Abstract
Photosynthetic dinoflagellates of the Family Symbiodiniaceae live symbiotically with many organisms that inhabit coral reefs and are currently classified into fifteen groups, including seven genera. Draft genomes from four genera, Symbiodinium, Breviolum, Fugacium, and Cladocopium, which have been isolated from corals, have been reported. However, no genome is available from the genus Durusdinium, which occupies an intermediate phylogenetic position in the Family Symbiodiniaceae and is well known for thermal tolerance (resistance to bleaching). We sequenced, assembled, and annotated the genome of Durusdinium trenchii, isolated from the coral, Favia speciosa, in Okinawa, Japan. Assembled short reads amounted to 670 Mb with ∼47% GC content. This GC content was intermediate among taxa belonging to the Symbiodiniaceae. Approximately 30,000 protein-coding genes were predicted in the D. trenchii genome, fewer than in other genomes from the Symbiodiniaceae. However, annotations revealed that the D. trenchii genome encodes a cluster of genes for synthesis of mycosporine-like amino acids, which absorb UV radiation. Interestingly, a neighboring gene in the cluster encodes a glucose–methanol–choline oxidoreductase with a flavin adenine dinucleotide domain that is also found in Symbiodinium tridacnidorum. This conservation seems to partially clarify an ancestral genomic structure in the Symbiodiniaceae and its loss in late-branching lineages, including Breviolum and Cladocopium, after splitting from the Durusdinium lineage. Our analysis suggests that approximately half of the taxa in the Symbiodiniaceae may maintain the ability to synthesize mycosporine-like amino acids. Thus, this work provides a significant genomic resource for understanding the genomic diversity of Symbiodiniaceae in corals.
Keywords: Symbiodiniaceae, Durusdinium trenchii, WGS, MAAs, GMC oxidoreductase
Significance
Dinoflagellates of the family Symbiodiniaceae include coral symbionts and have been well studied. Analyses from whole-genome sequencing of several genera have been reported, but no genome is available from the genus Durusdinium. Here, we report the draft genome of Durusdinium trenchii from the coral, Favia speciosa. The genomic analysis of this thermotolerant species shows that a cluster of genes for biosynthesis of mycosporine-like amino acids (MAAs), which absorb UV radiation, is conserved between Symbiodinium, an early-diverging lineage, and Durusdinium, which occupies an intermediate phylogenetic position in the Family Symbiodiniaceae. Both genera reportedly enhance thermal tolerance of corals. If coral bleaching is triggered by high solar radiation, a dinoflagellate capacity for MAA biosynthesis may contribute to bleaching resistance.
Introduction
Symbiotic dinoflagellates of the Family Symbiodiniaceae, are keystone photosynthetic organisms in coral reef ecosystems (LaJeunesse et al. 2018). The diversity of symbiotic dinoflagellate populations and their relationships with hosts have been analyzed and discussed (Baker 2003; Coffroth and Santos 2005; Coffroth et al. 2006; Pochon and Gates 2010; Green et al. 2014; Pochon et al. 2014). Dinoflagellate populations in stony corals, which form modern reefs, have attracted particular attention (Abrego et al. 2009; Thornhill et al. 2014; Shoguchi et al. 2020), because breakdown of coral-dinoflagellate symbiosis causes coral bleaching, decimating coral reef communities (coral holobionts) (Weis et al. 2008; Stat and Gates 2011).
Although many reasons have been discussed for collapse of this symbiotic relationship (Nakamura and van Woesik 2001; Iguchi et al. 2012), the main trigger is likely rising surface seawater temperatures (SSTs) caused by climate change (Weis et al. 2008). Possible bleaching has been also reported in other hosts, such as giant clams (Mies 2019). Recently, discussions of coral bleaching due to increasing SSTs have focused on heat-tolerant species of the genus Durusdinium (Symbiodiniaceae) (a member of previous clade D), because genetic variability of dinoflagellates in corals is thought to be a major factor in the bleaching phenomenon (Berkelmans and Van Oppen 2006; Stat and Gates 2011; Lesser 2019). Horizontal transmission types of symbiosis may be more adaptive than vertical transmission types (Weis et al. 2001; Yamashita et al. 2013; Hidaka 2016; Yuyama et al. 2018). Coral holobionts resulting from coral-Durusdinium symbiosis may be better adapted to rising SSTs than other types of coral holobionts.
Durusdinium includes heat-tolerant strains (Rowan 2004; Stat and Gates 2011). A metabolic analysis of cultured Symbiodiniaceae showed that D. trenchii has a low level of the sterol metabolite, C29 Stanol 2, suggesting metabolic differences among members of the family Symbiodiniaceae (Symbiodinium microadriaticum, Symbiodinium psygmophilum, and B. minutum) (Klueter et al. 2015). A recent report on effects of light and thermal stress indicates that the pan-tropical species, D. trenchii, is more thermotolerant than others so far examined (S. microadriaticum, B. minutum, and Cladocopium goreaui) (Lesser 2019). In addition, the heat-stress response of D. trenchii was compared between free-living and symbiotic cells and transcriptional activity in symbiotic dinoflagellates was drastically altered by thermal stress (Bellantuono et al. 2019).
Genomes of several taxa within the Symbiodiniaceae have been deciphered (Shoguchi et al. 2013, 2018; Aranda et al. 2016; Liu et al. 2018; Li et al. 2020) and genome evolution of this family has been discussed (González-Pech et al. 2019). However, no Durusdinium genome is available and the genetic basis for thermal tolerance remains unknown (Baker 2003; Weber and Medina 2012; Hidaka 2016). To provide a genomic resource for Durusdinium, we isolated D. trenchii from the coral, F. speciosa, in Okinawa, which will be useful for analyzing Durusdinium in coral holobionts.
One of the early-diverging lineages of the family Symbiodiniaceae is the genus Symbiodinium (LaJeunesse et al. 2018), which includes species having the ability to synthesize MAAs (Banaszak et al. 2000). Both Symbiodinium and Durusdinium have been known to enhance thermal tolerance of holobionts (Reynolds et al. 2008; Kemp et al. 2014; Aihara et al. 2016). The genome of Symbiodinium tridacnidorum has a cluster of genes for enzymes involved in MAA biosynthesis (Shoguchi et al. 2018). On the other hand, species in later-diverging groups (Breviolum and Cladocopium) appear not to have this metabolic pathway, as no MAA gene cluster has been found in their genomes. Therefore, a draft genome of Durusdinium, one of an intermediate group of seven genera in the family Symbiodiniaceae, may help to clarify when the ability to synthesize MAAs was lost during diversification of the Symbiodiniaceae. To explore the genetic background of the coral symbiont, Durusdinium, here we examined the genome and associated transcriptomes to determine whether Durusdinium also has this gene cluster.
Materials and Methods
Biological Materials
The culturable dinoflagellate, D. trenchii, is harbored by the coral, F. speciosa, in Okinawa, Japan. A single cell of D. trenchii was isolated using a glass-micropipette in May 2012. The Nagoya Protocol was not applicable to the dinoflagellate. The established culture strain is available as NIES-2907 in the Microbial Culture Collection at the National Institute for Environmental Studies (NIES) in Tsukuba (https://mcc.nies.go.jp). Cloned Durusdinium cells for nucleotide sequencing were basically maintained as previously described (Shoguchi et al. 2018). The culture medium included artificial seawater containing 1× Guillard’s (F/2) marine-water enrichment solution (Sigma–Aldrich) and soil extract (Provasoli et al. 1957). A 25 °C incubator for culturing was maintained on a 12 h-light/12 h-dark regime at an illumination of ∼20 μmol m−2 s−1 (Beedessee et al. 2015).
Nucleotide Sequencing and Assembly
Genomic DNA from clonal cultures was extracted using phenol-chloroform and cetyltrimethylammonium bromide (Shoguchi et al. 2013) and was used for Illumina library construction (supplementary table S1, Supplementary Material online). Libraries were sequenced using a HiSeq 2500 (Illumina) and paired-end reads were assembled de novo with Platanus (Kajitani et al. 2014) and Newbler. Assembled data were combined (Nishitsuji et al. 2020). Scaffolding with mate-pair information was carried out using SSPACE (ver. 3.0) (Boetzer et al. 2011). With Gapcloser, gaps inside scaffolds were closed with paired-end data. Finally, data were polished using Pilon (ver. 1.22) (Walker et al. 2014). The completeness of the assembled genome was evaluated by the recovery of 458 CEGMA and 303 BUSCO genes from the genome of D. trenchii (Parra et al. 2007; Simão et al. 2015; Beedessee et al. 2020). Total RNA for transcriptome sequencing was isolated from cultured cells at 25 °C, as described previously (Shoguchi et al. 2013). Two libraries (the difference of culture time at 0 and 3 days) were constructed following the manufacturer’s protocol and were sequenced using a HiSeq 2500. De novo assembly was performed using Trinity (Grabherr et al. 2011).
Gene Prediction and Annotation
RNA-seq reads were mapped to a soft-masked genome using STAR (Dobin et al. 2013) for passage to the BRAKER2 pipeline (Hoff et al. 2016). UTR and gene model prediction were performed with Augustus (v3.2.3) (Stanke et al. 2008). Intron and exon hints were generated with STAR (Dobin et al. 2013) and BLAT (Kent 2002), respectively, and were used to make final gene predictions using a modified version of Augustus (v3.2.3) (Stanke et al. 2008; Shoguchi et al. 2013). The final set of predicted proteins was annotated against UniProt (Magrane and UniProt Consortium 2011) and PFAM (Punta et al. 2012) where hits larger than 1e−5 were discarded. Putative contaminant sequences were identified basically following Chen et al. (2020). First, short scaffolds (<1 kb) were removed (Shoguchi et al. 2018). To find contaminant sequences, 45 scaffolds (>10 kb) with high GC content (>55%) were manually checked using a genome browser (Koyanagi et al. 2013). Twelve scaffolds predicted genes with introns that were supported by transcriptomes or had similarities to S. microadriaticum proteins (BlastX, E-value <10−20) in the NCBI database (https://pubmed.ncbi.nlm.nih.gov). About 33 scaffolds were removed as putative contaminant sequences.
Molecular Phylogenetic Tree and Protein Structure Predictions
Molecular phylogenetic analysis of the demethyl-4-deoxygadusol (DDG) synthase family was performed as described in our previous study (Shoguchi et al. 2018). Protein sequences of glucose–methanol–choline (GMC) oxidoreductases in the molecular phylogenetic analysis of Sorigué et al. (2017) and some proteins with GMC domains were collected from the NCBI database (https://pubmed.ncbi.nlm.nih.gov; last accessed July 21, 2020). Those and Symbiodiniaceae proteins with GMC domains were aligned with MAFFT (Katoh and Standley 2013). Molecular phylogenetic analysis was carried out using Bayesian inference with MrBayes v.3.2 (Ronquist et al. 2012), as previously described (Beedessee et al. 2019). Trees were visualized using Figtree (http://tree.bio.ed.ac.uk/software/figtree/). I-TASSER was used for 3 D prediction (Zhang 2008).
Results and Discussion
Draft Genome of Durusdinium
Three genomic libraries with insert sizes ranging from 500 bp to 19 kb were constructed from the cloned Durusdinium (supplementary table S1, Supplementary Material online). Short read sequencing (2 × 101 bp) produced ∼76 Gb of total sequencing data, which were assembled into a total length of 695 Mb. Thirty-three scaffolds, which were likely to be contaminant sequences, were removed from the initial assembly (supplementary table S2, Supplementary Material online). The final draft genome of D. trenchii (version 1.0) had a total length of 670.4 Mb with a scaffold N50 of 97.5 kb (table 1). Completeness of the D. trenchii genome was checked using CEGMA (Parra et al. 2007) and BUSCO (Simão et al. 2015). The 48% (145/303 BUSCO genes) hits on the D. trenchii proteins was comparable to other reported dinoflagellate genomes (44–71%) (supplementary fig. S1, Supplementary Material online). The GC content of the draft genome was 47.4%, comparable to GC contents of Symbiodinium (∼50%) and Cladocopium (∼44%) (table 1).
Table 1.
Genomic Compositions of Seven Genomes of the Family Symbiodiniaceae
| Durusdinium trenchii | Symbiodinium microadriaticum a | Symbiodinium tridacnidorum b | Breviolum minutum c | Fugacium kawagutii v3 d | Cladocopium goreaui e | Cladocopium sp. (C92)b | |
|---|---|---|---|---|---|---|---|
| A total assembled length of assembly (Mb) | 670.43 | 808.24 | 766.65 | 615.52 | 936.98 | 1,027.79 | 704.77 |
| G + C content (%) | 47.4 | 50.5 | 49.9 | 43.6 | 45.5 | 44.8 | 43.0 |
| No. of genes | 30,054 | 49,109 | 69,018 | 41,925 | 45,192 | 35,913 | 65,832 |
| Average length of genes (bp) | 15,030 | 12,898 | 8,834 | 11,959 | 7,242 | 6,967 | 8,192 |
| No. of exons per gene | 19.6 | 21.8 | 13.4 | 19.6 | 12.6 | 10.0 | 11.3 |
| Average length (bp) of exons | 90 | 110 | 105 | 100 | 126 | 176 | 130 |
| Average length (bp) of introns | 704 | 505 | 561 | 499 | 479 | 575 | 622 |
Aranda et al. (2016).
Shoguchi et al. (2018).
Shoguchi et al. (2013).
Li et al. (2020).
Liu et al. (2018).
Genome Annotations
Two RNA-seq libraries were constructed and sequenced. Reads of 2 × 134 bp produced ∼11 Gb of total sequencing data (supplementary table S1, Supplementary Material online). The de novo assembly produced 64,183 contigs with a GC content of ∼55%, similar to that of clade D from reported transcriptomes (González-Pech et al. 2017). Using transcriptome data as hints, 30,054 protein-coding genes were predicted (table 1), a number comparable to that of C. goreaui (Liu et al. 2018), but less than some other dinoflagellate genomes. A recent report indicated that a consistent gene-prediction approach is crucial for comparative genomic analysis, suggesting the difficulties of computational gene prediction for dinoflagellate genomes (Chen et al. 2020). Therefore, long-read transcriptomic data from various conditions are likely to be needed in future comparative genomic studies. Assembled genomic and transcriptomic data and annotation information are accessible from the following genome browser: https://marinegenomics.oist.jp/gallery (Koyanagi et al. 2013). The 28S rDNAs and ITS2 sequences (TRINITY_DN41397_c4_g2_i5) from the assembled sequences corresponded to the nuclear ribosomal ITS1/5.8S/ITS2 (KJ019889) in D. trenchii LaJeunesse sp. nov. (LaJeunesse et al. 2014, 2018), confirming that the clone is D. trenchii.
Gene Cluster for Sunscreen Biosynthesis
Analysis of the S. tridacnidorum (previously Symbiodinium sp. clade A3) genome identified a gene cluster for enzymes involved in MAA biosynthesis in Shoguchi et al. (2018). In addition, comparative analysis suggested that orthologs of these genes have been lost in the common ancestor of Breviolum and Cladocopium. To determine whether such losses occurred in the Durusdinium lineage, we performed BLAST and Pfam domain searches. Scaffold 2498 of the draft assembly contained a gene cluster for MAA biosynthesis, expression of which was supported by transcriptomic data (fig. 1a). Moreover, gene order was conserved between S. tridacnidorum and D. trenchii. The orthologous relationship between S. tridacnidorum and D. trenchii was confirmed in molecular phylogenetic analysis of the DDG synthase family (supplementary fig. S2, Supplementary Material online). In addition, we found that the neighboring gene to D-Ala D-Ala ligase homolog on the 3′ side of the cluster encoded an enzyme resembling the GMC oxidoreductase family. The homolog was also found in the genome of S. tridacnidorum and is located adjacent to the MAA gene cluster (fig. 1a), suggesting syntenic conservation of metabolic genes among members of the Symbiodiniaceae (Liu et al. 2018).
Fig. 1.
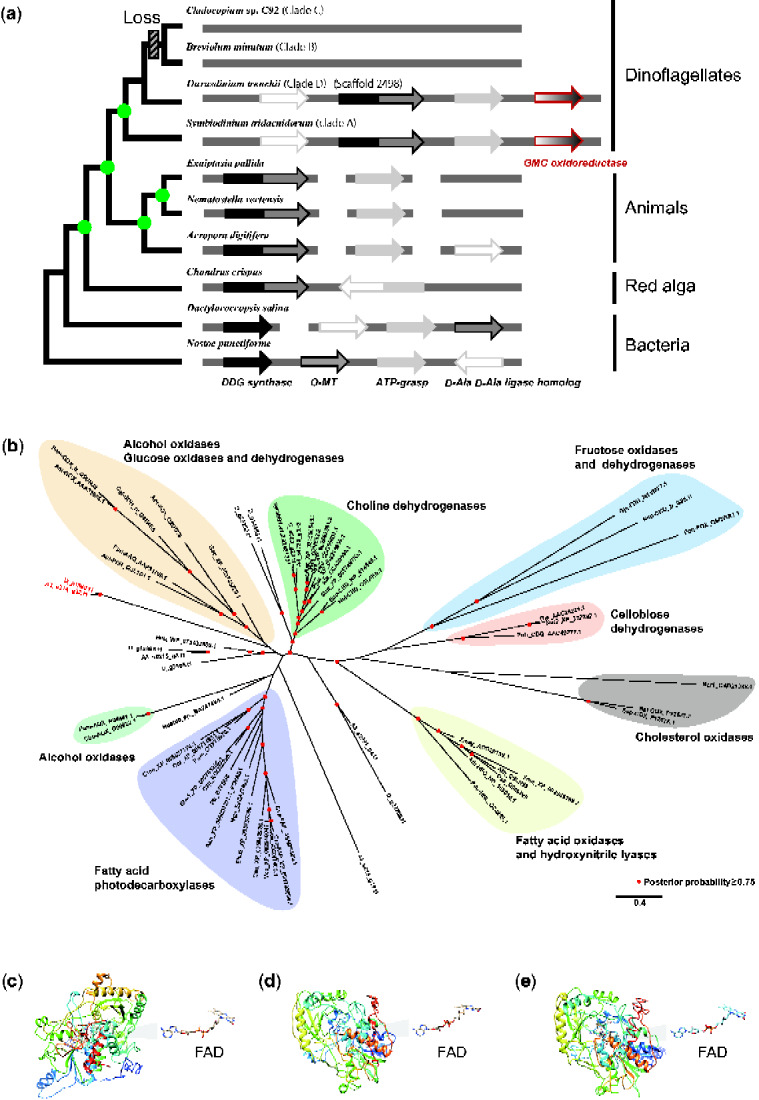
The dinoflagellates, Symbiodinium tridacnidorum and Durusdinium trenchii, both possess a probable gene cluster for MAA biosynthesis. (a) A gene cluster in the D. trenchii genome and a potential evolutionary scenario for MAA biosynthesis in the family Symbiodiniaceae. Topology of the tree is based on the phylogenetic tree of the DDG synthase family with bootstrap support >90%, as shown in green circle. The detail is shown in supplementary figure S2, Supplementary Material online. The positions of clades B and C with no MAA biosynthetic gene cluster are assumed based upon previous 28S rDNA phylogenies (Shoguchi et al. 2018). (b) A molecular phylogeny of GMC family enzymes showing evolutionary relationships of the proteins. Proteins from the neighboring GMC oxidoreductase in (a) the MAA biosynthetic gene cluster are shown in red. Genes for choline dehydrogenase are encoded in dinoflagellate genomes and others in the Symbiodiniaceae are unclassified enzymes in the GMC family. (c–e) 3D structures of the enzymes and their use of flavin adenine dinucleotide (FAD) as a cofactor were predicted using I-TASSER (Zhang 2008). (c) Fatty acid photodecarboxylase (FAP), a light-activated enzyme from Chlorella variabilis. (d) g1386 of D. trenchii. (e) s314_g32.t1 of S. tridacnidorum.
The predicted ligase had domains for the GMC oxidoreductase family (GMC_oxred_N of PF00732 and GMC_oxred_C of PF05199), which includes proteins having diverse catalytic activities. A molecular phylogeny of GMC oxidoreductases indicated that this one does not belong to a subfamily with known functions and that it may constitute a sister group of alcohol oxidases and glucose oxidases and dehydrogenases (fig. 1b). Recently, it has been shown that GMC oxidoreductases in algae include photoenzymes (Sorigué et al. 2017; Björn 2018), which have the light-capturing flavin adenine dinucleotide (FAD) as a cofactor (fig. 1c). Using I-TASSER software, prediction of the 3D structure of Symbiodiniaceae GMC oxidoreductases showed that they likely also carry FAD (fig. 1d and e), suggesting the possibility of a photoenzyme (Sorigué et al. 2017; Björn 2018). Future studies may clarify the relationship between light intensity and MAA biosynthesis. Coral genomes also have genes for MAAs (Shinzato et al. 2011, 2014), but they do not seem to have GMC oxidoreductase. If coral bleaching is triggered by high SSTs and insolation (Lesser 2019), the Symbiodiniaceae capacity for MAA biosynthesis may contribute to bleaching resistance.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank members of the DNA sequencing section (Drs Ryo Koyanagi and Miyuki Kanda) from the Okinawa Institute of Science and Technology (OIST) for conducting whole-genome sequencing and the Scientific Computing & Data Analysis section of OIST for IT support. We are grateful to Ms Maria Khalturina for sequencing library preparation and Drs Hiroya Yamano and Kaoru Sugihara for providing F. speciosa. We thank Dr Steven D. Aird for helpful comments and editing of the article. This work was supported in part by Japan Society for the Promotion of Science (No. 20K05798 to E.S. and No. 20H03235 to C.S.), Japan. We also greatly appreciate OIST support for the Marine Genomics Unit (N.S.).
Data Availability
This project has been deposited at DNA Data Bank of Japan (DDBJ) under the accession number PRJDB10306. Assembled genomic and transcriptomic data are accessible from the following site: https://marinegenomics.oist.jp/symbd/viewer/download?project_id=102.
Literature Cited
- Abrego D, van Oppen MJH, Willis BL. 2009. Onset of algal endosymbiont specificity varies among closely related species of Acropora corals during early ontogeny. Mol Ecol. 18(16):3532–3543. [DOI] [PubMed] [Google Scholar]
- Aihara Y, Takahashi S, Minagawa J. 2016. Heat induction of cyclic electron flow around photosystem I in the symbiotic dinoflagellate symbiodinium. Plant Physiol. 171(1):522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda M, et al. 2016. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci Rep. 6:39734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AC. 2003. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu Rev Ecol Evol Syst. 34(1):661–689. [Google Scholar]
- Banaszak AT, LaJeunesse TC, Trench RK. 2000. The synthesis of mycosporine-like amino acids (MAAs) by cultured, symbiotic dinoflagellates. J Exp Mar Bio Ecol. 249(2):219–233. [DOI] [PubMed] [Google Scholar]
- Beedessee G, et al. 2019. Diversified secondary metabolite biosynthesis gene repertoire revealed in symbiotic dinoflagellates. Sci Rep. 9(1):1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beedessee G, et al. 2020. Integrated omics unveil the secondary metabolic landscape of a basal dinoflagellate. BMC Biol. 18(1):139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beedessee G, Hisata K, Roy MC, Satoh N, Shoguchi E. 2015. Multifunctional polyketide synthase genes identified by genomic survey of the symbiotic dinoflagellate, Symbiodinium minutum. BMC Genomics 16:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellantuono AJ, Dougan KE, Granados‐Cifuentes C, Rodriguez‐Lanetty M. 2019. Free living and symbiotic lifestyles of a thermotolerant coral endosymbiont display profoundly distinct transcriptomes under both stable and heat stress conditions. Mol Ecol. 28(24):5265–5281. [DOI] [PubMed] [Google Scholar]
- Berkelmans R, Van Oppen MJH. 2006. The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc R Soc B. 273(1599):2305–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björn LO. 2018. Photoenzymes and related topics: an update. Photochem Photobiol. 94(3):459–465. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27(4):578–579. [DOI] [PubMed] [Google Scholar]
- Chen Y, González-Pech RA, Stephens TG, Bhattacharya D, Chan CX. 2020. Evidence that inconsistent gene prediction can mislead analysis of dinoflagellate genomes. J Phycol. 56(1):6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffroth MA, Lewis CF, Santos SR, Weaver JL. 2006. Environmental populations of symbiotic dinoflagellates in the genus Symbiodinium can initiate symbioses with reef cnidarians. Curr Biol. 16(23):R985–R987. [DOI] [PubMed] [Google Scholar]
- Coffroth MA, Santos SR. 2005. Genetic diversity of symbiotic dinoflagellates in the genus Symbiodinium. Protist 156(1):19–34. [DOI] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Pech RA, Bhattacharya D, Ragan MA, Chan CX. 2019. Genome evolution of coral reef symbionts as intracellular residents. Trends Ecol Evol. 34(9):799–806. [DOI] [PubMed] [Google Scholar]
- González-Pech RA, Ragan MA, Chan CX. 2017. Signatures of adaptation and symbiosis in genomes and transcriptomes of Symbiodinium. Sci Rep. 7(1):15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EA, Davies SW, Matz MV, Medina M. 2014. Quantifying cryptic Symbiodinium diversity within Orbicella faveolata and Orbicella franksi at the Flower Garden Banks, Gulf of Mexico. PeerJ 2:e386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka M. 2016. Life history and stress response of Scleractinian corals In: Kayanne H, editor. Coral reef science: strategy for ecosystem symbiosis and coexistence with humans under multiple stresses. Tokyo (Japan: ): Springer; p. 1–24. [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. 2016. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32(5):767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi A, et al. 2012. Effects of acidified seawater on coral calcification and symbiotic algae on the massive coral Porites australiensis. Mar Environ Res. 73:32–36. [DOI] [PubMed] [Google Scholar]
- Kajitani R, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24(8):1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp DW, Hernandez-Pech X, Iglesias-Prieto R, Fitt WK, Schmidt GW. 2014. Community dynamics and physiology of Symbiodinium spp. before, during, and after a coral bleaching event. Limnol Oceanogr. 59(3):788–797. [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12(4):656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klueter A, Crandall JB, Archer FI, Teece MA, Coffroth MA. 2015. Taxonomic and environmental variation of metabolite profiles in marine dinoflagellates of the genus symbiodinium. Metabolites 5(1):74–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi R, et al. 2013. MarinegenomicsDB: an integrated genome viewer for community-based annotation of genomes. Zoolog Sci. 30(10):797–800. [DOI] [PubMed] [Google Scholar]
- LaJeunesse TC, et al. 2014. Ecologically differentiated stress-tolerant endosymbionts in the dinoflagellate genus Symbiodinium (Dinophyceae) Clade D are different species. Phycologia 53(4):305–319. [Google Scholar]
- LaJeunesse TC, et al. 2018. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol. 28(16):2570–2580. [DOI] [PubMed] [Google Scholar]
- Lesser MP. 2019. Phylogenetic signature of light and thermal stress for the endosymbiotic dinoflagellates of corals (Family Symbiodiniaceae). Limnol Oceanogr. 64(5):1852–1863. [Google Scholar]
- Li T, et al. 2020. Genome improvement and core gene set refinement of Fugacium kawagutii. Microorganisms 8(1):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, et al. 2018. Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun Biol. 1:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M, UniProt Consortium. 2011. UniProt Knowledgebase: a hub of integrated protein data. Database 2011(0):bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mies M. 2019. Evolution, diversity, distribution and the endangered future of the giant clam–Symbiodiniaceae association. Coral Reefs. 38(6):1067–1084. [Google Scholar]
- Nakamura T, van Woesik R. 2001. Water-flow rates and passive diffusion partially explain differential survival of corals during the 1998 bleaching event. Mar Ecol Prog Ser. 212:301–304. [Google Scholar]
- Nishitsuji K, et al. 2020. Comparative genomics of four strains of the ediblebrown alga, Cladosiphon okamuranus. BMC Genomics 21(1):422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23(9):1061–1067. [DOI] [PubMed] [Google Scholar]
- Pochon X, Gates RD. 2010. A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai’i. Mol Phylogenet Evol. 56(1):492–497. [DOI] [PubMed] [Google Scholar]
- Pochon X, Putnam HM, Gates RD. 2014. Multi-gene analysis of Symbiodinium dinoflagellates: a perspective on rarity, symbiosis, and evolution. PeerJ 2:e394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provasoli L, Mclaughlin JJ, Droop MR. 1957. The development of artificial media for marine algae. Archiv Mikrobiol. 25(4):392–428. [DOI] [PubMed] [Google Scholar]
- Punta M, et al. 2012. The Pfam protein families database. Nucleic Acids Res. 40(D1):D290–D301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JM, Bruns BU, Fitt WK, Schmidt GW. 2008. Enhanced photoprotection pathways in symbiotic dinoflagellates of shallow-water corals and other cnidarians. Proc Natl Acad Sci U S A. 105(36):13674–13678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan R. 2004. Thermal adaptation in reef coral symbionts. Nature 430(7001):742–742. [DOI] [PubMed] [Google Scholar]
- Shinzato C, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476(7360):320–323. [DOI] [PubMed] [Google Scholar]
- Shinzato C, Mungpakdee S, Satoh N, Shoguchi E. 2014. A genomic approach to coral-dinoflagellate symbiosis: studies of Acropora digitifera and Symbiodinium minutum. Front Microbiol. 5:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoguchi E, et al. 2013. Draft Assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 23(15):1399–1408. [DOI] [PubMed] [Google Scholar]
- Shoguchi E, et al. 2018. Two divergent Symbiodinium genomes reveal conservation of a gene cluster for sunscreen biosynthesis and recently lost genes. BMC Genomics 19(1):458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoguchi E, et al. 2020. Correlation between organelle genetic variation and RNA editing in dinoflagellates associated with the coral Acropora digitifera. Genome Biol Evol. 12(3):203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Sorigué D, et al. 2017. An algal photoenzyme converts fatty acids to hydrocarbons. Science 357(6354):903–907. [DOI] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24(5):637–644. [DOI] [PubMed] [Google Scholar]
- Stat M, Gates RD. 2011. Clade D Symbiodinium in Scleractinian corals: a ‘Nugget’ of Hope, a selfish opportunist, an ominous sign, or all of the above? J Mar Biol. 2011:1–9. [Google Scholar]
- Thornhill DJ, Lewis AM, Wham DC, LaJeunesse TC. 2014. Host-specialist lineages dominate the adaptive radiation of reef coral endosymbionts. Evolution 68(2):352–367. [DOI] [PubMed] [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MX, Medina M. 2012. The role of microalgal symbionts (Symbiodinium) in holobiont physiology In: Advances in botanical research. Vol. 64 Elsevier; p. 119–140. [Google Scholar]
- Weis VM, Davy SK, Hoegh-Guldberg O, Rodriguez-Lanetty M, Pringle JR. 2008. Cell biology in model systems as the key to understanding corals. Trends Ecol Evol. 23(7):369–376. [DOI] [PubMed] [Google Scholar]
- Weis VM, Reynolds WS, deBoer MD, Krupp DA. 2001. Host-symbiont specificity during onset of symbiosis between the dinoflagellates Symbiodinium spp. and planula larvae of the scleractinian coral Fungia scutaria. Coral Reefs 20(3):301–308. [Google Scholar]
- Yamashita H, Suzuki G, Hayashibara T, Koike K. 2013. Acropora recruits harbor ‘rare’ Symbiodinium in the environmental pool. Coral Reefs 32(2):355–366. [Google Scholar]
- Yuyama I, Ishikawa M, Nozawa M, Yoshida M-A, Ikeo K. 2018. Transcriptomic changes with increasing algal symbiont reveal the detailed process underlying establishment of coral-algal symbiosis. Sci Rep. 8(1):16802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This project has been deposited at DNA Data Bank of Japan (DDBJ) under the accession number PRJDB10306. Assembled genomic and transcriptomic data are accessible from the following site: https://marinegenomics.oist.jp/symbd/viewer/download?project_id=102.


