Abstract
Untargeted metabolomics indicates that the number of unidentified small-molecule metabolites may exceed the number of protein-coding genes for many organisms, including humans, by orders of magnitude. Uncovering the underlying metabolic networks is essential for elucidating the physiological and ecological significance of these biogenic small molecules. Here we develop a click-chemistry-based enrichment strategy, DIMEN (deep interrogation of metabolism via enrichment), that we apply to investigate metabolism of the ascarosides, a family of signaling molecules in the model organism C. elegans. Using a single alkyne-modified metabolite and a solid-phase azide resin that installs a diagnostic moiety for MS/MS-based identification, DIMEN uncovered several hundred novel compounds originating from diverse biosynthetic transformations that reveal unexpected intersection with amino acid, carbohydrate, and energy metabolism. Many of the newly discovered transformations could not be identified or detected by conventional LC-MS analyses without enrichment, demonstrating the utility of DIMEN for deeply probing biochemical networks that generate extensive yet uncharacterized structure space.
Graphical Abstract
INTRODUCTION
Small molecules represent the chemical input, biosynthetic intermediates, and endproducts of cellular activity, and thus metabolomics is essential for understanding biological processes, complementing proteomics, transcriptomics, and genomics.1 In addition to their roles as building blocks and substrates in primary metabolism, many metabolites serve as signaling molecules, from the intra- and intercellular level, e.g., as hormones, to signaling at the interorganismal level via pheromones. Metabolomics faces challenges that do not apply to other members of the -omics family because the structural diversity of metabolites is not restricted by a limited number of templates and simple rules for assembly, as in the case of DNA, RNA, and proteins, and thus is vast and largely unpredictable. Correspondingly, LC-HRMS-based surveys of animal and plant metabolomes suggests that the number of distinct metabolites in a single species may exceed 100 000.2 Future improvements of analytical instrumentation may reveal even greater compound diversity. In parallel, studies of signaling networks in C. elegans and other systems indicate that biogenic small molecules can have profound biological activity even at femtomolar concentrations, providing a clear rationale for systematic structural and functional characterization of low abundance metabolites.3–6 Given these challenges, there exists a need for methods that facilitate prioritization of detection and structural characterization of specific compound families and biosynthetic pathways of interest.
We aimed to develop a click-chemistry-based strategy for targeted metabolomics that would enable in-depth exploration of specific metabolic pathways and compound families, beyond current detection limits, by (i) improving sensitivity through enrichment from overwhelming pools of unrelated metabolites, and (ii) facilitating recognition of relevant metabolites by means of a specific fragmentation signature in LC-MS/MS (Figure 1a). We envisioned that this could be accomplished by alkyne-labeling metabolic precursors or building blocks of interest, followed by incubation with the biological system and enrichment of labeled derivatives via click chemistry.7 Surprisingly, there are few examples of click chemistry-based enrichment of tagged metabolites to interrogate metabolism, whereas click chemistry has been employed extensively to enrich proteins and peptides.8,9
Figure 1.
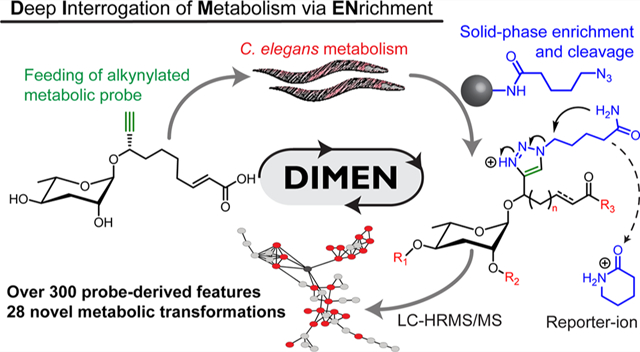
Development of DIMEN. (a) Schematic representation of metabolomes and purpose of DIMEN for the characterization of known, LC-MS visible (light gray) and LC-MS invisible (dark gray) molecules. (b) Glycosides of the dideoxy sugar ascarylose are combined with diverse primary metabolic building blocks in C. elegans. (c) Structure of solid-support, alkyne capture for enrichment and reporter-ion installation (ACER) resin (1), and alkyne probe ascr#3-YNE (3), which mimics ascr#3 (2), a precursor upstream of the biosynthesis of diverse signaling molecules. (d) Schematic overview of DIMEN analysis. C. elegans are grown with and without alkyne-labeled probe (blue star) and extracted. Probe is added to unlabeled samples before enrichment using the ACER resin. Enriched metabolomes are analyzed via LC-HRMS/MS to detect PDMs, e.g., ascr#3-LNK (4) and icas#3-LNK (5).
To demonstrate the utility of click chemistry-based enrichment for metabolomics, we selected a family of signaling molecules, the ascarosides, which play a central role in the life history of the model organism, C. elegans, and other nematodes (Figures 1b and S1a). The ascarosides form a large, structurally diverse family of compounds that is derived from modular assembly of building blocks from carbohydrate, amino acid, nucleoside, and cofactor metabolism around a core glycoside derived from the dideoxy sugar ascarylose and a variable fatty acid-like moiety.10–12
Ascarosides regulate development, aging, phenotypic plasticity, and many behaviors in C. elegans and other nematodes, and furthermore mediate multitrophic interactions of nematodes with fungi and plants.13–17 Biological activity has been observed down to femtomolar concentrations, and correspondingly ascaroside identification and metabolism has been extensively studied.4,12,18,19 Previous studies demonstrated uptake of synthetic ascarosides by C. elegans, and that introduction of an alkyne moiety in the fatty acid side chain proximal to the glycosidic linkage does not interfere with biological activity (Figure 1c).20–22 Feeding worms an alkyne-modified ascaroside probe may thus provide deep insight into ascaroside metabolism by enabling enrichment and recognition of derived metabolites, including derivatives of potential signaling molecules that have remained undetectable in conventional LC-HRMS.
RESULTS
Development of DIMEN.
Our workflow for deep interrogation of metabolism via enrichment (DIMEN) employs the copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC) to selectively bind probe-derived metabolites (PDMs) to an azide-modified solid support, using unlabeled metabolic samples with added probe as controls (Figure 1c,d). The resin-bound PDMs are extensively washed to remove unlabeled metabolites and cleaved to yield PDMs including a triazole moiety that confers a unique MS/MS signature or reporter-ion. PDMs are detected via comparative analysis of LC-HRMS data for the experimental and control samples. Finally, MS/MS fragmentation patterns are used to confirm the probe origin of the detected PDMs and further to discern structurally distinct families of PDMs via molecular networking software.23
As the solid support we selected a Rink-amide resin typically used for solid-phase peptide synthesis (SPPS) to which we attached a simple azido-valerate linker (“ACER resin” (1), Figure 1c).24 Initially we attempted to use more elaborate azide designs integrating a copper-chelating moiety in the azide linker attached to the solid support, which accelerate click chemistry and improve yields in some situations (Figure S2).25,26 However, in our case integration of a copper chelator in the azide linker caused extensive oxidative damage to the resin and, in some cases, also reduced stability of the linker (Figure S3). Lastly, the use of these more elaborate linkers confounded MS/MS analysis by saturating spectra with linker-associated fragments.
Screening different azide designs further demonstrated that exposure to CuAAC conditions can result in unexpected side reactions. To distinguish such CuAAC side products and probe-derived artifacts from compounds that are the result of biological transformations, DIMEN includes an incubation control arm in which the alkyne probe is added to an unlabeled metabolome sample (Figure 1d). Both the probe-treated and mock-treated metabolomes were then incubated with the ACER resin (1) and subjected to CuAAC, resulting in similar artifact profiles for the control and experimental arms and facilitating recognitions of “real” PDMs via comparative LC-HRMS analysis (Figure 1d).
Optimization of enrichment conditions was accomplished by doping concentrated C. elegans metabolome with a trace amount of the alkyne derivative of ascr#3 (2), ascr#3-YNE (3), and monitoring the recovery of the resulting linker-modified ascr#3-LNK (4). ascr#3 is one of the major ascarosides produced by C. elegans and can serve as a precursor for other known ascarosides.4,27 Different buffers, copper chelating ligands, reagent concentrations, and types of SPPS resins were tested (Figure S4). On the basis of these experiments we selected a polyethylene glycol acrylamide (PEGA) resin as solid support for the azide linker and 2-(4-((bis((1-(tert-butyl)-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)-1H-1,2,3-triazol-1-yl)acetic acid (BTTAA) as copper chelating ligand.28 Analysis of the resin supernatant following enrichment indicated that more than 99% of unlabeled metabolites had been removed and that ~80% of added ascr#3-YNE (3) had reacted (Figure 2a).
Figure 2.
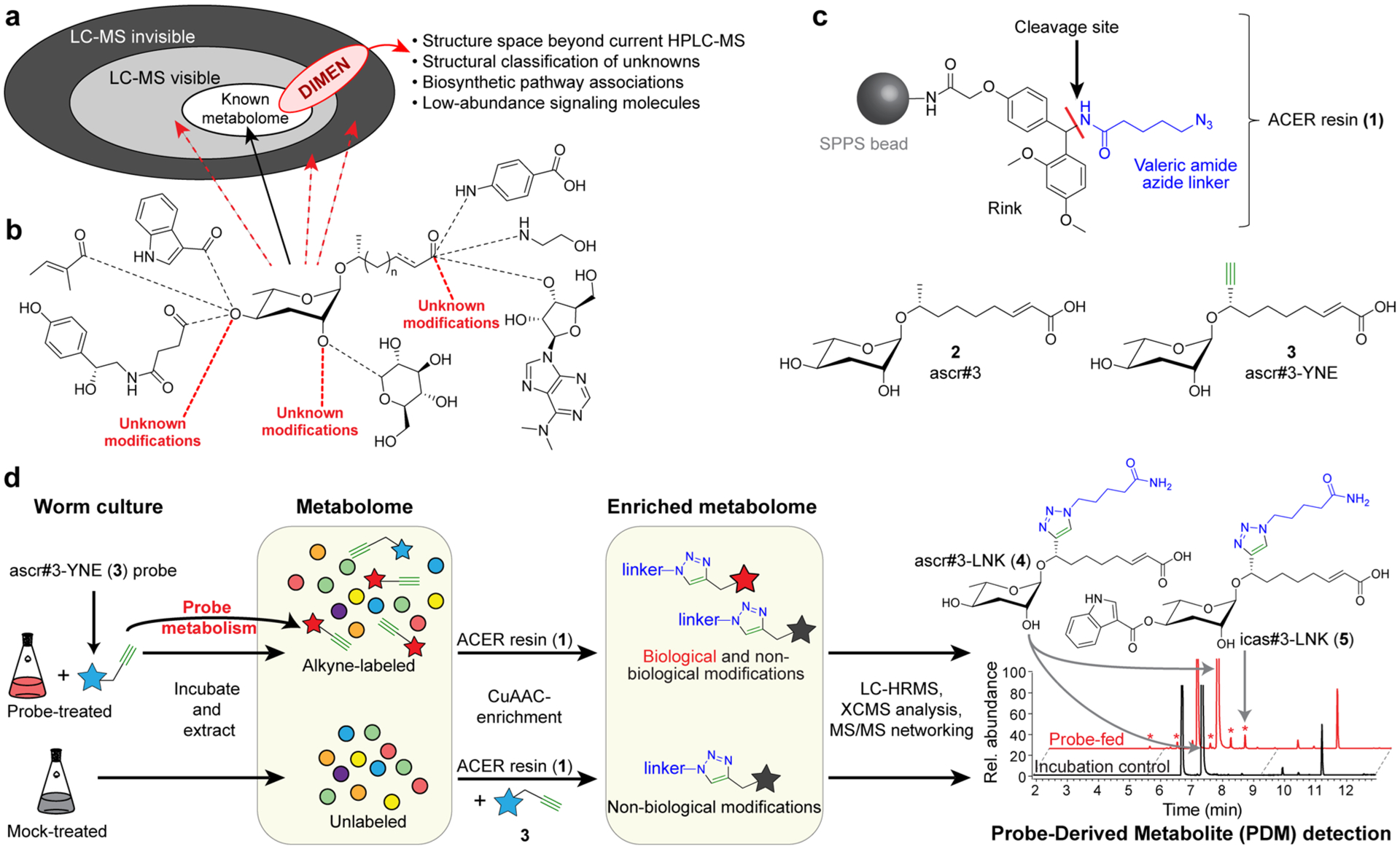
Enrichment of PDMs with DIMEN. (a) LC-HRMS chromatograms for a typical enrichment experiment. Peaks of highly abundant nontarget metabolites (red) are absent following enrichment and remain in the flow-through (intensities adjusted for sample volumes). (b) MS/MS of PDMs in positive mode produces a reporter-ion at m/z 100.0757. (c) Extracted-ion-chromatogram (EIC) for the reporter ion m/z 100.076 of a DIMEN-enriched sample, showing peaks representing known metabolic transformations (annotated in blue), novel transformations (examples annotated red), and unmodified probe and nonbiological cleavage products (annotated in green). See Figure S1 for structures.
Triazoles generally ionize well in positive-ion electrospray mode, facilitating mass spectrometric detection of PDMs (Figure S4d). Furthermore, use of the azido-valeryl modified rink resin resulted in PDMs that produce a diagnostic MS/MS fragment with a mass-to-charge of 100.0757 (Figures 2b and S5) that can be employed as a reporter-ion to automate PDM detection and corroborate that detected metabolites are in fact derived from ascr#3-YNE (Figure 2c and S6).
DIMEN Reveals Novel Metabolites.
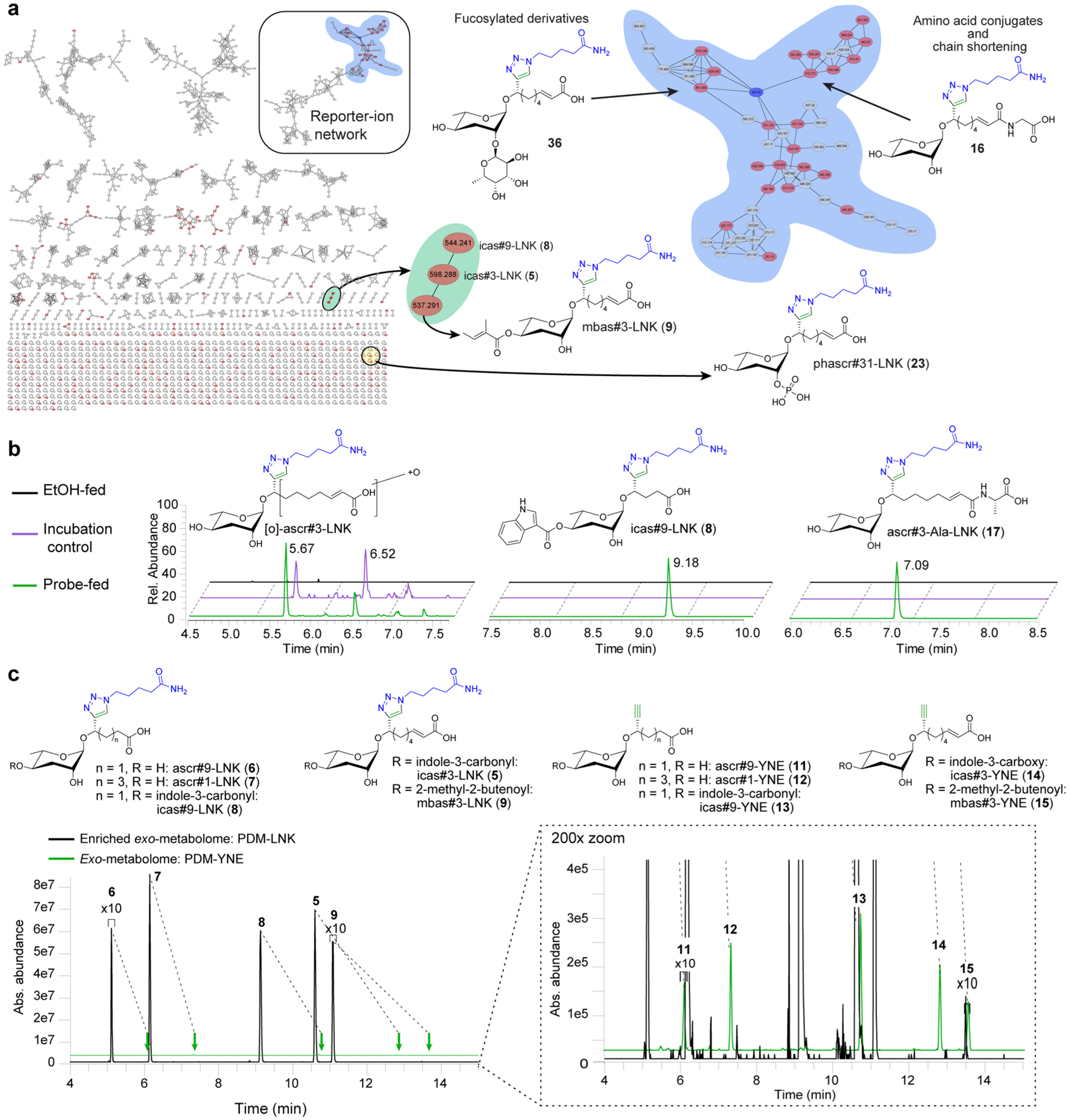
We then employed the optimized enrichment protocol to investigate metabolic transformations of ascr#3-YNE (3) in C. elegans. LC-HRMS/MS-based comparison of experimental and incubation control samples revealed more than 10 000 features, of which 585 were present only in the experimental samples and thus classified as likely PDMs (Figure 3a,b). Molecular networking revealed 27 clusters in which three or more nodes represent likely PDMs, most of which formed defined subclusters due to the shared reporter-ion fragment (Figure 3a).
Figure 3.
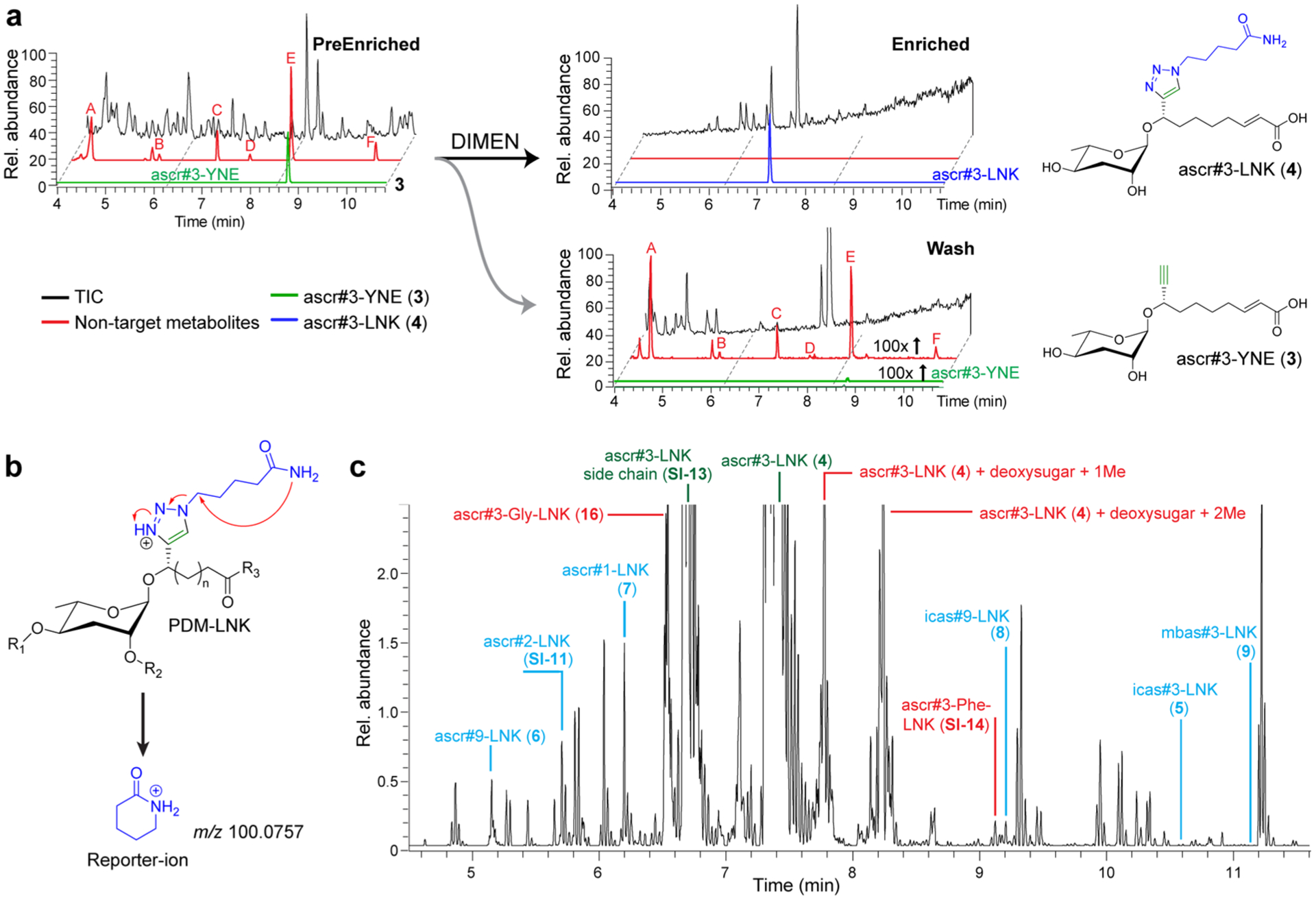
DIMEN of ascr#3-YNE-fed C. elegans. (a) Molecular network resulting from DIMEN analysis of ascr#3-YNE-treated C. elegans cultures. The blue node is ascr#3-LNK and the red nodes are differential between probe-fed and incubation control data sets. These include analogues of known ascarosides such as mbas#3-LNK (9) as well as several new compound families, e.g., a set of fucosylated PDMs, amino-acid ligated ascarosides, and phosphorylated ascarosides. (b) Example EICs for probe-derived artifacts detected in both the probe-fed and incubation control data sets that originate from abiological transformations (e.g., [O]-ascr#3-LNK, a product of nonenzymatic oxidation of ascr#3), PDMs only present in probe-fed data sets representing known ascaroside derivatives (e.g., icas#9-LNK (8), a derivative of the known icas#9), and PDMs only present in probe-fed data sets that represent novel metabolites (e.g., ascr#3-Ala-LNK (17)). (c) EICs filtered for alkyne-functionalized PDMs (PDM-YNE) in exo-metabolome (green), and linker-bound PDMs (PDM-LNK) in enriched exo-metabolome (black). DIMEN improves signal intensity >200-fold between pairs of PDM-YNE and PDM-LNK, due to enrichment and improvement of ionization efficiency (Figure S4d).
Detailed analysis of MS/MS spectra representing likely PDMs revealed expected derivatives of known metabolites as well as a large number of features that appeared to represent novel metabolic transformations. PDMs representing known metabolites (e.g., PDMs 5–9) include the linker analogues of side chain-shortened ascarosides, e.g., the linker derivative of the dauer pheromone component ascr#1 (10), ascr#1-LNK (7) (Figures 3c and S1, Table S1).29 Formation of ascr#1-LNK (4) and other chain-shortened derivatives of ascr#3-YNE (3) indicates that, similar to unmodified ascr#3 (2), the added ascr#3-YNE (3) enters peroxisomal β-oxidation, which is known to iteratively shorten ascaroside side chains, in two-carbon steps, e.g., from 9 carbons as in ascr#3 to 7 carbons as in ascr#1.12,30 Another example for a PDM representing a known metabolite is icas#9-LNK (8), the linker-analogue of the aggregation pheromone icas#9 (Figure 3b and S1).20,31 The detection of icas#9-LNK (8) suggests that metabolism of the probe ascr#3-YNE (3) closely mimics that of natural ascr#3 (2), which had previously been shown to undergo chain shortening and 4′-modification, e.g., attachment of an indolecarboxy moiety.4 Several other PDMs also represented known 4′-modified ascarosides, e.g., icas#3-LNK (5), mbas#3-LNK (9), and others (Figures 3a and S1, Table S1). Quantitation of PDMs representing known metabolites (5–9, 11–15) in a typical DIMEN experiment indicated a signal-to-noise gain of over 2 orders of magnitude, as a result of the enrichment and generally better ionization of PDM-LNK derivatives compared to the PDM-YNE precursors (Figure 3c and S4d).
Among PDMs representing novel metabolic transformations, we prioritized identification of the three largest subclusters in the MS/MS network, which revealed previously unreported ascarosides derived from amino acid ligation and phosphorylation, as well as di- and trisaccharides derived from attachment of fucose derivatives to the ascarylose. For each of these groups of PDMs, we proposed structures based on MS/MS analysis and subsequently reanalyzed the metabolome of unsupplemented C. elegans for candidate compounds that could constitute the corresponding naturally occurring ascaroside derivatives. To corroborate that such candidate compounds represent ascaroside derivatives, we confirmed their absence in the metabolome of daf-22 mutants, which are known to be defective in ascaroside biosynthesis.12,32,33 Finally, the structures of representative members for each of the identified new families of C. elegans metabolites were confirmed by synthesis (see SI Methods).
Amino Acid-Ligated Ascarosides.
Analysis of MS/MS spectra of one of the largest clusters of PDMs suggested that they represent amino acid conjugates of ascr#3-YNE (3) (Figure 4a). PDMs representing putative conjugates of glycine, alanine, and serine (16–18, Figures 3a,b, and 4a) were most abundant, although trace amounts of almost all proteinogenic amino acids could be detected (Figure S7). LC-HRMS analysis of the metabolome of unsupplemented C. elegans revealed several low abundance peaks for the expected m/z values that could plausibly represent amino acid conjugates of ascarosides as their production was abolished in daf-22 mutants (19–21, Figure 4b), but peak intensities were too low to obtain MS/MS data to confirm structures. We then surveyed available metabolic data for other ascaroside biosynthetic mutants, including mutants of acyl-CoA oxidase acox-1.1, in which production of saturated ascarosides with a side chain of 9 carbons is greatly increased (Figure S8).4,34 We found that acox-1.1 mutants produce larger amounts of compounds that have MS/MS spectra consistent with amino acid conjugates of saturated ascarosides with 9 carbon and longer side chains (19–21, Figures 4b,c and S9), e.g., ascr#10-Phe (22), whose structure was independently confirmed by synthesis (Figures 4c and S10). These data established amino acid conjugates as a new family of ascaroside derivatives which under standard laboratory conditions are produced only in very low concentrations in wild-type (Figure S9b).
Figure 4.
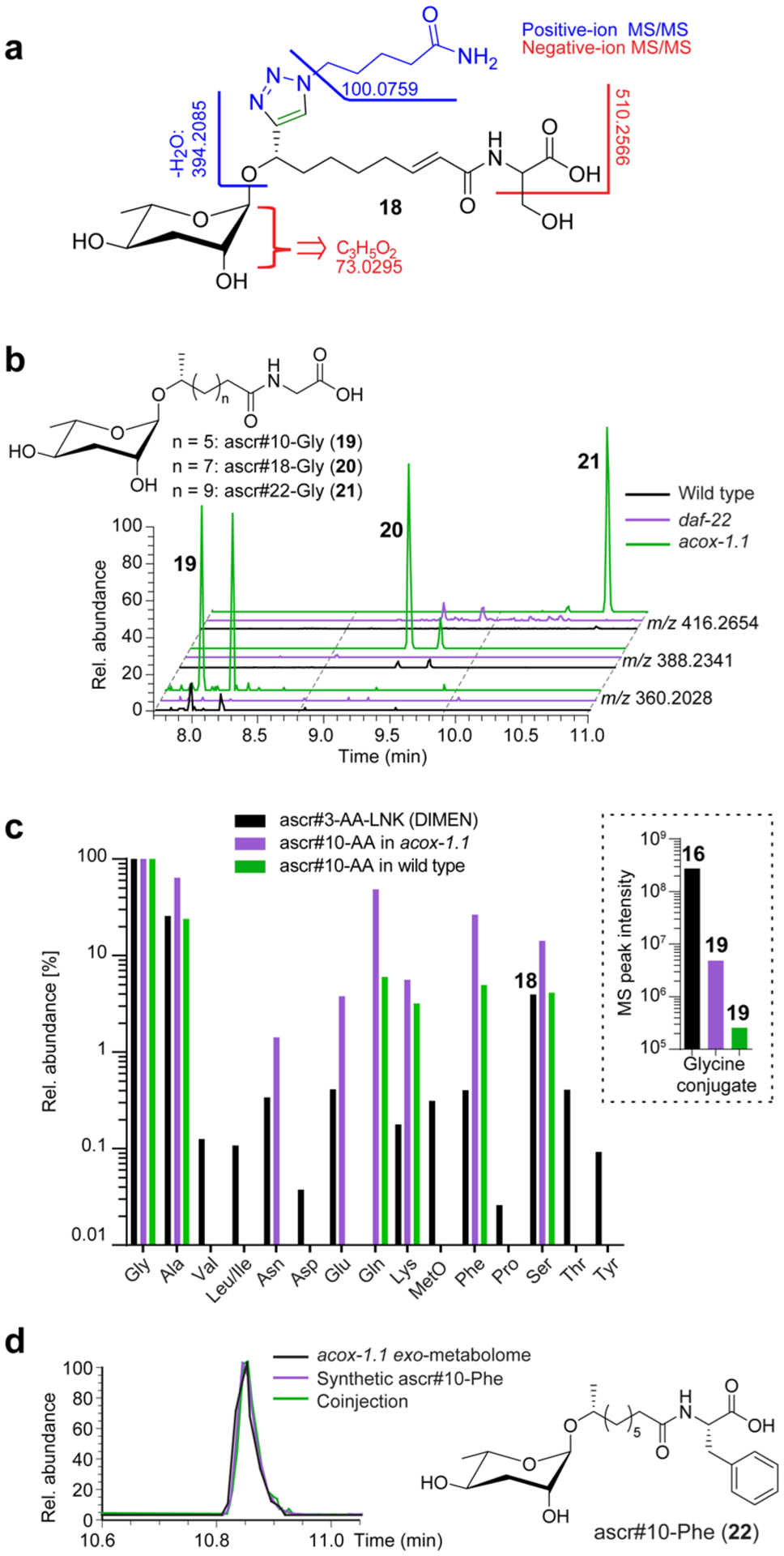
PDMs from amino acid ligation of ascr#3-YNE. (a) MS/MS fragmentation of ascr#3-Ser-LNK (19). (b) EICs [M–H]− for ascaroside glycine conjugates 19–22 in acox-1.1, wildtype, and daf-22 exo-metabolomes. (c) Relative abundances for PDMs representing ascr#3-amino acid-LNK conjugates as detected by DIMEN (black) as well as for natural ascr#10-amino acid conjugates in acox-1.1 mutants and wild-type worms (purple and green, respectively), normalized to the abundance of the corresponding glycine conjugate. Absolute abundances of the ascaroside-glycine conjugates are shown in the inset. (d) EICs for ascr#10-Phe (23) in acox-1.1 exo-metabolome, synthetic ascr#10-Phe, and coinjection confirm its identification.
Phosphorylated Ascarosides.
MS/MS spectra of a second family of PDMs suggested that they represent phosphorylated ascarosides, including phascr#31-LNK (23) and phicas#31-LNK (24) (Figures 3a, 5a,b). Their structures were inferred primarily from analysis of positive ion MS/MS spectra, since negative ion MS/MS spectra were dominated by phosphate fragments (Figures 5b and S11). LC-HRMS analysis of the metabolome of unsupplemented C. elegans revealed a daf-22-dependent compound whose MS/MS spectra and retention time were consistent with a phosphorylated ascr#3, (phascr#31, 25). Subsequent targeted analysis of the C. elegans metabolome revealed a series of abundant compounds whose MS/MS spectra and retention times suggested that they represent phosphorylated ascaroside derivatives (Figures 5c–f). To confirm these assignments and determine the site of phosphorylation, we synthesized the 2′- and 4′-phosphorylated derivatives of ascr#15 (26), phascr#151 (27), and phascr#152 (28), respectively (Figures 5e and S12). Comparison of retention times of natural and synthetic samples demonstrated that natural phosphorylated ascr#15 carries the phosphate in the 2′-position and further indicated that C. elegans produces a series of 2′-phosphorylated derivatives (29–32) of almost all known simple ascarosides (2, 10, 33–35) (Figure 5d).
Figure 5.
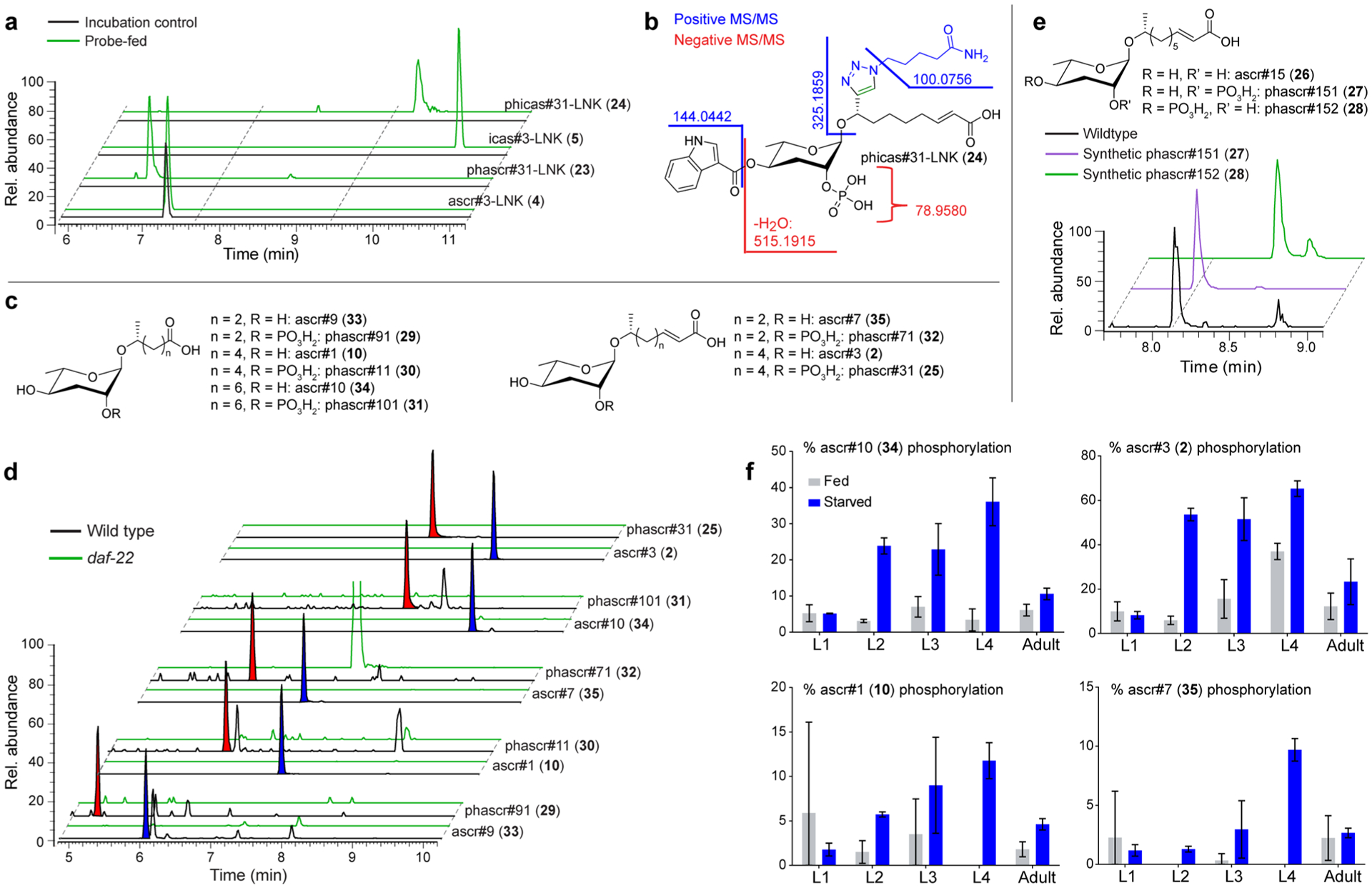
Identification of phosphorylated PDMs. (a) EICs for features representing phosphorylated PDMs in probe-fed and incubation control data sets. (b) MS/MS analysis of phicas#31-LNK (24). (c) Structures of simple ascarosides and their phosphorylated derivatives. (d) EICs for compounds in panel (c) in wildtype and daf-22 exo-metabolomes reveal extensive phosphorylation of ascarosides. Phosphorylated and corresponding nonphosphorylated ascaroside species are highlighted with red and blue shading, respectively. (e) EICs for synthetic phascr#151 (27), synthetic phascr#152) (28), and the phosphorylated ascr#15 in wild-type exo-metabolome indicates phosphorylation at the 2′-position. (f) Abundances of phosphorylated ascarosides relative to corresponding nonphosphorylated ascarosides in different life stages.
To estimate the concentrations of phosphorylated ascarosides relative to their nonphosphorylated counterparts, we established standard curves for ascr#15 (26) and phascr#151 (27) (Figure S13). Quantification across different life stages and different nutritional conditions revealed that production of phosphorylated ascarosides is greatly increased in L2, L3, and L4 larvae under starved conditions (Figure 5f). In fact, over 50% of some ascarosides, e.g., ascr#3 (2), were phosphorylated in starved larvae. Levels of ascaroside phosphorylation in adults do not appear to change significantly upon starvation; however, phosphorylated ascarosides still account for a significant fraction of total ascaroside production.
Although mass features we identified here as phosphorylated ascarosides have been previously shown to be daf-22 dependent, their uninformative MS/MS spectra prevented their recognition as an abundant class of ascarosides.12 As pointed out above, negative-ion MS/MS spectra of phosphorylated ascarosides are dominated by phosphate fragments and lack the m/z 73.0295 fragment that usually is diagnostic for ascarosides (Figure S14).4
Fucosylated Ascarosides.
A third family of PDMs was identified from the largest reporter-ion cluster for which MS/MS spectra suggested addition of one or two deoxyhexose moieties (Figure 6a,b, and S15–S17). These deoxy sugar moieties appeared to be decorated with up to two methyl groups each, resulting in a total of 11 observed combinations of methylated carbohydrate-modified PDMs (e.g., 36a–c, 37, also see Table S1).
Figure 6.
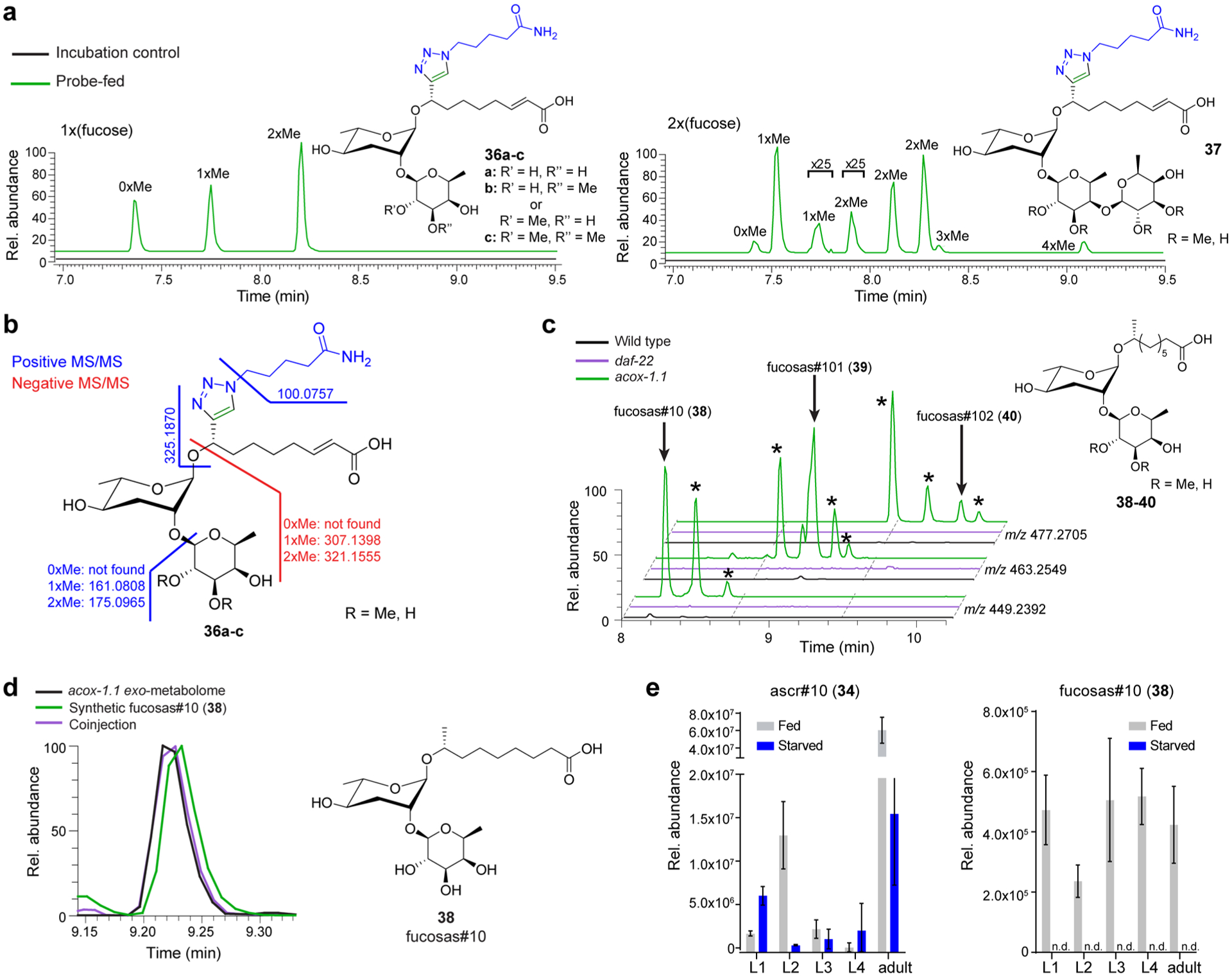
Identification of PDMs representing fucosylated ascarosides. (a) EICs for PDMs bearing one or two deoxyhexose moieties that are further decorated with additional methyl groups in enriched probe-fed and control data sets. (b) MS/MS analysis of fucosylated ascr#3-LNK decorated with up to two methyl groups (36a–c, also Figures S15–S17). (c) EICs for m/z values corresponding to fucosas#10 (38) and derivatives with one or two additional methyl groups (fucosas#101 (39) and fucosas#102 (40), respectively) in acox-1.1, wildtype, and daf-22 exo-metabolomes. Fucosylated ascr#10 derivatives are observed in wild-type C. elegans, are more abundant in acox-1.1 mutants, and not detected in daf-22 mutant worms. Additional isomers of fucosylated ascr#10 derivatives represent fucosyl-ascarosides with shorter side chains and mono- and di-O-methylation of the fucose, as suggested by MS/MS analysis (Figures S18–S20). (d) EICs for fucosas#10 (38) in acox-1.1 exo-metabolome (black), a synthetic sample (green), and in a sample where synthetic fucosas#10 is added to acox-1.1 exo-metabolome (purple) confirm identification of fucosas#10. (e) Ascr#10 (34) and fucosas#10 (38) were quantified in the exo-metabolome of wildtype worms. Production of fucosas#10 is dependent on nutritional conditions, but not strongly dependent on life stage, unlike ascr#10.
Similar to the amino acid conjugate PDMs, candidate metabolites corresponding to the deoxyhexosylated PDMs were detected only at very low abundances in wild-type C. elegans, with intensities that were too low for extensive MS/MS analysis. However, inspection of the exo-metabolome of acox-1.1 mutants revealed much larger amounts of putative deoxyhexosylated ascr#10 derivatives, likely because of the very high abundance of ascr#10 (34) in this mutant (Figure S8).4 MS/MS analysis indicated the presence of deoxyhexosylated ascr#10 with 0, 1, and 2 additional methyl groups (38–40, Figure 6c). Additionally, we detected several series of features that appeared to represent deoxyhexosylated derivatives of other ascarosides with different side chain lengths (Figures S18–S20).
On the basis of previous reports of mono- and di-O-methylated l-fucose as a component of C. elegans N-glycans,35 we hypothesized that the deoxyhexose in the ascr#3-YNE-derived PDMs may be fucose. To confirm the identity of the deoxyhexose, the mixture of PDMs obtained from the ascr#3-YNE (3) enrichment experiment was subjected to acidic hydrolysis to cleave all glycosidic bonds and then permethylated.36,37 The permethylated sample showed features with the same retention times and molecular weight as a synthetic mixture of the α- and β-isomers of permethylated fucose, supporting the assignment of the deoxyhexose as fucose (Figure S21). To confirm this assignment and to determine whether the fucose is attached to the 2′- or 4′-position of the ascarylose, we synthesized monofucosylated ascr#10 (fucosas#10, 38), one of the most abundant fucosylated ascarosides, using a thioglycosylation strategy (see SI Methods).38 Fucosas#10 (38) matched the retention time and MS/MS fragmentation pattern of the natural compound (Figures 6d and S22). Lastly, we quantified production of fucosylated ascarosides in the exo-metabolome in different life stages of wild-type C. elegans. Unlike ascr#10 (34), which is excreted in different amounts depending on life-stage, fucosas#10 (38) is excreted consistently at all life stages (Figure 6e). Additionally, we found that starvation reduced production of fucosas#10 to levels below the detection limit of LC-MS, suggesting that this compound may function in mediating starvation-associated metabolic responses (Figure 6e).
DIMEN in E. coli Bacteria.
To test applicability of DIMEN to other biological systems, we performed a simple experiment in E. coli OP50 bacteria, feeding 17-octadecynoic acid (C18-YNE) as a probe, using the conditions we had developed for C. elegans without further optimization. Analysis of MS/MS data for the reporter-ion at m/z 100.0757 revealed a series of chain shortened PDMs, indicating extensive metabolism of C18-YNE via β-oxidation (Figure S23). In-depth analysis of the data obtained for C18-YNE-treated E. coli is beyond the scope of this study; however, the preliminary results suggest that DIMEN is broadly applicable.
DISCUSSION
Untargeted metabolic studies between different mutants, life stages, or other biological conditions result in large and often overwhelming data sets that nonetheless capture only part of the structural diversity of metabolomes, given the limitations of current MS instrumentation and data processing capabilities.12,39–41 On the other hand, targeted studies, which often take advantage of reactive functional groups (i.e., acids, amines, thiols, etc.),42–44 may fail to discover novel metabolites with unexpected structural features. DIMEN enables discovery-oriented analyses in a highly sensitive, pathway-targeted manner by leveraging the advantages of both untargeted comparative metabolomics and enrichment strategies. As we demonstrated here, using a single alkyne-modified metabolite uncovered a large number of novel biosynthetic transformations that reveal unexpected intersection with amino acid metabolism, glycan chemistry, and phosphorylation.
Although click-chemistry mediated enrichment strategies have been used extensively for proteins, few studies employ click-chemistry-enabled enrichment of small molecules. Examples include a cleavable azide reactive resin (CARR) that was used to isolate azide-labeled fatty acids and phenylethylamines, as well as an azide-reactive cyclooctyne (ARCO) resin that was used to enrich azide-labeled peptides.8,9 In both cases, the strain-promoted azide–alkyne cycloaddition (SPAAC) click reaction was employed to enrich azide-labeled compounds.45 These strategies have the advantage of not requiring any copper, reducing agent, or chelating ligands when compared to CuAAC. However, the requirement of a strained alkyne for enrichment results in relatively large and bulky linkers that may confer undesirable chromatographic or MS/MS properties. One of few examples of CuAAC to isolate small molecules employed an azido-coumarin loaded resin to detect alkyne-containing natural products.46 The resulting fluorescence was then used to guide isolation of alkynated plant metabolites.
DIMEN’s use of CuAAC with the ACER resin results in a relatively small and polar linker-derived modification of the enriched PDMs, which benefits ionization and limits chromatographic perturbation. However, since we observed some undesired TFA-induced cleavage of the glycosidic bond in enriched ascr#3-LNK (4), future iterations of this DIMEN could benefit from implementation of other established chemo-orthogonal cleavage strategies (Figure 2c).42
Among the three newly identified compound classes, the amino acid conjugates reveal a direct connection between ascaroside signaling and amino acid abundance and thus nutritional status. Correspondingly, the previous identification of indole-carboxy- and p-hydroxybenzoyl-modified ascarosides (icas and hbas, respectively) connect ascaroside signaling to amino acid catabolism.11 Among the identified ascaroside amino acid conjugates, the glycine derivatives are by far the most abundant (Figure 4c), although free glycine is among the less abundant amino acids in C. elegans.47,48 This suggests that, like other types of modular ascarosides, amino acid-conjugated ascarosides are products of specific biosynthetic pathways and may serve specific signaling functions.19,49 Under laboratory conditions, the amino acid-conjugated ascarosides are barely observable by LC-HRMS in the exo-metabolome of wildtype C. elegans, whereas they accumulate in the acox-1.1 mutant (Figure 4b). Their identification highlights the potential of DIMEN for the discovery of low-abundance metabolites that may represent potent signaling molecules, even at low femtomolar concentrations, as shown for the case of hbas#3.4
In contrast, the phosphorylated ascarosides identified here are among the most abundant ascarosides in C. elegans, but had remained unidentified, despite the central role ascaroside signaling plays in the life history of this model organism. Although several compounds identified here as phosphorylated ascarosides had been observed previously via LC-MS based comparative metabolomics, they remained unidentified as their ascaroside origin could not be confirmed (Figure S14).12 DIMEN enabled structure elucidation by (i) demonstrating that these compounds must be derived from an ascaroside probe and (ii) providing robust fragmentation patterns that extended beyond phosphate fragments.
Small molecule phosphorylation regulates diverse biological processes, e.g., signal transduction through phosphorylated nucleosides, inositols, and lipid-linked inositides, and importantly, energy metabolism.50–52 Phosphorylation is often highly site-specific, e.g., in the case of inositol pyrophosphates.53 In the case of ascarosides, phosphorylation is specific for the 2′-position. Our observation that starvation increases the percentage of excreted phosphorylated ascarosides relative to their nonphosphorylated counterparts, particularly during the larval stages of C. elegans (Figure 5f), suggests that phosphorylated ascarosides have distinct signaling properties.
Like the phosphorylated and amino acid conjugated PDMs, fucosylated ascarosides produced an MS/MS subcluster that enabled their quick recognition as a novel compound family (Figures 3a and 6a). Fucose is obtained by mammals from food sources via a salvage pathway and through a highly conserved 3-step transformation of GDP mannose.54 Once acquired, fucose may be incorporated into N- and O-glycans or glycolipids through the activity of fucosyltransferases, with the most well-known example being the ABO antigens found on human blood cells.54 Because fucose is integral for cell–cell communication and signaling, disturbances of fucosylation result in developmental disease and autoimmune disorders.55 Fucosylation also plays an important role for chemical signaling involving bacteria, for example in host-commensal symbiosis in the human intestine.56,57 The C. elegans genome includes a partially annotated fucose biosynthetic pathway and over 30 predicted fucosyl transferases, indicating that fucose may play an important role in nematodes.58,59 Similar to other organisms, C. elegans incorporate fucose in the architecture of N- and O-glycans of glycoproteins. These fucose units can be O-methylated analogously to the enriched PDMs of this study.35,59,60 Surprisingly, there are few examples of fucosylated small molecules from other organisms, though our identification of fucosylated ascarosides and the prevalence of fucosyl transferases may suggest that fucosylated metabolites could be widespread.
In addition to the three compound classes we selected for follow up, the DIMEN MS/MS network (Figure 3a) contains many yet uncharacterized nodes that likely represent additional unknown structures and metabolic pathways. Using other alkyne-functionalized ascarosides—all data shown here were generated using a single probe—and application of DIMEN to biosynthesis mutants may reveal additional facets of ascaroside metabolism and help delineate biosynthetic pathways. Our partial analysis provides evidence for an extensive metabolic network beyond existing biochemical models in C. elegans and highlights the potential of click-chemistry-based enrichment and tagging approaches for uncovering hidden metabolites and biosyntheses in diverse biological systems.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Kiemle (SUNY-ESF) for assistance with NMR spectroscopy, and I. O’Doherty, D. Gentile, and R. Micikas (Cornell University) for synthetic starting materials. This work was partly supported by the NIH (T32 GM008500 to J.S.H. and J.A.B., and R01 GM088290 and R35 GM131877 to F.C.S.) and the Howard Hughes Medical Institute, with which F.C.S. is a Faculty Scholar.
ABBREVIATIONS
- ARCO
azide-reactive cyclooctyne
- BTTAA
2-(4-((Bis((1-(tert-butyl)-1H-1,2,3,-triazol-4-yl)methylamino)methyl)-1H-1,2,3-triazol-1-yl)acetic acid
- CARR
cleavable azide-reactive resin
- CuAAC
copper catalyzed azide alkyne cycloaddition
- DIMEN
deep interrogation of metabolism via enrichment
- GDP
guanosine diphosphate
- LC
liquid chromatography
- m/z
mass to charge ratio
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- PDM
probe-derived metabolite
- PEGA
polyethylene glycol acrylamide
- SPPS
solid phase peptide synthesis
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c06877.
DIMEN methods, synthetic procedures with NMR spectroscopic data, Supporting Figures S1–S24, and Tables S1–S7 (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.0c06877
The authors declare no competing financial interest.
Contributor Information
Joshua A. Baccile, Department of Chemistry, University of Tennessee, Knoxville, Tennessee 37996, United States.
Frank C. Schroeder, Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14853, United States.
REFERENCES
- (1).Dettmer K; Aronov PA; Hammock BD Mass Spectrometry-Based Metabolomics. Mass Spectrom. Rev 2007, 26 (1), 51–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Markley JL; Brüschweiler R; Edison AS; Eghbalnia HR; Powers R; Raftery D; Wishart DS The Future of NMR-Based Metabolomics. Curr. Opin. Biotechnol 2017, 43, 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Ludewig AH; Artyukhin AB; Aprison EZ; Rodrigues PR; Pulido DC; Burkhardt RN; Panda O; Zhang YK; Gudibanda P; Ruvinsky I; Schroeder FC An Excreted Small Molecule Promotes C. Elegans Reproductive Development and Aging. Nat. Chem. Biol 2019, 15 (8), 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).von Reuss SH; Bose N; Srinivasan J; Yim JJ; Judkins JC; Sternberg PW; Schroeder FC Comparative Metabolomics Reveals Biogenesis of Ascarosides, a Modular Library of Small-Molecule Signals in C. Elegans. J. Am. Chem. Soc 2012, 134 (3), 1817–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Echevarría-Machado I; Rosa RM; Larqué-Saavedra A Responses of Transformed Catharanthus Roseus Roots to Femtomolar Concentrations of Salicylic Acid. Plant Physiol. Biochem 2007, 45 (6–7), 501–507. [DOI] [PubMed] [Google Scholar]
- (6).Sowa G; Gekker G; Lipovsky MM; Shuxian H; Chao CC; Molitor TW; Peterson PK Inhibition of Swine Microglial Cell Phagocytosis of Cryptococcus Neoformans by Femtomolar Concentrations of Morphine. Biochem. Pharmacol 1997, 53 (6), 823–828. [DOI] [PubMed] [Google Scholar]
- (7).Lehmann J; Wright MH; Sieber SA Making a Long Journey Short: Alkyne Functionalization of Natural Product Scaffolds. Chem. - Eur. J 2016, 22 (14), 4666–4678. [DOI] [PubMed] [Google Scholar]
- (8).Péerez AJ; Wesche F; Adihou H; Bode HB Solid-Phase Enrichment and Analysis of Azide-Labeled Natural Products: Fishing Downstream of Biochemical Pathways. Chem. - Eur. J 2016, 22 (2), 639–645. [DOI] [PubMed] [Google Scholar]
- (9).Nessen MA; Kramer G; Back JW; Baskin JM; Smeenk LEJJ; De Koning LJ; Van Maarseveen JH; De Jong L; Bertozzi CR; Hiemstra H; De Koster CG Selective Enrichment of Azide-Containing Peptides from Complex Mixtures. J. Proteome Res 2009, 8 (7), 3702–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Von Reuss SH; Schroeder FC Combinatorial Chemistry in Nematodes: Modular Assembly of Primary Metabolism-Derived Building Blocks. Nat. Prod. Rep 2015, 32 (7), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Schroeder FC Modular Assembly of Primary Metabolic Building Blocks: A Chemical Language in C. Elegans. Chem. Biol 2015, 22 (1), 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Artyukhin AB; Zhang YK; Akagi AE; Panda O; Sternberg PW; Schroeder FC Metabolomic “Dark Matter” Dependent on Peroxisomal β-Oxidation in Caenorhabditis elegans. J. Am. Chem. Soc 2018, 140 (8), 2841–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Butcher RA Small-Molecule Pheromones and Hormones Controlling Nematode Development. Nat. Chem. Biol 2017, 13 (6), 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Butcher RA Decoding Chemical Communication in Nematodes. Nat. Prod. Rep 2017, 34 (5), 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Choe A; Von Reuss SH; Kogan D; Gasser RB; Platzer EG; Schroeder FC; Sternberg PW Ascaroside Signaling Is Widely Conserved among Nematodes. Curr. Biol 2012, 22 (9), 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Hsueh YP; Mahanti P; Schroeder FC; Sternberg PW Nematode-Trapping Fungi Eavesdrop on Nematode Pheromones. Curr. Biol 2013, 23 (1), 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Manosalva P; Manohar M; Von Reuss SH; Chen S; Koch A; Kaplan F; Choe A; Micikas RJ; Wang X; Kogel KH; Sternberg PW; Williamson VM; Schroeder FC; Klessig DF Conserved Nematode Signalling Molecules Elicit Plant Defenses and Pathogen Resistance. Nat. Commun 2015, 6, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zhou Y; Wang Y; Zhang X; Bhar S; Jones Lipinski RA; Han J; Feng L; Butcher R A Biosynthetic Tailoring of Existing Ascaroside Pheromones Alters Their Biological Function in C. Elegans. eLife 2018, 7, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Panda O; Akagi AE; Artyukhin AB; Judkins JC; Le HH; Mahanti P; Cohen SM; Sternberg PW; Schroeder FC Biosynthesis of Modular Ascarosides in C. Elegans. Angew. Chem., Int. Ed 2017, 56 (17), 4729–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Srinivasan J; von Reuss SH; Bose N; Zaslaver A; Mahanti P; Ho MC; O’Doherty OG; Edison AS; Sternberg PW; Schroeder FC A Modular Library of Small Molecule Signals Regulates Social Behaviors in Caenorhabditis elegans. PLoS Biol. 2012, 10 (1), e1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Ludewig AH; Izrayelit Y; Park D; Malik RU; Zimmermann A; Mahanti P; Fox BW; Bethke A; Doering F; Riddle DL; Schroeder FC Pheromone Sensing Regulates Caenorhabditis elegans Lifespan and Stress Resistance via the Deacetylase SIR-2.1. Proc. Natl. Acad. Sci. U. S. A 2013, 110 (14), 5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhang YK; Reilly DK; Yu J; Srinivasan J; Schroeder FC Photoaffinity Probes for Nematode Pheromone Receptor Identification. Org. Biomol. Chem 2020, 18 (1), 36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Wang M; Carver JJ; Phelan VV; Sanchez LM; Garg N; Peng Y; Nguyen DD; Watrous J; Kapono CA; Luzzatto-Knaan T; Porto C; Bouslimani A; Melnik AV; Meehan MJ; Liu WT; Crüsemann M; Boudreau PD; Esquenazi E; Sandoval-Calderón M; Kersten RD; Pace LA; Quinn RA; Duncan KR; Hsu C-C; Floros DJ; Gavilan RG; Kleigrewe K; Northen T; Dutton RJ; Parrot D; Carlson EE; Bertrand A; Michelsen CF; Jelsbak L; Sohlenkamp C; Pevzner P; Edlund A; McLean J; Piel J; Murphy BT; Gerwick L; Liaw C-C; Yang Y-L; Humpf H-U; Maansson M; Keyzers RA; Sims AC; Johnson AR; Sidebottom AM; Sedio BE; Klitgaard A; Larson CB; Boya PCA; Torres-Mendoza D; Gonzalez DJ; Silva DB; Marques LM; Demarque DP; Pociute E; O’Neill EC; Briand E; Helfrich EJN; Granatosky EA; Glukhov E; Ryffel F; Houson H; Mohimani H; Kharbush JJ; Zeng Y; Vorholt JA; Kurita KL; Charusanti P; McPhail KL; Fog Nielson K; Vuong L; Elfeki M; Traxler MF; Engene N; Koyama N; Vining OB; Baric R; Silva RR; Mascuch SJ; Tomasi S; Jenkins S; Macherla V; Hoffman T; Agarwal V; Williams PG; Dai J; Neupane R; Gurr J; Rodríguez AMC; Lamsa A; Zhang C; Dorrestein K; Duggan BM; Almaliti J; Allard P-M; Phapale P; Nothias L-F; Alexandrov T; Litaudon M; Wolfender J-L; Kyle JE; Metz TO; Peryea T; Nguyen D-T; VanLeer D; Shinn P; Jadhav A; Müller R; Waters KM; Shi W; Liu X; Zhang L; Knight R; Jensen PR; Palsson BO; Pogliano K; Linington RG; Gutiérrez M; Lopes NP; Gerwick WH; Moore BS; Dorrestein PC; Bandeira N Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol 2016, 34 (8), 828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Bernatowicz MS; Daniels SB; Köster H A Comparison of Acid Labile Linkage Agents for the Synthesis of Peptide C-Terminal Amides. Tetrahedron Lett. 1989, 30 (35), 4645–4648. [Google Scholar]
- (25).Uttamapinant C; Tangpeerachaikul A; Grecian S; Clarke S; Singh U; Slade P; Gee KR; Ting AY Fast, Cell-Compatible Click Chemistry with Copper-Chelating Azides for Biomolecular Labeling. Angew. Chem., Int. Ed 2012, 51 (24), 5852–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Bevilacqua V; King M; Chaumontet M; Nothisen M; Gabillet S; Buisson D; Puente C; Wagner A; Taran F Copper-Chelating Azides for Efficient Click Conjugation Reactions in Complex Media. Angew. Chem., Int. Ed 2014, 53 (23), 5872–5876. [DOI] [PubMed] [Google Scholar]
- (27).Srinivasan J; Kaplan F; Ajredini R; Zachariah C; Alborn HT; Teal P. E. a; Malik RU; Edison AS; Sternberg PW; Schroeder FC A Blend of Small Molecules Regulates Both Mating and Development in Caenorhabditis elegans. Nature 2008, 454 (7208), 1115–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Besanceney-webler C; Jiang H; Zheng T; Feng L; Soriano Del Amo D; Wang W; Klivansky LM; Marlow FL; Liu Y; Wu P Increasing the Efficacy of Bioorthogonal Click Reactions for Bioconjugation: A Comparative Study. Angew. Chem., Int. Ed 2011, 50 (35), 8051–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Jeong P-YY; Jung M; Yim Y-HH; Kim H; Park M; Hong E; Lee W; Kim YH; Kim K; Paik Y-KK Chemical Structure and Biological Activity of the Caenorhabditis elegans Dauer-Inducing Pheromone. Nature 2005, 433 (7025), 541–545. [DOI] [PubMed] [Google Scholar]
- (30).Izrayelit Y; Srinivasan J; Campbell SL; Jo Y; Von Reuss SH; Genoff MC; Sternberg PW; Schroeder FC Targeted Metabolomics Reveals a Male Pheromone and Sex-Specific Ascaroside Biosynthesis in Caenorhabditis elegans. ACS Chem. Biol 2012, 7 (8), 1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Butcher RA; Ragains JR; Clardy J An Indole-Containing Dauer Pheromone Component with Unusual Dauer Inhibitory Activity at Higher Concentrations. Org. Lett 2009, 11 (14), 3100–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Pungaliya C; Srinivasan J; Fox BW; Malik RU; Ludewig AH; Sternberg PW; Schroeder FC A Shortcut to Identifying Small Molecule Signals That Regulate Behavior and Development in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (19), 7708–7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Butcher RA; Ragains JR; Li W; Ruvkun G; Clardy J; Mak HY Biosynthesis of the Caenorhabditis elegans Dauer Pheromone. Proc. Natl. Acad. Sci. U. S. A 2009, 106 (6), 1875–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Zhang X; Wang Y; Perez DH; Jones Lipinski RA; Butcher RA Acyl-CoA Oxidases Fine-Tune the Production of Ascaroside Pheromones with Specific Side Chain Lengths. ACS Chem. Biol 2018, 13 (4), 1048–1056. [DOI] [PubMed] [Google Scholar]
- (35).Paschinger K; Gutternigg M; Rendić D; Wilson IBH The N-Glycosylation Pattern of Caenorhabditis elegans. Carbohydr. Res 2008, 343 (12), 2041–2049. [DOI] [PubMed] [Google Scholar]
- (36).Cheetham NWH; Sirimanne P Methanolysis Studies of Carbohydrates, Using H.P.L.C. Carbohydr. Res 1983, 112 (1), 1–10. [Google Scholar]
- (37).Ciucanu I; Costello CE Elimination of Oxidative Degradation during the Per-O-Methylation of Carbohydrates. J. Am. Chem. Soc 2003, 125 (52), 16213–16219. [DOI] [PubMed] [Google Scholar]
- (38).He H; Chen D; Li X; Li C; Zhao JH; Qin HB Synthesis of Trisaccharide Repeating Unit of Fucosylated Chondroitin Sulfate. Org. Biomol. Chem 2019, 17 (11), 2877–2882. [DOI] [PubMed] [Google Scholar]
- (39).Quinn RA; Melnik AV; Vrbanac A; Fu T; Patras KA; Christy MP; Bodai Z; Belda-Ferre P; Tripathi A; Chung LK; Downes M; Welch RD; Quinn M; Humphrey G; Panitchpakdi M; Weldon KC; Aksenov A; da Silva R; Avila-Pacheco J; Clish C; Bae S; Mallick H; Franzosa EA; Lloyd-Price J; Bussell R; Thron T; Nelson AT; Wang M; Leszczynski E; Vargas F; Gauglitz JM; Meehan MJ; Gentry E; Arthur TD; Komor AC; Poulsen O; Boland BS; Chang JT; Sandborn WJ; Lim M; Garg N; Lumeng JC; Xavier RJ; Kazmierczak BI; Jain R; Egan M; Rhee KE; Ferguson D; Raffatellu M; Vlamakis H; Haddad GG; Siegel D; Huttenhower C; Mazmanian SK; Evans RM; Nizet V; Knight R; Dorrestein PC Global Chemical Effects of the Microbiome Include New Bile-Acid Conjugations. Nature 2020, 579, 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Martín-Blázquez A; Díaz C; González-Flores E; Franco-Rivas D; Jiménez-Luna C; Melguizo C; Prados J; Genilloud O; Vicente F; Caba O; Pérez del Palacio J Untargeted LC-HRMS-Based Metabolomics to Identify Novel Biomarkers of Metastatic Colorectal Cancer. Sci. Rep 2019, 9 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Khizar M; Shi J; Saleem S; Liaquat F; Ashraf M; Latif S; Haroon U; Hassan SW; Rehman S. ur; Chaudhary HJ; Quraishi UM; Munis MFH Resistance Associated Metabolite Profiling of Aspergillus Leaf Spot in Cotton through Non-Targeted Metabolomics. PLoS One 2020, 15 (2), 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Carlson EE; Cravatt BF Chemoselective Probes for Metabolite Enrichment and Profiling. Nat. Methods 2007, 4 (5), 429–435. [DOI] [PubMed] [Google Scholar]
- (43).Conway LP; Garg N; Lin W; Vujasinovic M; Löhr JM; Globisch D Chemoselective Probe for Detailed Analysis of Ketones and Aldehydes Produced by Gut Microbiota in Human Samples. Chem. Commun 2019, 55 (62), 9080–9083. [DOI] [PubMed] [Google Scholar]
- (44).Garg N; Conway LP; Ballet C; Correia MSP; Olsson FKS; Vujasinovic M; Lohr J-M; Globisch D Chemoselective Probe Containing a Unique Bioorthogonal Cleavage Site for Investigation of Gut Microbiota Metabolism. Angew. Chem 2018, 130 (42), 14001–14005. [DOI] [PubMed] [Google Scholar]
- (45).Agard NJ; Prescher JA; Bertozzi CR A Strain-Promoted [3 + 2] Azide-Alkyne Cycloaddition for Covalent Modification of Biomolecules in Living Systems. J. Am. Chem. Soc 2004, 126 (46), 15046–15047. [DOI] [PubMed] [Google Scholar]
- (46).Jeon H; Lim C; Lee JM; Kim S Chemical Assay-Guided Natural Product Isolation via Solid-Supported Chemodosimetric Fluorescent Probe. Chem. Sci 2015, 6 (5), 2806–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Falk MJ; Zhang Z; Rosenjack JR; Nissim I; Daikhin E; Nissim I; Sedensky MM; Yudkoff M; Morgan PG Metabolic Pathway Profiling of Mitochondrial Respiratory Chain Mutants in C. Elegans. Mol. Genet. Metab 2008, 93 (4), 388–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Reinke SN; Hu X; Sykes BD; Lemire BD Caenorhabditis elegans Diet Significantly Affects Metabolic Profile, Mitochondrial DNA Levels, Lifespan and Brood Size. Mol. Genet. Metab 2010, 100 (3), 274–282. [DOI] [PubMed] [Google Scholar]
- (49).Falcke JM; Bose N; Artyukhin AB; Rödelsperger C; Markov GV; Yim JJ; Grimm D; Claassen MH; Panda O; Baccile JA; Zhang YK; Le HH; Jolic D; Schroeder FC; Sommer RJ Linking Genomic and Metabolomic Natural Variation Uncovers Nematode Pheromone Biosynthesis. Cell Chem. Biol 2018, 25 (6), 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Lee JY; Kim YR; Park J; Kim S Inositol Polyphosphate Multikinase Signaling in the Regulation of Metabolism. Ann. N. Y. Acad. Sci 2012, 1271 (1), 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Görke B; Stülke J Carbon Catabolite Repression in Bacteria: Many Ways to Make the Most out of Nutrients. Nat. Rev. Microbiol 2008, 6 (8), 613–624. [DOI] [PubMed] [Google Scholar]
- (52).Rose IA; O’Connell EL The Role of Glucose 6-Phosphate in the Regulation of Glucose Metabolism. J. Biol. Chem 1964, 239, 12–17. [PubMed] [Google Scholar]
- (53).Shears SB Intimate Connections: Inositol Pyrophosphates at the Interface of Metabolic Regulation and Cell Signaling. J. Cell. Physiol 2018, 233 (3), 1897–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Becker DJ; Lowe JB Fucose: Biosynthesis and Biological Function in Mammals. Glycobiology 2003, 13 (7), 41R. [DOI] [PubMed] [Google Scholar]
- (55).Li J; Hsu HC; Mountz JD; Allen JG Unmasking Fucosylation: From Cell Adhesion to Immune System Regulation and Diseases. Cell Chem. Biol 2018, 25 (5), 499–512. [DOI] [PubMed] [Google Scholar]
- (56).Pacheco AR; Curtis MM; Ritchie JM; Munera D; Waldor MK; Moreira CG; Sperandio V Fucose Sensing Regulates Bacterial Intestinal Colonization. Nature 2012, 492 (7427), 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Pickard JM; Maurice CF; Kinnebrew MA; Abt MC; Schenten D; Golovkina TV; Bogatyrev SR; Ismagilov RF; Pamer EG; Turnbaugh PJ; Chervonsky AV Rapid Fucosylation of Intestinal Epithelium Sustains Host-Commensal Symbiosis in Sickness. Nature 2014, 514 (7524), 638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Yan S; Serna S; Reichardt NC; Paschinger K; Wilson IBH Array-Assisted Characterization of a Fucosyltransferase Required for the Biosynthesis of Complex Core Modifications of Nematode N-Glycans. J. Biol. Chem 2013, 288 (29), 21015–21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Barrows BD; Haslam SM; Bischof LJ; Morris HR; Dell A; Aroian RV Resistance to Bacillus Thuringiensis Toxin in Caenorhabditis elegans from Loss of Fucose. J. Biol. Chem 2007, 282 (5), 3302–3311. [DOI] [PubMed] [Google Scholar]
- (60).Haslam SM; Dell A Hallmarks of Caenorhabditis elegans N-Glycosylation: Complexity and Controversy. Biochimie 2003, 85 (1–2), 25–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


