Abstract
Background
Epidermolysis bullosa (EB) is a rare genetic disease with widely different clinical manifestations, but the relationship between genotype and phenotype is not fully understood. In the present study, we recruited a Chinese family in which two members had been diagnosed with localized EB simplex (EBS), with clinical manifestation, including blisters and erosions on the soles of the feet since infancy.
Objective
To identify and confirm the genetic variation in a Chinese family diagnosed as localized EBS.
Methods
Our study included two patients, other healthy members of the family, and 100 normal controls. Genomic DNA samples were isolated from each participant, and then polymerase chain reaction (PCR) direct sequencing was performed.
Results
The results of PCR direct sequencing revealed a novel heterozygous missense mutation in codon 461 of exon 7 of KRT5 (c.1382T>C), which led to an amino acid change (p.L461P) in the patients with EBS but was absent in unaffected family members and 100 unrelated control samples.
Conclusion
The present study broadens the mutational spectrum of EBS, and this knowledge could be harnessed for prenatal screening, gene diagnosis, and gene therapy for localized EBS.
Keywords: Epidermolysis bullosa simplex, Keratin 5, Mutation
INTRODUCTION
Epidermolysis bullosa (EB) comprises a group of hereditary disorders, in which blisters occur spontaneously or after minor injury or friction. The phenotypic spectrum of EB is highly variable, and there are differences in severity and associated extraneous manifestations between different types of EB. However, all types of EB present with traumatic blistering and fragility1,2. Based on the level of the dermal-epidermal separation relative to the basement membrane, EB can be divided into four subtypes—simplex, junctional, dystrophic, and Kindler syndrome3. In the most common subtype, EB simplex (EBS), separation occurs at or above the basal layer of keratinocytes in the epidermis. Statistics from 1986 to 2002 showed that the prevalence of EB was about 11 cases per 1 million residents and the incidence rate was about 19 cases per 1 million live births4. EBS shows significant genetic and clinical heterogeneity. EBS can be subdivided into the generalized severe (Dowling-Meara), generalized intermediate (Koebner), and localized subtypes5. Localized EBS (OMIM 131800) is the mildest subtype, in which bullous lesions usually occur during the birth or infancy period, although they may also appear during adolescence or early adulthood. Clinically, the vesicles and bullae are primarily confined to the palmar and plantar regions without nail and mucosal damage6.
Almost all EBS mutations follow the pattern of autosomal dominant inheritance7. EBS is mainly associated with mutations in two genes, KRT5 and KRT14. KRT5 and KRT14 genes encode basal epidermal keratin 5 and 14, respectively8. Both keratin 5 and keratin 14 have a central rod-like alpha helix structure and a non-helix structure (head and tail) on both sides. At the beginning and end of the central rod-like region, there are two highly conserved amino acid sequences, called helix initiation peptide (HIP) and helix termination peptide (HTP)9. In most cases, the location of the mutation determines the severity of the clinical phenotype. For example, mutations in HIP and HTP that affect the assembly of keratin filaments lead to the most severe Dowling-Meara EBS (EBS-DM; OMIM 131760); Mutations often lead to localized EBS in nonhelical linker regions, while Koebner (EBS-K; OMIM 131900) mutations are more widely distributed along both keratin 5 and keratin 14 polypeptides10. To date, more than 150 different pathogenic mutations associated with these genes have been described10, most of them are associated with the more severe EBS-DM5.
In the present study, two patients from a Chinese family who had been diagnosed with localized EBS were evaluated, and a novel missense mutation c.1382T>C (p.L461P), which was located at the 2B segment of the KRT5, confirmed a diagnosis of localized EBS. Identification of new mutations and genotype-phenotype associations leads to a better understanding of the underlying pathophysiologic basis of localized EBS and is important for disease course prediction, gene diagnosis, genetic counseling, and gene therapy.
MATERIALS AND METHODS
Clinical evaluation and DNA sampling
In total, five members of the localized EBS pedigree (Fig. 1) were enrolled in the study. Members II:1 (42-years-old female) and III:1 (15-years-old female; proband) presented to our hospital. Our study also included proband's father, grandparents, and 100 normal control subjects. Written informed consent was obtained from all participating individuals (or their guardians), and 5 ml blood samples were drawn from each participant for DNA extraction. Clinical examinations of all participants were carried out by a dermatologist. The current research, which was performed in compliance with the principles of the Declaration of Helsinki, was authorized by the Institutional Review Board of Tongling People's Hospital. We received the patient's consent form about publishing all photographic materials. Ethical approval was gained from the Ethics Committee of Tongling People's Hospital (2020001).
Fig. 1. Family pedigree. The family tree shows two affected members across three generations. The unfilled symbols represent the unaffected members of the pedigree; the dark symbols indicate the affected members. Circles and squares represent females and males, respectively. The proband of pedigree has been marked with a red arrow.

Mutational analysis and multiple sequence alignment
We used the polymerase chain reaction (PCR) to amplify genomic DNA samples (Humphries et al. 1996)11, after which PCR products were sequenced with Sanger sequencing. Consulting the National Center for Biotechnology Information (NCBI) cDNA reference sequences NM_000424.3 for KRT5 and NM_000526.4 for KRT14, we identified a specific mutation present only in clinically affected family members. Multiple online bioinformatics software programs, including follows, PolyPhen2 (http://genetics.bwh.harvard.edu/pph2), SIFT (http://sift.bii.a-star.edu.sg), and Mutation Taster (http://www.mutationtaster.org/), were used to predict whether the resulting amino acid substitution could have an effect on the structure and function of the KRT5 and KRT14. We aligned the proteins of different species, including Bos taurus, Gallus gallus, Gorilla gorilla, Mus musculus, and Oryctolagus cuniculus. The change of structure between wild-type and mutation (p.L461P) was predicted by SWISS-MODEL (https://swissmodel.expasy.org, PBD: 3TNU). A structural comparison of the wild-type and mutation revealed notable differences between them.
RESULTS
Phenotypic features of the affected patients
There were two affected people in this family. The proband, a 15-year-old Chinese girl, was the only daughter of unrelated Chinese parents. She and her mother have similar clinical symptoms, and there is no other family history. Since birth, the proband and her mother have been prone to blisters after mechanical injury to the foot, but the blisters heal without scarring. The symptoms were worse in the summer and improved with age. Physical examinations were normal except the blisters (Fig. 2). Pathologic examination of the right plantar blister of the proband revealed hyperkeratosis of the epidermis, thickening of the granulosa and spinous layer, and vesicles form in the epidermis. The vessels in the dermal papillary layer were slightly dilated, with little inflammatory cell infiltration, and the perivascular stroma was slightly mucinous (Fig. 2).The diagnosis of this disease was based on the combination of typical clinical manifestations, family history and pathological examination, so it was diagnosed as localized EBS. The molecular tests we performed further confirmed the diagnosis.
Fig. 2. Clinical manifestations of the patients with localized epidermolysis bullosa simplex. The blisters in the figures has been marked with a red arrow. (A) Blisters and erosions are observed on the proband's right foot. (B) Similar blisters are found in the mother's left sole. (C) The epidermal epithelium is hyperkeratinized, the spinous layer is thickened, and the vesicles in the epidermis (H&E, ×40). (D) The fissure is located inside the epidermis (H&E, ×200).
Mutation analysis and protein structure modeling
Localized EBS is one of the most common clinical variant diseases of EBS, as well as the mildest type. According to published data, mutations in many genes are associated with EBS phenotype, including TGM5, PLEC, PKP1, KRT5, KRT14, DSP, JUP, DST, and EXPH5, but more than 75% of EBS cases are attributed to the mutation of KRT5 or KRT14 genes12. Thus, here we selected these two genes as the candidate causative genes to screen for mutation.
Molecular analysis of the proband's DNA indicated a heterozygous T to C transition at nucleotide position 1382 (c.1382T>C) in exon 7 of the KRT5 gene. The mutation leads to the substitution of proline residue for leucine at codon 461 (p. Leu461Pro). The identical mutation in KRT5 was detected in the proband's mother. The mutation is located at the 2B segment of the KRT5, confirming a diagnosis of localized EBS. No additional mutation was identified in KRT5 or KRT14. No mutation was found in unaffected family members or the 100 population-matched healthy controls (Fig. 3). PolyPhen2 (http://genetics.bwh.harvard.edu/pph2), SIFT (http://sift.bii.a-star.edu.sg), and Mutation Taster (http://www.mutationtaster.org/) all predicted that the mutation c.1382T>C (p.L461P) was likely to disrupt protein function, although proline and leucine have similar polarity.
Fig. 3. Mutation analysis of KRT5. The molecular analysis demonstrates a de novo heterozygous missense mutation c.1382T>C (p.Leu461Pro) in the KRT5 gene. (A) Unaffected members, (B) affected members.

We compared proteins from different species and obtained structural-based multi-sequence alignment of KRT5 from different species, showing that the mutation (p. l 461pro) is located in a highly conserved region (Fig. 4).
Fig. 4. Multiple-sequence alignment of KRT5 from different species. Structure-based multiple-sequence alignment of KRT5 from different species revealed that the mutation (p.Leu461Pro) was located within a highly conserved region.

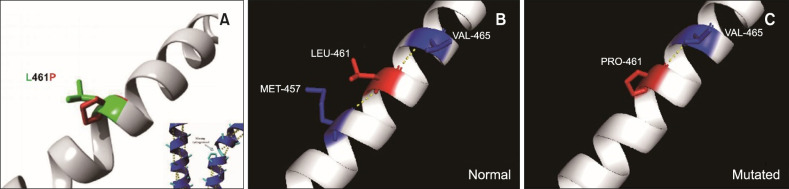
In wild-type KRT5, Leu461 forms two hydrogen bonds, with Met457 and Val465, while the mutant Pro461 only forms one hydrogen bond, with Val465. The reduction in hydrogen bonds may impact the stability of 2B, alpha-helical domains whose spatial structure is maintained by hydrogen bonds (Fig. 5). The c.1382T>C (p.L461P) mutation occurred in the non-helical junction region, and the proline of the same polarity replaced the leucine, which resulted in the existence of lightest EBS. Collectively, these results indicate that p.L461P in KRT5 is a causative mutation for localized EBS in this family.
Fig. 5. Structural model of the L461P mutation. The yellow dashed lines indicate the hydrogen bonds between two amino acids and demonstrate the difference between the structures. (A) LEU-461 and PRO-461 are in green and red, respectively. (B) In wild-type KRT5, LEU-461 forms two hydrogen bonds (with MET-457 and VAL-465). (C) The mutant PRO-461 only form sone hydrogen bond (with VAL-465).
Retrieved reported KRT5 mutation and analysis
To investigate whether there are any mutation-rich exons for KRT5 mutations, we retrieved all the reported mutation in KRT5. As shown in Table 18,10,13,14,15,16,17,18,19,20,21,22,23,24,25, about 28 novel KRT5 mutations responsible for EBS have been found since January 1, 2008. Mutations in the tail domain of KRT5 are often associated with EBS-migratory circinate erythema (EBS-Migr) and pigment forms of the disease, suggesting that this region may play a role in the regulation of pigmentation and inflammation. Also, as to the genetic diagnose, more attention should be paid to these exons coding for tail domains when screening for mutations causing EBS. We noticed that only a few mutations are associated with localized EBS, which are distributed in all regions of KRT5 but mainly gathered in the head and the non-helical linker regions. Examples include p.Val133Met, p.Asn146Lys, p.Met327Thr, p.Asp328Gly and the novel mutation (p.Leu 461Pro) in our study5,11,13. It revealed that if the patients are diagnosis as localized EBS, sequences coding for head and the non-helical linker regions of KRT5 should have propriety for the mutation screening. Because the process of gene expression is complicated and can be influenced by other genes and external factors, it may be cautious in predicting the relationship between genotype and phenotype14.
Table 1. Novel KRT5 mutations for localized epidermolysis bullosa simplex.
| Author (year) | Exon | Nucleotide change | Protein change | Keratin domain |
|---|---|---|---|---|
| Jerábková et al. (2010)15 | 1 | c.428T>C | p.V143A | Head |
| Wertheim-Tysarowska et al. (2016)13 | 1 | c.436A>T | p.N146Y | Head |
| Kim et al. (2017)16 | 1 | c.464T>C | p.L155P | Head |
| Kim et al. (2017)14 | 1 | c.502G>C | p.E168Q | Head |
| Minakawa et al. (2013)17 | 1 | c.505C>G | p.R169G | 1A |
| Arin et al. (2010)10 | 1 | c.514A>G | p.I172V | 1A, HIP |
| Kim et al. (2017)14 | 1 | c.535T>C | p.F179L | 1A |
| Glász-Bóna et al. (2009)18 | 1 | c.547A>G | p.I183V | 1A |
| García et al. (2011)19 | 1 | c.557T>A | p.V186E | 1A |
| Glász-Bóna et al. (2009)18 | 1 | c.570G>C | p.E190D | 1A |
| Bowden et al. (2009)20 | 1 | c.593C>G | p.T198S | 1A |
| Flohil et al. (2010)8 | 1 | c.596A>T | p.K199T | 1A |
| Gao et al. (2015)21 | 1 | c.605T>C | p.L202Q | 1A |
| Cho et al. (2014)22 | 1 | c.608T>C | p.L203P | L1 |
| García et al. (2011)19 | 3 | c.961A>C | p.T321P | L12 |
| Chiang et al. (2008)23 | 3 | c.971T>C | p.V324A | L12 |
| Wertheim-Tysarowska et al. (2016)13 | 3 | c.974T>C | p.L325P | L12 |
| Arin et al. (2010)10 | 3 | c.991C>A | p.R331S | L12 |
| Glász-Bóna et al. (2009)18 | 3 | c.991C>G | p.R331G | L12 |
| Lev-Tov et al. (2012)24 | 6 | c.1270G>C | p.A424P | 2B |
| García et al. (2011)19 | 6 | c.1283G>A | p.A428T | 2B |
| Minakawa et al. (2013)17 | 6 | c.1327A>G | p.K443E | 2B |
| Arin et al. (2010)10 | 6 | c.1362del4insAGCTGGTA | p.E455AfsX117 | 2B |
| Wertheim-Tysarowska et al. (2016)13 | 7 | c.1412G>A | p.R471H | 2B |
| Arin et al. (2010)10 | 7 | c.1438A>G | p.R480G | 2B, HTP |
| Arin et al. (2010)10 | 8 | c.1636C>A | p.L546I | Tail |
| Minakawa et al. (2013)17 | 9 | c.1644del4 | p.G550A | Tail |
| Bchetnia et al. (2012)25 | 9 | c.1675C>T | fsX82 p.R559X | Tail |
HIP: helix initiation peptide, HTP: helix termination peptide.
DISCUSSION
KRT5 and KRT14 genes encode the intermediate filament (IF) proteins in the basal layer of the epidermis and related complex epithelia26. The keratin intermediate filament (KIF) network is attached to desmosomes and hemi-desmosomes, connecting keratinocytes to adjacent cells and basement membranes, forming intercellular adhesions that protect epithelial cells from mechanical and other stresses27. EBS is closely related to mutations in KRT5 and KRT14, but the mechanism by which mutations cause the formation and collapse of keratin networks remains unclear28.
It is reported that disruption of KRT5/KRT14 filament network architecture can cause the fragility of basal keratinocytes, making the cells unable to bear mechanical pressure, then further leading to intracellular vacuoles, which eventually result in blistering of the skin. Sawant et al.29 found that threonine 150 (T150) KRT5 phosphorylation plays an important role in the formation of the KIF network, relating to the pathogenesis of EBS. In addition, other pathological mechanisms play important roles in the pathophysiology of EBS, including preinflammatory cytokines, the tumor necrosis factor, interleukin-1β (IL-1β), and signaling pathways which associated with them. Russell et al.30 found that the activation of extracellular signal-regulated kinase (ERK) and protein kinase B (PKB) signal transduction channels of ERK and PKB due to mechanical stresses were involved in EBS, leading to the resistance of keratin mutated cells to apoptosis after channel activation9. Beyond that, there are other mechanisms involved in the occurrence of disease, including impaired mechanical stress recovery, downregulation of cellular junction elements, and destruction of epithelial cell adhesion31. So far, we do not know how the mutation (p.L461P) causes clinical manifestation in this family; further studies are needed to provide insights into the detailed molecular pathogenesis of the mutations identified in the present study.
At present, there are no effective treatment methods for EBS; the main treatment is symptomatic management. For example, plantar injection of botulinum toxin, as a safe and long-lasting method, can effectively block the occurrence of blisters32. In addition, it has been reported that long-term oral erythromycin may be effective on EBS-DM33. A 1% diacerein ointment is considered to be a well-tolerated and safe targeted therapy that significantly reduces blisters in most EBS-DM patients34. Because of the overexpression of Th17 cytokines found in the blister roof and fluid in the skin of EBS-DM patients35, with anti-IL-17 agents treatment, a sharp reduction in the number of blisters in all patients has been observed36. The natural chemical sulforaphane produced by broccoli has been shown to treat blisters in KRT14 deficient mice by inducing the expression of KRT16 and KRT17 in the epidermal basal layer and improving the EBS phenotype37. Recently, Peking et al.27 successfully developed an ex vitro gene therapy using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 to correct a recurrent COL7A1 mutation of dystrophic EB in a xenograft mouse model. Clearly, considerable progress has been made in the treatment of EB with RNA, but there are limitations. Since it is a genetic disease, genome editing as state-of-the-art gene therapy approach may be harnessed for clinical treatment. Specifically, because the mutation is c.1382T>C in KRT5 gene, it could be corrected with cytidine base editor to convert C-to-T conversion38.
Taken together, this is the first study reporting the KRT5: p.L461P missense mutation at the 2B helix, or any mutation in codon 461 of KRT5. Identification of this novel mutation (p.L461P) in KRT5 would lead to a better understanding of the underlying pathophysiologic basis of localized EBS, which may be harnessed for gene diagnosis, genetic counseling, and gene therapy. Also, it provides more insight into the phenotype-genotype correlation in EBS.
ACKNOWLEDGMENT
We thank all the people for their partcipation in the project.
Footnotes
CONFLICTS OF INTEREST: The authors have nothing to disclose.
FUNDING SOURCE: This work was supported by grants from the Natural Science Foundation of China (81201181, F.G.), Zhejiang provincial & Ministry of Health research fund for medical sciences (WKJ2013-2-023, F.G.).
DATA SHARING STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Uitto J. Toward treatment and cure of epidermolysis bullosa. Proc Natl Acad Sci U S A. 2019;116:26147–26149. doi: 10.1073/pnas.1919347117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruckner-Tuderman L. Newer treatment modalities in epidermolysis bullosa. Indian Dermatol Online J. 2019;10:244–250. doi: 10.4103/idoj.IDOJ_287_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JYW, Liu L, Hsu CK, Aristodemou S, Ozoemena L, Ogboli M, et al. Mutations in KLHL24 add to the molecular heterogeneity of epidermolysis bullosa simplex. J Invest Dermatol. 2017;137:1378–1380. doi: 10.1016/j.jid.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Mellado F, Fuentes I, Palisson F, I Vergara J, Kantor A. Ophthalmologic approach in epidermolysis bullosa: a cross-sectional study with phenotype-genotype correlations. Cornea. 2018;37:442–447. doi: 10.1097/ICO.0000000000001525. [DOI] [PubMed] [Google Scholar]
- 5.Li JG, Feng J, Xiao SX, Ai YL, Wang JM, Peng ZH. A new mutation in the linker 12 domain of keratin 5 in a Chinese family with Weber-Cockayne epidermolysis bullosa simplex. Clin Exp Dermatol. 2004;29:539–541. doi: 10.1111/j.1365-2230.2004.01565.x. [DOI] [PubMed] [Google Scholar]
- 6.Stawczyk-Macieja M, Wertheim-Tysarowska K, Jakubowski R, Szczerkowska-Dobosz A, Krygier M, Wilkowska A, et al. A novel de novo mutation p.Ala428Asp in KRT5 gene as a cause of localized epidermolysis bullosa simplex. Exp Dermatol. 2019;28:1131–1134. doi: 10.1111/exd.13788. [DOI] [PubMed] [Google Scholar]
- 7.Kowalewski C, Hamada T, Wozniak K, Kawano Y, Szczecinska W, Yasumoto S, et al. A novel autosomal partially dominant mutation designated G476D in the keratin 5 gene causing epidermolysis bullosa simplex Weber-Cockayne type: a family study with a genetic twist. Int J Mol Med. 2007;20:75–78. [PubMed] [Google Scholar]
- 8.Flohil SC, Bolling MC, Kooi KA, Lemmink HH, Jonkman MF. A new pathogenic keratin 5 mutation in a Hindoestan family with localized epidermolysis bullosa simplex. Eur J Dermatol. 2010;20:27–79. doi: 10.1684/ejd.2010.0804. [DOI] [PubMed] [Google Scholar]
- 9.Khani P, Ghazi F, Zekri A, Nasri F, Behrangi E, Aghdam AM, et al. Keratins and epidermolysis bullosa simplex. J Cell Physiol. 2018;234:289–297. doi: 10.1002/jcp.26898. [DOI] [PubMed] [Google Scholar]
- 10.Arin MJ, Grimberg G, Schumann H, De Almeida, Chang YR, Tadini G, et al. Identification of novel and known KRT5 and KRT14 mutations in 53 patients with epidermolysis bullosa simplex: correlation between genotype and phenotype. Br J Dermatol. 2010;162:1365–1369. doi: 10.1111/j.1365-2133.2010.09657.x. [DOI] [PubMed] [Google Scholar]
- 11.Humphries MM, Mansergh FC, Kiang AS, Jordan SA, Sheils DM, Martin MJ, et al. Three keratin gene mutations account for the majority of dominant simplex epidermolysis bullosa cases within the population of Ireland. Hum Mutat. 1996;8:57–63. doi: 10.1002/(SICI)1098-1004(1996)8:1<57::AID-HUMU8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Fine JD, Bruckner-Tuderman L, Eady RA, Bauer EA, Bauer JW, Has C, et al. Inherited epidermolysis bullosa: updated recommendations on diagnosis and classification. J Am Acad Dermatol. 2014;70:1103–1126. doi: 10.1016/j.jaad.2014.01.903. [DOI] [PubMed] [Google Scholar]
- 13.Wertheim-Tysarowska K, Ołdak M, Giza A, Kutkowska-Kaźmierczak A, Sota J, Przybylska D, et al. Novel sporadic and recurrent mutations in KRT5 and KRT14 genes in Polish epidermolysis bullosa simplex patients: further insights into epidemiology and genotype-phenotype correlation. J Appl Genet. 2016;57:175–181. doi: 10.1007/s13353-015-0310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim EN, Harris AG, Bingham LJ, Yan W, Su JC, Murrell DF. A review of 52 pedigrees with epidermolysis bullosa simplex identifying ten novel mutations in KRT5 and KRT14 in Australia. Acta Derm Venereol. 2017;97:1114–1119. doi: 10.2340/00015555-2715. [DOI] [PubMed] [Google Scholar]
- 15.Jerábková B, Marek J, Bucková H, Kopecková L, Veselý K, Valícková J, et al. Keratin mutations in patients with epidermolysis bullosa simplex: correlations between phenotype severity and disturbance of intermediate filament molecular structure. Br J Dermatol. 2010;162:1004–1013. doi: 10.1111/j.1365-2133.2009.09626.x. [DOI] [PubMed] [Google Scholar]
- 16.Kim E, Harris A, Hyland V, Murrell DF. Digenic inheritance in epidermolysis bullosa simplex involving two novel mutations in KRT5 and KRT14. Br J Dermatol. 2017;177:262–264. doi: 10.1111/bjd.15053. [DOI] [PubMed] [Google Scholar]
- 17.Minakawa S, Nakano H, Nakajima K, Matsuzaki Y, Takiyoshi N, Akasaka E, et al. Mutational analysis on 16 Japanese population cases with epidermolysis bullosa simplex. J Dermatol Sci. 2013;72:330–332. doi: 10.1016/j.jdermsci.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Glász-Bóna A, Medvecz M, Sajó R, Lepesi-Benko R, Tulassay Z, Katona M, et al. Easy method for keratin 14 gene amplification to exclude pseudogene sequences: new keratin 5 and 14 mutations in epidermolysis bullosa simplex. J Invest Dermatol. 2009;129:229–231. doi: 10.1038/jid.2008.223. [DOI] [PubMed] [Google Scholar]
- 19.García M, Santiago JL, Terrón A, Hernández-Martín A, Vicente A, Fortuny C, et al. Two novel recessive mutations in KRT14 identified in a cohort of 21 Spanish families with epidermolysis bullosa simplex. Br J Dermatol. 2011;165:683–692. doi: 10.1111/j.1365-2133.2011.10428.x. [DOI] [PubMed] [Google Scholar]
- 20.Bowden PE, Knight AG, Liovic M. A novel mutation (p.Thr198Ser) in the 1A helix of keratin 5 causes the localized variant of epidermolysis bullosa simplex. Exp Dermatol. 2009;18:650–652. doi: 10.1111/j.1600-0625.2008.00820.x. [DOI] [PubMed] [Google Scholar]
- 21.Gao J, Wang X, Zheng F, Dong S, Qiu X. Novel keratin 5 mutation in a family with epidermolysis bullosa simplex. Exp Ther Med. 2015;10:2432–2436. doi: 10.3892/etm.2015.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho JW, Ryu HW, Kim SA, Nakano H, Lee KS. Weber-Cockayne type epidermolysis bullosa simplex resulting from a novel mutation (c. 608>C) in the keratin 5 gene. Ann Dermatol. 2014;26:739–742. doi: 10.5021/ad.2014.26.6.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang YY, Chao SC, Chen WY, Lee WR, Wang KH. Weber-Cockayne type of epidermolysis bullosa simplex associated with a novel mutation in keratin 5 and amyloid deposits. Br J Dermatol. 2008;159:1370–1372. doi: 10.1111/j.1365-2133.2008.08801.x. [DOI] [PubMed] [Google Scholar]
- 24.Lev-Tov H, Sivamani RK, Burrall B. A novel keratin 5 mutation in a familial cluster. Dermatol Online J. 2012;18:1. [PubMed] [Google Scholar]
- 25.Bchetnia M, Tremblay ML, Leclerc G, Dupérée A, Powell J, McCuaig C, et al. Expression signature of epidermolysis bullosa simplex. Hum Genet. 2012;131:393–406. doi: 10.1007/s00439-011-1077-7. [DOI] [PubMed] [Google Scholar]
- 26.Steinert PM. The two-chain coiled-coil molecule of native epidermal keratin intermediate filaments is a type I-type II heterodimer. J Biol Chem. 1990;265:8766–8774. [PubMed] [Google Scholar]
- 27.Peking P, Breitenbach JS, Ablinger M, Muss WH, Poetschke FJ, Kocher T, et al. An ex vivo RNA trans-splicing strategy to correct human generalized severe epidermolysis bullosa simplex. Br J Dermatol. 2019;180:141–148. doi: 10.1111/bjd.17075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bohnekamp J, Cryderman DE, Paululat A, Baccam GC, Wallrath LL, Magin TM. A drosophila model of epidermolysis bullosa simplex. J Invest Dermatol. 2015;135:2031–2039. doi: 10.1038/jid.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawant M, Schwarz N, Windoffer R, Magin TM, Krieger J, Mücke N, et al. Threonine 150 phosphorylation of keratin 5 is linked to epidermolysis bullosa simplex and regulates filament assembly and cell viability. J Invest Dermatol. 2018;138:627–636. doi: 10.1016/j.jid.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Russell D, Ross H, Lane EB. ERK involvement in resistance to apoptosis in keratinocytes with mutant keratin. J Invest Dermatol. 2010;130:671–681. doi: 10.1038/jid.2009.327. [DOI] [PubMed] [Google Scholar]
- 31.Homberg M, Ramms L, Schwarz N, Dreissen G, Leube RE, Merkel R, et al. Distinct impact of two keratin mutations causing epidermolysis bullosa simplex on keratinocyte adhesion and stiffness. J Invest Dermatol. 2015;135:2437–2445. doi: 10.1038/jid.2015.184. [DOI] [PubMed] [Google Scholar]
- 32.Swartling C, Karlqvist M, Hymnelius K, Weis J, Vahlquist A. Botulinum toxin in the treatment of sweat-worsened foot problems in patients with epidermolysis bullosa simplex and pachyonychia congenita. Br J Dermatol. 2010;163:1072–1076. doi: 10.1111/j.1365-2133.2010.09927.x. [DOI] [PubMed] [Google Scholar]
- 33.Chiaverini C, Fontas E, Vabres P, Bessis D, Mazereeuw J, Charlesworth A, et al. Oral erythromycin therapy in epidermolysis bullosa simplex generalized severe. Br J Dermatol. 2015;173:563–564. doi: 10.1111/bjd.13672. [DOI] [PubMed] [Google Scholar]
- 34.Ablinger M, Felder TK, Wimmer M, Zauner R, Hofbauer P, Lettner T, et al. Basal pharmacokinetic parameters of topically applied diacerein in pediatric patients with generalized severe epidermolysis bullosa simplex. Orphanet J Rare Dis. 2018;13:193. doi: 10.1186/s13023-018-0940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castela E, Tulic MK, Rozières A, Bourrat E, Nicolas JF, Kanitakis J, et al. Epidermolysis bullosa simplex generalized severe induces a T helper 17 response and is improved by apremilast treatment. Br J Dermatol. 2019;180:357–364. doi: 10.1111/bjd.16897. [DOI] [PubMed] [Google Scholar]
- 36.Mellerio JE. Potential therapeutic targeting of inflammation in epidermolysis bullosa simplex. Br J Dermatol. 2019;180:258–260. doi: 10.1111/bjd.17106. [DOI] [PubMed] [Google Scholar]
- 37.Kerns M, DePianto D, Yamamoto M, Coulombe PA. Differential modulation of keratin expression by sulforaphane occurs via Nrf2-dependent and -independent pathways in skin epithelia. Mol Biol Cell. 2010;21:4068–4075. doi: 10.1091/mbc.E10-02-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doman JL, Raguram A, Newby GA, Liu DR. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat Biotechnol. 2020;38:620–628. doi: 10.1038/s41587-020-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.