Abstract
COVID-19 pandemic results in record high deaths in many countries. Although a vaccine for SARS-CoV-2 is now available, effective antiviral drugs to treat coronavirus diseases are not available yet. Recently, EGCG, a green tea polyphenol, was reported to inhibit SARS-CoV-2 3CL-protease, however the effect of EGCG on coronavirus replication is unknown. In this report, human coronavirus HCoV-OC43 (beta coronavirus) and HCoV-229E (alpha coronavirus) were used to examine the effect of EGCG on coronavirus. EGCG treatment decreases 3CL-protease activity of HCoV-OC43 and HCoV-229E. Moreover, EGCG treatment decreased HCoV-OC43-induced cytotoxicity. Finally, we found that EGCG treatment decreased the levels of coronavirus RNA and protein in infected cell media. These results indicate that EGCG inhibits coronavirus replication.
Keywords: Coronavirus, 3CL-protease, EGCG, HCoV-OC43, HCoV-229E
Abbreviations: COVID-19, the 2019 novel coronavirus disease; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; 3CL-protease, 3C-Like protease; EGCG, (−)-epigallocatechin-3-gallate
Graphical abstract
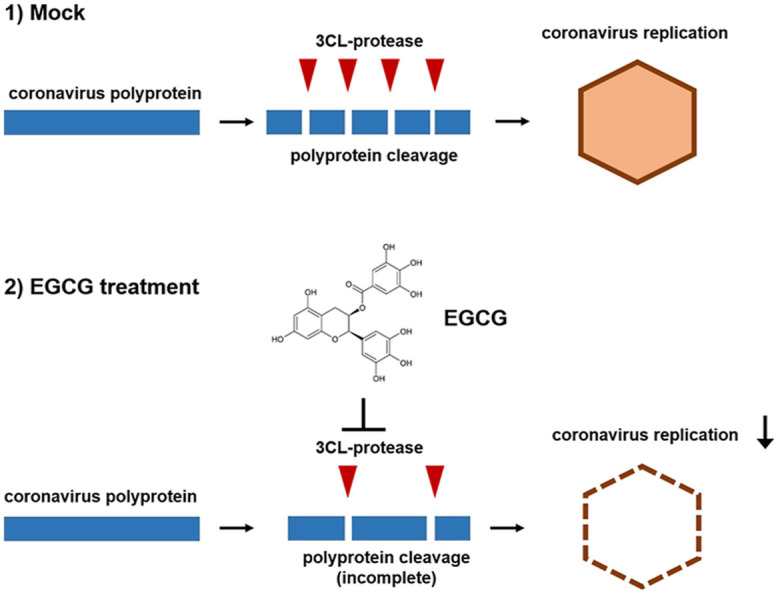
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a pandemic of acute respiratory disease, COVID-19, due to high transmission rate and pathogenicity of the virus [1]. Recently, COVID-19 vaccines have been developed, however effective antiviral drugs to treat coronavirus disease are not available [2]. Efficient antiviral therapy for coronavirus diseases should be established for immediate use and future safety.
Epigallocatechin-3-gallate (EGCG) is known to be the major catechin in green tea and accounts for 50%–80% of a brewed cup of green tea [3,4]. There is mounting evidence suggesting that EGCG is potentially useful to treat COVID-19. Because cleavage of viral polyproteins by proteases is a vital step for coronavirus replication, coronavirus 3CL-protease (Main protease or Mpro) is a promising drug target for coronavirus diseases [5,6]. EGCG is reported to inhibit SARS-CoV-2 3CL-protease using purified SARS-CoV-2 3CL-protease protein [7]. A computer simulation study also supports that EGCG can block the 3CL-protease activity of SARS-CoV-2 [[8], [9], [10], [11]]. In addition, statistical analysis demonstrated that the countries with high green tea consumption showed a striking reduction of COVID-19 risk including morbidity and mortality [12].
There are four coronavirus subfamilies: alpha, beta, gamma and delta; and SARS-CoV-2 belongs to beta coronaviruses [13]. In this report, we used human beta coronavirus, OC43 strain (HCoV-OC43) and human alpha coronavirus 229E strain (HCoV-229E) as a coronavirus model system to examine the effect of EGCG on coronavirus replication and production [14]. We found that EGCG treatment can efficiently reduce coronavirus production.
2. Materials and methods
2.1. Cell culture and infection
RD cells were maintained in MEM (Welgene, Seoul, Korea) containing 10% fetal bovine serum (FBS, Thermo Fisher Scientific, Waltham, MA, USA) and 1% penicillin-streptomycin solution (Welgene). RD cells were infected with HCoV-OC43 and HCoV-229E with the indicated dilutions of media from virus-infected cells, and the infected cells were maintained in MEM containing 2% FBS and 1% penicillin-streptomycin. Cell viability was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. HCoV-OC43 and HCoV-229E virus was obtained from ATCC (Rockville, MD, USA) and RD cells was obtained from Korean Cell Line Bank (KCLB, Seoul, Korea). (−)-Epigallocatechin gallate (EGCG) (E4134, purity ≥ 95%) was purchased from Sigma-Aldrich (Saint Louis, MO, USA) and MTT was purchased from USB Corporation (Cleveland, OH, USA).
2.2. Preparation of coronavirus and 3CL-protease assay
RD cells were infected with HCoV-OC43 and HCoV-229E and incubated for 7 days. The media from cell culture was collected and the virus fractions were isolated using an ultracentrifuge at 100,000 g for 4 h. Virus lysates were prepared using passive lysis buffer (Promega, Madison, WI, USA). A FRET-based protease assay was used to measure 3CL-protease activity [7,15]. The 3CL-protease activity was performed using 3CL-protein and FRET peptide in the reaction buffer (20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 5 mM EDTA, 5 mM DTT, 1% DMSO) for 5 h. For the inhibition assay, the purified 3CL-protease was incubated with EGCG for 1 h before the addition of substrate. Protease activity was calculated as the difference between the activity with virus particles and the activity with control at the indicated time.
2.3. Quantitative RT-PCR
For quantitative RT-PCR, cells and media were harvested and RNA was extracted using Trizol (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions and then subjected to RT-PCR using the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). HCoV-OC43 RdRp gene was amplified using the forward primer 5′-GAGTGTAGATGCCCGTCTCG-3′ and reverse primer 5′-TGTGGCACACGACTACCTTC-3’. HCoV-OC43 M gene was amplified using the forward primer 5′-ACGGTCACAATAATACGCGGT-3′ and reverse primer 5′-GGGTTGATGGCAGTCGGTAA-3’. HCoV-OC43 N gene was amplified using the forward primer 5′-AGGATGCCACCAAACCTCAG-3′ and reverse primer 5′-TGGGGAACTGTGGGTCACTA-3’. HCoV-229E RdRp gene was amplified using the forward primer 5′-CGCTGGTTGGCCCCTTAATA-3′ and reverse primer 5′-CGTGCACGTTCCTTACCAGA-3’. HCoV-229E M gene was amplified using the forward primer 5′-TTGGCCACTCGTACTTGCTT-3′ and reverse primer 5′-GTTCGAGCACGTCGGAAAAG-3’. HCoV-229E N gene was amplified using the forward primer 5′-ACAGGACCCCATAAAGCTGC-3′ and reverse primer 5′-CGCCTAACACCGTAACCTGT-3’.
2.4. Western blot and immunofluorescence assay
For Western blot, cells and media were harvested and resuspended in cell lysis buffer (150 mM NaCl, 50 mM HEPES (pH 7.5), and 1% NP40) containing a protease inhibitor cocktail (Roche, Basel, Switzerland). Cell lysates and virus lysates were resolved by SDS-PAGE and transferred to immune-blot PVDF membrane filters (Bio-Rad, Hercules, CA, USA). Virus proteins were detected with a 1:5000 dilution of primary HCoV-OC43 antibody using an ECL system (Dogen, Seoul, Korea). The images were acquired using the ImageQuant LAS 4000 System (GE Healthcare, Waukesha, WI, USA). The HCoV-OC43 antibody was purchased from Sigma-Aldrich. For immunofluorescence, RD cells were grown on sterilized glass coverslips. Cell were fixed with 4% paraformaldehyde. For immunostaining, cells were blocked with 3% bovine serum albumin in PBS, stained with a 1:1000 dilution of primary antibody in PBS, and stained with a 1:1000 dilution of Alexa Fluor 488–conjugated secondary antibody (Thermo Fisher Scientific). Finally, slides were washed 3 times with PBS, stained with DAPI, and mounted in mounting medium (Vector Laboratories, Burlingame, CA, USA). Images were captured with a Carl Zeiss LSM710 confocal microscope (Carl Zeiss, Oberkochen, Germany).
2.5. Statistical analysis
The results of 3CL-protease activity and MTT were evaluated by a 2-tailed Student’s t-test using Excel software (Microsoft, Redmond, WA, USA). A p-value of 0.05 was considered significant. For the calculation of half inhibitory concentration (IC50), AAT Bioquest website program was used (https://www.aatbio.com/tools/ic50-calculator-v1).
3. Results
3.1. EGCG inhibits the 3CL-protease activity of coronavirus
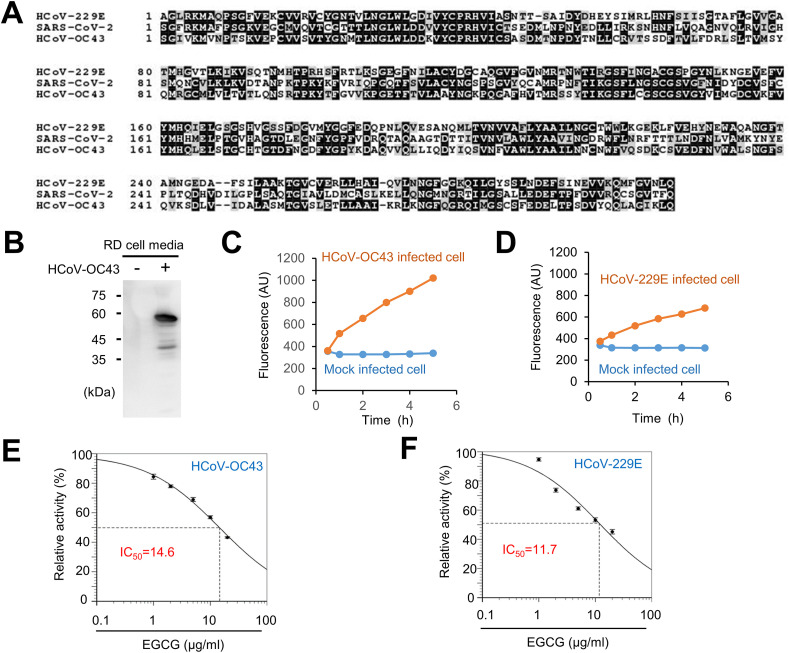
EGCG is reported to inhibit 3CL-protease activity of SARS-CoV-2 [7]. In order to determine the effect of EGCG on coronavirus replication, we used HCoV-OC43 and HCoV-229E virus strains as a proxy to study SARS-CoV-2. We examined and compared the sequence homology of 3CL-proteases between HCoV-OC43, HCoV-229E and SARS-CoV-2. We found that SARS-CoV-2 3CL-protease shows high similarity to HCoV-OC43 3CL-protease (sequence identity 48.18%) and HCoV-OC229E 3CL-protease (sequence identity 41.06%) (Fig. 1 A). Next, we examined HCoV-OC43 protease activity using virus particles of the infected cell media. Western blot with anti-HCoV-OC43 antibody showed the production of virus particles in the media (Fig. 1B). We showed 3CL-protease activity of HCoV-OC43 and HCoV-229E virus by FRET-based 3CL-protease assay (Fig. 1C and D). Next, we examined the effect of EGCG treatment on 3CL-protease activity of HCoV-OC43 and HCoV-229E. EGCG treatment decreased 3CL-protease activity of HCoV-OC43 and HCoV-229E in a dose-dependent manner, and the half maximal inhibitory concentration (IC50) was 14.6 μM (HCoV-OC43) and 11.7 μM (HCoV-229E), respectively (Fig. 1E and F). These results indicate that EGCG inhibits 3CL-protease activity of HCoV-OC43 and HCoV-229E.
Fig. 1.
EGCG inhibits 3CL-protease activity of HCoV-OC43 and HCoV-229E. (A) Amino acid sequence alignment of HCoV-229E 3CL-protease, HCoV-OC43 3CL-protease and SARS-CoV-2 3CL-protease. (B) Detection of HCoV-OC43 proteins in media. RD cells were infected with HCoV-OC43. Seven days after infection, media was collected and were probed with anti-HCoV-OC43 antibody. (C) HCoV-OC43 virus particles from infected cell media showed 3CL-protease activity in a FRET-based 3CL protease assay. (D) HCoV-229E virus particles showed 3CL-protease activity. (E) EGCG treatment inhibits 3CL-protease activity of HCoV-OC43 virus particles. (F) EGCG treatment inhibits 3CL-protease activity of HCoV-229E virus particles.
3.2. EGCG reduced coronavirus induced cytotoxicity
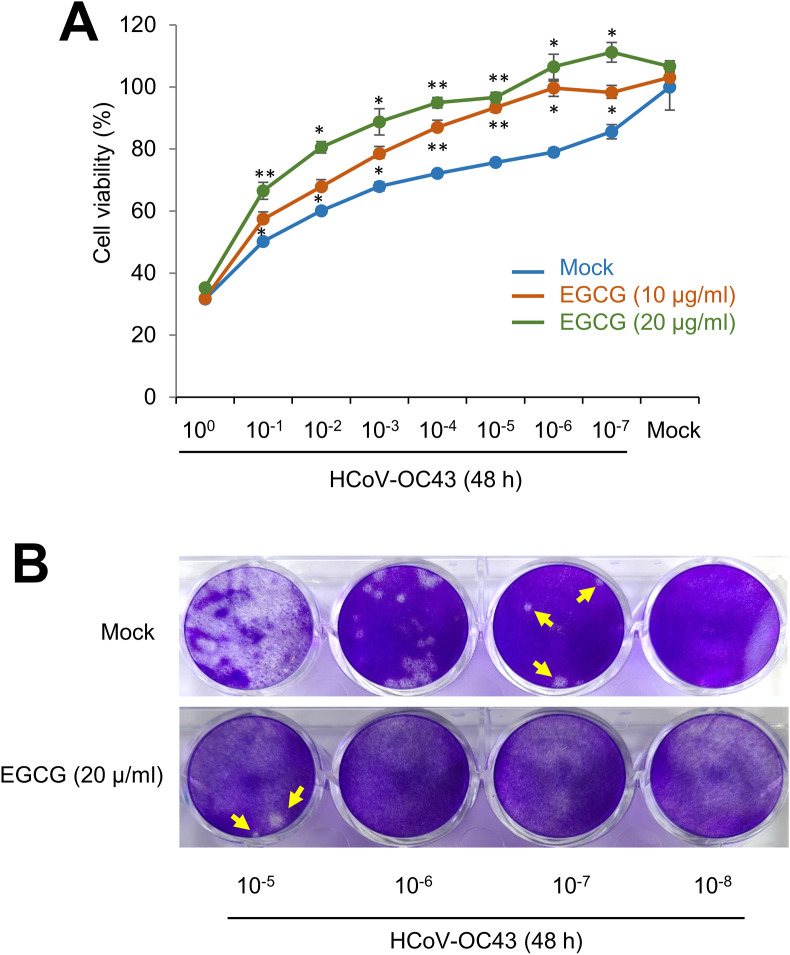
Because EGCG inhibits 3CL-protease activity, we hypothesized that EGCG reduces coronavirus induced cytotoxicity by inhibiting virus replication. We examined the effect of EGCG on coronavirus infection by measuring cell viability. While HCoV-229E did not show cytotoxicity on RD cells, HCoV-OC43 killed the RD cells (data not shown). RD cells were infected with various dilutions of HCoV-OC43 virus and incubated with mock or EGCG treatment. Cell viability assay showed that EGCG treatment decreases the cytotoxicity of HCoV-OC43 virus significantly (Fig. 2 A). Next, we evaluated the effect of EGCG on plaque formation. RD cells were treated with mock or EGCG, and the cells were infected with the indicated dilution of HCoV-OC43 conditioned media. EGCG treatment decreases plaque formation in host cells dramatically (Fig. 2B). We calculated the plaque forming unit (pfu) and found that the pfu of mock treatment was 2.875 ∗108 pfu/ml, while EGCG treatment had 1.25 ∗106 pfu/ml. These results indicate that EGCG treatment reduces HCoV-OC43 induced cytotoxicity and plaque formation.
Fig. 2.
EGCG treatment decreases HCoV-OC43 induced cytotoxicity. (A) EGCG treatment reduces the cytotoxicity of HCoV-OC43 infections. RD cells were infected with mock or HCoV-OC43 and cell viability was evaluated with MTT assay. The experiments were performed at least in triplicate. The graph shows mean and standard error. Control vs EGCG treatment: ∗p < 0.05, ∗∗p < 0.005. (B) RD cells were infected with mock or HCoV-OC43 and cells were overlaid with agarose. To visualize the plaque formation, the infected cells were stained with crystal violets. The number indicates dilution of conditioned media.
3.3. EGCG decreases coronavirus production and transmission
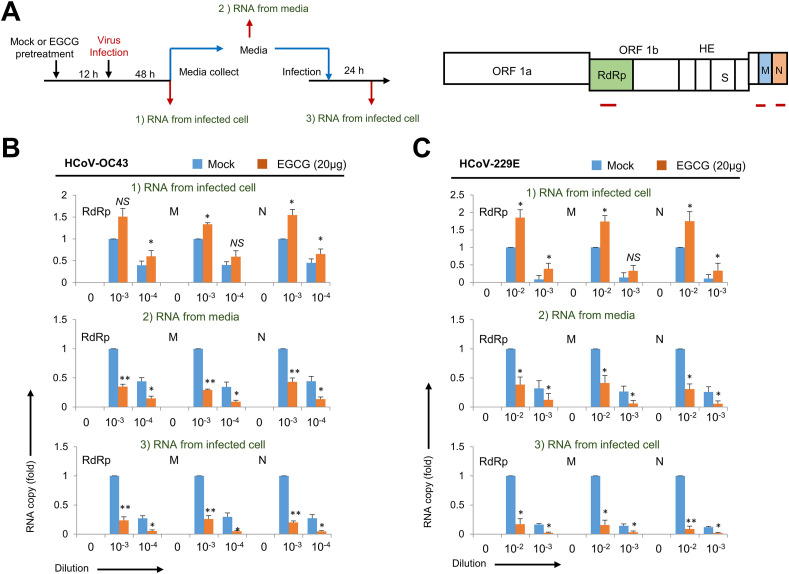
In order to determine the effect of EGCG on coronavirus replication, we evaluated the level of coronavirus RNA by quantitative RT-PCR. RD cells were treated with mock or EGCG and infected with coronavirus, and 48 h later, the copy number of coronavirus RNA was evaluated from infected cell and conditioned media. We also examined the level of produced coronavirus by allowing new RD cells to be infected by the conditioned media (Fig. 3 A). RNA-dependent RNA polymerase (RdRp) gene, M gene and N gene RNA were used to evaluate the level of coronavirus [16]. When we examined the RNA level of HCoV-OC43 in the infected cells, we unexpectedly found that EGCG treatment increases the levels of RdRp gene, M gene and N gene (Fig. 3B). However, the RNA levels of HCoV-OC43 genes were significantly reduced in the conditioned media by EGCG treatment (Fig. 3B). In addition, we quantified coronavirus RNA levels in cells infected with the conditioned media. The level of coronavirus RNA was significantly reduced when cells were infected with the virus which was produced in the presence of EGCG (Fig. 3B). When we examined the effect of EGCG treatment on the levels of coronavirus RNA with the HCoV-229E strain, we observed very similar trends (Fig. 3C). These results indicate that EGCG inhibits coronavirus production and transmission.
Fig. 3.
EGCG treatment decreases the infectious virus. (A) Experimental design and target locations. Schematic diagram of the experimental design illustrating HCoV-OC43 infections (left panel). Simplified genome structure of HCoV-OC43. The genomic locations of RdRp gene, M gene and N gene were indicated (right panel). (B) RD cells were infected HCoV-OC43, and RNA was collected from the infected cell and media. The RNA level of RdRp gene, M gene and N gene was evaluated by quantitative RT-PCR. The experiments were performed in triplicate, and the graph shows mean and standard error. (C) RD cells were infected HCoV-229E, and RNA was collected from the infected cell and media. The RNA level of RdRp gene, M gene and N gene was evaluated by quantitative RT-PCR. Control vs EGCG treatment: ∗p < 0.05, ∗∗p < 0.005, NS not significant.
3.4. EGCG reduces the expression of coronavirus protein
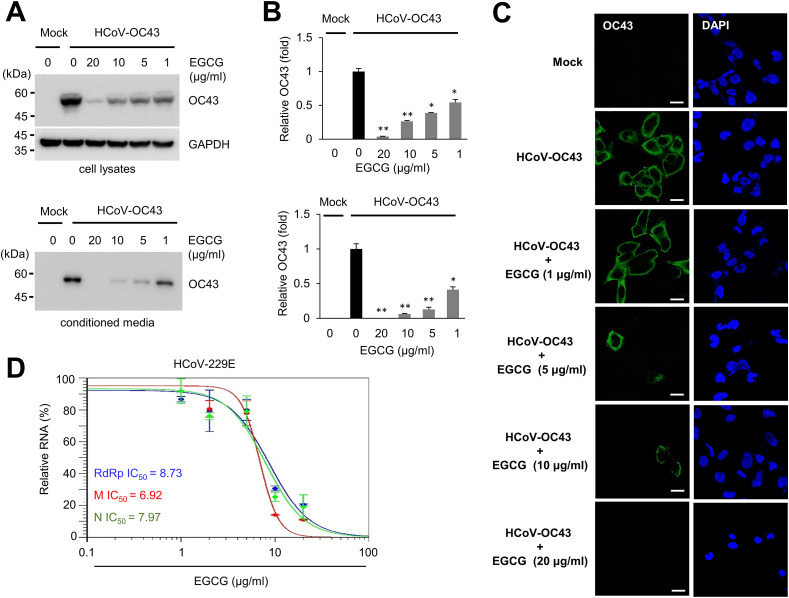
In order to confirm the reduced production of coronavirus by EGCG treatment, we examined the expression level of coronavirus proteins using anti-OC43 antibody. RD cells were treated with mock or EGCG and the cells were infected with HCoV-OC43. The infected cell and the conditioned media were used to evaluate coronavirus protein expression. EGCG treatment dramatically decreased the level of HCoV-OC43 proteins in the infected cells in a dose-dependent manner (Fig. 4 A and B). Reduction of OC43 protein is even more dramatic in the conditioned media. When the EGCG concentration is 20 μM, the expression of HCoV-OC43 proteins was not detected (Fig. 4A and B). These results indicate that EGCG treatment decreased the expression of coronavirus proteins.
Fig. 4.
EGCG inhibits coronavirus replication in a dose dependent manner. (A) EGCG treatment decreases the expression of HCoV-OC43 protein. RD cells were incubated with the indicated concentration of EGCG, and infected with HCoV-OC43 (10−4 dilution of conditioned media). Two days after infection, cell lysates (upper panel) and conditioned media (lower panel) were probed with anti-OC43 antibody. (B) The level of OC43 bands was quantified by Western blot. The experiments were performed in triplicate, and the graph shows mean and standard deviation. Control vs EGCG treatment: ∗p < 0.005, ∗∗p < 0.0005. (C) RD cells were infected with HCoV-OC43 (10−4 dilution of conditioned media) in the presence or absence of EGCG, and cells were immunostained with anti-OC43 antibody. Bars, 10 μM. (D) RD cells were infected with HCoV-229E (10−3 dilution of conditioned media), and the RNA levels of RdRp, M and N gene in the conditioned media were evaluated by quantitative RT-PCR.
Next, we examined the expression and localization of HCoV-OC43 protein in the infected cells. Immunofluorescent staining with anti-HCoV-OC43 antibody indicate that HCoV-OC43 protein is mainly localized around the plasma membrane (Fig. 4C). EGCG treatment decreased the number of positively stained cells and also decreased the intensity (Fig. 4C). We also examined the effect of EGCG treatment on HCoV-229E replication. Quantitative RT-PCR results showed that EGCG reduced the levels of HCoV-229E RNA from the conditioned media in a dose-dependent manner (Fig. 4D). We examined the quantity of RdRp, M and N RNA, and IC50 was 6–9 μg/ml in RdRp, M and N genes (Fig. 4D). These results indicate that EGCG treatment inhibits the coronavirus replication in a dose-dependent manner.
4. Discussion
Previously, we demonstrated that EGCG inhibits 3CL-protease activity of SARS-CoV-2 [7]. Because 3CL-protease is an essential enzyme for coronavirus, we expected that EGCG also inhibits coronavirus replication. We examined the effect of EGCG using low pathogenic human coronavirus HCoV-229E, and HCoV-OC43 as a proxy for SARS-CoV-2. EGCG treatment decreases viral RNA and viral protein production in the media indicating EGCG interferes with coronavirus replication. These results support that EGCG is a potentially useful mean for coronavirus diseases.
Our data suggests that EGCG protects host cells. EGCG treatment enhances the viability of infected cells and decreases plaque formation. In addition, EGCG decreases viral protein expression and viral RNA copy number. Since EGCG inhibits 3CL-protease, an essential enzyme for coronavirus, protein processing by enzyme cleavage is interfered, and other proteins will be inactive due to incomplete cleavage. EGCG is the major component of green tea and the safety of green tea has been proven. Therefore, we believe that the impacts of EGCG or green tea on the efficacy on coronavirus diseases should be studied further.
During our study, we found that EGCG treatment increases the copy number of coronavirus RNA in the infected cells (Fig. 3). However, the amount of coronavirus in the conditioned media was significantly lower, thus we speculate that the coronavirus RNA genome accumulates in the infected cells due to inefficient virus release from host cells by EGCG. However, it is clear that the elevated level of coronavirus RNA does not result in the increased expression of coronavirus protein (Fig. 4).
We also found that although the IC50 of EGCG on 3CL-protease is 11–15 μg/ml, we found that EGCG inhibits coronavirus replication even more efficiently (Fig. 4). We speculate that the coronavirus genome contains multiple cleavage sites for the 3CL-protease, and partial cleavage may also interfere with coronavirus replication [17]. In addition, we showed that the IC50 of EGCG on SARS-CoV-2 is lower than HCoV-OC43 and HCoV-229E. Therefore we speculate that EGCG or green tea should be more effective to treat SARS-CoV-2 [7]. Moreover, development of more potent derivatives of EGCG will be desirable to enhance the efficacy of EGCG. In addition, further study should be performed to examine the effect of EGCG on coronavirus disease in vivo.
Declaration of competing of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (2019R1A2C1006511).
References
- 1.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jomah S., Asdaq S.M.B., Al-Yamani M.J. Clinical efficacy of antivirals against novel coronavirus (COVID-19): a review. J Infect Public Health. 2020;13:1187–1195. doi: 10.1016/j.jiph.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham H.N. Green tea composition, consumption, and polyphenol chemistry. Prev. Med. 1992;21:334–350. doi: 10.1016/0091-7435(92)90041-f. [DOI] [PubMed] [Google Scholar]
- 4.Singh B.N., Shankar S., Srivastava R.K. Green tea catechin, epigallocatechin-3-gallate (EGCG): mechanisms, perspectives and clinical applications. Biochem. Pharmacol. 2011;82:1807–1821. doi: 10.1016/j.bcp.2011.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 6.Herold J., Gorbalenya A.E., Thiel V., Schelle B., Siddell S.G. Proteolytic processing at the amino terminus of human coronavirus 229E gene 1-encoded polyproteins: identification of a papain-like proteinase and its substrate. J. Virol. 1998;72:910–918. doi: 10.1128/jvi.72.2.910-918.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jang M., Park Y.I., Cha Y.E., Park R., Namkoong S., Lee J.I., Park J. Tea polyphenols EGCG and theaflavin inhibit the activity of SARS-CoV-2 3CL-protease in vitro. Evid Based Complement Alternat Med. 2020;2020 doi: 10.1155/2020/5630838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors - an in silico docking and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1779818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sagaama A., Brandan S.A., Ben Issa T., Issaoui N. Searching potential antiviral candidates for the treatment of the 2019 novel coronavirus based on DFT calculations and molecular docking. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e04640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mhatre S., Naik S., Patravale V. A molecular docking study of EGCG and theaflavin digallate with the druggable targets of SARS-CoV-2. Comput. Biol. Med. 2020;129:104137. doi: 10.1016/j.compbiomed.2020.104137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Storozhuk M. medRxiv; 2020. COVID -19: Could Green Tea Catechins Reduce the Risks? [Google Scholar]
- 13.Velavan T.P., Meyer C.G. The COVID-19 epidemic. Trop. Med. Int. Health. 2020;25:278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milewska A., Chi Y., Szczepanski A., Barreto-Duran E., Dabrowska A., Botwina P., Obloza M., Liu K., Liu D., Guo X., Ge Y., Li J., Cui L., Ochman M., Urlik M., Rodziewicz-Motowidlo S., Zhu F., Szczubialka K., Nowakowska M., Pyrc K. HTCC as a polymeric inhibitor of SARS-CoV-2 and MERS-CoV. J. Virol. 2021;95(4) doi: 10.1128/JVI.01622-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzym. Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hui R.K., Zeng F., Chan C.M., Yuen K.Y., Peiris J.S., Leung F.C. Reverse transcriptase PCR diagnostic assay for the coronavirus associated with severe acute respiratory syndrome. J. Clin. Microbiol. 2004;42:1994–1999. doi: 10.1128/JCM.42.5.1994-1999.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo C.J., Liang P.H. Characterization and inhibition of the main protease of severe acute respiratory syndrome coronavirus. Chembioeng Rev. 2015;2:118–132. [Google Scholar]