Abstract
Coronaviruses are a family of viruses that majorly cause respiratory disorders in humans. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a new strain of coronavirus that causes the coronavirus disease 2019 (COVID-19). WHO has identified COVID-19 as a pandemic as it has spread across the globe due to its highly contagious nature. For early diagnosis of COVID-19, the reverse transcription-polymerase chain reaction (RT-PCR) test is commonly done. However, it suffers from a high false-negative rate of up to 67% if the test is done during the first five days of exposure. As an alternative, research on the efficacy of deep learning techniques employed in the identification of COVID-19 disease using chest X-ray images is intensely pursued.
As pneumonia and COVID-19 exhibit similar/ overlapping symptoms and affect the human lungs, a distinction between the chest X-ray images of pneumonia patients and COVID-19 patients becomes challenging. In this work, we have modeled the COVID-19 classification problem as a multiclass classification problem involving three classes, namely COVID-19, pneumonia, and normal. We have proposed a novel classification framework which combines a set of handpicked features with those obtained from a deep convolutional neural network. The proposed framework comprises of three modules. In the first module, we exploit the strength of transfer learning using ResNet-50 for training the network on a set of preprocessed images and obtain a vector of 2048 features. In the second module, we construct a pool of frequency and texture based 252 handpicked features that are further reduced to a set of 64 features using PCA. Subsequently, these are passed to a feed forward neural network to obtain a set of 16 features. The third module concatenates the features obtained from first and second modules, and passes them to a dense layer followed by the softmax layer to yield the desired classification model. We have used chest X-ray images of COVID-19 patients from four independent publicly available repositories, in addition to images from the Mendeley and Kaggle Chest X-Ray Datasets for pneumonia and normal cases.
To establish the efficacy of the proposed model, 10-fold cross-validation is carried out. The model generated an overall classification accuracy of 0.974 0.02 and a sensitivity of 0.987 0.05, 0.963 0.05, and 0.973 0.04 at 95% confidence interval for COVID-19, normal, and pneumonia classes, respectively. To ensure the effectiveness of the proposed model, it was validated using an independent Chest X-ray cohort and an overall classification accuracy of 0.979 was achieved. Comparison of the proposed framework with state-of-the-art methods reveal that the proposed framework outperforms others in terms of accuracy and sensitivity. Since interpretability of results is crucial in the medical domain, the gradient-based localizations are captured using Gradient-weighted Class Activation Mapping (Grad-CAM). In summary, the results obtained are stable over independent cohorts and interpretable using Grad-CAM localizations that serve as clinical evidence.
Keywords: COVID-19, Machine Learning, Classification, Chest X-Rays, Deep Learning, Grad-CAM
1. Introduction
Soon after the onset of coronavirus disease 2019 (COVID-19) in late 2019 in Wuhan city of China, it started to spread globaly at an alarming rate [1]. Consequently, on March 11, 2020 WHO declared Covid-19 as a pandemic. The coronavirus disease causes severe respiratory problems in humans [2] and it has already accounted for more than 1.8 million deaths. The reverse transcription-polymerase chain reaction (RT-PCR) test is popularly used to detect the COVID-19 disease in humans. However, the test suffers from a high false negative rate, as high as 67%, if the test is done during early days of the onset of the disease. Moreover, a negative report of the test does not rule out COVID-19 even in case of patients showing strong symptoms [3]. Thus, the followup of COVID-19 patients becomes difficult.
Researchers have been finding alternative ways for identification of COVID-19 disease. Chest X-ray (CXR) images are popularly used to diagnose, evaluate, and monitor common respiratory and lung infections. Thus, recently, chest X-ray images have been used for early and improved detection of COVID-19 using machine learning techniques [4], [5]. Apart from medical diagnostics, machine learning techniques have found applications in diverse domains such as speech recognition, weather forecasting, and forecasting market trends [6], [7], [8], [9]. For example, Altan et al. [9] amalgamated long short-term memory (LSTM) neural network with empirical wavelet transform (EWT) decomposition using cuckoo search (CS) algorithm for forecasting the price of digital currency. Recently, Karasu et al. [8] applied support vector regression (SVR) for forecasting of crude oil prices in a multiobjective setting using particle swarm optimization (PSO). However, in this paper, we will focus on applying machine learning techniques for the detection of COVID-19. In order to automate the task of detecting various lung abnormalities, several researchers are applying deep learning techniques to identify the affected regions in a CXR image [10], [11], [12], [13]. Recently, the use of some well-known deep neural networks namely ImageNet (also known as ’AlexNet’) [14], VGGNet [15], GoogLeNet [16], ResNet [17], and their variations has been explored for identification of COVID-19 using CXR images [18], [19], [20], [21], [22], [23].
Several researchers have approached the COVID-19 detection problem as a binary classification problem to distinguish between COVID-19 and Non-COVID/Normal classes. Based on experimentatios with VGG16, VGG19, ResNet18, ResNet50, and ResNet101, Ismael and Şengür [24] used a pretrained ResNet50 model for extracting features from CXR images from a data set having 180 COVID-19 and 200 normal patients’ images. Their classification model achieved the best accuracy of 92.63% using support vector machine (SVM) with linear kernel. Similarly, Afshar et al. [25] proposed a capsule-network (CapsNet) based framework which exploited spatial information to overcome an inability of CNNs for recognizing the same object when it undergoes various transformations. They experimented with a dataset having 183 COVID-19 Chest X-ray images and achieved an initial accuracy of 95.7% and sensitivity of 90% which was improved to 98.3% and 98.6% respectively after pre-training the model with another dataset. However, the above works do not attempt to distinguish the pneumonia patients from COVID-19 and normal classes. As Pneumonia, a respiratory illness, shares several closely related symptoms with COVID-19 [26], it is important to distinguish COVID-19 cases from several forms of pneumonia cases.
In the multi-class classification domain, most of the researchers have incorporated Convolutional Neural Networks (CNNs) for the classification task to identify COVID-19 using CXR images. Bukhari et al. [27] used a ResNet-50 architecture [17] for assessment of COVID-19 using 278 CXR images that belonged to three classes, namely COVID-19 (89), normal (93), and pneumonia (96), and obtained an overall accuracy of 98.24% and F1-score equal to 0.98. Apostolopoulos et al. [22] experimented with different CNNs on two datasets, each comprising 224 CXR images of COVID-19. They obtained highest classification accuracy of 93.48% and 92.85% on one dataset using VGG19 and MobileNetv2 respectively, and 96.78% using MobileNet on another dataset. Similarly, Khan et al. [18] proposed a deep CNN architecture based on Xception architecture [28] and achieved an overall accuracy of 90.21% on a dataset that comprises 284 Covid-19 CXR images. Mahmud et al. [20] proposed a CNN-CovXNet in which variation in the dilation rate is introduced to identify significant features in CXR images and obtained an overall accuracy of 90.2%. Rajaraman et al. [29] proposed iteratively pruned deep learning ensemble that pruned the neurons to the maximum of 50% in each convolutional layer to obtain a accuracy of 99.01%. They experimented on a dataset that comprises 313 COVID-19 instances. Basu et al. [30] based their model on domain extension transfer learning (DETL). Using a pre-trained network on National Institutes of Health (NIH) CXR image dataset [10], they obtained an overall accuracy by fine-tuning 12 layer CNN. Wang et al. [23] proposed an evolving deep convolutional architecture, called COVID-Net, inpired by the design of FermiNets [31] – a generative-synthesis approach that leads to the evolution of efficient neural network designs that satisfy the specified optimization criteria. Using CXR images having 358 COVID-19 images, they achieved an accuracy of 93.39%. Aslan et al. [12] proposed two deep learning architectures which takes segmented lung area as an input and yielded better results with hybrid architecture that comprise pretrained Alexnet architecture followed by additional BiLSTM (Bidirectional Long Short-Term Memory) layer leveraged for identification of sequential and temporal properties. They experimented on dataset that comprise only 219 samples of COVID-19 and achieved an accuracy of 98.70%. Loey et al. [13] used generative adversarial nets [32] for dataset augmentation and experimented with AlexNet, GoogLeNet, and ResNet and reported maximum accuracy of 85.19% using AlexNet. Soares et al. [19] used VGG16 network [15] to obtain 97.30% accuracy on a set of for 375 images.
Significant efforts have been made to explore the applicability of handcrafted features for the detection of COVID-19 [4], [5], [33], [34]. Khuzani et al. [4] used multilayer neural networks (MNN) to classify COVID-19 positives from CXR images. Using FFT, textural analysis, DWT, GLCM, and GLDM methods, they obtained a pool of 252 features. From this feature pool, they extracted a set of 64 features using Principal Component Analysis (PCA) which was input to a multi-layer Neural network to attain an accuracy of 94.04%. In contrast, Altan et al. [33] used two-dimensional (2D) curvelet transformations to improve their model accuracy for classifying the CXR images into three classes, namely, COVID-19, normal, and viral pneumonia. The 2D curvelet transformation captures a high degree of directionality and parabolic scaling relationship to the Fourier coefficients obtained from CXR images. Finally, inverse 2D Fourier transform was applied to obtain the curvelet coefficients. Subsequently, the chaotic salp swarm algorithm (CSSA) was applied to minimize the number of features and maximize the classification accuracy. The model achieved an accuracy of 99.69% using EfficientNet-B0 model. Sergio et al. [34] proposed a classifier that used texture features of CXR images with feed-forward neural networks and convolutional neural networks and achieved the best accuracy accuracy of 96.83% using a former model on a set of 255 COVID-19 CXR images.
A deep learning based model’s performance depends on the amount of data used for training the model. However, most of the above cited works used a small number of COVID-19 images [12], [13], [18], [27], [30], [34]. Another important aspect, validation on an independent cohort to establish the efficacy of a model, is missing from several works [13], [18], [19], [20], [27], [34]. Again, interpretability of results based on localizations used to mark the affected lung regions, a crucial issue in the medical domain, has not been properly addressed [12], [13], [18], [33]. To address the above short comings, the proposed framework uses a balanced data set that comprise 520 COVID-19 patients’ CXR images and an equal number of normal and pneumonia CXR images. This results in an improved classification model. To establish the stability and performance of the proposed framework, 10-fold cross-validation is done. Again, an independent cohort having 471 CXR images with equal number of images for the three classes is used to validate the model. Finally, Grad-CAM based localizations are captured to serve as interpretable clinical evidence.
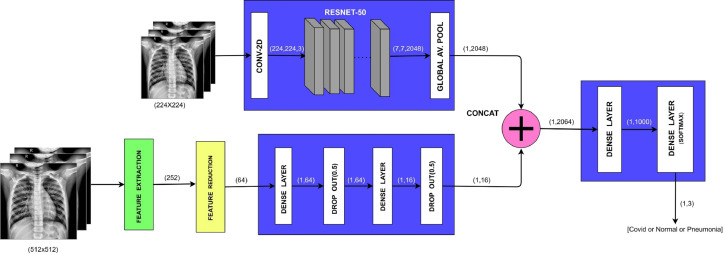
Fig. 1 presents the workflow of the proposed approach. As a part of preprocessing, a CXR image is transformed to gray scale, resized, and normalized. For an improved detection of COVID-19 subjects from datasets that comprise of COVID-19, normal, and pneumonia subjects, we combine a set of features obtained from ResNet-50 [17] with another set of handpicked features obtained from conventional image processing methods. The CXR datasets used in this work are collected from publicly available repositories [10], [21], [35], [36], [37], [38]. The proposed framework comprises of three modules. In the first module, we exploit the strength of transfer learning using ResNet-50 by training the network on a set of preprocessed images and thereby obtaining a vector of 2048 features. The second module exploits the texture and frequency domain features of the CXR images to yield a set of 252 handpicked features which is further reduced to a set of 64 features using Principal Component Analysis (PCA). Subsequently, the feature set is passed to a feed forward neural network to finally obtain a set of 16 features. The features obtained from first and second module are combined to serve as input to the dense layer of the third module resulting in a feature set of size 1000. Finally, these are fed to a softmax layer to yield the desired classification model. The entire model is trained end-to-end. The proposed model yeilds an overall accuracy of 0.974 0.02 at 95% confidence interval. Comparison of the proposed framework with state-of-the-art methods reveal that the proposed framework outperforms others in terms of accuracy and sensitivity. Also, the results obtained are validated using an independent cohort.
Fig. 1.
Workflow of the Proposed Approach.
To summarize, in this paper we have targeted the problem of improved detection of COVID-19 using CXR images and towards this end we have:
-
1.
proposed a three-module multi-class classification framework to distinguish between COVID-19, normal and pneumonia CXR images.
-
2.
combined a set of handpicked features obtained from conventional image processing methods with those obtained using the ResNet-50 based transfer learning.
-
3.
employed 10-fold cross validation on our data set having 1560 images, 520 images belonging to each of the three classes.
-
4.
validated the proposed framework on an independent chest X-ray cohort having 471 images having equal number of images belonging to each of the classes.
-
5.
applied Grad-CAM to obtain localizations that serve as interpretable clinical evidence.
The rest of the paper is outlined as follows: in section 2, we describe the datasets, preprocessing steps, and the architectural details, in section 3, we discuss the outcome of the experiments, and in section 4, we present the conclusions and mention the scope for future work.
2. Materials and methods
In this section, we describe our datasets, followed by the data preprocessing steps and a detailed description of the network architecture.
2.1. Dataset description
To begin with, we enumerate the class wise data distribution of some of the popular publicly available COVID-19 datasets:
-
1.
COVID-19 Radiography Database (Kaggle) [35]. It comprises 2905 samples, COVID-19: 219, normal: 1341, Viral pneumonia: 1345.
-
2.
COVID-19 Image Data Collection [21]. It comprises 760 samples, COVID-19: 538, ARDS: 14, Other Diseases: 222.
-
3.
Fig. 1-COVID-chestxray-dataset [36]. It comprises 53 COVID-19 samples.
-
4.
Actualmed-COVID-chestxray-dataset [37]. It comprises 150 COVID-19 samples.
In order to maintain uniformity among the chest X-ray images obtained from different sources, we only consider images having frontal views, namely, Poster anterior (PA) and Erect anteroposterior (AP) views. The first two databases mentioned above contain 520 COVID-19 images corresponding to PA and AP views. In order to construct a balanced data set for training the network, an equal number of pneumonia and normal chest X-ray images are included. We select an equal number of CXR images related to viral and bacterial pneumonia from Kaggle database [35] and Mendeley database [38]. Thus, our dataset comprises 260 images of each of the type bacterial pneumonia and viral pneumonia, randomly selected from Kaggle and Mendeley databases. Similarly, we randomly select 520 images for normal class from Kaggle and Mendeley databases. The final dataset having 1560 chest X-ray images is now randomized and used for 10-fold cross-validation.
Further, an independent cohort is created using a new dataset that was never used during 10-fold cross-validation. This cohort comprises of 157 unique COVID-19 samples collected from the cohorts at serial numbers 3 and 4 mentioned above, and also has an equal number of normal and pneumonia images taken from the Mendeley database.
2.2. Preprocessing
Since the CXR images are collected from various sources, they are resized and transformed from RGB to grayscale to ensure uniformity across different datasets. The resultant images are then subjected to min-max normalization [39] to speed up the convergence process.
2.3. Architectural details
The proposed framework (Fig. 2) uses a set of handpicked image features in conjunction with the features obtained from ResNet-50 for improved detection of COVID-19 from chest X-ray images. In the first module, preprocessed chest X-ray images of size are passed as an input to the 2D convolutional layer which transforms images to for ResNet-50 model. The resultant images are passed to ResNet-50 [17] – pre-trained on ImageNet. The output of ResNet-50 is connected to a global average pooling (GAP) layer to generate a vector of 2048 features. During the training, the images are subjected to random projective transformations such as scaling, shearing, zooming, and horizontal flip to enhance the robustness and generalization capability of the network.
Fig. 2.
Architecture of the Proposed Network.
In the second module, we incorporate a feature extraction unit which yields 252 features obtained from CXR images rescaled to . This set of 252 features so obtained is processed using principal component analysis (PCA) to obtain a vector of 64 features. This feature vector of size 64 is further passed to a feed forward neural network having a pair of dense layer and a dropout layer, followed by another pair of dense layer and a dropout layer to finally obtain a vector of 16 features. Based on experimentations, we have employed the ReLU activation function in dense layers and dropout regularization factor of 0.50 so as to prevent the network from overfitting.
The feature extraction unit involves a process of extracting textual and frequency domain features (widely used in image processing works [40], [41], [42]). The texture feature set is generated using CXR image in spatial domain, gray-level co-occurrence matrix (GLCM) [43], [44] and gray-level difference matrix (GLDM) [4], [45]. For each category of texture features, the computation of 14 statistical values – area, mean, standard deviation, skewness, kurtosis, energy, entropy, max, min, mean absolute deviation, median, range, root mean square, and uniformity is carried out. In this manner, we extract 14 spatial domain features, 56 GLCM features, and 56 GLDM features generating a combined texture feature set of size 126.
Drawing inspiration from Zargari et al. [46] (used statistical features for predicting chemotherapy response in ovarian cancer patients), we also obtain frequency features by computing the aforementioned statistical values on the transformed region resulted by applying Fast Fourier Transform (FFT) [42] and two-level Discrete Wavelet Transform (DWT) [47]. These features are concatenated with the texture feature pool constructed above. We extract 14 FFT features and 112 DWT features. Thus, the texture and frequency features so obtained comprise of 252 features.
In the third module of the proposed network, the outputs from both the modules are concatenated to form a vector of features which are passed to a dense layer employing ReLU activation function to obtain a reduced set of 1000 features. These features serve as input to a softmax layer that assigns classification probabilities of the input image belonging to one of three classes, i.e., COVID-19, pneumonia, and normal.
3. Results and discussion
In this section, we present the classification results obtained using the proposed framework. We compare the performance of the proposed framework with other state-of-the-art methods. Further, we validate the performance of proposed framework on an independent cohort. For clinical interpretation of the results, the gradient-based localizations are captured using Gradient-weighted Class Activation Mapping (Grad-CAM). We carried out all the experiments using Python 3.6.9 on the NVIDIA Tesla K80 GPU in Google Colaboratory environment. For implementing the proposed framework, we have used NumPy, SciPy, Scikit-learn, Keras, and Scikit-image libraries.
3.1. Classification results
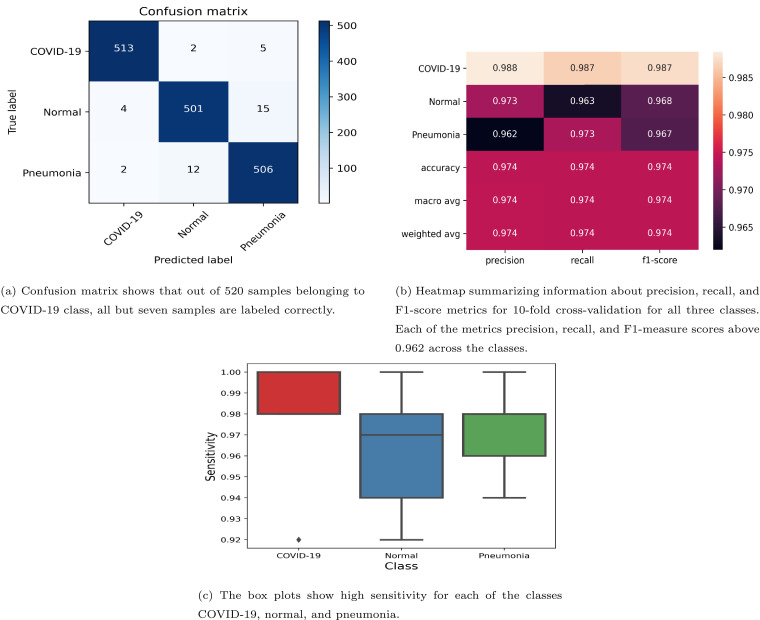
Using the proposed framework described in the previous section, we classify the chest X-ray images into three classes: normal, pneumonia, and COVID-19 by training the network end-to-end. To assess the effectiveness of the proposed model in terms of variance over different test data, we perform 10-fold cross-validation. To avoid overfitting during model construction, out of the 90% of the data set being used for training in a fold, we reserved 10% of this data as the hold-out validation set. The results of the 10-fold cross-validation are summarized in a confusion matrix (see Fig. 3(a)). The diagonal entries in the confusion matrix indicate the number of samples correctly classified for each class, while off-diagonal entries indicate the number of samples wrongly assigned to each class. We note that out of 520 COVID-19 patients, 513 are correctly identified, five are misclassified as pneumonia and two are labeled as normal. Similarly, pneumonia and normal subjects are also labeled by the proposed classifier with high accuracy. Thus, we obtain an overall accuracy of 0.974 0.02 and a high sensitivity of 0.987 0.05, 0.963 0.05, and 0.973 0.04 at 95% confidence interval for COVID-19, normal, and pneumonia classes, respectively. This is also evident from the heatmap in Fig. 3(b) which summarizes information about precision, recall, and F1-score metrics for 10-fold cross-validation for all three classes. Note that the proposed framework is able to label almost all COVID-19 patients correctly, thus achieving high average values of the precision, recall, and F1-score () across 10 different folds (heatmap in Fig. 3(b)). For the Normal subtype, the framework yields high values (greater than 0.963) of precision, recall, and F1-measure. Similarly, for the Pneumonia subtype, the framework yields precision, recall, and F1-measure scores greater than 0.962.
Fig. 3.
10-fold cross-validation classification results using proposed model.
The box plots in Fig. 3(c) show the robustness of the proposed framework in distinguishing the COVID-19 subjects from pneumonia and normal cases. Indeed, the sensitivity values for the COVID-19 class lie in the close range [0.98, 1.00]. Thus, the classification results attest the robustness of the proposed framework in distinguishing COVID-19 patients from others when the 10-fold cross-validation is carried out.
3.2. Comparison with other state-of-the-art works
To evaluate the relative effectiveness of the proposed framework, we compare the results obtained using the proposed framework with related work in literature. Table 1 shows the accuracy and sensitivity of the proposed framework along with other state-of-the-art classifiers in detecting COVID-19 from chest X-ray images. It may be noted that the proposed system performs better than the other state-of-the-art approaches in terms of accuracy and sensitivity. Although Bukharia et al. [27] report a slightly higher value of accuracy (0.98), they experimented on relatively small-sized datasets and did not report the sensitivity of their classifiers. Further, while majority of the related works have modeled the problem as binary classification problem (COVID-19 vs. Normal), we have included the closely related pneumonia class in our the multi-class classification model. Also, effect of variability in data sets has not been studied as most of these works do not report their cross-validation results. While, validation on an independent cohort is essential to establish the effectiveness of a model, somehow, this aspect has been missing in a majority of related works. Most importantly, interpretability of results in terms of localization is crucial in medical domain. However, we could not find such interpretations in several works.
Table 1.
Comparison of the proposed network with other state-of-the-art machine learning and deep learning algorithms, in terms of overall accuracy and sensitivity value to detect COVID-19.
| Research group | Dataset used | Technique | Overall accuracy (%) |
COVID-19 sensitivity (%) |
|---|---|---|---|---|
| Proposed Network | COVID-19: 520 normal: 520 pneumonia (Viral/Bacterial):520 |
ResNet50 + NN | 0.974 0.02 | 0.987 0.05 |
| Bukharia et al. [27] | COVID-19: 89 normal: 93 pneumonia (Viral/Bacterial):96 |
ResNet50 | 0.982 | —– |
| Apostolopoulos et al. [22] | COVID-19: 224 normal: 504 pneumonia (Viral/Bacterial): 700 |
VGGNet19 MobileNet v2 |
0.935 0.928 |
0.86 0.94 |
| Khan et al. [18] | COVID-19: 290 normal: 1203 pneumonia (Viral/Bacterial):1591 |
Xception | 0.902 | 0.89 |
| Khuzani et al. [4] | COVID-19: 140 normal: 140 pneumonia (Viral/Bacterial):140 |
Multilayer NN | 0.94 | 0.100 |
| Loey et al. [13] | COVID-19: 69 normal: 79 pneumonia (Viral/Bacterial):158 |
AlexNet GoogLeNet ResNet18 |
0.852 0.815 0.845 |
0.100 0.818 0.100 |
| Soares et al. [19] | COVID-19: 175 normal: 100 pneumonia (Viral/Bacterial): 100 |
VGG16 | 0.973 | —– |
| Wang et al. [10] | COVID-19: 358 normal: 8066 pneumonia (Viral/Bacterial): 5538 |
VGGNet-19 ResNet50 COVID-Net |
0.83 0.906 0.933 |
0.587 0.83 0.91 |
3.3. Gradient-based Localization using Grad-CAM
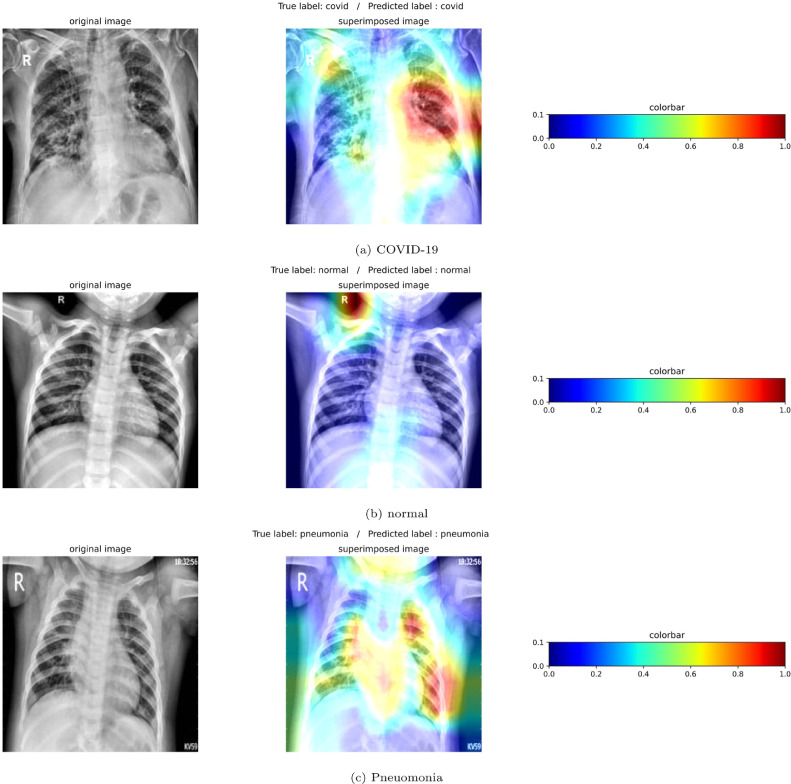
Although effectiveness of the proposed architecture in detecting COVID-19 from chest X-ray images is evident from the results tabulated in previous section, in order to be useful in clinical practice, it is necessary to relate the classification results to clinical evidence. For this purpose, we used Gradient-weighted class activation mapping (Grad-CAM) technique. Grad-CAM is a popular tool used to produce a localization map highlighting the most important regions which assist the proposed network in predicting a class. Using this tool, we obtained gradient-based localization (Fig. 4 ) for the images that we processed. The red-colored region marks the region of interest (RoI) responsible for activating the final convolutional layer of the proposed network. The blue-colored region acts as an evidence [48] of the class identified by the proposed network. Radiological validation was done by a radiologist, who confirmed that lung regions marked in red colour in the figure relate to those regions of the lung which are predominantly affected in COVID-19.
Fig. 4.
The gradient-based localization of COVID-19, normal, and pneumonia case using Grad-CAM.
3.4. Validation of proposed framework on an independent cohort
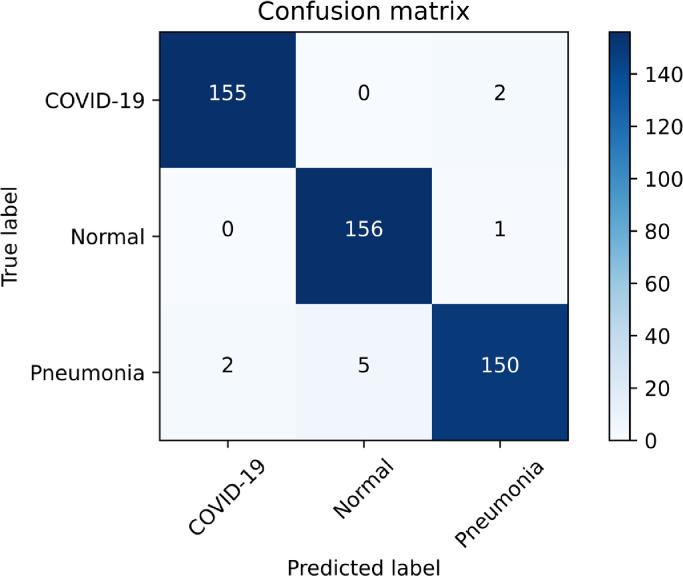
We also evaluate the effectiveness of the proposed framework using an independent cohort of the CXR images that is not used during 10-fold cross-validation. For this purpose, we use 157 unique CXR images from two COVID-19 datasets ([36] and [37]). The equal number of CXRs for pneumonia and normal are taken from Mendeley dataset [38]. Classification results are depicted through confusion matrix in Fig. 5 . The diagonal entries in the confusion matrix indicate the number of samples correctly classified for each class, while off-diagonal entries indicate the number of samples wrongly assigned to each class. As seen in the confusion matrix, out of 157 COVID-19 patients, only two cases of COVID-19 are misclassified (as pneumonia). In case of pneumonia, out of 157 subjects, 150 are correctly classified and seven are wrongly classified (two as COVID-19 and five as normal). Out of 157 normal subjects, only one is misclassified (as pneumonia). Thus, the proposed framework achieves an overall accuracy of 0.979 on the unseen data which validates the generalization capability of the model. Results obtained establishes the strength of proposed framework on an independent cohort.
Fig. 5.
Performance of the proposed framework on unseen CXR images gathered from [36],[37], and [38]. Note that only two cases of COVID-19 out of 157 patients are labelled wrongly as pneumonia and an equal number of pneumonia cases are labeled as COVID-19.
4. Conclusion
Given the low sensitivity of the RT-PCR test in detecting COVID-19, coupled with delayed availability of the results, there is an urgent need to devise alternative and more efficient methods for diagnosing COVID-19. Recently, chest X-ray images have been used for early and improved detection of COVID-19. In this work, we have examined the feasibility of using a set of handpicked features in conjunction with those obtained from ResNet-50. For this purpose, we proposed a framework having three modules. While the first module, comprising of ResNet-50 based transfer learning, yields 2048 features, the second module exploits frequency and texture domain features to identify 252 handpicked features, which are finally reduced to a set of 16 features using PCA and a feed-forward neural network. The concatenated feature set is passed to the dense layer followed by softmax layer of the third module for the classification task. The use of handpicked features in conjunction with those obtained via transfer learning using ResNet50 has enhanced the learning ability of the model. To establish the stability and performance of the proposed end-to-end model, 10-fold cross-validation is carried out.
Experiments on the dataset having 1560 chest X-ray images (including 520 images of each of the three classes: COVID-19, pneumonia, and normal) reveal that the proposed model (trained end-to-end) yields an overall accuracy of 0.974 0.02. For the 10-fold cross-validation, we obtain sensitivity of 0.987 0.05, 0.963 0.05, and 0.973 0.04 at 95% confidence interval for COVID-19, normal, and pneumonia classes, respectively. The results of the experiments demonstrate the efficacy of the proposed framework, and also establish that the hybrid approach of concatenating the handpicked features with the features obtained from the deep neural network enhance the detection of COVID-19. Further, validation of the framework on an independent Chest X-ray cohort results in an overall accuracy of 0.979. Comparison of the proposed framework with state-of-the-art methods reveals that the proposed framework outperforms others in terms of accuracy and sensitivity. Since interpretability of results is important in the medical domain, the use of Grad-CAM has enabled us to mark the lung regions affected in COVID-19, thereby generating useful clinical evidence.
As part of the future work, we propose to evaluate the effect of segmentation on localizing the lung regions affected in COVID-19. Further, the clinical data such as data regarding symptoms, age, and treatment history of patients may be integrated for improved detection of the COVID-19 disease.
CRediT authorship contribution statement
Sheetal Rajpal: Writing - original draft, Conceptualization, Methodology, Software. Navin Lakhyani: Visualization, Validation. Ayush Kumar Singh: Data curation, Software. Rishav Kohli: Data curation, Software. Naveen Kumar: Model Evaluation, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.WHO. Archived: WHO Timeline - COVID-19, 2020. https://www.who.int/news-room/detail/27-04-2020-who-timeline---covid-19.
- 2.Ng M.-Y., Lee E.Y., Yang J., Yang F., Li X., Wang H., et al. Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiology: Cardiothoracic Imaging. 2020;2(1):e200034. doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson J., Whiting P.F., Brush J.E. Interpreting a covid-19 test result. BMJ. 2020;369 doi: 10.1136/bmj.m1808. [DOI] [PubMed] [Google Scholar]
- 4.Khuzani A.Z., Heidari M., Shariati S.A. COVID-Classifier: An automated machine learning model to assist in the diagnosis of COVID-19 infection in chest x-ray images. medRxiv. 2020 doi: 10.1038/s41598-021-88807-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajpal S, Kumar N, Rajpal A. Cov-elm classifier: An extreme learning machine based identification of covid-19 using chest-ray images, 2020. arXiv preprint arXiv:200708637.
- 6.Deng L., Li X. Machine learning paradigms for speech recognition: An overview. IEEE Transactions on Audio, Speech, and Language Processing. 2013;21(5):1060–1089. [Google Scholar]
- 7.Holmstrom M., Liu D., Vo C. Machine learning applied to weather forecasting. Stanford University. 2016:2–4. [Google Scholar]
- 8.Karasu S., Altan A., Bekiros S., Ahmad W. A new forecasting model with wrapper-based feature selection approach using multi-objective optimization technique for chaotic crude oil time series. Energy. 2020;212:118750. [Google Scholar]
- 9.Altan A., Karasu S., Bekiros S. Digital currency forecasting with chaotic meta-heuristic bio-inspired signal processing techniques. Chaos, Solitons & Fractals. 2019;126:325–336. [Google Scholar]
- 10.Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; pp. 2097–2106. [Google Scholar]
- 11.Nanni L., Lumini A., Brahnam S. Local binary patterns variants as texture descriptors for medical image analysis. Artificial intelligence in medicine. 2010;49(2):117–125. doi: 10.1016/j.artmed.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Aslan M.F., Unlersen M.F., Sabanci K., Durdu A. CNN-based transfer learning-BiLSTM network: A novel approach for COVID-19 infection detection. Applied Soft Computing. 2020:106912. doi: 10.1016/j.asoc.2020.106912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loey M., Smarandache F., M Khalifa N.E. Within the Lack of Chest COVID-19 X-ray Dataset: A Novel Detection Model Based on GAN and Deep Transfer Learning. Symmetry. 2020;12(4):651. [Google Scholar]
- 14.Krizhevsky A., Sutskever I., Hinton G.E. Advances in neural information processing systems. 2012. Imagenet classification with deep convolutional neural networks; pp. 1097–1105. [Google Scholar]
- 15.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition, 2014. arXiv preprint arXiv:14091556.
- 16.Szegedy C., Liu W., Jia Y., Sermanet P., Reed S., Anguelov D., et al. Proceedings of the IEEE conference on computer vision and pattern recognition. 2015. Going deeper with convolutions; pp. 1–9. [Google Scholar]
- 17.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 18.Khan A.I., Shah J.L., Bhat M.M. Coronet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Computer Methods and Programs in Biomedicine. 2020:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soares LP, Soares CP. Automatic Detection of COVID-19 Cases on X-ray images Using Convolutional Neural Networks, 2020. arXiv preprint arXiv:200705494.
- 20.Mahmud T., Rahman M.A., Fattah S.A. CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Computers in Biology and Medicine. 2020:103869. doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M. COVID-19 Image Data Collection: Prospective Predictions Are the Future, 2020. arXiv preprint arXiv:200611988.
- 22.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Wong A. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-Ray Images, 2020. arXiv preprint arXiv:200309871. [DOI] [PMC free article] [PubMed]
- 24.Ismael A.M., Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Systems with Applications. 2020;164:114054. doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. Covid-caps: A capsule network-based framework for identification of covid-19 cases from x-ray images. Pattern Recognition Letters. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao D., Yao F., Wang L., Zheng L., Gao Y., Ye J., et al. A comparative study on the clinical features of covid-19 pneumonia to other pneumonias. Clinical Infectious Diseases. 2020 doi: 10.1093/cid/ciaa247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bukhari S.U.K., Bukhari S.S.K., Syed A., SHAH S.S.H. The diagnostic evaluation of Convolutional Neural Network (CNN) for the assessment of chest X-ray of patients infected with COVID-19. medRxiv. 2020 [Google Scholar]
- 28.Chollet F. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Xception: Deep learning with depthwise separable convolutions; pp. 1251–1258. [Google Scholar]
- 29.Rajaraman S, Siegelman J, Alderson PO, Folio LS, Folio LR, Antani SK. Iteratively Pruned Deep Learning Ensembles for COVID-19 Detection in Chest X-rays, 2020. arXiv preprint arXiv:200408379. [DOI] [PMC free article] [PubMed]
- 30.Basu S, Mitra S. Deep Learning for Screening COVID-19 using Chest X-Ray Images, 2020. arXiv preprint arXiv:200410507.
- 31.Wong A, Shafiee MJ, Chwyl B, Li F. Ferminets: Learning generative machines to generate efficient neural networks via generative synthesis, 2018. arXiv preprint arXiv:180905989.
- 32.Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., et al. Advances in neural information processing systems. 2014. Generative adversarial nets; pp. 2672–2680. [Google Scholar]
- 33.Altan A., Karasu S. Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm and deep learning technique. Chaos, Solitons & Fractals. 2020;140:110071. doi: 10.1016/j.chaos.2020.110071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varela-Santos S., Melin P. A new approach for classifying coronavirus COVID-19 based on its manifestation on chest X-rays using texture features and neural networks. Information sciences. 2020;545:403–414. doi: 10.1016/j.ins.2020.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahman T, Chowdhury M, Khandakar A. COVID-19 Radiography Database | Kaggle, ; 2020. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database.
- 36.Chung AG. GitHub - agchung/Figure1-COVID-chestxray-dataset: Figure 1 COVID-19 Chest X-ray Dataset Initiative, 2020a. https://github.com/agchung/Figure1-COVID-chestxray-dataset.
- 37.Chung AG. GitHub - agchung/Actualmed-COVID-chestxray-dataset: Actualmed COVID-19 Chest X-ray Dataset Initiative, 2020b. https://github.com/agchung/Actualmed-COVID-chestxray-dataset.
- 38.Kermany D., Zhang K., Goldbaum M. Large dataset of labeled optical coherence tomography (oct) and chest x-ray images. Mendeley Data, v3. 2018;3 [Google Scholar]; http://dx doi org/1017632/rscbjbr9sj.
- 39.Jain A., Nandakumar K., Ross A. Score normalization in multimodal biometric systems. Pattern recognition. 2005;38(12):2270–2285. [Google Scholar]
- 40.Haralick R.M., Shanmugam K., Dinstein I.H. Textural features for image classification. IEEE Transactions on systems, man, and cybernetics. 1973;6:610–621. [Google Scholar]
- 41.Pardeshi R., Patil R., Ansingkar N., Deshmukh P.D., Biradar S. Ambient Communications and Computer Systems. Springer; 2020. DWT-LBP Descriptors for Chest X-Ray View Classification; pp. 381–389. [Google Scholar]
- 42.Leibstein J.M., Nel A.L. Detecting tuberculosis in chest radiographs using image processing techniques. University of Johannesburg. 2006 [Google Scholar]
- 43.Zare M.R., Seng W.C., Mueen A. Automatic classification of medical x-ray images. Malaysian Journal of Computer Science. 2013;26(1):9–22. [Google Scholar]
- 44.MathWorks. Texture Analysis Using the Gray-Level Co-Occurrence Matrix (GLCM) - MATLAB & Simulink, 2020. https://www.mathworks.com/help/images/texture-analysis-using-the-gray-level-co-occurrence-matrix-glcm.html#::text=More-,Texture%20Analysis%20Using%20the%20Gray%2DLevel%20Co%2DOccurrence%20Matrix%20(,gray%2Dlevel%20spatial%20dependence%20matrix.
- 45.Kim J.K., Park H.W. Statistical textural features for detection of microcalcifications in digitized mammograms. IEEE transactions on medical imaging. 1999;18(3):231–238. doi: 10.1109/42.764896. [DOI] [PubMed] [Google Scholar]
- 46.Zargari A., Du Y., Heidari M., Thai T.C., Gunderson C.C., Moore K., et al. Prediction of chemotherapy response in ovarian cancer patients using a new clustered quantitative image marker. Physics in Medicine & Biology. 2018;63(15):155020. doi: 10.1088/1361-6560/aad3ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parveen N., Sathik M.M. Detection of pneumonia in chest X-ray images. Journal of X-ray Science and Technology. 2011;19(4):423–428. doi: 10.3233/XST-2011-0304. [DOI] [PubMed] [Google Scholar]
- 48.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE international conference on computer vision. 2017. Grad-cam: Visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]