Abstract
Glucocorticoids are widely used for therapy of hematological malignancies. Unfortunately, chronic treatment with glucocorticoids commonly leads to adverse effects including skin and muscle atrophy and osteoporosis. We found recently that REDD1 (regulated in development and DNA damage 1) plays central role in steroid atrophy. Here we tested whether REDD1 suppression makes glucocorticoid-based therapy of blood cancer safer. Unexpectedly, ~50% of top putative REDD1 inhibitors selected by bioinformatics screening of LINCS database were PI3K/Akt/mTOR inhibitors. We selected Wortmannin, LY294002 and AZD8055 for our studies, and showed that they blocked basal and glucocorticoid-induced REDD1 expression. Moreover, all PI3K/mTOR/Akt inhibitors modified glucocorticoid receptor function shifting it towards therapeutically important transrepression. PI3K/Akt/mTOR inhibitors enhanced anti-lymphoma effects of Dexamethasone in vitro and in vivo, in lymphoma xenograft model. The therapeutic effects of PI3K inhibitor+Dexamethasone combinations ranged from cooperative to synergistic, especially in case of LY294002 and Rapamycin, used as a previously characterized reference REDD1 inhibitor. We found that co-administration of LY294002 or Rapamycin with Dexamethasone protected skin against Dexamethasone-induced atrophy, and normalized RANKL/OPG ratio indicating a reduction of Dexamethasone-induced osteoporosis. Together, our results provide foundation for further development of safer and more effective glucocorticoid-based combination therapy of hematological malignancies using PI3K/Akt/mTOR inhibitors.
Keywords: glucocorticoids, glucocorticoid receptor, PI3K/Akt/mTOR inhibitors, REDD1, hematological malignancies, combination therapy
Introduction
Though the treatment of hematological malignancies has evolved over the past decades, glucocorticoids (Gcs) remain the essential component of combination therapies, and the glucocorticoid receptor (GR) is a well-recognized target for blood cancers (1,2). Unfortunately patients chronically treated with Gcs develop numerous metabolic and atrophic adverse effects including osteoporosis, muscle waste, skin atrophy that strongly affect their life quality (2–4). Thus, there is a great need for the development of safer GR-targeted therapies.
GR is a transcription factor that regulates gene expression via transactivation (TA) that requires GR binding to the responsive elements in gene promoters, and transrepression (TR) that is mediated, among other mechanisms, by binding and blocking other TFs including pro-inflammatory NF-kB and AP-1 (2,5,6). GR TR is an important mechanism underlying therapeutic anti-inflammatory and anti-cancer effects of Gcs, and it usually does not require GR DNA-binding (7). GR TA regulates GR signaling linked to gluconeogenesis and catabolism, and often mediates steroid atrophic effects in different tissues (2,8). Thus, for years the major efforts in the field have been directed towards the development of selective GR agonists/modulators (SEGRAM) that shift GR activity towards TR (9). The alternative approach could be the combination of Gcs with compounds protecting tissues against steroid adverse effects.
Recently mTOR/Akt inhibitor REDD1 (Regulated in development and DNA damage 1) was identified as the key mediator of Gcs-induced atrophy in different tissues. Indeed, in REDD1 KO animals muscle and skin were protected against Gcs atrophy (10,11). At the same time in REDD1 KO mice the anti-inflammatory potential of Gcs was completely preserved (10), suggesting that inhibitors of REDD1 expression could dissociate therapeutic and adverse atrophogenic effects of Gcs.
As REDD1 pharmacological inhibitors are not known, we applied drug repurposing approach. Using bioinformatics screening of Library of Integrated Network-Based Cellular Signatures (LINCS) database of ~ 20,000 transcriptional signatures induced by FDA-approved and experimental drugs (http://lincsproject.org/LINCS/), we identified a significant number of putative inhibitors of REDD1 expression among PI3K/Akt/mTOR inhibitors (12,13) including classical experimental inhibitors such as LY294002 and Wortmannin (WM), and drugs in clinical trials such as Rapamycin (Rapa) and AZD8055. This was completely unexpected as PI3K/Akt/mTOR inhibitors block major pro-proliferative signaling in cells. We demonstrated that compounds from this pharmacological class indeed inhibited REDD1 in keratinocytes and attenuated skin atrophy induced by topical Gcs but did not interfere with their anti-inflammatory effects (12,13).
The goal of this work was to evaluate whether the same approach (Gcs+PI3K/Akt/mTOR inhibitor) is applicable to increase safety of systemic Gcs used for blood cancer. We analyzed the effects of PI3K/Akt/mTOR inhibitors AZD8055, WM and LY294002 on REDD1 expression and GR function in leukemia and lymphoma cells. We assessed atrophogenic effects of systemic Dexamethasone (Dex)+/−LY294002 or Rapa, previously validated as REDD1 inhibitor (13), using osteoporosis and skin atrophy mouse models. We also monitored anti-lymphoma effects of this combination therapy in human lymphoma xenograft model to assure that the regimens chosen for tissues protection remain optimal for anti-cancer activity. We found that LY294002 and Rapa prevented development of Dex-induced osteoporosis and skin atrophy, and enhanced Dex anti-lymphoma effects.
Materials and methods
Chemicals.
LY294002, Wortmannin (WM), AZD8055 were from LC Laboratories (Woburn, MA), Dexamethasone (Dex) and other chemicals were from Sigma-Aldrich (St. Louis, MO).
Computational screen of REDD1 inhibitors.
Putative REDD1 inhibitors were identified by computational screening of LINCS library as described in (12). The molecular signature of each compound in each experiment is presented at LINCS as a list of DEGs - differentially expressed genes, ordered by descending expression fold-change. The top putative REDD1 inhibitors were selected according to the number of LINCS experiments in which REDD1 was within 100 most down-regulated genes in treated cells. For statistical computing, we used the R project version 3.2.5 (https://www.r-project.org/). Selected compounds are presented in Supplementary Table 1
Cell cultures.
Acute lymphoblastic leukemia cell line CEM and mantle cell lymphoma cell line Granta were obtained from ATCC and cultured in RPMI-1640 (Thermo Fischer) with 10% FBS (HyClone), sodium piruvate (10 mM), HEPES (10 mM) and antibiotics (referred thereafter as complete medium; Thermo Fischer) as described (14). Peripheral blood samples were collected from healthy volunteers. The ethical Committee of the Blokhin Cancer Research Center approved this study, written informed consents were provided. Mononuclear cell fraction was isolated by 1.077 g/ml Ficoll-Isopaque density-gradient centrifugation and cultured in the complete medium. Cells were pretreated with Rapa (0,1–5 uM), LY294002 (10–100 uM), WM (1–10 uM) and AZD8055 (0,1–2 uM) or vehicle for 6 h, then were treated for Dex (1–10 uM) or vehicle for 24–144 h as indicated.
Western blot analysis.
Western blot analysis was performed as described in (10). The following antibodies were used: GR (H-300) (Santa Cruz Biotechnology, Dallas, TX), phospho-GR (Ser211), phospho-4E-BP1 (Thr37/46), phospho-Akt (Ser473), Akt, 4E-BP1, cleaved and full-length PARP, GAPDH, HDAC1 (Cell Signaling, San Jose, CA), REDD1 (Proteintech Group, Rosemont, IL).
Analysis of cell viability and apoptosis induction.
Cell viability was assessed using the MTT assay as follows: cells were incubated at 37°C for 3 h with 20 ul of MTT solution (5 mg/ml). Supernatant was discarded and 150 μL of 100% DMSO was added to each well. Optical density was measured at 495 nm using a microplate reader MultiScan MCC 340 (Labsystems). The IC50 values was calculated using Quest Graph IC50 Calculator free software. Isobolograms were constructed as described previously (15). In brief, IC50 values of PI3K inhibitors or Dex as single agents were plotted as 1 on the x and y axes, respectively. Various concentrations of PI3K inhibitors and for Dex were combined. The IC50 values for each compound in the combination were calculated and plotted as a percentage of each single drug. The disposition of data points below, above or on the line of additivity indicates synergy, antagonism or additivity, correspondingly.
Apoptosis induction was evaluated using Western blot analysis of PARP cleavage and propidium iodine (PI) staining as described in (14).
RNA extraction and Q-PCR.
Total RNA isolation, reverse transcription and Q-PCR were performed as described in (10). Primers were designed with NCBI Primer-BLAST (Supplementary Table 2). Results were normalized to the expression of the housekeeping RPL27 gene (10)
Luciferase assay.
GRE.Luc, NF-κB.Luc and control mCMV.Luc reporter cells were generated as described (13) using viral stocks obtained from Northwestern University SDRC DNA/RNA delivery Core. Luciferase activity was measured as described (10).
Xenograft tumor growth study.
10 million Granta cells in 200 ul BD Matrigel Matrix were subcutaneously injected into the right flank of 7 weeks old female nu/nu mice (Taconic). Animals were randomized and treated every 48 h with LY294002 (20 mg/kg), Rapamycin (5 mg/kg) of vehicle (30% PEG3350, 4% DMSO, 5% Tween 20 in PBS) followed by Dex (1 mg/kg) of vehicle (5% DMSO in PBS) 6 h later. Body weight was recorded twice a week. Tumor size was measured twice a week by digital calipers, drug interaction mode was calculated based on combination index method described in (16,17). Combination index (CI) of Dex+Rapa/LY294002 was calculated as ratio (observed fraction value of combination group)/(expected fraction value of combination group). The observed fractions were calculated as ratio (mean value of experiment)/(mean value of control), and the expected fractions values of combination effect between combined groups and single drug groups were calculated as expected fraction=(mean observed fraction of Dex)×(mean observed fraction of Rapa/LY294002). CI<1, CI=1 and CI>1 indicates a synergy, additive effect and antagonism, respectively.
Glucocorticoid-induced osteoporosis (GIOP).
GIOP was induced as described (18) by i.p. injection of Dex (10 mg/kg) in 12 weeks old BALB/c female mice (“Stolbovaya”, Moscow region, Russia) every 48 h for 5 weeks. 6 h prior to Dex, animals were treated i.p. with LY294002 (20 mg/kg), Rapamycin (5 mg/kg) of vehicle (30% PEG3350, 4% DMSO, 5% Tween-20 in PBS). Body weight was recorded twice a week.
All animal experiments were in compliance with Northwestern University ACUC and Blokhin NMRCO approved protocols.
Histological analysis and immunostaining.
Xenograft and skin samples were fixed in formaldehyde, embedded in paraffin, sections were stained with hematoxylin and eosin. The bone samples after formaldehyde fixation were decalcinated in decalcination buffer (0,5 M EDTA, 0,7 mM NaOH, pH=7.0) for 14 days, then washed with PBS twice and further processed according to standard histology protocol. Ki67 and Caspase 3 were detected by immunostaining with antibodies to Ki-67 and Caspase-3 (Cell Signaling, San Jose, CA).
Morphometric analysis.
Quantification of the epidermal width, dermal adipose thickness, bone thickness and cellularity of xenograft tissue was performed in sections stained with hematoxylin and eosin. At least 5 individual fields per slide of each individual sample/treatment (at least 50 images/treatment group) were analyzed using Axioplan2 microscope software (Carl Zeiss). The epidermal, dermal adipose, or bone thickness and cell density in treated animals is presented as % to control animals.
Statistical analysis.
Mean and standard deviation values were calculated using Microsoft Excel software. The treatment effects in each experiment were compared by one-way ANOVA or t-test. Differences between groups were considered significant at P <0.05. All in vitro experiments were repeated three times. In animal experiments, we used 10 animals/experimental group.
Results
PI3K inhibitors suppressed REDD1 expression in lymphoid cells
REDD1 pharmacological inhibitors are not known. Thus, we searched for the inhibitors of REDD1 expression using bioinformatics screening of a large LINCS library of transcriptomic signatures of ~20,000 small molecule drugs tested at large range of concentrations in ~50 human cell lines (http://lincsproject.org/LINCS/). We scored compounds according to the number of LINCS experiments in which REDD1 expression was significantly inhibited (REDD1 was within top 100 down-regulated genes), and found that inhibitors of the PI3K/mTOR/Akt pathway represented the major pharmacological class among the top inhibitors of REDD1 expression. Even though, it was a rather unexpected outcome, we have recently validated REDD1 suppressive effects of several PI3K/Akt/mTOR inhibitors in human keratinocytes (12,13).
For this study we selected three compounds: classical experimental PI3K inhibitors Wortmannin (WM) and LY294002; and dual mTOR inhibitor AZD8055 that recently entered clinical trials as anti-cancer drug (19) (Supplementary Table 1). We also used mTOR inhibitor, Rapamycin (Rapa), as a reference REDD1 inhibitor characterized previously (13). It is known that PI3K activation leads to the phosphorylation and activation of Akt, which in turn activates mTOR in both mTORC1 and mTORC2 complexes (20). Thus, we expected that the selected inhibitors could affect to some degree all branches of PI3K/ Akt/mTOR signaling.
The anti-REDD1 effects of these compounds were tested in mantle cell lymphoma Granta and acute lymphoblastic leukemia CEM cells, which are widely used for the screening of novel chemotherapeutics for blood cancer treatment (1,2,14).
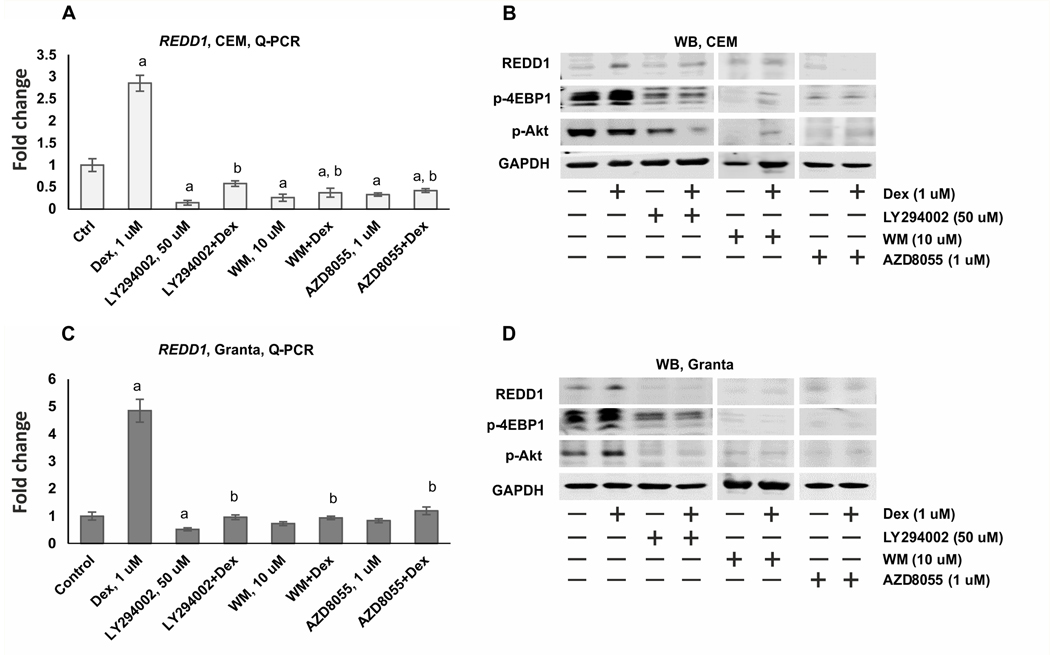
The optimal regimen of cell treatment: pre-treatment with REDD1 inhibitors for 6 h followed by Dex treatment for 24 h was chosen as described (12,13). Dex alone significantly induced REDD1 expression at mRNA/protein levels in both Granta and CEM cells. All tested compounds, LY294002 (50 uM), WM (10 uM) and AZD8055 (1 uM) prevented REDD1 induction (Figure 1, Supplementary Figure 1).
Figure 1. PI3K/mTOR/Akt inhibitors suppress the expression of REDD1 in CEM and Granta cells.
CEM (A-B) and Granta cells (C-D) were pretreated with solvent (Control), LY294002 (50 uM), AZD8055 (1 uM) and WM (10 uM) for 6 h and treated with either solvent or glucocorticoid Dex (1 uM) for 24 h. (A, C) Q-PCR analysis of REDD1 mRNA expression in CEM (A) and Granta (C) cells. The Q-PCR results were normalized to the expression of housekeeping gene RPL27, and presented as fold change compared to control. The mean +/− SD was calculated for three individual RNA samples/condition. Statistically significant differences as compared to: a-control; b-Dex (p<0.001) as and where indicated. (B, D) Western blot analysis of REDD1 protein levels, phosphorylation of mTOR downstream substrate 4-EBP1 and Akt phosphorylation at Thr308 in CEM (B) and Granta (D) cells. GAPDH was used as the loading controls.
The selected regimen and concentrations of REDD1 inhibitors strongly down-regulated PI3K/Akt/mTOR signaling monitored by phosphorylation of mTOR substrate 4E-BP1 (eukaryotic initiation factor 4E binding protein 1), and Akt at Ser473, the major site regulating Akt activity (Figure 1 B, D). PI3K/Akt/mTOR inhibitors also significantly reduced basal expression of REDD1 mRNA (in CEM cells, Figure 1A), and protein (more pronounced in Granta cells, Figure 1D).
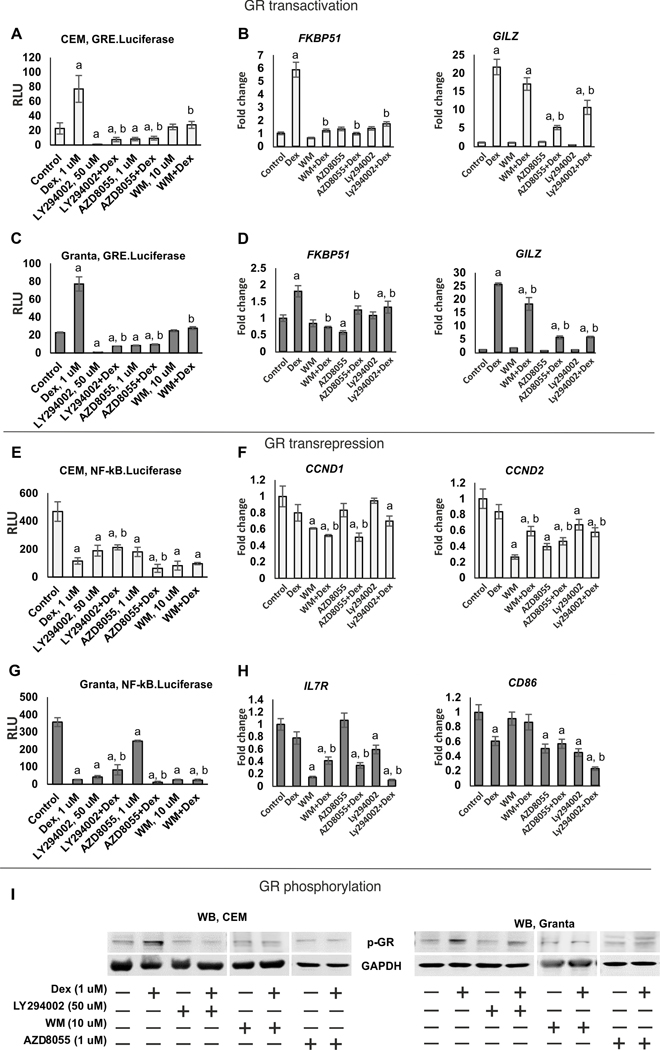
PI3K inhibitors decreased GR phosphorylation, nuclear translocation, and shifted GR function towards transrepression.
REDD1 is a GR target gene (10,21). To assess whether REDD1 inhibitors could modify GR function, we evaluated LY294002, WM and AZD8055 effect on GR TA and TR using CEM and Granta cells transduced with GRE.Luc and NF-kB.Luc reporters. All three tested compounds inhibited Dex-induced GRE.Luciferase activity in both cell lines (Figure 2A, C). Moreover, they also negatively affected basal GR activity when applied individually, with LY294002 being the most effective. Attenuation of GR TA was further confirmed by Q-PCR analysis of known GR-target genes including GR chaperone FKBP51 (12,14), mediator of Gcs anti-inflammatory effects GILZ (22,23), transcriptional activator KLF9 (24) and inhibitor of cytokine production MKP-1 (25) (Figure 2B, D and Supplementary Figure 2A, B).
Figure 2. PI3K/mTOR/Akt inhibitors shift GR function towards transrepression.
CEM (A, E) and Granta (C, G) cells stably expressing GRE.Luciferase and NF-κB.Luciferase were pretreated with LY294002 (50 uM), AZD8055 (1 uM) and WM (10 uM) for 6 h and treated with either solvent or glucocorticoid Dex (1 uM) for 24 h. (A, C, E, G) Luciferase assay. Luciferase activity is presented as mean +/− SD for three individual wells/point. (B, D, F, H) Q-PCR analysis of the expression of GR- and NF-kB regulated endogenous genes. Q-PCR results for repressed genes (CCND1, CCND2, IL7R, CD86) and activated genes (FKBP51, GILZ) were normalized to the expression of housekeeping gene RPL27, and presented as fold change compared to control. Cells were treated as above. The mean +/− SD was calculated for three individual RNA samples/condition. Statistically significant differences as compared to: a-control; b-Dex (p<0.001) as and where indicated. (I) Western blot analysis of GR phosphorylation. Cells were treated as above. GAPDH was used as loading control.
NF-kB activity was strongly reduced according to kB.Luciferase test by Dex as well as by PI3K/Akt/mTOR inhibitors during individual treatment, and in case of Dex combination with AZD8055 or WM, we observed additive inhibitory effect in both cell lines (Figure 2E, G). This finding was further corroborated by Q-PCR analysis of negative regulation of several endogenous genes such as cell cycle regulators cyclins CCND1/D2, and IL7R involved in lymphoid cell survival (26), and to the lesser extent, a marker of malignant lymphoid cells CD86 (Figure 2F, H).
To identify the molecular mechanisms underlying GR functional changes induced by PI3K/Akt/mTOR inhibitors, we assessed how these compounds affected major steps in GR activation: GR phosphorylation at Ser211 and nuclear translocation that are critical for GR TA (27,28).
All studied compounds prevented GR phosphorylation induced by Dex in both cell types to a different degree (Figure 2I). Two out of 3 tested PI3K inhibitors, LY294002 and AZD8055, in addition to the effect on GR phosphorylation significantly prevented GR nuclear import (Supplementary Figure 2C).
Cooperative anti-lymphoma effects of Dex and PI3K/Akt/mTOR inhibitors in vitro.
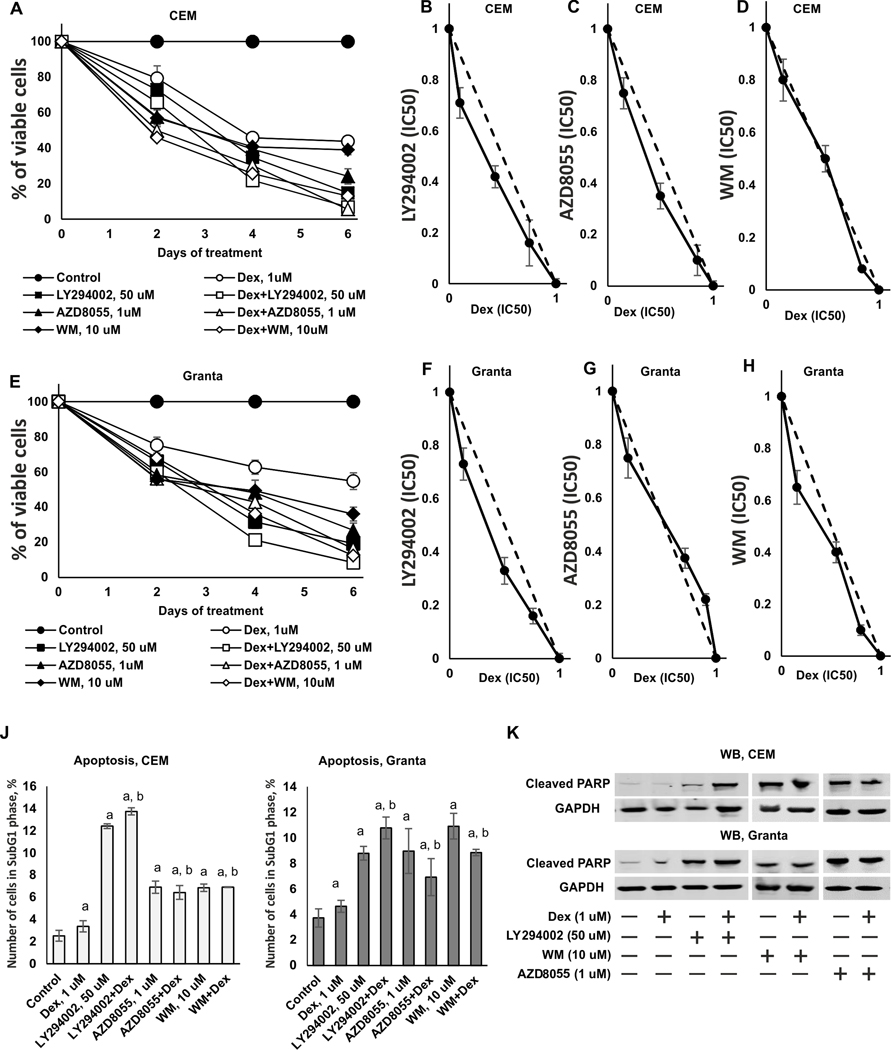
PI3K/Akt/mTOR inhibitors were developed as anti-cancer drugs. To assess whether and how PI3K/Akt/mTOR inhibitors interacted with Dex anti-lymphoma effects we first evaluated the efficacy of the combined treatment in vitro using such readouts as cell viability (MTT test) and apoptosis (PARP cleavage and sub-G1-phase cell population determined by FACS). We tested cytotoxic effect of Dex and PI3K inhibitors in wide concentration range (LY294002 1–500 uM, WM 0,1–25 uM, AZD8055 0,05–5 uM) (Supplementary Figure 3) and determined IC50 values after 24 h treatment (Supplementary Table 3). PI3K inhibitors at optimal, close to IC50 concentrations (50 uM for LY294002, 1 uM for AZD8055, and 10 uM for WM) decreased CEM and Granta survival by 70–90%, and Dex (1 uM) by 50–55% when used alone for 6 days (Figure 3A, E). At the same time, normal lymphocytes from healthy volunteers were significantly more resistant to cytotoxic effects of combinational treatments (Supplementary Figure 4).
Figure 3. Synergistic and additive cytotoxic effects of PI3K/mTOR/Akt inhibitors and Dexamethasone in CEM and Granta cells.
(A-E) Cytotoxic effect was determined using MTT assay. CEM (A) and Granta (E) cells were pretreated with solvent (Control), LY294002 (50 uM), AZD8055 (1 uM) and WM (10 uM) for 6 h and treated with either solvent or glucocorticoid Dex (1 uM) for 24–144 h. (B-D) Isobologram analysis in CEM cells for the combination of LY294002 (B), AZD8055 (C), WM (D) and Dex. (F-H) Isobologram analysis in Granta cells for the combination of LY294002 (F), AZD8055 (G), WM (H) and Dex. The concentration, which resulted in 50% cell growth inhibition (IC50), is expressed as 1.0 on X and Y axis of the isobologram. Y-axis: LY294002, AZD8055 or WM (IC50); X-axis: Dex (IC50). (J, K) Apoptosis induction was evaluated by flow cytometry using PI staining (J) and by Western blot analysis of cleaved PARP level (K). GAPDH was used as loading control. The mean +/− SD was calculated for three individual samples/condition. Statistically significant differences as compared to: a-control; b-Dex (p<0.001) as and where indicated.
To assess the type of interactions between Dex and PI3K/Akt/mTOR inhibitors in anti-lymphoma tests, we used determined IC50 values for inhibitors and Dex after 24 h treatment (Supplementary Table 3), generated isobolograms and used statistical models to determine synergy, additivity or antagonism between Dex and PI3K inhibitors (Figure 3B-D, F–H). Isobologram analysis revealed synergy in antiproliferative effects of Dex and LY294002 in both cell types (Figure 3B, F). Synergy was also determined for Dex and Rapa, used as reference REDD1 inhibitor (Supplementary Figure 5). The effects of Dex and AZD8055 or WM on lymphoma cell survival were mostly additive (Figure 3C, D, G, H).
Next, we examined effect of Dex combined with LY294002, AZD8055, WM or reference inhibitor Rapa on apoptosis. When used alone, Dex and all three PI3K inhibitors as well as Rapa stimulated apoptosis in CEM and Granta cells as assessed by increase in sub-G1-phase cell population (Figure 3J and Supplementary Figure 5). LY294002 cooperated with Dex in apoptosis induction; other inhibitors increased apoptosis development compared to Dex only in both cell types. These findings were confirmed by Western blot analysis of PARP cleavage (Figure 3K), that revealed the especially pronounced cooperative pro-apoptotic effects between LY294002 and Dex in CEM cells.
PI3K inhibitors enhance anti-lymphoma effects of Dexamethasone in vivo
To assess whether REDD1 inhibitors from PI3K/Akt/mTOR pharmacological class enhance Dex anti-lymphoma activity in vivo, we used previously established s.c. Granta xenograft model in athymic nu/nu female mice (2). We selected LY294002 that displayed synergistic anti-lymphoma effects with Dex in vitro. In addition, we used as a positive control mTOR inhibitor Rapamycin whose synergistic anti-cancer effects with Dex in vitro were reported (29). We previously characterized Rapa as REDD1 inhibitor and tissue protector against atrophic effects of topical glucocorticoids in skin (13).
Animals were randomized to different treatment groups after tumor volume reached 50 mm3. All compounds were delivered via i.p. injections of LY29400 (20mg/kg), Rapa (5 mg/kg) or vehicle in the morning followed by Dex treatment (1 mg/kg) or vehicle 6 h later following the optimal regimens for REDD1 inhibition developed in vitro (2,13). All treatment regiments were only minimally toxic according to the steady increase in body weight monitored twice weekly (Supplementary Figure 6). Tumor volume was measured also twice a week (Supplementary Figure 7).
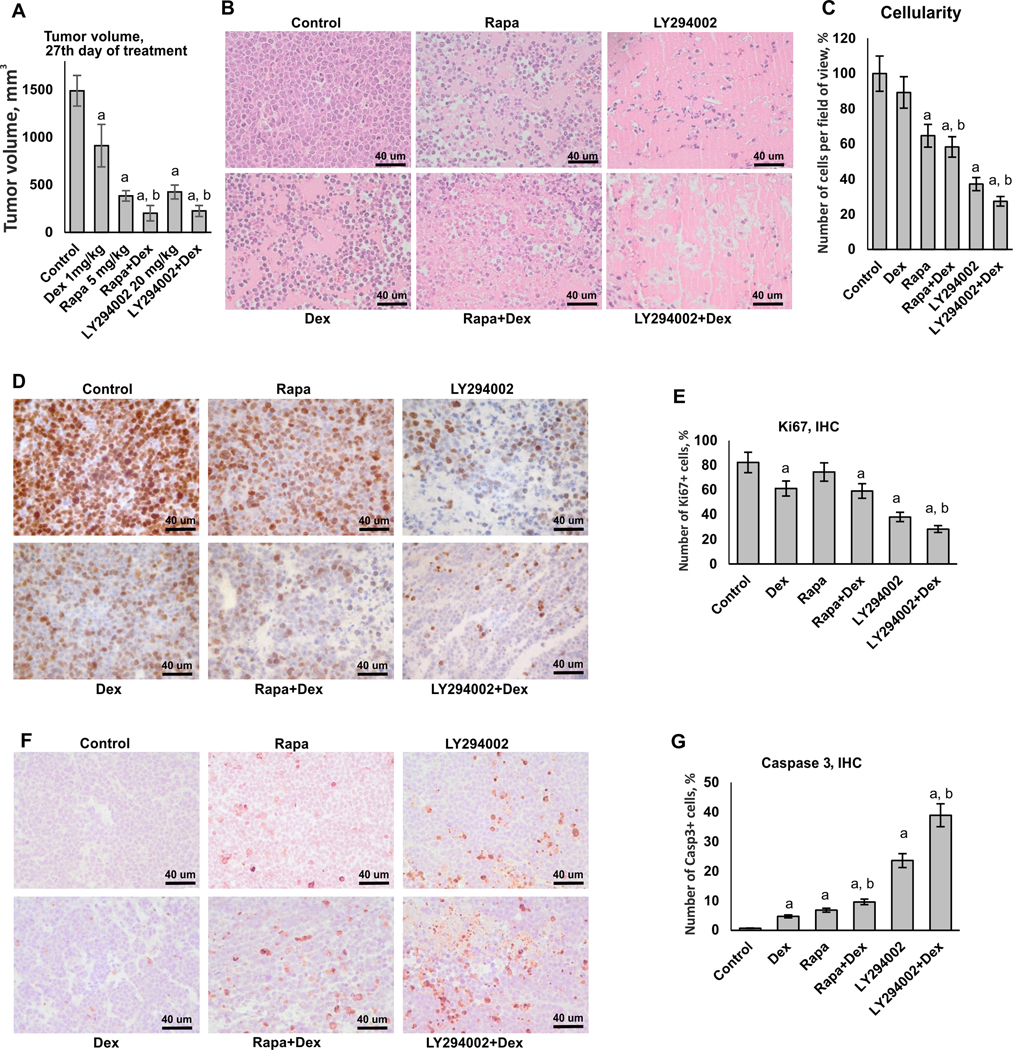
At the end-point of experiment (27th day of treatment), the average tumor volume in control was 1489±161,3 mm3. In groups treated with Dex and LY294002 individually, the tumor volume decreased by ~ 40% and ~70% correspondingly (Figure 4A). We observed the remarkable 85% decrease in tumor volume in LY294002+Dex group. Similar cooperation in anti-cancer effects was observed for Rapa and Dex (~75% decrease in tumor volume in Rapa-treated group and ~85% in Rapa+Dex group), which was in line with published data (29–32). To determine the mode of combined action of Dex and LY294002/Rapa, we used the combination index (CI) approach based on the difference between the expected and observed mean tumor volumes in Dex+LY294002 or Dex+Rapa groups compared to control. Calculated CI values of 0.87 for Dex+LY294002 and 0.85 for Dex+Rapa indicated synergistic mode of interaction.
Figure 4. Cooperative anti-cancer effects of PI3K/mTOR/Akt inhibitors and Dexamethasone in lymphoma xenograft model.
Subcutaneous xenografts were established using Granta cells. Mice (10 animals/group) were treated every 48 h with i.p. injections of LY294002 (20 mg/kg), Rapa (5 mg/kg) or vehicle (30% PEG3350, 4% DMSO, 5% Tween-20 in PBS) followed by i.p. injections of Dex (1 mg/kg) of vehicle (5% DMSO in PBS) 6 h later. Control mice were monitored until the tumor size reached 1500 mm3 at which point they were euthanized. (A) Tumor volume on the 27th day of treatment. (B) H&E staining. (C) Quantification of cellularity in xenografts. (D, F) Immunohistochemical staining of xenograft samples for Ki67 (D), and caspase 3 (F). Scale bars: 40 um. (E, G) Quantification of Ki67- (E) and caspase 3- (G) positive cells. At least five individual fields per slide of each individual sample/treatment (at least 50 images/treatment group) were analyzed using Axioplan2 microscope software (Carl Zeiss). The cell density of xenografts and the number of Ki67- and caspase 3-positive cells in treated animals is presented as % to control. In A, C, E and G bar graphs the mean +/− SD is presented. Statistically significant differences as compared to: a-control; b-Dex (p<0.001) as and where indicated.
The reduction in xenograft tumor volume in Dex+LY294002 and Dex+Rapa groups reflected drastic decrease of tumor cellularity, by 50–70% in groups with combined treatments with superior effect of Dex+LY294002 (Figure 4B, C). It is known that the major effects of Gcs in transformed lymphoid cells are growth inhibition and apoptosis (33,34). Thus, we evaluated proliferative activity of the cells in xenografts as well as apoptosis by immunostaining with proliferation marker Ki67 and apoptosis marker caspase 3. We observed the lowest number of Ki67-positive cells and the highest percentage of apoptotic cells in the xenografts from LY294002+Dex treated mice (Figure 4D-G) that further confirmed that LY294009 co-treatment enhanced anti-lymphoma effects of Dex. Effects of Rapa on antiproliferative and proapoptotic Dex activity were less pronounced, but still statistically significant for increased apotosis induction.
Protective effect of LY294002 and Rapamycin on systemic atrophic effects of Dex in osteoporosis and skin atrophy models
We selected LY294002 that most significantly modulated GR function, and displayed synergistic anti-lymphoma effects with Dex both in vitro and in vivo, to test its tissue protective effect against atrophy induced by systemic exposure to Dex.
The mechanisms of Gcs atrophic effects are complex, and depend on the tissue. In skin and muscle, the GR molecular signature appeared to be similar (10), and in both tissues Gcs significantly shift metabolism to catabolic processes that affect lipids, sphingolipids, proteins (10,35). The steroid-induced skin atrophy involves all skin compartments and results in overall skin thinning, changes in collagen synthesis, atrophy of epidermis and dermal adipose (8,36,37).
In bone, Gcs induce the shift of bone cell populations towards bone-resorbing osteoclasts via increased osteoclast formation and inhibition of proliferation of bone-forming osteoblasts leading to an increased risk of fragility and bone fractures (38). One of the major signaling pathways that regulates osteoclast formation from their precursors as well as their activation and survival during normal bone remodeling and in a variety of pathologic conditions is mediated by RANK (receptor activator of nuclear factor κB)/RANKL (RANK ligand) signaling (39). Another important regulator of osteoclasts is cytokine Osteoprotegerin (OPG) that acts as a decoy receptor for RANKL. Thus, OPG protects the skeleton from excessive bone resorption, and RANKL/OPG ratio is frequently used as an important determinant of bone mass and skeletal integrity (39). Collagen family proteins (especially Col1a1 whose polymorphisms are associated with age-related osteoporosis) also play a key role in bone formation, structure and mineralization (40,41).
To test whether REDD1 inhibitors protect against systemic atrophic effects of Gcs, we used previously described model of Gcs-induced osteoporosis in Balb/c female mice (18), and assessed atrophy in both bone and skin induced by high-dose i.p. Dex injections (10 mg/kg) every 48 h for 5 weeks. LY294002 (20 mg/kg), Rapa (5 mg/kg) or vehicle were injected 6 h prior animal treatment with Dex. Over the course of the experiment general animal health, as determined by cage-side observations and animal body weight, was similar between control and experimental groups; moreover, we saw a normal steady increase in animal body weight in all groups during the experiment (Supplementary Figure 8).
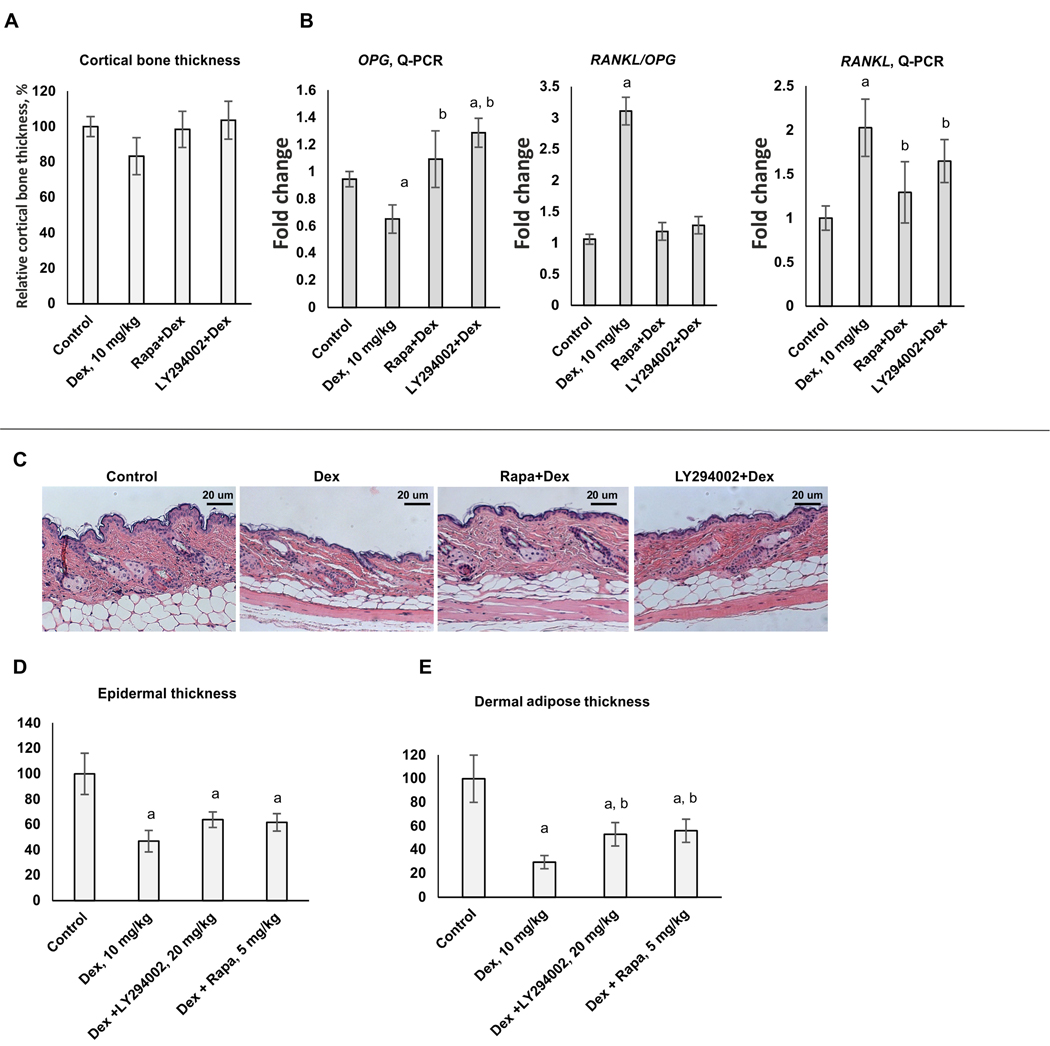
We revealed the tendency in decrease of femoral bone thickness between control and Dex-treated animals (Figure 5A). Unfortunately, this tendency was not statistically significant, possibly due to the duration of treatment. However, changes in widely accepted osteoporosis markers: RANKL (associated with bone resorption), OPG (protector from bone resorption) and RANKL/OPG ratio clearly indicated that in Dex-treated group femur bones started to be affected by osteoporosis: RANKL/OPG ratio was significantly increased after 5-week treatment (Figure 5B). Importantly, both LY294002 and Rapa induced down-regulation of RANKL expression, up-regulation of OPG expression, and, correspondingly, the significant decrease of RANKL/OPG ratio towards control level. Moreover, expression of Col1a1 and especially Col2a1 decreased after Dex chronic treatment, was restored in bone samples from animals treated with LY294002 or Rapa (Supplementary Figure 9)
Figure 5. Protective effect of LY294002 and Rapamycin against atrophic effects of systemic chronic Dex treatment in bone and skin.
12 weeks old female BALB/c mice (10/group) were treated every 48 h with i.p. injections of LY294002 (20 mg/kg), Rapamycin (5 mg/kg) or vehicle (30% PEG3350, 4% DMSO, 5% Tween-20 in PBS) followed by i.p. injections of Dex (10 mg/kg) of vehicle (5% DMSO in PBS) 6 h later for 5 weeks. (A) Quantification of cortical bone thickness. (B) Q-PCR analysis of Rankl and Opg mRNA expression in bone tissue. The Q-PCR results (for three individual RNA samples/condition) were normalized to the expression of housekeeping gene RPL27, and presented as fold change compared to control. (C) H&E skin staining. (D, E) Quantification of the epidermal (D) and dermal adipose (E) thickness as described in Materials and Methods. In bar graphs the mean +/− SD is presented. Statistically significant differences as compared to: a-control; b-Dex (p<0.001) as and where indicated.
To test whether LY294002 and Rapamycin could prevent development of skin atrophy induced by systemic Dex delivery, we assessed morphological changes in skin of Balb/c female mice. In agreement with previously published results (8), chronic treatment with systemic Dex led to a very significant cutaneous atrophy: epidermal thickness was reduced by ~50%, and dermal adipose tissue was reduced by ~ 70% (Figure 5C-E). Remarkably, LY294002 and Rapamycin partially protected skin from these hypoplastic effects of Dex: LY294002+Dex-treated skin displayed only 35 % and 45% reduction in epidermal and dermal adipose thickness accordingly. The protective effects of Rapamycin were similar.
Discussion
Significant success in cancer treatment is turning many cancers including blood cancer into chronic diseases. Thus, there has been a growing interest to the treatment-related adverse effects impacting health and quality of life during/following cancer therapy. The focus of studies presented here is on the use of tissue protectors to mitigate adverse effects of systemic Gcs without impact on their anti-lymphoma potential.
Gcs are very effective against a number of hematological malignancies. However, their significant atrophic and metabolic adverse effects motivated development of novel steroidal and non-steroidal GR activators and modulators (SEGRAM) with reduced side effects. Numerous SEGRAM have been synthesized and tested by industry and academia, however, the success was rather limited (42). There also have been efforts to limit systemic side effects of Gcs using short-living Gcs-based pro-drugs that act locally and then undergo deactivation (42). Unfortunately, only few SEGRAM reached clinical trials but have never been approved for wide-scale clinical use.
We propose here an alternative approach to safer GR-targeted therapies via combination of glucocorticoids with tissue protectors which we selected by the bioinformatics screening of small molecule drugs based on their negative effect on atrophogene REDD1. As far as we know, this is the first successful attempt to use drug repurposing approach to seek for compounds preventing atrophy.
We tested here anti-atrophogenic compounds from the pharmacological class of PI3K/Akt/mTOR inhibitors. The unexpected finding that PI3K/Akt/mTOR inhibitors can potently inhibit the expression of Akt/mTOR genetic regulator REDD1, confirms the existence of a little known feedback loop in Akt/mTOR signaling. As shown here and in our recent work (13), this loop tightly regulates REDD1 expression at RNA/protein levels. However, the molecular mechanisms underlying transcriptional regulation of REDD1 at basal level by mTOR have yet to be identified. Interestingly, mTOR also regulates REDD1 proteasomal degradation and thus, mTOR inhibitors can reduce REDD1 protein stability (43).
One of our most intriguing findings was the capacity of numerous PI3K/Akt/mTOR inhibitors to modify GR function shifting overall GR activity towards therapeutically important TR both in lymphoid cell types (Figure 2), and also in epithelial cells as assessed by Luciferase test and by global changes in Gcs transcriptome (12,13). These effects could underlie the feedback loop in PI3K/Akt/mTOR signaling after the exposure of Gcs.
The inhibition of GR TA followed inhibition of GR phosphorylation and decreased GR nuclear import. It is known that GR is phosphorylated on multiple residues by mitogen-activated protein kinases (MAPKs), cyclin-dependent kinases (CDKs), casein kinase II (CK2) and glycogen synthase kinase-3β (GSK-3β) (27,44). Interestingly, some of PI3K inhibitors such as LY294002 do have off-target effects mediated by binding to BET proteins (Bromodomain and Extra-Terminal motif proteins) regulating the histone acetylation. As GR is also involved into epigenetic regulation of gene expression, possible cross-talk between Gcs/GR and BET signaling could affect the expression of GR target genes, especially cytokines and chemokines implicated into anti-inflammatory Gcs response (45–47).
Interestingly, Akt itself can be directly involved in GR phosphorylation and GR function tuning (48). These literature data suggest that PI3K/Akt/mTOR inhibitors affected GR phosphorylation status via GR kinases as mTOR and Akt crosstalk with signaling pathways involving p38, GSK-3beta is known (27,44). At the same time, the exaggeration of GR TR branch (associated with anti-inflammatory and anti-cancer effects of Gcs) in part reflects profound negative effect of PI3K inhibitors on NF-kB activation (12,13).
The directional changes in GR function (down-regulation of GR TA) and blockage of atrophogene REDD1 suggested that PI3K/Akt/mTOR indeed could protect tissues against steroid atrophy. Using two different models of steroid atrophy – skin atrophy and osteoporosis, we validated protective effects of PI3K inhibitors when they and Gcs were delivered systemically. Even though the causative role of REDD1 in skin and muscle atrophy was proven experimentally (10,11), the mechanisms underlying REDD1 effects in bone will be an interesting direction for future investigation.
The PI3K/AKT/mTOR pathway plays a central role in regulating growth and survival of lymphoid cells, and constitutive PI3K activation is frequently observed in different types of lymphoid malignancies (49,50). Importantly, in our work, combination of Gcs with PI3K inhibitors enhanced anti-lymphoma effects of Dex in vitro and in vivo, in human lymphoma xenograft model, and the therapeutic effects of PI3K inhibitor + Dex combinations ranged from cooperative to synergistic (especially in case of Rapamycin and LY294002). The mechanism underlying cooperative cytotoxicity of Gcs and PI3K inhibitors could involve the attenuation of Gcs-induced pro-survival autophagy and shift to Gcs-dependent apoptosis in cells lacking REDD1 (51). Interestingly, pharmacologic inhibition of PI3K, mTOR and Akt could overcome resistance/increase sensitivity of leukemia cells to Gcs (52).
Overall, our findings provide a proof of principle for using compounds from PI3K/Akt/mTOR inhibitors class to prevent atrophic effects induced by chronic systemic treatment with Gcs. This combination also appeared to be exceptionally promising due to enhanced anti-lymphoma activity of Gcs. In addition to atrophy induced in different tissues, chronic treatment with Gcs results in metabolic, gastrointestinal and cardiovascular side effects (53). As some of those adverse effects including glucose metabolism also strongly depend on TA branch of GR signaling (54), it is conceivable that PI3K/Akt/mTOR inhibitors could potentially ameliorate some metabolic adverse effects of Gcs, suggesting clinical applications for wide range of different diseases and disorders treated with Gcs.
Supplementary Material
Acknowledgements:
Work is supported by R01GM112945, R01AI125366 (to IB and JTD), HESI-THRIVE grant (to IB), Russian Science Foundation grant 17-75-20124 (to EL). The authors would like to thank University of Chicago Genomics Facility, and Northwestern University SDRC (5 P30 AR057216) morphology and phenotyping, skin tissue engineering, and DNA/RNA delivery Cores for technical support.
Financial Information: Work is supported by R01GM112945, R01AI125366 (to IB and JTD), HESI-THRIVE grant (to IB), Russian Science Foundation grant 17-75-20124 (to EL).
Abbreviations:
- CCND1
cyclin D1
- CCND3
cyclin D3
- CD86
Cluster of Differentiation 86
- Col1a1
Collagen, type I, alpha 1 Chain
- Col2a1
Collagen Type II Alpha 1 Chain
- DEG
differentially expressed gene
- Dex
Dexamethasone
- FACS
Fluorescence-activated cell sorting
- FKBP51
FK506-binding protein 5
- 4E-BP1
eukaryotic initiation factor 4E binding protein 1
- Gcs
glucocorticoids
- GILZ
Glucocorticoid-induced Leucine Zipper
- GIOP
glucocorticoid-induced osteoporosis
- GR
Glucocorticoid receptor
- GRE
glucocorticoid responsive element
- GSK-3β
glycogen synthase kinase-3β
- IL7R
Interleukin 7 Receptor
- KLF9
Kruppel-like factor 9
- Luc
Luciferase
- MKP-1
Mitogen-activated protein kinase (MAPK) phosphatase 1
- mTOR
mammalian target of Rapamycin
- NF-κB
nuclear factor kappa B
- OPG
Osteoprotegerin
- PI3K
Phosphoinositide 3-kinase
- RANKL
Receptor activator of nuclear factor kappa-Β ligand
- Rapa
Rapamycin
- REDD1
regulated in development and DNA damage response 1
- rpS6
ribosomal protein S6
- Q-PCR
quantitative polymerase chain reaction
- SEGRAM
selective glucocorticoid receptor agonist or modulator
- TA
transactivation
- TF
transcription factor
- TR
transrepression
- WM
Wortmannin
Footnotes
Conflict of Interest Disclosures
The authors declare no competing financial interests.
References
- 1.Scheijen B. Molecular mechanisms contributing to glucocorticoid resistance in lymphoid malignancies. Cancer Drug Resist. 2019; 2:647–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lesovaya E, Yemelyanov A, Swart AC, Swart P, Haegeman G, Budunova I. Discovery of Compound A--a selective activator of the glucocorticoid receptor with anti-inflammatory and anti-cancer activity. Oncotarget. 2015;6:30730–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hachemi Y, Rapp AE, Picke A-K, Weidinger G, Ignatius A, Tuckermann J. Molecular mechanisms of glucocorticoids on skeleton and bone regeneration after fracture. J Mol Endocrinol. 2018;61:R75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conaway HH, Henning P, Lie A, Tuckermann J, Lerner UH. Activation of dimeric glucocorticoid receptors in osteoclast progenitors potentiates RANKL induced mature osteoclast bone resorbing activity. Bone. 2016;93:43–54. [DOI] [PubMed] [Google Scholar]
- 5.Kleiman A, Tuckermann JP. Glucocorticoid receptor action in beneficial and side effects of steroid therapy: lessons from conditional knockout mice. Mol Cell Endocrinol. 2007;275:98–108. [DOI] [PubMed] [Google Scholar]
- 6.Ramamoorthy S, Cidlowski JA. Ligand-induced repression of the glucocorticoid receptor gene is mediated by an NCoR1 repression complex formed by long-range chromatin interactions with intragenic glucocorticoid response elements. Mol Cell Biol. 2013;33:1711–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Bosscher K, Vanden Berghe W, Haegeman G. The interplay between the glucocorticoid receptor and nuclear factor-kappaB or activator protein-1: molecular mechanisms for gene repression. Endocr Rev. 2003;24:488–522. [DOI] [PubMed] [Google Scholar]
- 8.Schoepe S, Schäcke H, May E, Asadullah K. Glucocorticoid therapy-induced skin atrophy. Exp Dermatol. 2006;15:406–20. [DOI] [PubMed] [Google Scholar]
- 9.De Bosscher K, Beck IM, Haegeman G. Classic glucocorticoids versus non-steroidal glucocorticoid receptor modulators: survival of the fittest regulator of the immune system? Brain Behav Immun. 2010;24:1035–42. [DOI] [PubMed] [Google Scholar]
- 10.Baida G, Bhalla P, Kirsanov K, Lesovaya E, Yakubovskaya M, Yuen K, et al. REDD1 functions at the crossroads between the therapeutic and adverse effects of topical glucocorticoids. EMBO Mol Med. 2015;7:42–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Britto FA, Begue G, Rossano B, Docquier A, Vernus B, Sar C, et al. REDD1 deletion prevents dexamethasone-induced skeletal muscle atrophy. Am J Physiol Endocrinol Metab. 2014;307:E983–93. [DOI] [PubMed] [Google Scholar]
- 12.Agarwal S, Mirzoeva S, Readhead B, Dudley JT, Budunova I. PI3K inhibitors protect against glucocorticoid-induced skin atrophy. EBioMedicine. 2019;41:526–37. A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lesovaya E, Agarwal S, Readhead B, Vinokour E, Baida G, Bhalla P, et al. Rapamycin Modulates Glucocorticoid Receptor Function, Blocks Atrophogene REDD1, and Protects Skin from Steroid Atrophy. J Invest Dermatol. 2018;138:1935–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lesovaya E, Yemelyanov A, Kirsanov K, Popa A, Belitsky G, Yakubovskaya M, et al. Combination of a selective activator of the glucocorticoid receptor Compound A with a proteasome inhibitor as a novel strategy for chemotherapy of hematologic malignancies. Cell Cycle. 2013;12:133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin C-J, Lee C-C, Shih Y-L, Lin T-Y, Wang S-H, Lin Y-F, et al. Resveratrol enhances the therapeutic effect of temozolomide against malignant glioma in vitro and in vivo by inhibiting autophagy. Free Radic Biol Med. 2012;52:377–91. [DOI] [PubMed] [Google Scholar]
- 16.Chou T-C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–81. [DOI] [PubMed] [Google Scholar]
- 17.Yang H, Wang J, Fan J-H, Zhang Y-Q, Zhao J-X, Dai X-J, et al. Ilexgenin A exerts anti-inflammation and anti-angiogenesis effects through inhibition of STAT3 and PI3K pathways and exhibits synergistic effects with Sorafenib on hepatoma growth. Toxicol Appl Pharmacol. 2017;315:90–101. [DOI] [PubMed] [Google Scholar]
- 18.McLaughlin F, Mackintosh J, Hayes BP, McLaren A, Uings IJ, Salmon P, et al. Glucocorticoid-induced osteopenia in the mouse as assessed by histomorphometry, microcomputed tomography, and biochemical markers. Bone. 2002;30:924–30. [DOI] [PubMed] [Google Scholar]
- 19.Naing A, Aghajanian C, Raymond E, Olmos D, Schwartz G, Oelmann E, et al. Safety, tolerability, pharmacokinetics and pharmacodynamics of AZD8055 in advanced solid tumours and lymphoma. Br J Cancer. 2012;107:1093–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dennis MD, Coleman CS, Berg A, Jefferson LS, Kimball SR. REDD1 enhances protein phosphatase 2A-mediated dephosphorylation of Akt to repress mTORC1 signaling. Sci Signal. 2014;7:ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Kubica N, Ellisen LW, Jefferson LS, Kimball SR. Dexamethasone represses signaling through the mammalian target of rapamycin in muscle cells by enhancing expression of REDD1. J Biol Chem. 2006;281:39128–34. [DOI] [PubMed] [Google Scholar]
- 22.Yang N, Baban B, Isales CM, Shi X-M. Role of glucocorticoid-induced leucine zipper (GILZ) in inflammatory bone loss. PLoS One. 2017;12:e0181133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan H, Kao W, Yang YH, Gu R, Harris J, Fingerle-Rowson G, et al. Macrophage migration inhibitory factor inhibits the antiinflammatory effects of glucocorticoids via glucocorticoid-induced leucine zipper. Arthritis Rheumatol (Hoboken, NJ). 2014;66:2059–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leigh R, Mostafa MM, King EM, Rider CF, Shah S, Dumonceaux C, et al. An inhaled dose of budesonide induces genes involved in transcription and signaling in the human airways: enhancement of anti- and proinflammatory effector genes. Pharmacol Res Perspect. 2016;4:e00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vollmer TR, Stockhausen A, Zhang J-Z. Anti-inflammatory effects of mapracorat, a novel selective glucocorticoid receptor agonist, is partially mediated by MAP kinase phosphatase-1 (MKP-1). J Biol Chem. 2012;287:35212–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ligons DL, Tuncer C, Linowes BA, Akcay IM, Kurtulus S, Deniz E, et al. CD8 lineage-specific regulation of interleukin-7 receptor expression by the transcriptional repressor Gfi1. J Biol Chem. 2012;287:34386–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen W, Dang T, Blind RD, Wang Z, Cavasotto CN, Hittelman AB, et al. Glucocorticoid receptor phosphorylation differentially affects target gene expression. Mol Endocrinol. 2008;22:1754–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galliher-Beckley AJ, Cidlowski JA. Emerging roles of glucocorticoid receptor phosphorylation in modulating glucocorticoid hormone action in health and disease. IUBMB Life. 2009;61:979–86. [DOI] [PubMed] [Google Scholar]
- 29.Zhang C, Ryu Y-K, Chen TZ, Hall CP, Webster DR, Kang MH. Synergistic activity of rapamycin and dexamethasone in vitro and in vivo in acute lymphoblastic leukemia via cell-cycle arrest and apoptosis. Leuk Res. 2012;36:342–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei G, Twomey D, Lamb J, Schlis K, Agarwal J, Stam RW, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10:331–42. [DOI] [PubMed] [Google Scholar]
- 31.Hall CP, Reynolds CP, Kang MH. Modulation of Glucocorticoid Resistance in Pediatric T-cell Acute Lymphoblastic Leukemia by Increasing BIM Expression with the PI3K/mTOR Inhibitor BEZ235. Clin Cancer Res. 2016;22:621–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silveira AB, Laranjeira ABA, Rodrigues GOL, Leal PC, Cardoso BA, Barata JT, et al. PI3K inhibition synergizes with glucocorticoids but antagonizes with methotrexate in T-cell acute lymphoblastic leukemia. Oncotarget. 2015;6:13105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ploner C, Schmidt S, Presul E, Renner K, Schröcksnadel K, Rainer J, et al. Glucocorticoid-induced apoptosis and glucocorticoid resistance in acute lymphoblastic leukemia. J Steroid Biochem Mol Biol. 2005;93:153–60. [DOI] [PubMed] [Google Scholar]
- 34.Amaral JD, Solá S, Steer CJ, Rodrigues CMP. Role of nuclear steroid receptors in apoptosis. Curr Med Chem. 2009;16:3886–902. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu N, Yoshikawa N, Ito N, Maruyama T, Suzuki Y, Takeda S, et al. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 2011;13:170–82. [DOI] [PubMed] [Google Scholar]
- 36.Hengge UR, Ruzicka T, Schwartz RA, Cork MJ. Adverse effects of topical glucocorticosteroids. J Am Acad Dermatol. 2006;54:1–15; quiz 16–8. [DOI] [PubMed] [Google Scholar]
- 37.Kimura T, Doi K. Dorsal skin reactions of hairless dogs to topical treatment with corticosteroids. Toxicol Pathol. 27:528–35. [DOI] [PubMed] [Google Scholar]
- 38.Frenkel B, White W, Tuckermann J. Glucocorticoid-Induced Osteoporosis. Adv Exp Med Biol. 2015;872:179–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyce BF, Xing L. Biology of RANK, RANKL, and osteoprotegerin. Arthritis Res Ther. 2007;9 Suppl 1:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clancy BM, Johnson JD, Lambert AJ, Rezvankhah S, Wong A, Resmini C, et al. A gene expression profile for endochondral bone formation: oligonucleotide microarrays establish novel connections between known genes and BMP-2-induced bone formation in mouse quadriceps. Bone. 2003;33:46–63. [DOI] [PubMed] [Google Scholar]
- 41.Boskey AL, Imbert L. Bone quality changes associated with aging and disease: a review. Ann N Y Acad Sci. 2017;1410:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petta I, Peene I, Elewaut D, Vereecke L, De Bosscher K. Risks and benefits of corticosteroids in arthritic diseases in the clinic. Biochem Pharmacol. 2019;165:112–25. [DOI] [PubMed] [Google Scholar]
- 43.Tan CY, Hagen T. mTORC1 dependent regulation of REDD1 protein stability. PLoS One. 2013;8:e63970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ismaili N, Garabedian MJ. Modulation of glucocorticoid receptor function via phosphorylation. Ann N Y Acad Sci. 2004;1024:86–101. [DOI] [PubMed] [Google Scholar]
- 45.Sacta MA, Tharmalingam B, Coppo M, Rollins DA, Deochand DK, Benjamin B, et al. Gene-specific mechanisms direct glucocorticoid-receptor-driven repression of inflammatory response genes in macrophages. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sacta MA, Chinenov Y, Rogatsky I. Glucocorticoid Signaling: An Update from a Genomic Perspective. Annu Rev Physiol. 2016;78:155–80. [DOI] [PubMed] [Google Scholar]
- 47.Dai J, Zhou S, Ge Q, Qin J, Li J, Ju H, et al. Recruitment of Brd3 and Brd4 to acetylated chromatin is essential for proinflammatory cytokine-induced matrix-degrading enzyme expression. J Orthop Surg Res. 2019;14:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Habib T, Sadoun A, Nader N, Suzuki S, Liu W, Jithesh PV, et al. AKT1 has dual actions on the glucocorticoid receptor by cooperating with 14-3-3. Mol Cell Endocrinol. 2017;439:431–43. [DOI] [PubMed] [Google Scholar]
- 49.Polak R, Buitenhuis M. The PI3K/PKB signaling module as key regulator of hematopoiesis: implications for therapeutic strategies in leukemia. Blood. 2012;119:911–23. [DOI] [PubMed] [Google Scholar]
- 50.Curran E, Smith SM. Phosphoinositide 3-kinase inhibitors in lymphoma. Curr Opin Oncol. 2014;26:469–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molitoris JK, McColl KS, Swerdlow S, Matsuyama M, Lam M, Finkel TH, et al. Glucocorticoid elevation of dexamethasone-induced gene 2 (Dig2/RTP801/REDD1) protein mediates autophagy in lymphocytes. J Biol Chem. 2011;286:30181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corsello SM, Bittker JA, Liu Z, Gould J, McCarren P, Hirschman JE, et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat Med. 2017;23:405–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adcock IM, Mumby S. Glucocorticoids. Handb Exp Pharmacol. 2017;237:171–96. [DOI] [PubMed] [Google Scholar]
- 54.Robertson S, Allie-Reid F, Vanden Berghe W, Visser K, Binder A, Africander D, et al. Abrogation of glucocorticoid receptor dimerization correlates with dissociated glucocorticoid behavior of compound a. J Biol Chem. 2010;285:8061–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.