) (all from Jackson ImmunoResearch Laboratories, Inc.) were added and samples were incubated at 37°C for 1h. Wells were washed and bound antibodies were detected by the addition of 100 μL of 3,3′,5,5′-tetramethylbenzimidine (TMB, ScyTek Laboratories, Logan, UT). The enzymatic reaction was stopped by the addition of 100 μL 1 N H2SO4 and the absorbance was measured at 450/630 nm within 5 minutes.
SARS-CoV-2 RBD ELISA
The presence of SARS-CoV-2 RBD-specific antibodies in the plasma from 25 recovered COVID-19 patients before and after depletion was measured using an ELISA adapted from a recently described protocol (Beaudoin-Bussières et al., 2020; Perreault et al., 2020; Prévost et al., 2020). Recombinant RBD proteins were produced in transfected FreeStyle 293F cells (Invitrogen, Carlsbad, CA, USA) and purified by nickel affinity chromatography. Recombinant RBD was diluted to 2.5 μg/mL in PBS (Thermo Fisher Scientific, Waltham, MA, USA) and 100 μl of the dilution was distributed in the wells of flat-bottom 96-well microplates (Immulon 2HB; Thermo Scientific). The plates were placed overnight at 2-8°C for antigen adsorption. For the assay, the plates were emptied and a volume of 300 μl/well of blocking buffer (PBS-0.1% Tween (Sigma)-2% BSA (Sigma)) was added. The microplates were incubated for one hour at room temperature (RT) followed by washing four times (ELx405 microplate washer, Bio-Tek) with 300 μL/well of washing solution (PBS-0.1% Tween). Because the reaction is time sensitive, samples, negative and positive controls were prepared in triplicates in a plate, then transferred in the RBD coated plate by reverse multi-pipetting. The negative control was prepared from a pool of 23 COVID negative plasmas while the positive control was a characterized plasma from a recovered patient. After transfer, the plates were incubated for 60 minutes at 20-24°C. After four washes, 100 μL of either goat anti-human IgA+G+M (H+L) HRP conjugate (1/30 000) for the detection of all isotypes, goat anti-human IgM (μ-chain specific) HRP conjugate (1/15 000), F(ab’)2 fragment goat anti-human IgA (α chain specific) HRP conjugate (1/4500) (all from Jackson Immunosearch Laboratories, Inc.) or goat anti-human IgG (γ-chain specific) HRP conjugate (1/50 000) (Invitrogen) were added and samples were incubated at 20-24°C for 60 minutes. Wells were washed four times and bound antibodies were detected by the addition of 100 μL of 3,3′,5,5′-tetramethylbenzimidine (ScyTek Laboratories). The enzymatic reaction was stopped by the addition of 100 μL 1 N H2SO4 and the absorbance was measured at 450/630 nm within 5 minutes.
Flow cytometry analysis of cell-surface staining
293T cells stably expressing SARS-CoV-2 Spike with a C-GFP tag (293T-Spike) were mixed at a 1:1 ratio with non-transduced 293T cells and were stained with plasma from SARS-CoV-2-infected individuals (1:250 dilution). Plasma binding to cell-surface Spike was revealed using fluorescent secondary antibodies able to detect all Ig isotypes (anti-human IgM+IgG+IgA; Jackson ImmunoResearch Laboratories, Inc.) or specific to IgG isotype (Biolegend), IgM isotype (Jackson ImmunoResearch Laboratories, Inc.) or IgA isotype (Jackson ImmunoResearch Laboratories, Inc.). The living cell population was gated on the basis of a viability dye staining (Aqua Vivid, Invitrogen). Samples were acquired on a LSRII cytometer (BD Biosciences, Mississauga, ON, Canada) and data analysis was performed using FlowJo v10.5.3 (Tree Star, Ashland, OR). The signal obtained with 293T (GFP- population) was subtracted from the signal obtained with 293T-Spike (GFP+ population) to remove unspecific signal.
Neutralization assay using pseudoviral particles
Target cells were infected with single-round luciferase-expressing lentiviral particles as described previously (Prévost et al., 2020). Briefly, 293T cells were transfected by the calcium phosphate method with the lentiviral vector pNL4.3 R-E- Luc (NIH AIDS Reagent Program) and a plasmid encoding for SARS-CoV-2 Spike at a ratio of 5:4. Two days post-transfection, cell supernatants were harvested and stored at –80°C until use. 293T-ACE2 target cells were seeded at a density of 1 × 104 cells/well in 96-well luminometer-compatible tissue culture plates (Perkin Elmer) 24h before infection. Recombinant viruses in a final volume of 100 μL were incubated with the indicated plasma dilutions (1/50; 1/250; 1/1250; 1/6250; 1/31 250) for 1h at 37°C and were then added to the target cells followed by incubation for 48h at 37°C; cells were lysed by the addition of 30 μL of passive lysis buffer (Promega) followed by one freeze-thaw cycle. An LB942 TriStar luminometer (Berthold Technologies) was used to measure the luciferase activity of each well after the addition of 100 μL of luciferin buffer (15mM MgSO4, 15mM KPO4 [pH 7.8], 1mM ATP, and 1mM dithiothreitol) and 50 μL of 1mM d-luciferin potassium salt (Prolume). The neutralization half-maximal inhibitory dilution (ID50) represents the sera dilution to inhibit 50% of the infection of 293T-ACE2 cells by recombinant viruses.
Microneutralization assay using live SARS-CoV-2 viral particles
A microneutralization assay for SARS-CoV-2 serology was performed as previously described (Amanat et al., 2020). The assay was conducted with the person blinded to the sample identity. Experiments were conducted with the SARS-CoV-2 USA-WA1/2020 virus strain. This reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281. One day prior to infection, 2x104 Vero E6 cells were seeded per well of a 96 well flat bottom plate and incubated overnight (37°C/5% CO2) to permit Vero E6 cell adherence. On the day of infection, all plasma samples were heat inactivated at 56°C for one hour. Non-depleted plasma from each donor was also included in this assay. Plasma dilutions were performed in a separate 96 well culture plate using MEM supplemented with penicillin (100 U/mL), streptomycin (100 μg/mL), HEPES, L-Glutamine (0.3 mg/mL), 0.12% sodium bicarbonate, 2% FBS (all from Thermo Fisher Scientific) and 0.24% BSA (EMD Millipore Corporation). Plasma dilutions ranged from 1:50 to 1:31 250. In a Biosafety Level 3 laboratory (ImPaKT Facility, Western University), 103 TCID50/mL SARS-CoV-2 USA-WA1/2020 virus strain was prepared in MEM + 2% FBS and combined with an equivalent volume of respective plasma dilution for one hour at room temperature. After this incubation, all media was removed from the 96 well plate seeded with Vero E6 cells and virus:plasma mixtures were added to each respective well at a volume corresponding to 600 TCID50 per well and incubated for one hour further at 37°C. Both virus only and media only (MEM + 2% FBS) conditions were included in this assay. All virus:plasma supernatants were removed from wells without disrupting the Vero E6 monolayer. Each plasma dilution (100 μL) was added to its respective Vero E6-seeded well in addition to an equivalent volume of MEM + 2% FBS and was then incubated for 48 hours. Media was then discarded and replaced with 10% formaldehyde for 24 hours to cross-link Vero E6 monolayer. Formaldehyde was removed from wells and subsequently washed with PBS. Cell monolayers were permeabilized for 15 minutes at room temperature with PBS + 0.1% Triton X-100 (BDH Laboratory Reagents), washed with PBS and then incubated for one hour at room temperature with PBS + 3% non-fat milk. An anti-mouse SARS-CoV-2 nucleocapsid protein (Clone 1C7, Bioss Antibodies) primary antibody solution was prepared at 1 μg/mL in PBS + 1% non-fat milk and added to all wells for one hour at room temperature. Following extensive washing with PBS, an anti-mouse IgG HRP secondary antibody solution was formulated in PBS + 1% non-fat milk. One hour post-room temperature incubation, wells were washed with PBS, SIGMAFAST OPD developing solution (Millipore Sigma) was prepared as per manufacturer’s instructions and added to each well for 12 minutes. Dilute HCl (3.0 M) was added to quench the reaction and the optical density at 490 nm of the culture plates was immediately measured using a Synergy LX multi-mode reader and Gen5 microplate reader and imager software (BioTek®).
Quantification and statistical analysis
Statistics were analyzed using GraphPad Prism version 8.0.2 (GraphPad, San Diego, CA, (USA). Every dataset was tested for statistical normality and this information was used to apply the appropriate (parametric or nonparametric) statistical test. P values < 0.05 were considered significant; significance values are indicated as ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Acknowledgments
We thank Stefan Pöhlmann for kindly providing the plasmid expressing the SARS-CoV-2 S glycoprotein. This work was supported by Ministère de l’Économie et de l’Innovation du Québec “Programme de soutien aux organismes de recherche et d’innovation,” the Fondation du CHUM, Canada’s COVID-19 Immunity Task Force (CITF) in collaboration with the Canadian Institutes of Health Research (CIHR), and a CIHR foundation grant 352417 to A.F. Funding was also provided by an operating grant from CIHR from the Canadian 2019 Novel Coronavirus (COVID-19) Rapid Research Funding Opportunity (FRN440388 to J.D.D. and G.A.D.) and an Infrastructure Grant from CFI for the Imaging Pathogens for Knowledge Translation (ImPaKT) Facility (36287 to J.D.D. and G.A.D.). A.F. is the recipient of a Canada Research Chair on Retroviral Entry (RCHS0235 950-232424). R.G. is supported by a MITACS Accélération postdoctoral fellowship. J.P. is supported by a CIHR graduate fellowship. The graphical abstract was prepared using images from BioRender.com.
Author contributions
R.G., M.C., J.P., R.B., and A.F. designed the studies. R.G. and S.D. performed neutralization experiments with pseudoviral particles. J.P. performed flow cytometry experiments. C.F., G.A.D., and J.D.D. performed microneutralization assays with infectious wild-type SARS-CoV-2 and analyzed the results. M.C., E.D., N.D., P.L., A.L.-L., and T.T. depleted plasma samples and performed the ELISA. J.R. provided new reagents. A.L. performed statistical analysis. C.L. provided scientific and clinical input. R.G., M.C., R.B., and A.F. wrote the manuscript with input from the others. Every author read, edited, and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: February 10, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108790.
Supplemental information
References
- Amanat F., White K.M., Miorin L., Strohmeier S., McMahon M., Meade P., Liu W.-C., Albrecht R.A., Simon V., Martinez-Sobrido L. An in vitro microneutralization assay for SARS-CoV-2 serology and drug screening. Curr. Protoc. Microbiol. 2020;58:e108. doi: 10.1002/cpmc.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S.P., Prévost J., Richard J., Perreault J., Tremblay T., Drouin M., Fournier M.-J., Lewin A., Bazin R., Finzi A. High-throughput detection of antibodies targeting the SARS-CoV-2 Spike in longitudinal convalescent plasma samples. bioRxiv. 2020 doi: 10.1101/2020.10.20.346783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin-Bussières G., Laumaea A., Anand S.P., Prévost J., Gasser R., Goyette G., Medjahed H., Perreault J., Tremblay T., Lewin A. Decline of humoral responses against SARS-CoV-2 spike in convalescent individuals. MBio. 2020;11:e02590-20. doi: 10.1128/mBio.02590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann-Leitner E.S., Mease R.M., Duncan E.H., Khan F., Waitumbi J., Angov E. Evaluation of immunoglobulin purification methods and their impact on quality and yield of antigen-specific antibodies. Malar. J. 2008;7:129. doi: 10.1186/1475-2875-7-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov J.D., Lacroix-Desmazes S., Kaveri S.V., Vassilev T.L. Transition towards antigen-binding promiscuity of a monospecific antibody. Mol. Immunol. 2007;44:1854–1863. doi: 10.1016/j.molimm.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Ladinsky M., Cho A., Jankovic M., Schaefer-Babajew D. Evolution of antibody immunity to SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.11.03.367391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzelak L., Velay A., Madec Y., Gallais F., Staropoli I., Schmidt-Mutter C., Wendling M.-J., Meyer N., Planchais C., Rey D. Sex differences in the decline of neutralizing antibodies to SARS-CoV-2. medRxiv. 2020 doi: 10.1101/2020.11.12.20230466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilpain P., Le Bihan C., Foulongne V., Taourel P., Pansu N., Maria A.T.J., Jung B., Larcher R., Klouche K., Le Moing V. Rituximab for granulomatosis with polyangiitis in the pandemic of covid-19: lessons from a case with severe pneumonia. Ann. Rheum. Dis. 2020;80:e10. doi: 10.1136/annrheumdis-2020-217549. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes R., Pedotti R., Koendgen H. COVID-19 in persons with multiple sclerosis treated with ocrelizumab—a pharmacovigilance case series. Mult. Scler. Relat. Disord. 2020;42:102192. doi: 10.1016/j.msard.2020.102192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kridin K., Ahmed A.R. Post-rituximab immunoglobulin M (IgM) hypogammaglobulinemia. Autoimmun. Rev. 2020;19:102466. doi: 10.1016/j.autrev.2020.102466. [DOI] [PubMed] [Google Scholar]
- Lodge R., Lalonde J.P., Lemay G., Cohen E.A. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Looney R.J., Srinivasan R., Calabrese L.H. The effects of rituximab on immunocompetency in patients with autoimmune disease. Arthritis Rheum. 2008;58:5–14. doi: 10.1002/art.23171. [DOI] [PubMed] [Google Scholar]
- Newell K.L., Clemmer D.C., Cox J.B., Kayode Y.I., Zoccoli-Rodriguez V., Taylor H.E., Endy T.P., Wilmore J.R., Winslow G. Switched and unswitched memory B cells detected during SARS-CoV-2 convalescence correlate with limited symptom duration. medRxiv. 2020 doi: 10.1101/2020.09.04.20187724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R.A., Mok C.K., Tsang O.T., Lv H., Ko R.L., Wu N.C., Yuan M., Leung W.S., Chan J.M., Chik T.S. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro Surveill. 2020;25:2000421. doi: 10.2807/1560-7917.ES.2020.25.16.2000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault J., Tremblay T., Fournier M.-J., Drouin M., Beaudoin-Bussières G., Prévost J., Lewin A., Bégin P., Finzi A., Bazin R. Waning of SARS-CoV-2 RBD antibodies in longitudinal convalescent plasma samples within four months after symptom onset. Blood. 2020;136:2588–2591. doi: 10.1182/blood.2020008367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J., Gasser R., Beaudoin-Bussières G., Richard J., Duerr R., Laumaea A., Anand S.P., Goyette G., Benlarbi M., Ding S. Cross-sectional evaluation of humoral responses against SARS-CoV-2 spike. Cell Rep. Med. 2020:100126. doi: 10.1016/j.xcrm.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavi F., Nourbakhsh B., Azimi A.R. B-cell depleting therapies may affect susceptibility to acute respiratory illness among patients with multiple sclerosis during the early COVID-19 epidemic in Iran. Mult. Scler. Relat. Disord. 2020;43:102195. doi: 10.1016/j.msard.2020.102195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Koops H., Krueger K., Vallbracht I., Hasseli R., Skapenko A. Increased risk for severe COVID-19 in patients with inflammatory rheumatic diseases treated with rituximab. Ann. Rheum. Dis. 2020 doi: 10.1136/annrheumdis-2020-218075. Published online June 26, 2020. [DOI] [PubMed] [Google Scholar]
- Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., Hemmings O., O’Byrne A., Kouphou N., Galao R.P. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmeen S., Elghawy A., Zarlasht F., Yao Q. COVID-19 in rheumatic disease patients on immunosuppressive agents. Semin. Arthritis Rheum. 2020;50:680–686. doi: 10.1016/j.semarthrit.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormani M.P., De Rossi N., Schiavetti I., Carmisciano L., Cordioli C., Moiola L., Radaelli M., Immovilli P., Capobianco M., Trojano M. Social Science Research Network; 2020. Disease Modifying Therapies and COVID-19 Severity in Multiple Sclerosis. [Google Scholar]
- St-Amour I., Laroche A., Bazin R., Lemieux R. Activation of cryptic IgG reactive with BAFF, amyloid beta peptide and GM-CSF during the industrial fractionation of human plasma into therapeutic intravenous immunoglobulins. Clin. Immunol. 2009;133:52–60. doi: 10.1016/j.clim.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Sterlin D., Fadlallah J., Adams O., Fieschi C., Parizot C., Dorgham K., Rajkumar A., Autaa G., El-Kafsi H., Charuel J.-L. Human IgA binds a diverse array of commensal bacteria. J. Exp. Med. 2020;217:e20181635. doi: 10.1084/jem.20181635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Viant C., Gaebler C., Cipolla M., Hoffmann H.-H., Oliveira T.Y., Oren D.A. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2020;13:eabf1555. doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate new code.