Abstract
Coronavirus disease (Covid-19) has been spreading all over the world and its diagnosis is attracting more research every moment. It is need of the hour to develop automated methods, which could detect this disease at its early stage, in a non-invasive way and within lesser time. Currently, medical specialists are analyzing Computed Tomography (CT), X-Ray, and Ultrasound (US) images or conducting Polymerase Chain Reaction (PCR) for its confirmation on manual basis. In Pakistan, CT scanners are available in most hospitals at district level, while X-Ray machines are available in all tehsil (large urban towns) level hospitals. Being widely used imaging modalities to analyze chest related diseases, produce large volume of medical data each moment clinical environments. Since automatic, time efficient and reliable methods for Covid-19 detection are required as alternate methods, therefore an automatic method of Covid-19 detection using Convolutional Neural Networks (CNN) has been proposed. Three publically available and a locally developed dataset, obtained from Department of Radiology (Diagnostics), Bahawal Victoria Hospital, Bahawalpur (BVHB), Pakistan have been used. The proposed method achieved on average accuracy (96.68 %), specificity (95.65 %), and sensitivity (96.24 %). Proposed model is trained on a large dataset and is being used at the Radiology Department, (BVHB), Pakistan.
Keywords: Covid-19 detection, Chest radiology images, Machine learning
1. Introduction
The novel coronavirus (COVID-19) is a new virus that has not been previously identified or diagnosed in human beings. Severe Acute Respiratory Syndrome (SARS) and Middle Eastern Respiratory Syndrome (MERS) are also caused by coronavirus [1]. Coronaviruses are a large family of viruses [1], transferred to humans from animals. This outbreak started in December 2019, from Wuhan, China, and has spread over the entire globe. Now it has affected almost all countries of the world. It has since been declared as major outbreak by World Health Organization (WHO). According to recent reports of WHO, there are almost 108,315,631 confirmed cases of coronavirus reported globally. As a result of this pandemic, almost 2,379,264 patients died across the world due to coronavirus. To diagnose Covid-19 disease, Polymerase Chain Reaction (PCR) test is mostly being used. Although, PCR is a gold standard method, it also has some key limitations. It is too sensitive and any sort of sample contamination, even DNA trace amounts can generate false result. In order to design its primers, prior sequence information is also needed. Resultantly, it can identify the presence or absence of known genes or pathogens [2]. The concept of Computer Aided Diagnostics (CAD) has clinically proved itself a trustworthy tool for assisting the medical practitioners. It is helping in almost every department of medical healthcare units, and its demand is increasing day by day due to its time efficiency, non-invasiveness, accuracy, and ease of use.
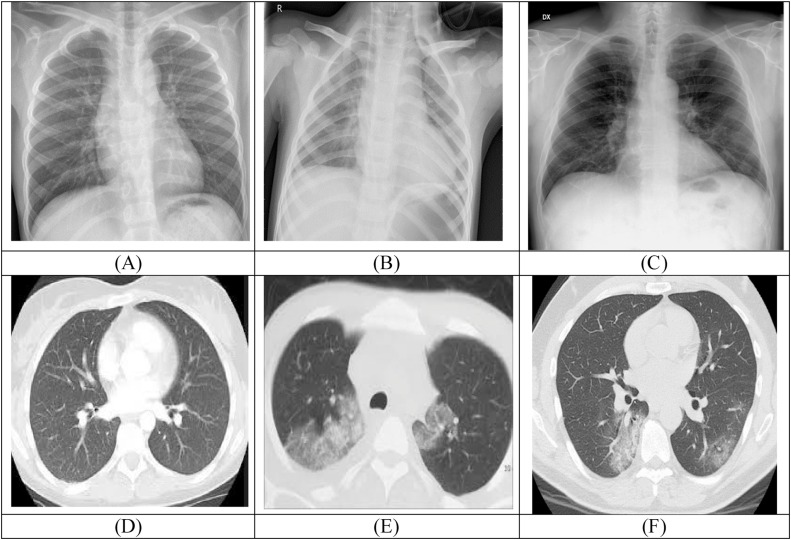
In this study, we focused on the global problem of coronavirus disease detection from chest radiology images. Early investments in characterizing this viral infection can handsomely help in improving the epidemic response, in terms of quick & continuous monitoring, and evaluation of the subjects. Computer Tomography (CT), X-Ray and Ultrasound (US) techniques are being applied to scan the patient to determine the presence and severity of Covid-19 [3,4]. Therefore, in the proposed research activity, normal, pneumonia, and Covid-19 chest images have been used and shown in Fig. 1 .
Fig. 1.
Chest radiography images (A) Normal (X-Ray), (B) Pneumonia (X-Ray), (C) infected with Covid-19 (X-Ray), (D) Normal (CT), (E) Pneumonia (CT), (F) infected with Covid-19 (CT).
In a research [5], the authors introduced deep convolutional neural network based model for the detection of Covid-19 from chest radiography images. In their proposed model, human machine collaborative design strategy has been adopted, where human driven network design prototyping is combined with machine driven design exploration to produce a network architecture tailored for the identification of Covid-19, yielding 83.5 % accuracy.
In another study [6], the authors detected Covid-19 using 150 CT images. Different feature extraction techniques, including grey level co-occurrence matrix, local directional patterns, grey level run length matrix, grey level size zone matrix, and discrete wavelet transform have been applied to extract features. During this process, they used 2-fold, 5-fold, and 10-fold cross validation. After feature extraction, they used SVM as a classifier. Their reported method obtained 99.68 % classification accuracy.
In a study [7], authors used a neural network based model (COVNet) for Covid-19 detection using CT images and they also classified pneumonia and other lung related diseases. This model is able to extract 2D local and 3D global features. The COVNet is based on RestNet50, which takes a series of CT images as input and generates features for those images and then all extracted features are combined by max poling operation. Finally, features are mapped to connected layers and SoftMax activation function, which distinguishes between Covid-19, pneumonia and other lung diseases. The dataset includes 4536 3D volumetric CT images and the results achieved were, sensitivity as 90 % and specificity as 96 %.
In another study [8], authors proposed a deep learning based models for detection and quantification of Covid-19. The proposed model comprises of CT images analyses at two levels, i.e., 3D images used for the analysis of nodules and focal opacities and 2D images used to localized and detected large size opacities clinically representative of Covid-19. In 3D images, they used off-the-shelf software that detected nodules, small opacities and also detected lung pathology and provide quantitative measurements. In 2D images, firstly region of interest has been extracted using lung segmentation module. In next step, they used RestNet50, 2D deep CNN to detected Covid-19. They used a total of 270 slices, 150 normal and 120 Covid-19 suspected and achieved 98.2 % sensitivity and 92.2 % specificity. In a research [9], the authors proposed a model to automatically detect Covid-19 using deep convolutional neural network from chest X-Ray images. They used pre-trained models, i.e., ResNet50, InceptionV3, and Inception-ResNetV2 but these models provided high detection accuracy for small datasets only. Their dataset was of 100 images comprising of 50 images of confirm Covid-19 patients, and the rest of 50 images were of normal patients. They concluded that ResNet50 achieved highest accuracy as 98 % on 5-fold cross validation.
In the study [10], the authors have been proposed a CNN based model namely CoroNet, which uses Xception architecture for automatically detection of Covid-19 from chest X-Ray images. The proposed model classifies three classes, i.e., bacterial pneumonia, viral pneumonia, and Covid-19. Further, it identifies how Covid-19 is different from other pneumonia. They implemented their proposed model for two and three class classification on two publically available databases. The model is trained and tested on a small dataset, which contains normal (310), bacterial pneumonia (330), viral pneumonia (327), and Covid-19 (284) images. The overall accuracy of their proposed model is 89.6 %. Another method of Covid-19 detection from chest X-Ray images has been proposed in a research [11], in which Covid-19, normal and pneumonia have been classified. DarkCovidNet, a deep learning model is used with 17 convolutional layers. They used a total of 1125 images out of that 125 images of Covid-19, 500 images of normal and 500 pneumonia images for experiments. Accuracy of their reported model for binary class classification is 98.08 %, while 87.02 % for multiclass classification. However, their dataset is also, which is unable to deal with multiformility and variability present in chest X-Ray images.
Above literature reveals that most of the work has been performed either on CT images or X-Ray images. Moreover, they trained their models on small datasets. Considering this motivation, CNNs were employed for Covid-19 detection using both CT and X-Ray images for the proposed study. The main contribution of this study is as follows:
-
1.
Considering the mentioned limitations of PCR method, CNN architectures were studied to address this problem and a model for Covid-19 detection was proposed which avoids tedious feature extraction process and learns features from images automatically.
-
2.
The proposed model achieved reasonable accuracy and hence is suitable for the detection of Convid-19 at early stage.
2. Material and methods
Nowadays, majority of researchers are using machine learning methods for diagnosis. Most of them use traditional approaches for detection, classification and grading different abnormalities, which include feature selection, extraction, reduction, and classification through these features. The main issue with these methods is the time consumption for feature engineering. Further to this, these traditional methods have low performance measures. To cope with such issues, deep learning architectures were explored. The potential of deep features motivated us towards the investigation of CNNs architectures.
2.1. Dataset
Chest images have been obtained from three publicly available datasets and a locally developed dataset from the Department of Radiology (Diagnostics), Bahawal Victoria Hospital, Bahawalpur (BVHB), Pakistan. It is radiologically documented that there is difference in medical traits of locals [12], so, this serves the reason, why a locally developed dataset has been used? The detail of these datasets have been shown in Table 1 . The datasets under experiment consist of 7021 images of both normal, and pneumonia, while 1066 images of Covid-19 infection.
Table 1.
Datasets used for experiments.
2.2. Preprocessing
It is radiologically documented that the intensity distribution of a tissue type varies even if image of the same subject is acquired using same scanner in different time frames. Therefore, to have the intensity ranges and contrast similar across acquisitions and subjects, intensity normalization proposed by [16] has been applied on each image. Resultantly, histogram of each image is similar across subjects.
2.3. The proposed CNN architecture
In this research, in order to classify images into normal, pneumonia, and Covid-19, CNN has been used, because CNN is a state-of-the-art area of machine learning inspired by human brain. CNN works like a human visual system and is designed based upon the assumption that raw data consists of two dimensional images, which enables certain properties to be encoded. So, CNN has been used, which works by convolving images with kernels to get feature maps. In a feature map, units are connected to previous layers through kernel weights and these weights are tweaked during training through a backpropagation process. Because same kernels have been used by all units, so, fewer weights have been trained by convolutional layer. Following are the apartments used with CNN to achieve the target of chest image classification.
1) Activation Function: This function has been used to transform the data into non-linear form. The activation function used is rectifier linear unit(ReLU), and is represented by Eq. (1).
| (1) |
2) Pooling: This layer has been used to combine spatially neighbor features present in feature maps. Average-pooling or max-pooling is used to join features, however, in this work, max-pooling has been used.
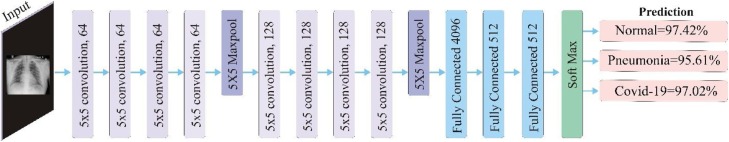
3) Architecture: Since, the visual clues of pneumonia have much similarity with the visual clues of Covid-19, which makes classification a challenging task in such case. This complexity has been reduced by tuning the proposed CNN based model on intensity normalization transformation of each image. Invariance was achieved and irrelevant detail was eliminated using pooling. However, pooling has also eliminated some important details, therefore, overlapped pooling with 3 × 3 receptive fields and 2 × 2 stride has been applied to keep information of a location. In convolutional layers, represented by the Eq. (2), feature maps have been padded before convolution. Use of padding ensured feature maps of same dimensions. The proposed CNN architecture has been depicted in Fig. 2 .
| (2) |
where I is an 2D array containing segmented brain tumor and K is a kernel convolution function.
Fig. 2.
The proposed CNN based architecture.
The details of the proposed architecture for normal, pneumonia, and Covid-19 classification has been presented in Table 2 .
Table 2.
Architecture of the CNN deigned for normal, pneumonia, and Covid-19 image classification.
| Layer # | Type of Layer | Filter size | Stride | No. of filters | FC units | Input |
|---|---|---|---|---|---|---|
| Layer-1 | Convolution Layer | 5 × 5 | 2 × 2 | 64 | – | 4 × 512 × 512 |
| Layer-2 | Convolution Layer | 5 × 5 | 2 × 2 | 64 | – | 64 × 512 × 512 |
| Layer-3 | Convolution Layer | 5 × 5 | 2 × 2 | 64 | – | 64 × 512 × 512 |
| Layer-4 | Convolution Layer | 5 × 5 | 2 × 2 | 64 | – | 64 × 512 × 512 |
| Layer-5 | Max Pooling | 5 × 5 | 3 × 2 | – | – | 64 × 512 × 512 |
| Layer-6 | Convolution Layer | 5 × 5 | 2 × 2 | 128 | – | 64 × 256 × 256 |
| Layer-7 | Convolution Layer | 5 × 5 | 2 × 2 | 128 | – | 128 × 256 × 256 |
| Layer-8 | Convolution Layer | 5 × 5 | 2 × 2 | 128 | – | 128 × 256 × 256 |
| Layer-9 | Convolution Layer | 5 × 5 | 2 × 2 | 128 | – | 128 × 256 × 256 |
| Layer-10 | Max Pooling | 5 × 5 | 3 × 2 | – | – | 128 × 256 × 256 |
| Layer-11 | Fully Connected | – | – | – | 512 | 4096 |
| Layer-12 | Fully Connected | – | – | – | 512 | 512 |
| Layer-13 | Fully Connected | – | – | – | 512 | 512 |
| Layer-14 | Fully Connected | – | – | – | 4 | 512 |
Hyper-parameters of the proposed architecture and their values have been listed in Table 3 and were tuned empirically. There are three classes (normal, pneumonia and covid-19) to classify. The CNNs were developed using MATLAB 2018b.
Table 3.
Hyper-parameters of the proposed CNN architecture.
| Stage | Hyper-parameter | Value |
|---|---|---|
| Initialization | bias | 0.1 |
| weights | Random | |
| Dropout | p | 0.3 |
| Training | Maximum epochs | 25 |
| v | 0.9 | |
| Initial ꞓ | 0.0002 | |
| Final ꞓ | 0.0002 | |
| Batch | 128 |
3. Results
Classification of images containing normal, Pneumonia, and Covid-19 required several steps from preprocessing to recognition. Dataset has been divided into 60 %, 20 %, and 20 % splits for training, cross validation, and testing sets, respectively. The results obtained with the proposed method have also been compared with state-of-the-art methods.
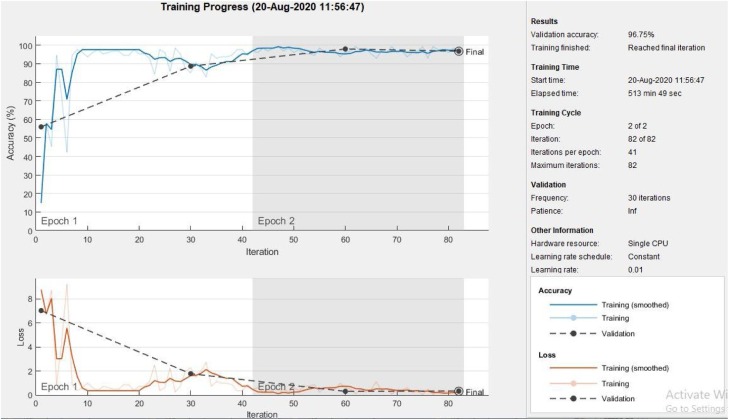
As shown in the confusion matrix represented as Table 4 , the accuracy achieved for normal is 97.42 %. Similarly, the accuracies for Pneumonia, and Covid-19 are 95.61 %, and 97.02 %, respectively. Overall, average accuracy achieved through the proposed model is 96.68 %. Performance plot representing accuracy and loss is shown in Fig. 3 .
Table 4.
Confusion matrix representing accuracy obtained through the proposed method.
| – | Normal | Pneumonia (%) | Covid-19 (%) |
|---|---|---|---|
| Normal | 97.42 | 1.68 | 0.90 |
| Pneumonia | 1.07 | 95.61 | 3.32 |
| Covid-19 | 0.91 | 2.07 | 97.02 |
| Overall accuracy of the proposed system | 96.68 % | ||
Fig. 3.
Performance plot showing accuracy and loss of the proposed model.
4. Discussion
Comparison of the results obtained through the proposed method with the results obtained from the state-of-the-art methods has been presented in Table 5 . The research activity [5] has classified the images into normal, bacterial pneumonia, non Covid-19 viral pneumonia and Covid-19 images. Their proposed model has attained the accuracy of 92.4 %, which is low. In another study [8] Covid-19 and non-Covid-19 images of Chinese and USA patients have been classified. Similarly, the study [6] has also classified Covid-19 and non-Covid-19 images with overall achieved accuracy of 99.68 %, which is the highest reported so far. In another study [9], normal and Covid-19 images have been classified, with reported highest accuracy as 98 % (using RestNet50 pre-trained model). However, the last three studies (above) have conducted their experiments on considerably lesser number of images, which possibly could not have enough variability and multiformity. Another study [7] classified Covid-19, non-pneumonia images with overall accuracy of 96 %, but their dataset was small. Similarly, the study [10], consisting of small dataset, i.e., normal (310), bacterial pneumonia (330), viral pneumonia (327), and Covid-19 (284) achieved overall accuracy of 89.6 %, which is lower than other so far conducted studies. In another study [11], X-Ray images have been used for Covid-19, pneumonia, and normal images classification with two class accuracy = 98.08 % and three class accuracy = 87.02 % on very small dataset, i.e., Covid-19 (127), pneumonia (500), and normal (500). Hence, this study may also suffer with variability and multiformity issues.
Table 5.
Comparison with state-of-the-art methods.
| Sr. No. | Method | Dataset | Evaluation measures |
|---|---|---|---|
| 1. | COVID-NET [5] | X-Ray Images | Accuracy = 92.4 % |
| Covid-19 Images = 68 | |||
| Normal Images = 1203 | |||
| Bacterial Pneumonia Images = 931 | |||
| Non-Covid-19 viral Pneumonia Images = 660 | |||
| 2. | Deep Convolutional Neural Network + Resnet-50 [8] | CT Images | AUC = 0.99 % |
| Covid-19 Patients = 56 | Sensitivity = 98.2 % | ||
| Non-Covid-19 Chinese Patients = 51 | Specificity = 92.2 % | ||
| Non-Covid-19 USA Patients = 49 | |||
| 3. | SVM, GLCM | CT Images | Accuracy = 99.68 % |
| LDP, GLRLM, | Covid-19 Images = 53 | ||
| GLSZM | Non-Covid-19 Images = 97 | ||
| Discrete Wavelet Transform [6] | |||
| 4. | Convolutional Neural Network based models (ResNet50, InceptionV3, and Inception-ResNetV2) [9] | X-Ray images | ResNet50 |
| Covid-19 Images = 50 | Accuracy = 98 % | ||
| Normal Images = 50 | Specificity = 100 % | ||
| InceptionV3 | |||
| Accuracy = 97 % | |||
| Specificity = 100 % | |||
| Inception-ResNetV2 | |||
| Accuracy = 87 % | |||
| Specificity = 90 % | |||
| 5. | COVNet (RestNet50) [7] | CT Images | AUC = 0.96 |
| Covid-19 Images = 1296 | Sensitivity = 90 % | ||
| Community Acquired Pneumonia Images = 1735 | Specificity = 96 % | ||
| Non-Pneumonia Images = 1325 | |||
| 6. | CNN CoroNet Xception architecture based model [10] | X-Ray images | Accuracy = 89.6 % |
| Normal = 310 | |||
| Bacterial pneumonia = 330 | |||
| Viral pneumonia = 327 | |||
| Covid-19 = 284 | |||
| 7. | DarkCovidNet [11] | X-Ray images | Binary class |
| Covid-19 = 127 | accuracy = 98.08 % | ||
| Pneumonia CXR = 500 | Multiclass accuracy = 87.02 % | ||
| Normal CXR = 500 | |||
| 8. | The Proposed Method | CT and X-Ray Images | |
| Normal Images = 7021 | Accuracy = 96.68 % | ||
| Pneumonia Images = 7021 | Sensitivity = 96.24 % | ||
| Covid-19 Images = 1066 | Specificity = 95.65 % |
Last row of Table 5 shows no. images for each of normal, pneumonia, and Covid-19 images used for experiments, and the accuracies obtained from the proposed system. The proposed model has achieved 96.68 %, 96.24 %, and 95.65 %, accuracy, sensitivity and specificity respectively. The proposed model has been trained and tested on a large number of images for each category as compared to the existing state-of-the-art methods. The average accuracy of our proposed model is 96.68 %.
5. Conclusion
The proposed model has been trained on a large dataset consisting of both CT and X-Ray images. Three publicly available and one locally developed dataset has been used for research and experiments. Locally developed dataset has been used to enable the proposed model to deal with medical traits of locals. The, Printed Circuit Board (PCB) is also designed as a protocol to integrated with exiting X-Ray and CT scanners. Currently, the proposed model is helping in the Radiology Department, Bahawal Victoria Hospital, Bahawalpur, Pakistan in decision-making.
CRediT authorship contribution statement
Ghulam Gilanie: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Writing - original draft, Visualization, Project administration. Usama Ijaz Bajwa: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - review & editing, Supervision. Mustansar Mahmood Waraich: Conceptualization, Validation, Investigation, Resources, Supervision. Mutyyba Asghar: Software, Data curation, Visualization. Rehana Kousar: Software, Data curation, Visualization. Adnan Kashif: Software, Data curation, Visualization. Rabab Shereen Aslam: Software, Data curation. Muhammad Mohsin Qasim: Software, Data curation. Hamza Rafique: Software, Data curation.
Declaration of Competing Interest
The authors report no declarations of interest.
References
- 1.Health, N.I.o. COVID-19. 2020 [cited 2020 05 April 2020]; Available from: https://www.niaid.nih.gov/diseases-conditions/covid-19.
- 2.Garibyan L., Avashia N. Research techniques made simple: polymerase chain reaction (PCR) J. Invest. Dermatol. 2013;133(3):e6. doi: 10.1038/jid.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buonsenso D., et al. novel coronavirus disease-19 pnemoniae: a case report and potential applications during COVID-19 outbreak. Eur. Rev. Med. Pharmacol. Sci. 2020;24:2776–2780. doi: 10.26355/eurrev_202003_20549. [DOI] [PubMed] [Google Scholar]
- 4.Mullis K.B. The unusual origin of the polymerase chain reaction. Sci. Am. 1990;262(4):56–65. doi: 10.1038/scientificamerican0490-56. [DOI] [PubMed] [Google Scholar]
- 5.Wang L., Wong A. 2020. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest Radiography Images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barstugan M., Ozkaya U., Ozturk S. 2020. Coronavirus (COVID-19) Classification Using CT Images by Machine Learning Methods. arXiv preprint arXiv:2003.09424. [Google Scholar]
- 7.Li L., et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020:200905. [Google Scholar]
- 8.Gozes O., et al. 2020. Rapid Ai Development Cycle for the Coronavirus (covid-19) Pandemic: Initial Results for Automated Detection & Patient Monitoring Using Deep Learning Ct Image Analysis. arXiv preprint arXiv:2003.05037. [Google Scholar]
- 9.Narin A., Kaya C., Pamuk Z. 2020. Automatic Detection of Coronavirus Disease (COVID-19) Using X-ray Images and Deep Convolutional Neural Networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan A.I., Shah J.L., Bhat M.M. Coronet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozturk T., et al. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., et al. 2020. Abnormal Respiratory Patterns Classifier May Contribute to Large-scale Screening of People Infected With COVID-19 in an Accurate and Unobtrusive Manner. arXiv preprint arXiv:2002.05534. [Google Scholar]
- 13.Dao, J.P.C.a.P.M.a.L. COVID-19 image data collection. 2020 [cited 2020 20 March 2020]; Available from: https://github.com/ieee8023/covid-chestxray-dataset.
- 14.America, R.S.o.N. RSNA pneumonia detection challenge. 2019 [cited 2020 01 April 2020]; Available from: https://www.kaggle.com/c/rsnapneumonia-detection-challenge/data.
- 15.Radiopaedia.org. Radiopaedia. 2020 [cited 2020 04 April 2020]; Available from: https://radiopaedia.org/articles/covid-19-3?lang=gb#article-images.
- 16.Nyúl L.G., Udupa J.K., Zhang X. New variants of a method of MRI scale standardization. IEEE Trans. Med. Imaging. 2000;19(2):143–150. doi: 10.1109/42.836373. [DOI] [PubMed] [Google Scholar]