Abstract
Local adaptation can drive diversification of closely related species across environmental gradients and promote convergence of distantly related taxa that experience similar conditions. We examined a potential case of adaptation to novel visual environments in a species flock (Great Lakes salmonids, genus Coregonus) using a new amplicon genotyping protocol on the Oxford Nanopore Flongle and MinION. We sequenced five visual opsin genes for individuals of Coregonus artedi, Coregonus hoyi, Coregonus kiyi, and Coregonus zenithicus. Comparisons revealed species-specific differences in a key spectral tuning amino acid in rhodopsin (Tyr261Phe substitution), suggesting local adaptation of C. kiyi to the blue-shifted depths of Lake Superior. Ancestral state reconstruction demonstrates that parallel evolution and “toggling” at this amino acid residue has occurred several times across the fish tree of life, resulting in identical changes to the visual systems of distantly related taxa across replicated environmental gradients. Our results suggest that ecological differences and local adaptation to distinct visual environments are strong drivers of both evolutionary parallelism and diversification.
Keywords: genomics, genotyping, nanopore, long-read sequencing, parallel evolution, toggling
Significance
Previous research has shown parallel evolution of vision genes across the fish tree of life. Two key questions are what role do they play in ecological diversification across light availability gradients and whether these changes can be reversible. This study demonstrates one vision gene in particular, rhodopsin, is involved in local adaptation across a depth gradient in Great Lakes salmonids. Additionally, we provide evidence that several species of salmonids have undergone parallel reversals to an ancestral rhodopsin allele that was last seen in this lineage over 175 Ma, showcasing an extreme example of evolutionary reversal over deep time.
Introduction
Local adaptation to novel environments can drive genetic and phenotypic differentiation among closely related organisms. Diversification may occur as populations become locally adapted to distinct conditions, leading to the evolution of divergent traits that are beneficial in each lineage’s preferred environment. Conversely, a trait may be sufficiently advantageous in a particular environment that multiple distantly related taxa converge upon it, sometimes due to the same mutation or amino acid substitution occurring independently (i.e., parallel evolution; Zhang and Kumar 1997; Futuyma and Kirkpatrick 2017). For example, parallel substitutions have occurred in myoglobin in pinnipeds and cetaceans (Romero-Herrera et al. 1978), lysozyme in ruminants and colobine monkeys (Stewart et al. 1987), and rhodopsin in fishes colonizing brackish or freshwater ecosystems (Hill et al. 2019). In this study, we examined a specific case of local adaptation in the teleost visual system that has led to diversification among similar taxa and parallel evolution among distantly related fishes.
Due to their importance in ecological interactions and their dynamic evolutionary history, the evolution of visual pigment genes (i.e., opsins) in marine and freshwater fishes has received considerable attention (Lin et al. 2017; reviewed in Rennison et al. 2012; Carleton et al. 2020). The vertebrate visual opsin system is divided into five subgroups—one rod opsin (rhodopsin) responsible for vision under low light conditions and four cone opsins (long-wave sensitive, short-wave sensitive 1, short-wave sensitive 2, and rhodopsin 2) responsible for color vision. These subgroups are based in part on differences in their peak absorbance spectra, with each group resulting from a series of gene duplications and subsequent amino acid substitutions, leading to small but measurable functional differentiation among the five opsin types (Okano et al. 1992; Yokoyama 2000). Opsin genes can shape the evolution of vision via several mechanisms. First, single nucleotide polymorphisms (SNPs) can cause nonsynonymous substitutions in key spectral tuning residues, in some cases driving adaptation to different light environments (Terai et al. 2002; Marques et al. 2017). Alternatively, copy number variants (CNVs) may result from tandem duplications and/or whole genome duplications and undergo neofunctionalization to maximally absorb at a new wavelength (e.g., expansions of rod opsins in deep-sea fishes [Musilova et al. 2019] and cone opsins in shallow-water fishes [Weadick and Chang 2007]), although gene conversion may sometimes work to homogenize newly formed paralogs, inhibiting this process (Carleton et al. 2020). Vision can also be modified by variation in opsin expression patterns based on both ontogenetic and environmental cues (Shand et al. 2008; Hofmann et al. 2009).
The cisco species flock (genus Coregonus) of the Laurentian Great Lakes presents a well-suited opportunity to study local adaptation of visual opsins to novel light environments based on depth differences among species (Harrington et al. 2015). The four extant cisco species in Lake Superior show generally low levels of interspecific variation across the genome (Turgeon and Bernatchez 2003; Turgeon et al. 2016; Ackiss et al. 2020) despite considerable differences in depth preferences (Rosinski et al. 2020). Coregonus artedi is typically epilimnetic (10–80 m), whereas Coregonus hoyi and Coregonus zenithicus are both found at intermediate depths (40–160 m), and Coregonus kiyi can be found at depths of 80–>200 m (Rosinski et al. 2020). Based on the observed ecological differentiation, we hypothesize that divergent selection may have acted to fine-tune opsins for maximum absorption of wavelengths of light that penetrate to each species’ preferred depth. Here we assess the evolution of five visual opsin genes in the Coregonus species flock to better understand mechanisms underlying their divergence across a depth gradient.
Materials and Methods
Oxford Nanopore sequencing is contributing to a rapidly expanding toolkit for DNA sequencing, owing to low up-front costs, enhanced ability to detect DNA or RNA base modifications, and read lengths limited only by the quality and quantity of input nucleic acids. Nanopore sequencing also allows for straightforward haplotyping, as whole molecules can be sequenced for each amplicon with no need for assembly. This approach has been successfully applied to microbial metabarcoding and pathogen identification (Shin et al. 2016; Moon et al. 2018; Rames and Macdonald 2018), as well as human genotyping (Cornelis et al. 2017, 2019). As flow cell quality and base-calling algorithms have improved, the accuracy and functionality of nanopore amplicon sequencing have rapidly improved. However, its application to SNP genotyping in nonhuman eukaryotes with large and complex genomes remains relatively unexplored. It is particularly relevant to understand whether accurate genotypes can be obtained, and, if so, what coverage depth is needed to do so.
In the present study, we sequenced amplicons of five teleost opsin genes in a total of 74 samples on the Oxford Nanopore Flongle device. In combination with the PCR Barcoding Expansion 1–12 (Oxford Nanopore Technologies), we sequenced and genotyped 12 individuals simultaneously on a single Flongle flow cell, following the pipeline shown in figure 1 (for a complete protocol, see Supplemental Protocol). This study is one of the first to demonstrate the accuracy and utility of amplicon sequencing with the Oxford Nanopore Flongle for SNP genotyping of eukaryotic samples.
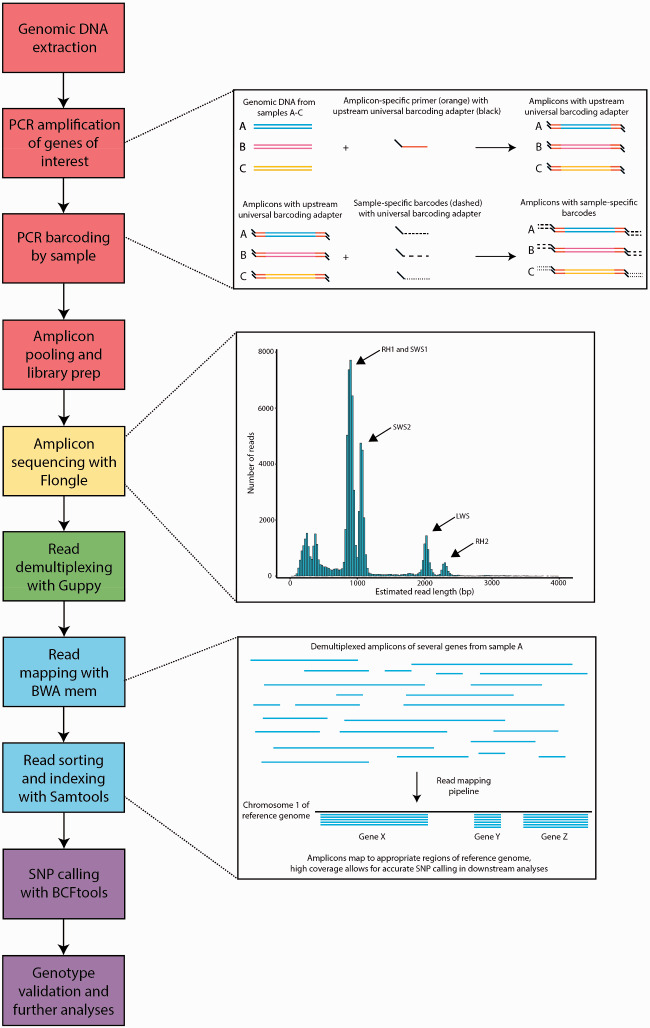
Fig. 1.
Summary of steps for amplicon sequencing and bioinformatic analyses. Boxes on the left represent individual steps, color-coded based on their phase: red represents sample preparation, yellow represents nanopore sequencing, green represents sample demultiplexing, blue represents read mapping, and purple represents genotyping and analysis. Larger boxes to the right show additional information for each of the steps: the simplified mechanisms by which amplicons are generated and barcoded (top); frequency histogram with read length on the x-axis and number of reads in the y-axis (middle); and how reads are mapped to the reference genome (bottom).
A preliminary assembly of the de novo transcriptome of C. artedi (NCBI BioProject #PRJNA659559; Bernal et al. 2020) was used as a reference to extract gene sequences of long-wave sensitive (LWS), short-wave sensitive 1 (SWS1), short-wave sensitive 2 (SWS2), rhodopsin (RH1), and rhodopsin 2 (RH2), representing one gene from each teleost visual opsin subfamily. For each of the five genes of interest, a fragment ∼700–2,100 bp in length was amplified for 18 samples of C. artedi, 19 C. hoyi, 21 C. kiyi, and 16 C. zenithicus (supplementary tables S1–S3, Supplementary Material online). All amplicons from a single individual were assigned a specific barcode and were pooled into a library containing amplicons from 12 samples, which were sequenced simultaneously on a single Flongle flow cell (fig. 1). This process was then repeated until amplicons from all samples were sequenced. Raw reads were deposited in SRA under NCBI BioProject #PRJNA664981. Sample-specific barcodes were detected and trimmed using Guppy v3.2.4 (Oxford Nanopore Technologies) with the command guppy_barcoder, and reads from each sample were mapped with BWA v0.7.17 using the command bwa mem, with version one of the Coregonus sp. “balchen” genome assembly as a reference (De-Kayne et al. 2020; GCA_902810595.1). To verify the accuracy of nanopore amplicon genotyping, we performed a rarefaction analysis in which SNPs were called at various levels of coverage (i.e., maximum possible coverage; 2,000; 1,000; 500; 250; 100; 75; 50; and 25×). An annotated bash script detailing the entire bioinformatic pipeline is available from GitHub (https://github.com/KrabbenhoftLab/rhodopsin; last accessed December 3, 2020).
Results and Discussion
On average, Flongle sequencing runs yielded 206.13 Mb (SD ± 166.64 Mb; min–max: 26.84–471.50 Mb), with an average of 184,958 reads (±154,877 reads; 23,468–435,138 reads), though yield varied based on flow cell quality (flow cells used were earlier versions with low number of active pores). The average sequence N50 was 1,117 bp (±305 bp; 897–1,852 bp), with read length abundances peaking at the approximate lengths of our amplicons (fig. 1). After resequencing genes with low coverage following first-round sequencing, the average coverage was 3,199.58× across all five genes (±4,804.24×, 10.47–31,158.31×; supplementary table S1, Supplementary Material online). Coverage varied slightly by species, but this is likely an artifact of stochastic differences in PCR efficiency and sequencing yield (supplementary table S4, Supplementary Material online). Amplicon reads mapped uniquely (i.e., one genomic region per amplicon) to the C. sp. “balchen” genome, confirming that each amplicon does indeed correspond to a sequence of a single gene. However, based on a BLAST search of our opsins against the C. sp. “balchen” genome, multiple functional copies of LWS, RH2, and SWS1 exist in the Coregonus sp. genome, providing another potential avenue for opsin evolution (supplementary table S5, Supplementary Material online). Additionally, it was determined that the copy of LWS analyzed here is partially pseudogenized and perhaps nonfunctional.
The SNP calls from nanopore data were then compared with Sanger sequences of RH1 for the same individuals. Although accuracy remained high at all sequencing depths (>90%), we found incongruencies in a small proportion of samples between 10× and 75×. Only when reaching 100× coverage were genotypes called with complete accuracy for all individuals, in relation to Sanger sequences. Considering that small errors can impact the results of analyses involving amplicons with few variant sites, we recommend a minimum per-amplicon coverage of 100× for future work.
We used a conservative genotyping approach, with the goal of assessing the coverage needed for accurate genotyping on a Flongle flow cell. Based on our findings, this approach could be used for higher throughput sequencing, which could involve more amplicons, more individuals, or a combination of both. Considering that we generated ∼200 Mb of sequence data per run, the number of individuals and amplicons that can be sequenced simultaneously at 100× coverage can be estimated as follows:
200,000,000 bp=100*A*N*NA,
where A is the amplicon size (in bp), N is the number of samples to be sequenced simultaneously, and NA is the number of amplicons to be sequenced per sample. To optimize throughput for the maximum number of samples, the PCR Barcoding Expansion 1–96 (EXP-PBC096, Oxford Nanopore Technologies) can be used to generate sequence data for 96 samples simultaneously. Assuming an average amplicon size of 1,000 bp, one could sequence 20 amplicons across 96 samples in a single 24-h Flongle sequencing run for ∼$300, or $0.16 per genotype (supplementary table S6, Supplementary Material online). The use of a MinION flow cell (not analyzed here) would increase output by a factor of ∼16× (based on differences in number of total pores) and further reduce the cost per genotype. With the growth of nanopore sequencing, these conservative cost estimates are expected to drop in upcoming years.
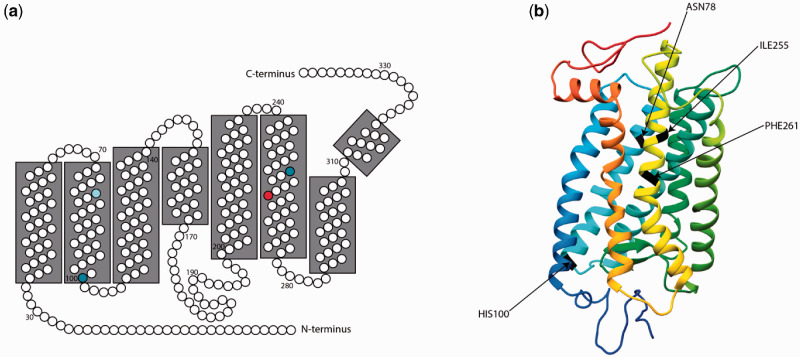
The average fixation index (FST) of the detected SNPs in the five opsins analyzed across all four species was 0.11 (supplementary table S7, Supplementary Material online). The only large differences (FST > 0.4) were found in four SNPs detected within the coding sequence of rhodopsin, with no highly differentiated SNPs among the four cone opsins. This suggests that differences in dim-light vision and changes in rhodopsin could be driving local adaptation by depth. Of the four high FST SNPs, one (FST = 0.44) was a synonymous SNP at amino acid residue 78. One SNP (FST = 0.44) resulted in a shift from asparagine to histidine at amino acid residue 100, which is located near the C-terminal end of transmembrane helix two, possibly in the extracellular matrix (fig. 2a and b; see also Yokoyama 2000). Another (FST = 0.44) resulted in a change from valine to isoleucine at residue 255, which is located in transmembrane helix six, facing away from the retinal binding pocket (fig. 2a and b, see also Baldwin 1993; Hunt et al. 1996). Neither residue 100 nor 255 are known to be key spectral tuning sites in rhodopsin (Yokoyama 2000), although Yokoyama and Jia (2020) recently reported that Ile255Val may have an effect on spectral tuning of RH2. The three aforementioned SNPs possess the exact same FST and changes in genotype were completely consistent across all samples, suggesting that these sites are tightly linked.
Fig. 2.
(a) 2D model of Coregonus artedi rhodopsin based on a 3D model generated using PHYRE v2.0 (Kelley et al. 2015) and visualized in UCSF Chimera (Pettersen et al. 2004). Amino acid residues are numbered in order from N-terminus to C-terminus. Amino acid residue 78 (the location of a synonymous, high FST SNP) is colored in light blue, residues 100 and 255 (locations of nonsynonymous, high FST SNPs) are colored in dark blue, and residue 261 (location of a high FST SNP known to be involved in spectral tuning) is colored in red. The 2D model was constructed for the specific model obtained for C. artedi, following Yokoyama (2000) and Musilova et al. (2019). (b) 3D model of bovine rhodopsin, colored blue (N-terminus) to red (C-terminus). Amino acid residues 78, 100, 255, and 261 have been colored in black, and are labeled accordingly.
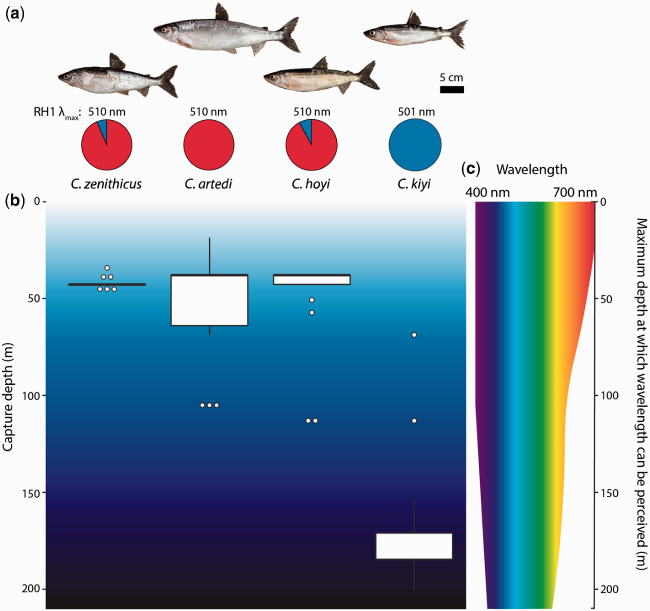
The most strongly segregating SNP (FST = 0.88) occurred at amino acid residue 261 of rhodopsin, which is located in transmembrane helix six, facing the retinal binding pocket of the protein (fig. 2a and b, see also Baldwin 1993; Hunt et al. 1996; Yokoyama 2000). Coregonus artedi, C. hoyi, and C. zenithicus, inhabitants of a relatively shallow, broad-spectrum light environment, were primarily homozygous for tyrosine at this locus (fig. 3). Meanwhile, C. kiyi, which inhabits the blue-shifted deeper waters of Lake Superior, was completely homozygous for phenylalanine. The presence of phenylalanine at this site is known to cause an 8 nm blue-shift in the absorbance spectrum, likely due to the change in polarity associated with this substitution (i.e., polar to nonpolar Y261F) (Yokoyama et al. 1995). Genotypic associations at this locus vary consistently with depth (fig. 3), providing evidence that C. kiyi is adapted to life in deep water after evolving from shallow-water ancestors. This hypothesis is further corroborated by phenotypic data, as C. kiyi have significantly larger eye diameters (as a proportion of total head length) than C. artedi (P < 0.001), C. zenithicus (P < 0.001), and C. hoyi (P < 0.001; supplementary fig. S1, Supplementary Material online). The predictable variation of both genetic and morphological traits provides key evidence that local adaptation to depth accompanies diversification of Great Lakes ciscoes (supplementary fig. S2, Supplementary Material online).
Fig. 3.
(a) The four cisco species included in this study: Coregonus zenithicus (n = 16), Coregonus artedi (n = 18), Coregonus hoyi (n = 19), and Coregonus kiyi (n = 21). For each species, the approximate λmax of the most common allele for rhodopsin is shown. Pie charts below each photo indicate the allele frequency at residue 261 of rhodopsin, where red represents tyrosine and blue represents phenylalanine. (b) Boxplots indicating the approximate capture depths of samples from each of the four species. (c) An approximation of the visible light spectrum in Lake Superior, from 400 to 700 nm wavelength. The narrowing of the spectrum with increased depth shows how the ability of organisms to perceive certain wavelengths of light diminishes with increasing depth, particularly with red and orange light (following Harrington et al. 2015).
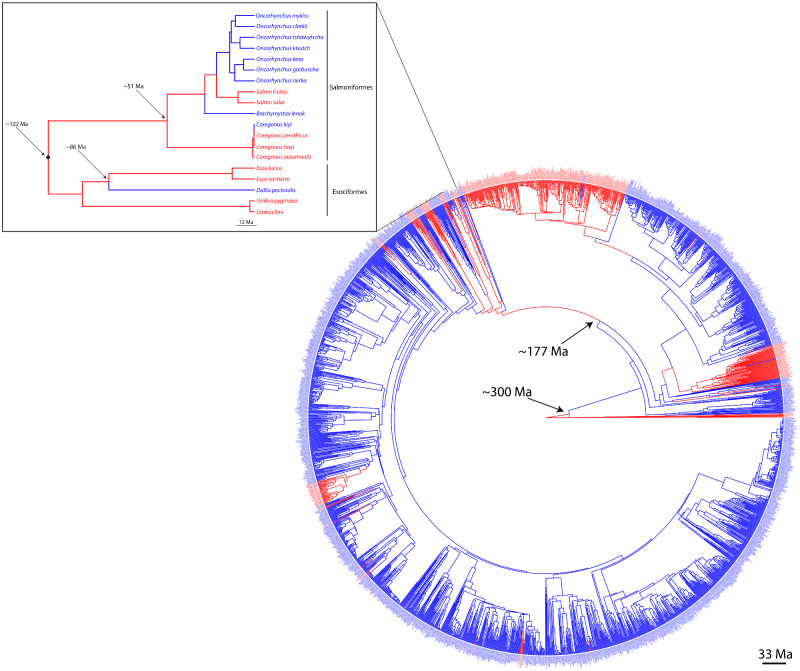
The shift between the two aforementioned amino acids at rhodopsin residue 261 was recently analyzed in a deep phylogenetic context. Hill et al. (2019) showed that fish lineages that have undergone a habitat change from blue-shifted marine waters to red-shifted brackish or freshwater have independently converged on the red-shift-associated 261Tyr phenotype over 20 times across the fish tree of life. We performed an ancestral state reconstruction at this amino acid residue to further examine these evolutionary changes. Our results show that parallel reversals and “toggling” (i.e., switching back and forth several times between two alleles in a single lineage; sensu Delport et al. 2008; Porubsky et al. 2020) are common occurrences at this site across the fish tree of life (fig. 4), consistent with the findings of Musilova et al. (2019). Indeed, it appears that C. kiyi has undergone a reversal to the blue-shifted 261Phe after ∼175 Myr of differentiation from its marine ancestors (fig. 4). Whether toggling is due to true re-evolution of an identical mutation or through re-emergence of a low-frequency allele following incomplete lineage sorting is unknown. Interestingly, this reversal to a deep ancestral state over millions of years has also occurred in several other lineages across the fish tree of life, including in the channel catfish (Ictalurus punctatus), rainbow trout (Oncorhynchus mykiss), Kessler’s sculpin (Leocottus kesslerii), and European flounder (Platichthys flesus), among others, and can occur in both directions (i.e., Phe261Tyr or Tyr261Phe; fig. 4). The Salmoniformes and Esociformes in particular appear to have undergone a high degree of parallel reversal at this site, with multiple taxa (Oncorhynchus spp., Brachymystax lenok, C. kiyi, and Dallia pectoralis) undergoing independent parallel reversals to the deep ancestral blue-shifted allele (fig. 4). These findings indicate that rhodopsin residue 261 may be able to toggle between these two amino acids depending on what is advantageous in a particular light environment, even across incredibly long time scales.
Fig. 4.
The time-calibrated fish tree of life (Rabosky et al. 2018) colorized according to the allele possessed by each species at rhodopsin residue 261, with red indicating the red-shifted tyrosine and blue indicating the blue-shifted phenylalanine (following Hill et al. 2019). Ancestral states were reconstructed using RAxML (Stamatakis 2014) and plotted using the R package ggtree (Yu et al. 2017). Arrows indicate divergence times of clades of interest, in millions of years (Chang et al. 2019). The enhanced clade showcases the Salmoniformes (including Coregonus) and Esociformes; this group shows several parallel reversals to the phylogenetically deep “blue” ancestral state (last seen in the ancestor to this group ∼175 Ma).
Conclusions
Our results indicate that local adaptation to distinct visual environments is associated with genetic and morphological differentiation among the closely related ciscoes of Lake Superior. The identification of several high FST SNPs in rhodopsin, including Phe261Tyr, is particularly relevant, as the shifts between these two amino acids at residue 261 are identical to those observed across similar light availability gradients in phylogenetically distant fishes (Hill et al. 2019). Adaptation to similar light conditions therefore seems to lead to phenotypic convergence due to parallel single-nucleotide changes at this site. Additionally, the discovery of independent reversals to the ancient ancestral state in C. kiyi (and other lineages), provides evidence of genetic toggling, whereby organisms transition bidirectionally between different alleles in response to environmental pressures. This result is striking because the genetic background is presumably very different across these taxa after more than ∼175 Myr of divergence. In this case, the conserved nature of rhodopsin protein function likely potentiates evolutionary reversals over deep time periods. The observation of amino acid toggling in Great Lakes Coregonus species and across the fish tree of life is at the extreme end of the time scale of parallel evolution and evolutionary reversals, given the generally held prediction that the likelihood of evolutionary reversal diminishes as time progresses (Storz 2016; Blount et al. 2018).
Long-read amplicon sequencing using the Oxford Nanopore Flongle is highly amenable to genotyping complex eukaryotes. The methodology described here is simple and reliable, and offers the promise of rapid, low-cost genotyping in nonmodel organisms. This protocol has many potential applications—nanopore amplicon genotyping could be used for CRISPR validation, screening for inherited genetic disorders, and eukaryotic eDNA metabarcoding, among others.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the US Geological Survey Research Vessel Kiyi Captain Joe Walters, First Mate Keith Peterson, and Engineer Charles Carrier, as well as Lake Superior Biological Station staff Mark Vinson and Lori Evrard for assistance with sample collections. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the US Government. All sampling and handling of fish were carried out in accordance with guidelines for the care and use of fishes by the American Fisheries Society (Jenkins et al. 2014). Jessie Pelosi provided critical assistance with data analysis. Christopher Loretz, Kelly Harrington, Mary Alice Coffroth, Daniel MacGuigan, Tianying Lan, Wendylee Stott, Amanda Ackiss, Andrew Muir, Thomas Dowling, Hannah Waterman, Victor Albert, Vincent Lynch, and Omer Gokcumen provided valuable assistance or feedback on the study. Jessica Poulin coordinated the University at Buffalo Honors Program in Biological Sciences that facilitated this research. Finally, we wish to thank the editors and anonymous reviewers for providing thoughtful, constructive, and timely feedback, particularly in the midst of a global pandemic. This study was supported by the Great Lakes Fishery Commission (Award #2018_KRA_44073 to T.J.K.), the University at Buffalo Department of Biological Sciences (Philip G. Miles Fellowship to K.M.E.), and the University at Buffalo Honors College (Award to K.M.E.).
Data Availability
Raw sequence reads generated on the Oxford Nanopore Flongle were deposited in SRA, under the BioProject #PRJNA664981. BAM alignments of these sequences to the Coregonus sp. balchen genome V1 were deposited in Dryad (doi:10.5061/dryad.crjdfn32s), along with the VCF file used to call SNPs. Raw Sanger sequencing files were also deposited in the aforementioned Dryad repository.
Literature Cited
- Ackiss AS, Larson WA, Stott W. 2020. Genotyping-by-sequencing illuminates high levels of divergence among sympatric forms of coregonines in the Laurentian Great Lakes. Evol Appl. 13(5):1037–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin JM. 1993. The probable arrangement of the helices in G protein-coupled receptors. EMBO J. 12(4):1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal MA, et al. 2020. Transcriptomic divergence predicts morphological and ecological variation underlying an adaptive radiation. BioRxiv. [Google Scholar]
- Blount ZD, Lenski RE, Losos JB. 2018. Contingency and determinism in evolution: replaying life’s tape. Science 362(6415):eaam5979. [DOI] [PubMed] [Google Scholar]
- Carleton KL, Escobar-Camacho D, Stieb SM, Cortesi F, Marshall NJ. 2020. Seeing the rainbow: mechanisms underlying spectral sensitivity in teleost fishes. J Exp Biol. 223(8):jeb193334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Rabosky DL, Smith SA, Alfaro ME. 2019. An R package and online resource for macroevolutionary studies using the ray-finned fish tree of life. Methods Ecol Evol. 10(7):1118–1124. [Google Scholar]
- Cornelis S, et al. 2019. Forensic tri-allelic SNP genotyping using nanopore sequencing. Forensic Sci Int Genet. 38:204–210. [DOI] [PubMed] [Google Scholar]
- Cornelis S, Gansemans Y, Deleye L, Deforce D, Van Nieuwerburgh F. 2017. Forensic SNP genotyping using Nanopore MinION sequencing. Sci Rep. 7(1):41759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-Kayne R, Zoller S, Feulner PGD. 2020. A de novo chromosome-level genome assembly of Coregonus sp. “Balchen”: one representative of the Swiss Alpine whitefish radiation. Mol Ecol Resour. 20(4):1093–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delport W, Scheffler K, Seoighe C. 2008. Frequent toggling between alternative amino acids is driven by selection in HIV-1. PLoS Pathog. 4(12):e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futuyma D, Kirkpatrick M. 2017. Evolution. 4th ed Sunderland (MA: ): Sinauer. [Google Scholar]
- Harrington KA, Hrabik TR, Mensinger AF. 2015. Visual sensitivity of deepwater fishes in Lake Superior. PLoS One 10(2):e0116173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J, et al. 2019. Recurrent convergent evolution at amino acid residue 261 in fish rhodopsin. Proc Natl Acad Sci USA. 116(37):18473–18478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann CM, et al. 2009. The eyes have it: regulatory and structural changes both underlie cichlid visual pigment diversity. PLoS Biol. 7(12):e1000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt DM, Fitzgibbon J, Slobodyanyuk SJ, Bowmaker JK. 1996. Spectral tuning and molecular evolution of rod visual pigments in the species flock of cottoid fish in Lake Baikal. Vis Res. 36(9):1217–1224. [DOI] [PubMed] [Google Scholar]
- Jenkins JA, et al. 2014. Guidelines for the use of fishes in research—revised and expanded, 2014. Fisheries 39(9):415–416. [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction, and analysis. Nat Protoc. 10(6):845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J-J, Wang F-Y, Li W-H, Wang T-Y. 2017. The rises and falls of opsin genes in 59 ray-finned fish genomes and their implications for environmental adaptation. Sci Rep. 7(1):15568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques DA, et al. 2017. Convergent evolution of SWS2 opsin facilitates adaptive radiation of threespine stickleback into different light environments. PLoS Biol. 15(4):e2001627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, et al. 2018. Diagnosis of Haemophilus influenzae pneumonia by nanopore 16S amplicon sequencing of sputum. Emerg Infect Dis. 24(10):1944–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musilova Z, et al. 2019. Vision using multiple distinct rod opsins in deep-sea fishes. Science 364(6440):588–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano T, Kojima D, Fukada Y, Shichida Y, Yoshizawa T. 1992. Primary structures of chicken cone visual pigments: vertebrate rhodopsins have evolved out of cone visual pigments. Proc Natl Acad Sci USA. 89(13):5932–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 25(13):1605–1612. [DOI] [PubMed] [Google Scholar]
- Porubsky D, et al. 2020. Recurrent inversion toggling and great ape genome evolution. Nat Genet. 52(8):849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL, et al. 2018. An inverse latitudinal gradient in speciation rate for marine fishes. Nature 559(7714):392–395. [DOI] [PubMed] [Google Scholar]
- Rames E, Macdonald J. 2018. Evaluation of MinION nanopore sequencing for rapid enterovirus genotyping. Virus Res. 252:8–12. [DOI] [PubMed] [Google Scholar]
- Rennison DJ, Owens GL, Taylor JS. 2012. Opsin gene duplication and divergence in ray-finned fish. Mol Phylogenet Evol. 62(3):986–1008. [DOI] [PubMed] [Google Scholar]
- Romero-Herrera AE, Lehmann H, Joysey KA, Friday AE. 1978. On the evolution of myoglobin. Philos Trans R Soc Lond B Biol Sci. 283(995):61–163. [DOI] [PubMed] [Google Scholar]
- Rosinski CL, Vinson MR, Yule DL. 2020. Niche partitioning among native ciscoes and nonnative rainbow smelt in Lake Superior. Trans Am Fish Soc. 149(2):184–203. [Google Scholar]
- Shand J, et al. 2008. The influence of ontogeny and light environment on the expression of visual pigment opsins in the retina of the black bream, Acanthopagrus butcheri. J Exp Biol. 211(9):1495–1503. [DOI] [PubMed] [Google Scholar]
- Shin J, et al. 2016. Analysis of the mouse gut microbiome using full-length 16S rRNA amplicon sequencing. Sci Rep. 6(1):29681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CB, Schilling JW, Wilson AC. 1987. Adaptive evolution in the stomach lysozymes of foregut fermenters. Nature 330(6146):401–404. [DOI] [PubMed] [Google Scholar]
- Storz JF. 2016. Causes of molecular convergence and parallelism in protein evolution. Nat Rev Genet. 17(4):239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai Y, Mayer WE, Klein J, Tichy H, Okada N. 2002. The effect of selection on a long wavelength-sensitive (LWS) opsin gene of Lake Victoria cichlid fishes. Proc Natl Acad Sci USA. 99(24):15501–15506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon J, Bernatchez L. 2003. Reticulate evolution and phenotypic diversity in North American ciscoes, Coregonus ssp. (Teleostei: Salmonidae): implications for the conservation of an evolutionary legacy. Conserv Genet. 4(1):67–81. [Google Scholar]
- Turgeon J, et al. 2016. Morphological and genetic variation in Cisco (Coregonus artedi) and Shortjaw Cisco (C. zenithicus): multiple origins of Shortjaw Cisco in inland lakes require a lake-specific conservation approach. Conserv Genet. 17(1):45–56. [Google Scholar]
- Weadick CJ, Chang BSW. 2007. Long-wavelength sensitive visual pigments of the guppy (Poecilia reticulata): six opsins expressed in a single individual. BMC Evol Biol. 7(Suppl. 1):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Knox BE, Yokoyama S. 1995. Rhodopsin from the fish, Astyanax: Role of tyrosine 261 in the red shift. Invest Ophthalmol Vis Sci. 36(5):939–945. [PubMed] [Google Scholar]
- Yokoyama S. 2000. Molecular evolution of vertebrate visual pigments. Prog Retin Eye Res. 19(4):385–419. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Jia H. 2020. Origin and adaptation of green-sensitive (RH2) pigments in vertebrates. FEBS Open Bio. 10(5):873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 8(1):28–36. [Google Scholar]
- Zhang J, Kumar S. 1997. Detection of convergent and parallel evolution at the amino acid sequence level. Mol Biol Evol. 14(5):527–536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence reads generated on the Oxford Nanopore Flongle were deposited in SRA, under the BioProject #PRJNA664981. BAM alignments of these sequences to the Coregonus sp. balchen genome V1 were deposited in Dryad (doi:10.5061/dryad.crjdfn32s), along with the VCF file used to call SNPs. Raw Sanger sequencing files were also deposited in the aforementioned Dryad repository.