Abstract
Light-inducible optogenetic systems offer precise spatiotemporal control over a myriad of biologic processes. Unfortunately, current systems are inherently limited by their dependence on external light sources for their activation. Further, the utility of laser/LED-based illumination strategies are often constrained by the need for invasive surgical procedures to deliver such devices and local heat production, photobleaching and phototoxicity that compromises cell and tissue viability. To overcome these limitations, we developed a novel BRET-activated optogenetics (BEACON) system that employs biologic light to control optogenetic tools. BEACON is driven by self-illuminating bioluminescent-fluorescent proteins that generate “spectrally tuned” biologic light via bioluminescence resonance energy transfer (BRET). Notably, BEACON robustly activates a variety of commonly used optogenetic systems in a spatially restricted fashion, and at physiologically relevant time scales, to levels that are achieved by conventional laser/LED light sources.
Keywords: optogenetics, chemogenetics, bioluminescence, BRET, NanoLuciferase, LumiFluor
Graphical Abstract
Optogenetics was first extensively applied in the field of neuroscience for the light-activated control of neuronal action potentials in an effort to map neural circuits in the brain.1 However, the past decade has seen an explosion in both the identification of novel proteins responsive to varying wavelengths of light across the visible and near-infrared spectrum, and their use by the broader scientific community.1–3 Consequently, this has led to the creation of a wide variety of unique optogenetic systems that can be leveraged to precisely control a myriad of biologic processes, and in a spatiotemporally defined manner.2,4–8 Indeed, these diverse optogenetic tool kits have been adopted by fields ranging from chemistry (drug uncaging)9 and synthetic biology (cell signaling circuits),10–12 to molecular biology (protein-protein interactions, recruitment, signaling and transcription)3,4,13,14 and cancer biology.15,16 However, despite the rapidly expanding adoption and application of optogenetics across diverse disciplines, there are several hurdles inherent to currently available technologies. In particular, supplying adequate light intensity and of an appropriate wavelength is a major issue that often requires expensive light sources (lasers/LED boards) that are specially engineered to deliver specific light requirements (wavelength, intensity, and duration) needed for different photosensitive proteins.17 This requirement for powerful, externally delivered light for extended intervals, via microscope illumination paths for in vitro studies, or using surgically implanted optical fibers for in vivo experiments, can provoke undesirable photobleaching, photoinactivation, and phototoxicity, all of which compromise cell viability and experimental outcomes.18,19 Thus, there is a clear need to develop noninvasive actuators in the form of spectrally tuned “in situ” light sources that can be deployed, either as surrogates or as a complementary activation paradigm, to conventionally used external light sources to enable spatial and temporal control over optogenetic systems. To address this need, we adapted self-illuminating biologic light sources with tunable emission intensity and wavelengths (LumiFluor)20 to develop and validate a chemo-optogenetic activation paradigm, coined BEACON (BRET-activated optogenetics). Here we report that the in cellulo biologic light generated with BEACON serves as a robust and tunable light source suitable for activating various light-responsive optogenetic systems.
RESULTS AND DISCUSSION
We recently reported the creation of novel bioluminescent-fluorescent proteins (LumiFluor)20 generated by fusing an ATP-independent luciferase (NanoLuc) to the enhanced Green Fluorescent Protein (eGFP) (Figure 1A). Using a similar strategy, additional LumiFluor-like spectral variants employing NanoLuc were subsequently generated.21 While several bioluminescence resonance energy transfer (BRET)-based molecules conceptually similar to LumiFluors have recently emerged, these have employed conventional luciferases like Renilla, Gaussia, or Firefly as the donor moiety.22–28 Compared to luciferases that have low quantum yield (QY) and consume 10- to 26-fold more substrate per photon emitted (e.g., Renilla, RLuc; QY = 0.02−0.1),29,30 or are subject to catalytic byproduct autoinhibition (e.g., Firefly),31 NanoLuc is resistant to autoinhibition and displays a stable, glow-type light emission with naturally high QY (≥0.34) per molecule of substrate catalyzed (Table S1).32,33 Importantly, LumiFluor molecules display even greater QY due to the natural resonance energy transfer phenomenon of BRET.34,35 Upon the addition of its catalytic substrate, furimazine (FZ), NanoLuc generates bioluminescent light that excites a proximally located acceptor molecule (eGFP), which then re-emits light according to its own emission spectra (Emmax ~ 510 nm), thereby effectively “tuning” the spectral light output (Figure 1A). We previously demonstrated that this green LumiFluor (dubbed GpNLuc) exhibits a >10-fold increase in light output compared to NanoLuc alone and is nearly 100× brighter than previously reported BRET fusion proteins that employ a “conventional” luciferase like RLuc (ex. NanoLantern).20,22 As expected, the enhanced quantum yield offered by LumiFluor results in a significant ~3-fold increase (p < 0.0005) in overall power output compared to NanoLuc alone (Figure 1B). Thus, our optimized BRET-based biologic light sources can readily generate photon power outputs in the mW/m2 range, when expressed at physiologically relevant levels (μM). Given that most optogenetic systems can be activated by power densities within the mW/m2 to W/m2 range, and the liabilities associated with externally delivered light sources for the activation of optogenetic systems, we sought to evaluate the utility and robustness of our LumiFluors toward a BRET-activated optogenetics (BEACON) paradigm.
Figure 1.
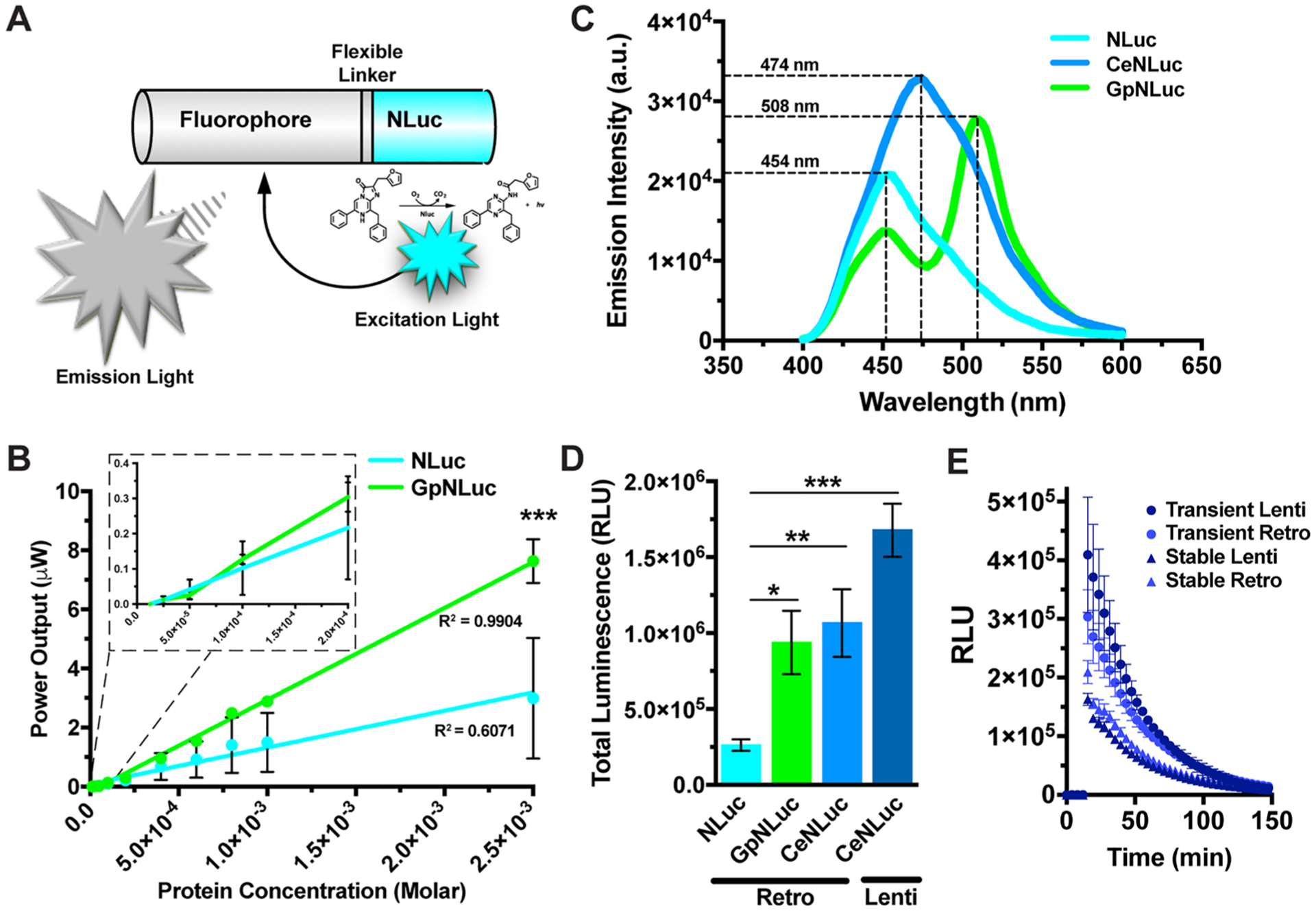
Development and characterization of the LumiFluor biologic light sources. (A) Schematic of bioluminescent-fluorescent (“LumiFluor”) fusion molecules. The C-terminus of fluorescent proteins such as eGFP or mCerulean3 were fused to the N-terminus of NanoLuciferase using a short flexible 5 amino acid linker (DISGG). (B) Surface power densities generated by varying concentration of purified recombinant GpNLuc protein versus unmodified NLuc, (FZ = 100 μM, n = 4). (C) Spectral emission scans of transiently transfected HEK293T cells expressing NLuc, CeNLuc, and GpNLuc biologic light sources at equimolar levels (FZ = 50 μM). (D) Total luminescence intensities of transiently transfected HEK293T cells expressing NLuc, GpNLuc, and CeNLuc at equimolar levels (FZ = 20 μM, n = 2). (E) Characterization of the luminescence intensities of LumiFluor-expressing HEK293 cells harboring the CeNLuc retroviral or lentiviral expression vectors in a transiently transfected versus stably integrated format (FZ = 20 μM, n = 2). Stable cells were selected by FACS or Puromycin, respectively. Data are expressed as mean ± SEM, n = biologic replicates, *p < 0.05, **p < 0.005, ***p < 0.0005 as determined by a Student’s t-test. All experiments were performed with at least 2–4 technical replicates each.
Optogenetic proteins responsive to a wide range of wavelengths ranging from the violet to red/far-red spectrum have been developed, yet optogenetic systems responsive to the blue-green spectrum (Ex 420–520 nm) continue to dominate the field. While this green light source (GpNLuc, Figure S1B right) offers peak spectral emission in the 510 nm range, it suffers from less than optimal output in the 450–470 nm range (Figure 1C). Thus, we first sought to develop a “cyan” biologic light source that would be compatible with the most commonly used blue-green sensitive optogenetic systems. We created this cyan-based LumiFluor (CeNLuc, Figure S1B left) by fusing NanoLuc in-frame with a cyan fluorescent protein, mCerulean3, separated by a flexible five amino acid linker (DISGG). Due to its improved quantum yield (0.87), superior photostability, enhanced brightness and pH insensitivity, mCerulean3 was chosen as the BRET acceptor over other cyan fluorescent proteins like mTurquoise.36 Further, mCerulean3 lacks the photoswitching behavior manifest with mTurquoise and other cyan fluorescent proteins,36 an important variable given that photosensitive proteins can be inactivated by exposure to other wavelengths.3 Analysis of the spectral emission profiles of NLuc donor emission and mCerulean3 acceptor excitation predicts robust CeNLuc BRET signal due to the high J(λ) overlap integral between donor and acceptor.20 As expected, we confirmed high CeNLuc BRET efficiency as evidenced by a single, wide emission peak centered around the mCerulean3 emission maxima (~474 nm) (Figure 1C). Equimolar expression of each biologic light source from a retroviral based plasmid (using a PGK promoter, Figure S1A, top) revealed that while the total light output from GpNLuc and CeNLuc are nearly identical, they are both ~4 fold brighter than NanoLuc alone (Figure 1D).
To further characterize this “cyan” biologic light source we transiently transfected or stably transduced cells with lentiviral- or retroviral-based constructs that express CeNLuc at varying levels. The stably integrated cells achieved lower total light emissions compared to the transiently transfected conditions due to fewer reporter templates, which is also manifest as reduced CeNLuc mRNA expression levels (Figure 1E and Figure S1C). To define the temporal kinetics of light emission from our “cyan” biologic light source, we performed a furimazine dose titration using the CeNLuc-expressing stable cell lines generated in Figure 1E. While a sustained light output is observed when using higher doses of furimazine, lower dose conditions displayed a shorter-lived, flash-type light emission profile (Figure S1D,E and Figure S3B), indicating that the kinetic profile for light emission can be readily fine-tuned by modulating the concentration of the luciferase substrate. Furthermore, total photon output exhibits a strong linear relationship (R2 = 0.99) with cell number (Figure S1F–H), indicating that total light emission can also be fine-tuned by modulating both the number of CeNLuc expressing cells present in the system (Figure S1F–H) and the CeNLuc expression level per cell (Figure S1D,E).
Prior to testing the utility of a CeNLuc-activated optogenetic paradigm, we next characterized the total photon power output and the photon power densities achieved by this biologic light source. Not surprisingly, the total photon power output displays a strong linear relationship (R2 = 0.97) with cell number (Figure S1I). Although CeNLuc expressing cells can only emit power densities in the low mW/m2 range, at a distance of ~2 cm (Figure S1J), light power densities are inversely related to the square of the distance from a point source. Therefore, the power densities generated in the immediate vicinity of CeNLuc (100s nm range) are orders of magnitude higher, and should therefore activate commonly used blue-green optogenetic systems. While the CeNLuc expressing cells can collectively generate photon power densities in the mW/m2 range, the per cell photon output is only ~450 fW (femtoWatt) (Figure S1K). Thus, not only can cells expressing CeNLuc readily achieve photon emissions in the mW/m2 range, but the total light output and kinetic profile of light emission can be reliably fine-tuned by modulating both cell number and the amount of furimazine added to the system, respectively.
To assess the feasibility and range of applications of the BEACON system, we rigorously tested the ability of the CeNLuc LumiFluor to activate a diverse collection of blue-green optogenetic systems, including the cryptochrome-based CRY2-CIBN, LOV-based FKF1-Gi and iLID, and the VVD-based pMagnet systems. We first applied BEACON to several light-activated transcriptional systems (Figure 2A), wherein the addition of furimazine leads to robust biologic light production (cytoplasmic), and subsequent activation of the optogenetic system (nuclear) to induce the expression of a reporter gene. We found that both GpNLuc and CeNLuc can equivalently activate the split Cre recombinase CRY2-CIBN dimerization system37 (Figure S2A) in a dose dependent fashion (Figure 2B), in keeping with the previously established spectral profile of CRY2-CIBN photoactivation.37,38 Notably, CeNLuc can potently activate the CRY2-CIBN system at levels comparable to conventionally used LED (~37 W/m2) light sources (Figure 2C, p < 0.0005). We next evaluated the utility of BEACON on a Gal4-DBD based FKF1-GI dimerization system39 (Figure S2B) and found that the “cyan” biologic light source, CeNLuc, activated this optogenetic system to higher levels (~2-fold, p < 0.05) than GpNLuc (Figure 2D). Moreover, additional optimization of experimental variables (Figure S2C) demonstrated that CeNLuc-based activation of FKF1-GI can reliably achieve ~40-fold gene induction (Figure 2E), which is superior to that seen with many other Gal4 fusion transcription factors such as CREB-Gal4.40 We also confirmed that beyond conventional optogenetic systems, BEACON is applicable and robust for activating newer, nascently emerging optogenetic systems. This includes a dCas9-gRNA based CRY2-CIBN dimerization system41 (Figure S2D) that takes advantage of gRNA targeting specificity for the precise and targeted activation of gene expression. Optimization of the experimental parameters (Figure S2E) revealed that CeNLuc-based activation of this system leads to robust (~20-fold, p < 0.0005) induction of reporter gene expression (Figure 2F), comparable to what others have observed for constitutively active dCas9-CRISPR transcription activation systems.42–44 We next applied BEACON to a recently described enhanced split Cre recombinase system based on a newly optimized, VVD-based, light-responsive optogenetic system (pMagnet).45,46 While CeNLuc mediated activation of pMagnet only results in a modest induction of the reporter gene (~4-fold), subcellular colocalization of CeNLuc (2xNLS-CeNLuc) and pMagnet leads to significantly enhanced (~10-fold, p < 0.0005) induction of the reporter gene (Figure 2G). Thus, the LumiFluor-based BEACON platform can potently activate a variety of widely used conventional blue-green optogenetic systems and does so to levels comparable to that achieved by traditional LED light sources. Moreover, the potency of CeNLuc-mediated activation of these optogenetic systems can be further enhanced by colocalizing the biologic light source and the light responsive proteins within the same subcellular compartment.
Figure 2.
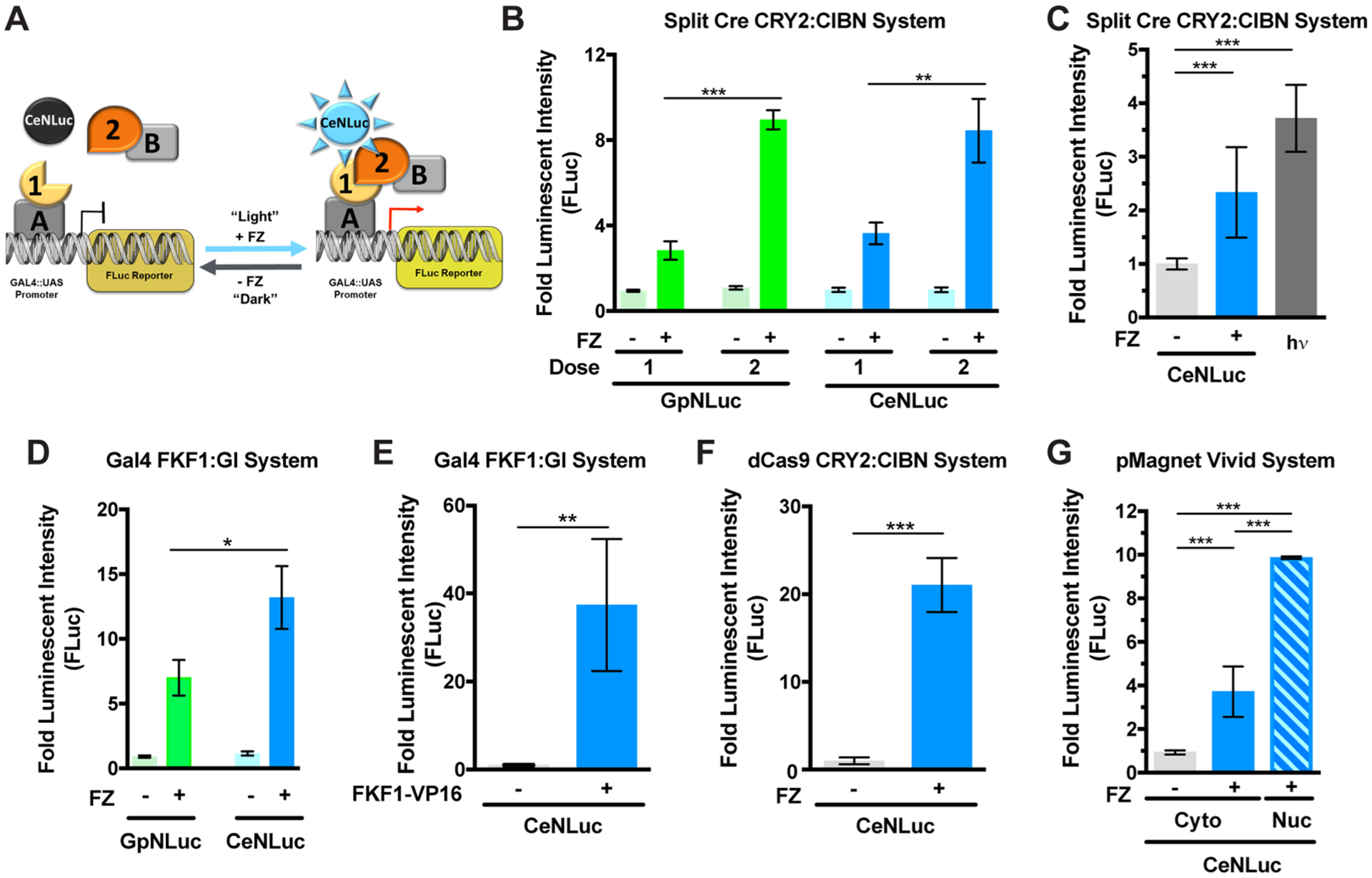
LumiFluors can activate diverse optogenetic systems. (A) Schematic of optogenetic transcription systems wherein biologic light generated by CeNLuc upon FZ administration (left) promotes interactions between light-responsive proteins partners (right). Biologic light-mediated optogenetic control is measured with a firefly luciferase reporter before (“Dark”) and after (“Light”) FZ-induced illumination (right). (B) Both the “green” (GpNLuc) and “cyan” (CeNLuc) LumiFluor light sources comparably activate the split Cre recombinase CRY2-CIBN system, and in a dose dependent fashion (FZ = 20 μM, n = 2). (C) CeNLuc (2 pulses of FZ) activates the CRY2-CIBN system at levels comparable to those seen when using a ~37 W/m2 LED light source (2 × 5 min pulses, n = 3). (D) CeNLuc (Emmax = 474 nm) is significantly more potent at activating the FKF1-GI system than the GpNLuc (Emmax = 508 nm) light source (FZ = 20 μM, n = 2). (E) Optimization of the FKF1-GI plasmid ratios reveals that CeNLuc mediated activation can achieve ~40-fold transcriptional induction (FZ = 20 μM, n = 2). (F) CeNLuc robustly activates a dCas9-based CRY2-CIBN transcriptional activation system (FZ = 20 μM, n = 2). (G) While CeNLuc only modestly activates the pMagnet optogenetic system, subcellular colocalization of CeNLuc with this optogenetic system robustly enhances pMagnet activation. All experiments were conducted using 20 μM FZ. All experiments were performed in transiently transfected HEK293T cells as indicated in the SI Materials and Methods. Data are expressed as mean ± SEM, n = biologic replicates, *p < 0.05, **p < 0.005, ***p < 0.0005 as determined by a Student’s t-test. All experiments were performed with at least 3–4 technical replicates each.
While optogenetic systems have widespread application as transcriptional activation systems, their true power lies in the ability to tightly regulate specific cellular processes in a spatiotemporally defined manner.37,47–49 Moreover, their unique ability to be rapidly switched on/off in a targeted, subcellular fashion has made them an indispensable tool for modern cell biologists.2 With this in mind, we tested the ability of our LumiFluor-based BEACON platform to activate optogenetic systems at physiologically relevant spatiotemporal scales. For this, we developed a modified version of the recently described LOV2 domain-based iLID system,5,50 wherein light stimulation induces a conformational change that exposes the SsrA peptide located on an outer mitochondrial membrane (OMM)-anchored iLID-SsrA fusion (iLID-mito, charcoal-purple), which then promotes SsrA binding to SspB and recruitment of an otherwise cytoplasmically diffuse SspBR73Q-iRFP670 (iRFPmicro, magenta-red) to the mitochondrial surface via direct interaction of the SsrA-SspB peptides (Figure 3A). Interestingly, when expressed in a nontargeted manner, CeNLuc is incapable of reliably activating iLID-mito (Figure 3B and Figure S3A). In contrast, mitochondrial colocalization of CeNLuc (OMMCeNLuc) results in rapid and robust activation of the iLID-mito system (Figure 3B and Figure S3A). Most importantly, OMMCeNLuc can activate iLID-mito to levels nearly identical to those observed when using powerful conventional light sources (Figure 3B; hν = 680 W/m2 and Figure S3A; hν = 37 W/m2).
Figure 3.
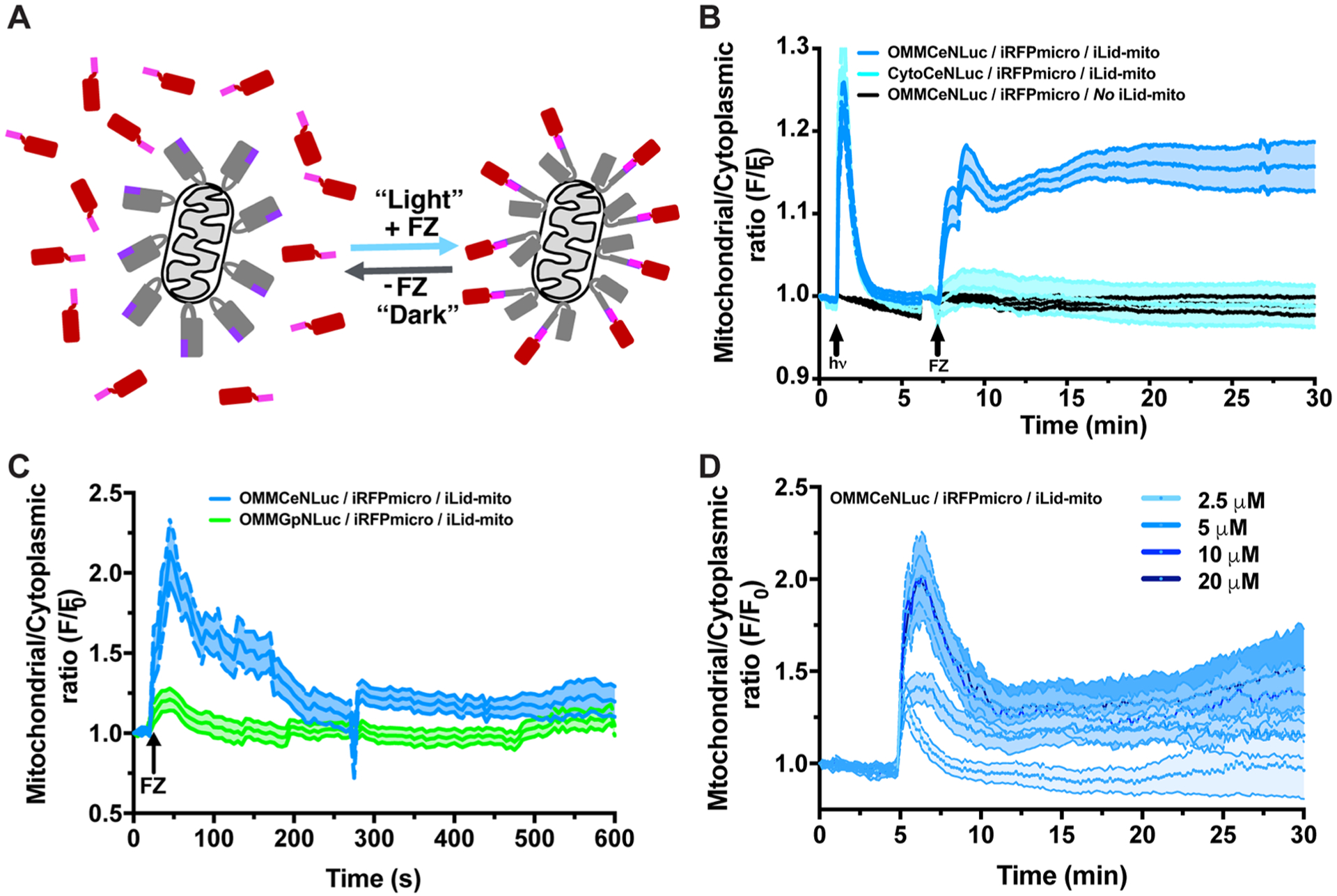
Tunable wavelengths and power output coupled with targeted subcellular LumiFluor activation enables robust spatial control of optogenetic systems. (A) Schematic of the iLID-mito recruitment system that is comprised of an outer mitochondrial membrane-targeted Venus-iLID-SsrA (charcoal-purple) and a diffusely expressed, cytoplasm-localized iRFP670-SspB (red-magenta). Under dark conditions (− FZ), the LOV2-based iLID protein sterically blocks (“cages”) the SsrA peptide from interacting with its natural obligate binding partner SspB. Exposure of iLID to light (+ FZ) induces a conformational change in the LOV2 Jα helix that exposes the SsrA peptide, enabling it to bind SspB, which promotes the rapid and robust recruitment of iRFP670-SspB to the mitochondrial surface. (B) Subcellular colocalization of the CeNLuc biologic light source and iLID-mito system leads to a rapid and robust activation of iLID, at levels comparable to a powerful light pulse provided by a traditional (680 W/m2) light source. Cytoplasm-localized CeNLuc (CytoCeNluc, n = 6 cells) or mitochondrially targeted CeNLuc (OMM, n = 9 cells) were cotransfected with the iLID-mito system into HeLa cells. Cells lacking Venus-iLID were used as negative controls (n = 6 cells). Experiments were conducted using 20 μM FZ and data are expressed as mean ± SEM. (C) While both CeNLuc and GpNLuc light sources can activate various optogenetic systems (Figure 2), only the cyan light source (CeNLuc) can activate the iLID-mito system. Experiments were conducted using 10 μM FZ and data are expressed as mean ± SEM (OMM-CeNLuc = 6 cells, OMM-GpNLuc = 3 cells). (D) Differential iLID-mito activation profiles can be generated by titrating the amounts of FZ to “tune” CeNLuc-based biologic light output levels. Data shown are the average of 3–6 cells per condition expressed as mean ± SEM. All experiments were performed in transiently transfected HeLa cells as indicated in the SI Materials and Methods.
Given that GpNLuc and CeNLuc activate the CRY2-CIBN and FKF1-GI optogenetic systems with different levels of potency, we directly compared their ability to activate the iLID-mito system. We observed a strong preference (~2-fold, p < 0.0005) of the iLID-mito system for OMMCeNLuc compared to OMMGpNLuc (Figure 3C). While the apparent discrepancy between CeNLuc-versus GpNLuc-based activation of the iLID system seems counterintuitive, this is expected based on the absorbance/activation profile of the flavin-based LOV2 optogenetic system. Although peak absorbance for the LOV2 domain is ~450 nm, there is a precipitous drop in absorbance starting ~480–490 nm and light >500 nm has negligible effects in activating LOV2 domains. Importantly, CeNLuc has a broad emission peak centered at ~470 nm, which is ideally suited to activate the LOV2 optogenetic system compared to the narrower GpNLuc emission peak (centered ~510 nm). Thus, this apparent discrepancy of the LumiFluors toward activating iLID (CeNLuc ≫ GpNLuc) and the CRY2/CIBN (CeNLuc ~ GpNLuc) optogenetic systems is expected based on their previously published photoactivation profiles.51–54 Next, we explored the ability of CeNLuc to dynamically activate iLID-mito translocation by performing a furimazine dose titration experiment intended to generate long-lasting versus pulsatile kinetics. Differential temporal kinetics of iLID-mito activation are readily achieved, wherein a low dose of furimazine (2.5 μM) generates a short-lived pulse of iLID-mito activation, a subsaturating dose (5 μM) generates a more sustained and long lasting activation, and saturating doses (10–20 μM) generate a large peak of initial activation followed by a modest decay but sustained and long lasting-steady state of iLID-mito activation (Figure 3D). Thus, CeNLuc activates the iLID-mito system in a spatially restricted fashion and modulating the experimental parameters (i.e., FZ dosing) enables one to generate dynamic kinetic temporal activation profiles.
Apart from being able to modulate activation in a spatiotemporally defined fashion, most neurobiology and cell biology applications of optogenetics require repeated, and often pulsatile, stimulation. Unlike other luciferases, NanoLuc (the BRET donor in LumiFluors) is resistant to autoinhibition by its catalytic byproducts.33 To test if LumiFluors are amenable to repeated pulsatile activation paradigms without significant loss in overall light output, we designed studies based on the light signal decay kinetics from the FZ dose titrations performed on HEK293 cells stably expressing the retroviral CeNLuc construct (Figure S3B). We postulated that low doses of FZ (0.625–2.5 μM) could be repeatedly applied to generate pulsatile patterns of light output since this dosing regimen generates more acute bursts of light with a short half-life of 2–5 min (Figure S3B). Indeed, repeated pulses of FZ can be used to generate intense, but short-lived (5–10 min), pulsatile biologic light output without significant loss in overall light output over the course of the experiment (Figure S3C). Consequently, repeated 15 or 5 min pulsing of cells with FZ (see Figure S4 for experimental setup) leads to robust activation of the iLID-mito system, and this can subsequently be rapidly turned off by withdrawing FZ from the system (Figure 4A,B and Video S1). Notably, repeated pulses of FZ enables rapid and robust activation of iLID-mito with minimal signal loss over time. Thus, the BEACON platform can be applied for spatially restricted activation of optogenetic systems and leveraged for the repeated and pulsatile activation of optogenetic systems at physiologically relevant time scales (seconds to minutes).
Figure 4.
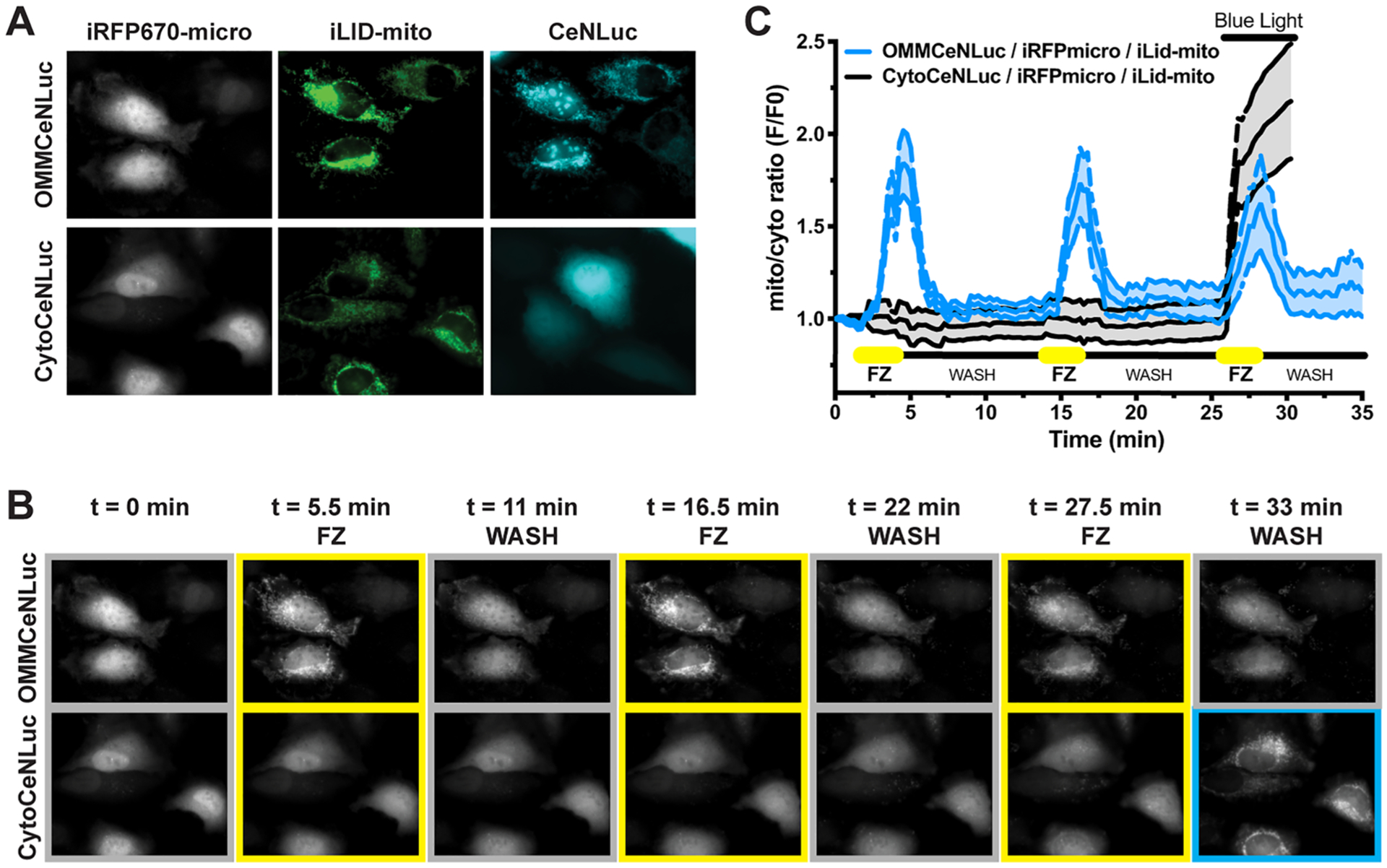
BEACON platform enables dynamic spatial and temporal control of optogenetic systems. (A) Representative images of HeLa cells cotransfected with the iLID-mito:iRFP670-micro system and OMM-CeNLuc (n = 4 cells) compared to control cells cotransfected with Cyto-CeNLuc (n = 4 cells). (B,C) Representative images (B) and quantification (C) of biologic light-induced iRFP670-micro translocation to iLID-mito on mitochondria. Pulsatile addition of FZ enables repeated activation of the iLID-mito system, while washing out the FZ results in rapid inactivation of iLID-mito. A 680 W/m2 pulse of 474 nm blue light source was used as a positive control for Cyto-CeNLuc. All experiments were conducted using 10 μM FZ. Data shown are the average of 3–6 cells per condition expressed as mean ± SEM. All experiments were performed in transiently transfected HeLa cells as indicated in the SI Materials and Methods.
Advantages of the BEACON system stem from the tunable nature of the “cold” biologic light that is produced, where the light intensity and wavelength generated by the BRET-based LumiFluors in response to a chemical substrate enables precise spatiotemporal modulation of molecular and cellular processes. Importantly, this ability to dynamically control light in a remote manner creates opportunities to noninvasively interrogate complex biological processes in vitro and in vivo. Similar chemo-optogenetic approaches have been recently described, but these generally employ direct fusions of the luciferase to an optogenetic actuator of interest (Luminopsins), or require that the luciferase is in a molecular complex with the photosensitive protein (SPARK2).55–59 In contrast, we demonstrate that robust expression or colocalization of the LumiFluor with photosensitive proteins is sufficient to achieve robust activation across a variety of optogenetic systems, thus eliminating the need for additional engineering and functional validation of each new protein fusion pairs. These data suggest that the BEACON paradigm relies on a radiative excitation-emission process known as fluorescence by unbound excitation from luminescence (FUEL) capable of activating distant light sensitive proteins, as opposed to a direct intermolecular BRET based activation.60 On the basis of our results (iLID, Figure 3 and Figure 4), the BEACON paradigm provides an opportunity to functionally dissect the importance of spatially localized signaling events,61 where LumiFluors allow one to selectively activate subcellular pools of light sensitive proteins while retaining the ability to stimulate systemic (whole cell) pools using conventional light sources within the same experiment/cell. Thus, BEACON offers a unique tool for synthetic biology approaches that can be multiplexed with orthogonal chemogenetic-based protein recruitment systems such as FKBP-rapamycin-FRB62 or receptor activation systems such as DREADD.12,63–65 For example, this could be accomplished by combining the BEACON platform with FKBP-rapamycin-FRB or eDHFR-TMP, enabling one to shuttle the LumiFluor light source between multiple subcellular compartments for selectively activating photosensitive proteins of choice in a spatially defined manner, while still preserving the ability to perform whole cell stimulations using conventional illumination approaches.66–69 Finally, similar LumiFluor-like molecules with additional spectral profiles have been recently described.21 Thus, one could theoretically multiplex “biologic light” sources (e.g., GLuc + LumiFluor)70 to selectively activate distinct pools of colocalized optogenetic systems (e.g., Luminopsin + iLID, respectively) due to the advent of ultrapotent and highly specific inhibitors for NanoLuc.71
While biologic light is not envisioned to replace all conventional light sources in optogenetics, we submit that BEACON offers a complementary noninvasive, affordable, and facile chemo-optogenetic approach to activate optogenetic systems that can be rapidly adopted toward a diverse variety of blue-green optogenetic systems. Moreover, we envision this optogenetic toolbox also including the rapidly expanding palette of novel luciferases (e.g., AkaLuc)72 and BRET-based “LumiFluor”-like molecules (e.g., Antares),73,74 which could be leveraged to activate additional red-shifted optogenetic systems. Finally, these LumiFluor tools could also be applied to the field of photochemistry,9,75 where light sensitive small molecules can be switched into an active state or uncaged/ photoreleased from pro-drug carriers76 to regulate diverse processes, including cell signaling, mitosis, motility, and proteolysis, or be activated as chemotherapeutic agents (photostatins).77 Thus, the BEACON platform expands both the optogenetic and the chemogenetic tool set to include the equivalent of small molecule-regulated molecular on/off light switches that enable one to dissect complex cellular processes involved in physiology and disease.
Supplementary Material
Methods and Materials; Figure S1: Characterization of power and photon output for the cyan-tuned LumiFluor biologic light source (CeNLuc);
Figure S2: Illustration of the various light-activated transcriptional reporter systems used, and optimization of plasmid ratios for maximal BEACON activation;
Figure S3: Representative images for CeNLuc mediated activation of iLID-iRFP670 recruitment to the outer mitochondrial membrane, and half-life analyses of CeNLuc mediated pulsatile light production;
Figure S4: Live cell imaging perfusion system setup;
Figure S5: Amino acid sequences of the various LumiFluor proteins evaluated for cellular optogenetic regulation;
Figure S6: Nucleotide sequence and plasmid map of 8× gRNA promoter for cellular optogenetic regulation of a Firefly luciferase reporter (PDF)
Table S1: Summary of previously described RLuc BRET-based biologic light sources compared to LumiFluors (PDF)
Video S1: Live cell imaging of CeNLuc mediated pulsatile activation of iLID-micro optogenetic system (AVI)
ACKNOWLEDGMENTS
The authors thank James Bear, Brian Kuhlman, Garret D. Stuber, Klaus K. Hahn, Paul M. Johnson and members of the Amelio Lab for reagents, helpful discussions, suggestions, and/ or scientific review of this manuscript. This work was supported by a NIH/NCI grant R01-CA167093 (to J.L. Cleveland), by a NIH/NIGMS T32-GM007092 training grant (to A.M. Musicant), NIH/NINDS and NIH/NCI grants R01-NS103486 and U01-CA207160 (to D.S. Lawrence), University Cancer Research Fund (UCRF; to A.L. Amelio), UNC Lineberger Tier 3 Developmental Award (to A.L. Amelio), the Cortner-Couch Endowed Chair for Cancer Research of the University of South Florida School of Medicine (to J.L. Cleveland) and by a NIH/NCI Howard Temin Pathway to Independence Award in Cancer Research R00-CA157954 (to A.L. Amelio). This work was also supported in part by NCI Comprehensive Cancer Center Grants P30-CA076292 and P30-CA016806 awarded to the H. Lee Moffitt Cancer Center & Research Institute and to the UNC Lineberger Comprehensive Cancer Center, respectively.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.9b00277.
The authors declare no competing financial interest.
REFERENCES
- (1).Deisseroth K (2015) Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci 18, 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Müller K, and Weber W (2013) Optogenetic tools for mammalian systems. Mol. BioSyst 9, 596–608. [DOI] [PubMed] [Google Scholar]
- (3).Tischer D, and Weiner OD (2014) Illuminating cell signalling with optogenetic tools. Nat. Rev. Mol. Cell Biol 15, 551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Beyer HM, Naumann S, Weber W, and Radziwill G (2015) Optogenetic control of signaling in mammalian cells. Biotechnol. J 10, 273–283. [DOI] [PubMed] [Google Scholar]
- (5).Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, and Kuhlman B (2015) Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. U. S. A 112, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW, and Gardner KH (2014) An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol 10, 196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Yamada M, Suzuki Y, Nagasaki SC, Okuno H, and Imayoshi I (2018) Light Control of the Tet Gene Expression System in Mammalian Cells. Cell Rep. 25, 487–500. [DOI] [PubMed] [Google Scholar]
- (8).Pathak GP, Vrana JD, and Tucker CL (2013) Optogenetic control of cell function using engineered photoreceptors. Biol. Cell 105, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lindberg E, Angerani S, Anzola M, and Winssinger N (2018) Luciferase-induced photoreductive uncaging of small-molecule effectors. Nat. Commun 9, 3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Lim WA (2010) Designing customized cell signalling circuits. Nat. Rev. Mol. Cell Biol 11, 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Liu Z, Zhang J, Jin J, Geng Z, Qi Q, and Liang Q (2018) Programming Bacteria With Light-Sensors and Applications in Synthetic Biology. Front. Microbiol 9, 2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Toettcher JE, Voigt CA, Weiner OD, and Lim WA (2011) The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nat. Methods 8, 35–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Toettcher JE, Weiner OD, and Lim WA (2013) Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Johnson HE, and Toettcher JE (2018) Illuminating developmental biology with cellular optogenetics. Curr. Opin. Biotechnol 52, 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kiełbus M, Czapiński J, Odrzywolski A, Stasiak G, Szymańska K, Kałafut J, Kos M, Giannopoulos K, Stepulak A, and Rivero-Müller A (2018) Optogenetics in cancer drug discovery. Expert Opin. Drug Discovery 13, 459–472. [DOI] [PubMed] [Google Scholar]
- (16).Bugaj LJ, Sabnis AJ, Mitchell A, Garbarino JE, Toettcher JE, Bivona TG, and Lim WA (2018) Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science 361, No. eaao3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Yizhar O, Fenno LE, Davidson TJ, Mogri M, and Deisseroth K (2011) Optogenetics in neural systems. Neuron 71, 9–34. [DOI] [PubMed] [Google Scholar]
- (18).Kale RP, Kouzani AZ, Walder K, Berk M, and Tye SJ (2015) Evolution of optogenetic microdevices. Neurophotonics 2, 031206–031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Stockley JH, Evans K, Matthey M, Volbracht K, Agathou S, Mukanowa J, Burrone J, and Káradóttir RT (2017) Surpassing light-induced cell damage in vitro with novel cell culture media. Sci. Rep 7, 849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Schaub FX, Reza MS, Flaveny CA, Li W, Musicant AM, Hoxha S, Guo M, Cleveland JL, and Amelio AL (2015) Fluorophore-NanoLuc BRET Reporters Enable Sensitive In Vivo Optical Imaging and Flow Cytometry for Monitoring Tumorigenesis. Cancer Res. 75, 1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Suzuki K, Kimura T, Shinoda H, Bai G, Daniels MJ, Arai Y, Nakano M, and Nagai T (2016) Five colour variants of bright luminescent protein for real-time multicolour bioimaging. Nat. Commun 7, 13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Saito K, Chang Y-F, Horikawa K, Hatsugai N, Higuchi Y, Hashida M, Yoshida Y, Matsuda T, Arai Y, and Nagai T (2012) Luminescent proteins for high-speed single-cell and whole-body imaging. Nat. Commun 3, 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Takai A, Nakano M, Saito K, Haruno R, Watanabe TM, Ohyanagi T, Jin T, Okada Y, and Nagai T (2015) Expanded palette of Nano-lanterns for real-time multicolor luminescence imaging. Proc. Natl. Acad. Sci. U. S. A 112, 201418468–201414356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Pfleger KDG, Seeber RM, and Eidne KA (2006) Bioluminescence resonance energy transfer (BRET) for the real-time detection of protein-protein interactions. Nat. Protoc 1, 337–345. [DOI] [PubMed] [Google Scholar]
- (25).Borghei G, and Hall EAH (2014) BRET-linked ATP assay with luciferase. Analyst 139, 4185–4192. [DOI] [PubMed] [Google Scholar]
- (26).Mezzanotte L, Iljas JD, Que I, Chan A, Kaijzel E, Hoeben R, and Löwik C (2017) Optimized Longitudinal Monitoring of Stem Cell Grafts in Mouse Brain Using a Novel Bioluminescent/Near Infrared Fluorescent Fusion Reporter. Cell Transplant 26, 1878–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hall MP, Woodroofe CC, Wood MG, Que I, Van’t Root M, Ridwan Y, Shi C, Kirkland TA, Encell LP, Wood KV, Löwik C, and Mezzanotte L (2018) Click beetle luciferase mutant and near infrared naphthyl-luciferins for improved bioluminescence imaging. Nat. Commun 9, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Mezzanotte L, Blankevoort V, Löwik CWGM, and Kaijzel EL (2014) A novel luciferase fusion protein for highly sensitive optical imaging: from single-cell analysis to in vivo whole-body bioluminescence imaging. Anal. Bioanal. Chem 406, 5727–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Inouye S, and Shimomura O (1997) The use of Renilla luciferase, Oplophorus luciferase, and apoaequorin as bioluminescent reporter protein in the presence of coelenterazine analogues as substrate. Biochem. Biophys. Res. Commun 233, 349–353. [DOI] [PubMed] [Google Scholar]
- (30).Loening AM, Fenn TD, Wu AM, and Gambhir SS (2006) Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng., Des. Sel 19, 391–400. [DOI] [PubMed] [Google Scholar]
- (31).Leitao JMM, and Esteves da Silva JCG (2010) Firefly luciferase inhibition. J. Photochem. Photobiol., B 101, 1–8. [DOI] [PubMed] [Google Scholar]
- (32).Shimomura O, Masugi T, Johnson FH, and Haneda Y (1978) Properties and reaction mechanism of the bioluminescence system of the deep-sea shrimp Oplophorus gracilorostris. Biochemistry 17, 994–998. [DOI] [PubMed] [Google Scholar]
- (33).Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, and Wood KV (2012) Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol 7, 1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ward WW, and Cormier MJ (1976) In vitro energy transfer in Renilla bioluminescence. J. Phys. Chem 80, 2289–2291. [Google Scholar]
- (35).Ward WW, and Cormier MJ (1979) An energy transfer protein in coelenterate bioluminescence. Characterization of the Renilla green-fluorescent protein. J. Biol. Chem 254, 781–788. [PubMed] [Google Scholar]
- (36).Markwardt ML, Kremers G-J, Kraft CA, Ray K, Cranfill PJC, Wilson KA, Day RN, Wachter RM, Davidson MW, and Rizzo MA (2011) An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One 6, No. e17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, and Tucker CL (2010) Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Repina NA, Rosenbloom A, Mukherjee A, Schaffer DV, and Kane RS (2017) At Light Speed: Advances in Optogenetic Systems for Regulating Cell Signaling and Behavior. Annu. Rev. Chem. Biomol. Eng 8, 13–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Yazawa M, Sadaghiani AM, Hsueh B, and Dolmetsch RE (2009) Induction of protein-protein interactions in live cells using light. Nat. Biotechnol 27, 941–945. [DOI] [PubMed] [Google Scholar]
- (40).Amelio AL, Miraglia LJ, Conkright JJ, Mercer BA, Batalov S, Cavett V, Orth AP, Busby J, Hogenesch JB, and Conkright MD (2007) A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc. Natl. Acad. Sci. U. S. A 104, 20314–20319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Polstein LR, and Gersbach CA (2015) A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol 11, 198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, and Vale RD (2014) A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, and Jaenisch R (2013) Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23, 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Agne M, Blank I, Emhardt AJ, Gäbelein CG, Gawlas F, Gillich N, Gonschorek P, Juretschke TJ, Krämer SD, Louis N, Müller A, Rudorf A, Schäfer LM, Scheidmann MC, Schmunk LJ, Schwenk PM, Stammnitz MR, Warmer PM, Weber W, Fischer A, Kaufmann B, Wagner HJ, and Radziwill G (2014) Modularized CRISPR/dCas9 effector toolkit for target-specific gene regulation. ACS Synth. Biol 3, 986–989. [DOI] [PubMed] [Google Scholar]
- (45).Kawano F, Okazaki R, Yazawa M, and Sato M (2016) A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nat. Chem. Biol 12, 1059–1064. [DOI] [PubMed] [Google Scholar]
- (46).Kawano F, Suzuki H, Furuya A, and Sato M (2015) Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat. Commun 6, 6256. [DOI] [PubMed] [Google Scholar]
- (47).Hallett RA, Zimmerman SP, Yumerefendi H, Bear JE, and Kuhlman B (2016) Correlating in Vitro and in Vivo Activities of Light-Inducible Dimers: A Cellular Optogenetics Guide. ACS Synth. Biol 5, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, and Glotzer M (2012) TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 9, 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Pathak GP, Strickland D, Vrana JD, and Tucker CL (2014) Benchmarking of optical dimerizer systems. ACS Synth. Biol 3, 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Lungu OI, Hallett RA, Choi EJ, Aiken MJ, Hahn KM, and Kuhlman B (2012) Designing photoswitchable peptides using the AsLOV2 domain. Chem. Biol 19, 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Wang H, and Hahn KM (2016) LOVTRAP: A Versatile Method to Control Protein Function with Light. Curr. Protoc Cell Biol 73, 21.10.21–21.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Gauden M, Crosson S, van Stokkum IHM, van Grondelle R, Moffat K, and Kennis JTM (2004) Low-temperature and time-resolved spectroscopic characterization of the LOV2 domain of Avena sativa phototropin 1. Proc. SPIE 5463, 97–104. [Google Scholar]
- (53).Swartz TE, Corchnoy SB, Christie JM, Lewis JW, Szundi I, Briggs WR, and Bogomolni RA (2001) The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem 276, 36493–36500. [DOI] [PubMed] [Google Scholar]
- (54).Durr H, Salomon M, and Rudiger W (2005) Chromophore Exchange in the LOV2 Domain of the Plant Photoreceptor Phototropin1 from Oat. Biochemistry 44, 3050–3055. [DOI] [PubMed] [Google Scholar]
- (55).Berglund K, Clissold K, Li HE, Wen L, Park SY, Gleixner J, Klein ME, Lu D, Barter JW, Rossi MA, Augustine GJ, Yin HH, and Hochgeschwender U (2016) Luminopsins integrate opto- and chemogenetics by using physical and biological light sources for opsin activation. Proc. Natl. Acad. Sci. U. S. A 113, E358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Berglund K, Birkner E, Augustine GJ, and Hochgeschwender U (2013) Light-emitting channelrhodopsins for combined optogenetic and chemical-genetic control of neurons. PLoS One 8, No. e59759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Kim CK, Cho KF, Kim MW, and Ting AY (2019) Luciferase-LOV BRET enables versatile and specific transcriptional readout of cellular protein-protein interactions. eLife 8, 1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Berglund K, Tung JK, Higashikubo B, Gross RE, Moore CI, and Hochgeschwender U (2016) Combined Optogenetic and Chemogenetic Control of Neurons, pp 207–225, Humana Press, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Tung JK, Gutekunst C-A, and Gross RE (2015) Inhibitory luminopsins: genetically-encoded bioluminescent opsins for versatile, scalable, and hardware-independent optogenetic inhibition. Sci. Rep 5, 14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Dragavon J, Sinow C, Holland AD, Rekiki A, Theodorou I, Samson C, Blazquez S, Rogers KL, Tournebize R, and Shorte SL (2014) A step beyond BRET: Fluorescence by Unbound Excitation from Luminescence (FUEL). J. Visualized Exp, No. e51549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Naim N, White AD, Reece JM, Wankhede M, Zhang X, Vilardaga J-P, and Altschuler DL (2019) Luminescence-activated nucleotide cyclase regulates spatial and temporal cAMP synthesis. J. Biol. Chem 294, 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Putyrski M, and Schultz C (2012) Protein translocation as a tool: The current rapamycin story. FEBS Lett. 586, 2097–2105. [DOI] [PubMed] [Google Scholar]
- (63).Roth BL (2016) DREADDs for Neuroscientists. Neuron 89, 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Xie M, and Fussenegger M (2018) Designing cell function: assembly of synthetic gene circuits for cell biology applications. Nat. Rev. Mol. Cell Biol 19, 507–525. [DOI] [PubMed] [Google Scholar]
- (65).Lim WA (2010) Designing customized cell signalling circuits. Nat. Rev. Mol. Cell Biol 11, 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Lin Y-C, Nihongaki Y, Liu T-Y, Razavi S, Sato M, and Inoue T (2013) Rapidly reversible manipulation of molecular activity with dual chemical dimerizers. Angew. Chem., Int. Ed 52, 6450–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Liu P, Calderon A, Konstantinidis G, Hou J, Voss S, Chen X, Li F, Banerjee S, Hoffmann JE, Theiss C, Dehmelt L, and Wu YW (2014) A Bioorthogonal Small-Molecule-Switch System for Controlling Protein Function in Live Cells. Angew. Chem., Int. Ed 53, 10049–10055. [DOI] [PubMed] [Google Scholar]
- (68).Schifferer M, Feng S, Stein F, and Schultz C (2017) Reversible Chemical Dimerization by rCD1. Methods Enzymol. 583, 173–195. [DOI] [PubMed] [Google Scholar]
- (69).O’Banion CP, Vickerman BM, Haar L, and Lawrence DS (2019) Compartmentalized cAMP Generation by Engineered Photoactivated Adenylyl Cyclases. Cell Chem. Biol 26, 1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Wires ES, Henderson MJ, Yan X, Bäck S, Trychta KA, Lutrey MH, and Harvey BK (2017) Longitudinal monitoring of Gaussia and Nano luciferase activities to concurrently assess ER calcium homeostasis and ER stress in vivo. PLoS One 12, No. e0175481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Walker JR, Hall MP, Zimprich CA, Robers MB, Duellman SJ, Machleidt T, Rodriguez J, and Zhou W (2017) Highly Potent Cell-Permeable and Impermeable NanoLuc Luciferase Inhibitors. ACS Chem. Biol 12, 1028–1037. [DOI] [PubMed] [Google Scholar]
- (72).Iwano S, Sugiyama M, Hama H, Watakabe A, Hasegawa N, Kuchimaru T, Tanaka KZ, Takahashi M, Ishida Y, Hata J, Shimozono S, Namiki K, Fukano T, Kiyama M, Okano H, Kizaka-Kondoh S, McHugh TJ, Yamamori T, Hioki H, Maki S, and Miyawaki A (2018) Single-cell bioluminescence imaging of deep tissue in freely moving animals. Science 359, 935–939. [DOI] [PubMed] [Google Scholar]
- (73).Chu J, Oh Y, Sens A, Ataie N, Dana H, Macklin JJ, Laviv T, Welf ES, Dean KM, Zhang F, Kim BB, Tang CT, Hu M, Baird MA, Davidson MW, Kay MA, Fiolka R, Yasuda R, Kim DS, Ng H-L, and Lin MZ (2016) A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat. Biotechnol 34, 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Yeh H-W, Karmach O, Ji A, Carter D, Martins-Green MM, and Ai H. w. (2017) Red-shifted luciferase-luciferin pairs for enhanced bioluminescence imaging. Nat. Methods 14, 971–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Lee H-M, Larson DR, and Lawrence DS (2009) Illuminating the chemistry of life: design, synthesis, and applications of ″caged″ and related photoresponsive compounds. ACS Chem. Biol 4, 409–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Shell TA, and Lawrence DS (2015) Vitamin B12: A Tunable, Long Wavelength, Light-Responsive Platform for Launching Therapeutic Agents, pp 2866–2874, ACS Publications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Borowiak M, Nahaboo W, Reynders M, Nekolla K, Jalinot P, Hasserodt J, Rehberg M, Delattre M, Zahler S, Vollmar A, Trauner D, and Thorn-Seshold O (2015) Photoswitchable Inhibitors of Microtubule Dynamics Optically Control Mitosis and Cell Death. Cell 162, 403–411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods and Materials; Figure S1: Characterization of power and photon output for the cyan-tuned LumiFluor biologic light source (CeNLuc);
Figure S2: Illustration of the various light-activated transcriptional reporter systems used, and optimization of plasmid ratios for maximal BEACON activation;
Figure S3: Representative images for CeNLuc mediated activation of iLID-iRFP670 recruitment to the outer mitochondrial membrane, and half-life analyses of CeNLuc mediated pulsatile light production;
Figure S4: Live cell imaging perfusion system setup;
Figure S5: Amino acid sequences of the various LumiFluor proteins evaluated for cellular optogenetic regulation;
Figure S6: Nucleotide sequence and plasmid map of 8× gRNA promoter for cellular optogenetic regulation of a Firefly luciferase reporter (PDF)
Table S1: Summary of previously described RLuc BRET-based biologic light sources compared to LumiFluors (PDF)
Video S1: Live cell imaging of CeNLuc mediated pulsatile activation of iLID-micro optogenetic system (AVI)


