Abstract
The latent reservoir of HIV-1 in resting CD4+ T cells is a major barrier to cure. It is unclear whether the latent reservoir of HIV-1 resides principally in particular subsets of CD4+ T cells, a finding that would have implications for understanding its stability and developing curative therapies. Recent work has shown that proliferation of HIV-1–infected CD4+ T cells is a major factor in the generation and persistence of the latent reservoir and that latently infected T cells that have clonally expanded in vivo can proliferate in vitro without production of virions. In certain CD4+ memory T cell subsets, the provirus may be in a deeper state of latency, allowing the cell to proliferate without producing viral proteins, thus permitting escape from immune clearance. To evaluate this possibility, we used a multiple stimulation viral outgrowth assay to culture resting naïve, central memory (TCM), transitional memory (TTM), and effector memory (TEM) CD4+ T cells from 10 HIV-1–infected individuals on antiretroviral therapy who showed viral suppression. On average, only 1.7% of intact proviruses across all T cell subsets were induced to transcribe viral genes and release replication-competent virus after stimulation of T cells. We found no consistent enrichment of intact or inducible proviruses in any T cell subset. Furthermore, we observed notable plasticity among the canonical memory T cell subsets after activation in vitro and saw substantial person-to-person variability in the inducibility of infectious virus release after T cell stimulation. This finding complicates the vision for a targeted cure approach for HIV-1 based on T cell memory subsets.
One sentence summary:
Latent replication-competent HIV-1 proviruses are widely distributed among the memory subsets with no significant differences in inducibility within any particular subset.
INTRODUCTION
The major barrier to HIV-1 cure is the latent reservoir (LR) (1, 2), comprised of resting CD4+ T-cells harboring latent, replication-competent proviruses (3-7). The LR was first demonstrated using a quantitative viral outgrowth assay (QVOA) in which virus production from resting CD4+ T-cells was induced upon T-cell activation (4, 7-10). The slow decay of the LR necessitates lifelong antiretroviral therapy (ART) to prevent viral rebound (1, 11, 12). The reservoir is maintained in part by memory cell proliferation (13-16) driven by cytokines, antigen, or effects related to the site of proviral integration (13-27). Latently infected cells carrying replication-competent proviruses can persist through clonal expansion in vivo (13-15, 28).
Cellular markers of latent infection (29-31) would allow selective reservoir targeting. Specific subpopulations of CD4+ T-cells may be enriched in cells carrying latent, replication-competent proviruses and may differ in propensity for clonal expansion or latency reversal (8-11, 13-16). However, specific markers of latent infection have not been confirmed (32-37), and persistent HIV-1 has been found in all CD4+ memory subsets and at lower levels in naïve CD4+ T-cells (29, 38-44). Three subsets of memory CD4+ T-cells are defined by the cell surface proteins CD45, CCR7, and CD27 (45-47): central memory (TCM: CD45RO+CCR7+CD27+), transitional memory (TTM: CD45RO+CCR7−CD27+), and effector memory cells (TEM: CD45RO+CCR7−CD27−) (48, 49). Naïve cells (TN: CD45RO−CCR7+CD27+) home to secondary lymphoid organs where they encounter antigen and differentiate into memory cells (48-54). TCM are long-lived cells that home to secondary lymphoid organs and mediate recall responses (46, 55, 56). TTM are in a transient state between TCM and TEM phenotypes (47, 57). TEM cells home to inflammatory sites for rapid effector function (48, 57-59).
Early studies demonstrated higher frequencies of HIV-1 in memory CD4+ T-cells than in TN (4, 60), consistent with models of reservoir formation (4, 61). Proviral DNA has been measured in TCM, TTM, and TEM (38-40) as well as Th1 cells (62, 63), T follicular helper cells (TfH) (64), and stem cell-like memory cells (TSCM) (41, 42). Several studies have shown that TCM harbor the majority of latent HIV-1 proviruses and have the longest half-life of memory subsets (38, 40, 56, 58, 59, 65-68). However, another study showed that TEM harbor intact HIV-1 proviruses at a higher frequency (69).
One factor contributing to disparate results is the assay used to measure infected cells (70). The QVOA provides a definitive minimal estimate of LR size but misses replication-competent proviruses not induced by a single round of activation (4, 8-10, 28). HIV-1 DNA measurements using qPCR dramatically overestimate LR size since they do not discriminate between intact and defective proviruses (71, 72). Near-full-length sequencing can distinguish between intact and defective proviruses, but is not quantitative and does not provide information on inducibility (69, 73, 74). A recently developed intact proviral DNA assay (IPDA) can distinguish between intact proviruses and those with overt fatal defects (75).
It is important to understand the relative inducibility as well as the distribution of proviruses in different subsets. Genes involved in T-cell activation, migration, and transcriptional regulation are more poised (present in an open chromatin state without active transcription) in resting memory cells than in TN (76, 77). DNA methylation decreases within genes that regulate differentiation toward a memory state (78). Because these genes are less transcriptionally restricted in TEM, this subset displays a higher proliferative response to IL-7 and IL-15 than less differentiated subsets (38, 50, 51, 79).
Initial evidence for differential inducibility of latent HIV-1 proviruses came from multiple stimulation viral outgrowth assays (MSVOA) demonstrating that many replication-competent proviruses are induced in vitro only after multiple rounds of T-cell activation ((14, 28). One hypothesis is that inducibility depends on the memory subset harboring the latent virus. Repeated activation may drive differentiation to a TEM phenotype with a transcriptional landscape that facilitates proviral transcription. We describe here simultaneous analysis of the distribution and inducibility of intact proviruses in CD4+ T-cell subsets.
RESULTS
Intact proviruses are similarly distributed across memory subsets
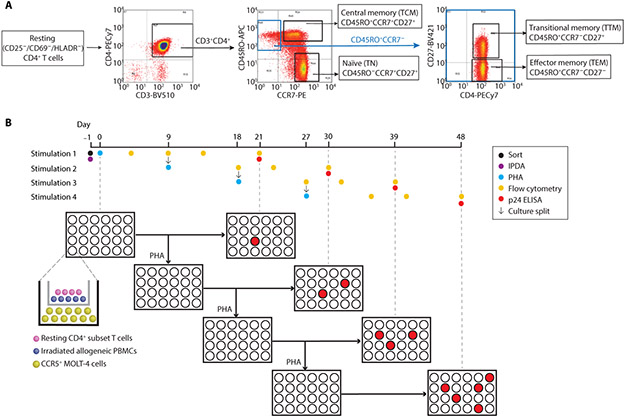
To understand the distribution and inducibility of latent HIV-1 proviruses in T-cell subsets, we isolated resting CD4+ T-cells from 10 individuals who had started ART during chronic infection and maintained viral suppression for >6 months (Table S1). Resting cells were stained for the subset-defining surface markers (CD27, CCR7, and CD45RO), CD3, and CD4. Cells were then sorted and analyzed as described in Figures 1A and B.
Figure 1. Subset sorting strategy and MSVOA culture schematic.
(A) Resting CD4+ T-cells were isolated from leukapheresis products from 10 individuals on ART as described in Methods. Purified resting cells were sorted into the respective subsets as shown. (B) Timeline for MSVOA. A small aliquot of the sorted resting cells was set aside for IPDA analysis and calculation of intact and defective provirus frequencies (Fig. 2B). The remaining cells from each subset were seeded in the MSVOA as previously described (14). Cells were activated with PHA on day 0, then both top and bottom chambers of each cultures were split in half on day 9. One set of split wells was incubated without further stimulation until day 21 for p24 ELISA. These results provided the viral outgrowth data in Fig. 4. The other set of split wells was activated again with PHA. This process was repeated every 9 days for 4 total stimulations. Analysis of p24 in the supernatants was carried out 21 days after each PHA stimulation (Fig. 5). Flow cytometric analysis of subset marker expression was done every 5-7 days (Fig. 3).
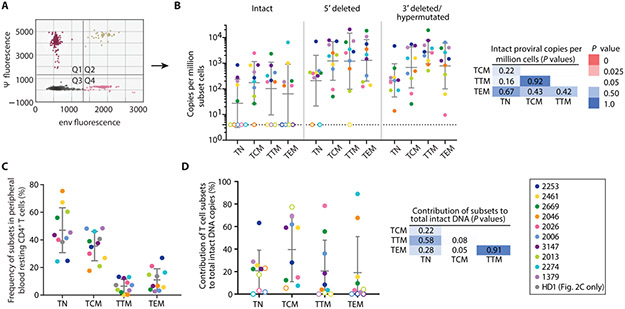
The IPDA separately quantitates intact and defective proviruses (Fig. 2A) (75). Consistent with previous observations (4, 60, 80), TN contained nearly 10-fold fewer proviral copies (both intact and defective) than memory cells (Fig. 2B). Intact proviral frequency in different subsets varied widely between individuals, but mean values were similar (Fig. 2B). Similarly, no significant differences across the memory subsets were observed in the frequencies of 5′ deleted proviruses or 3′ deleted/hypermutated proviruses (Fig. 2B). As expected based on previous studies with unfractionated resting CD4+ T-cells (28, 75, 81), there were ~10-fold more defective proviruses than intact proviruses.
Fig. 2. Frequencies and proviral DNA content of CD4+ T-cell subsets in peripheral blood.
(A) Representative IPDA results from sorted resting CD4+ T-cells. Droplets in quadrant 1 (Q1) contain proviruses with a 3΄ deletion in the env gene and/or APOBEC3G-induced hypermutation. Q4 contains proviruses with deletions at the 5՛ end of the genome encompassing the packaging signal (ψ). Q2 contains intact provirus. Most droplets (Q3) do not contain a provirus. (B) IPDA analysis on resting CD4+ T-cells from each subset. Geometric means ± SD are shown. For samples in which no provirus was detected (open symbols), the limit of detection (LOD, 4 copies/106 cells) was used in calculations. All subsets contained at least one type of provirus (intact or defective). Mann-Whitney tests were performed to compare ICPM between each subset. P-values are two-tailed. (C) Frequency of each subset in leukapheresis samples from 10 individuals and 1 healthy donor (HD1). Bars show mean ± SD. (D) Contribution of each subset to the circulating pool of intact proviruses determined from the frequency of intact proviruses found in each subset (Fig. 2B) and subset frequency in peripheral blood (Fig. 2C).
To determine the contribution of each subset to the total pool of intact proviruses, we calculated the frequency of each subset in peripheral blood (Fig. 2C). As expected, TN were the most abundant, followed by TCM, TEM, and TTM. The contribution of each subset to the pool of intact proviruses was then calculated. We observed large person-to-person variation (Fig. 2D) with no consistent major contribution from any subset.
Subsets differentiate towards an effector phenotype following multiple stimulations
To understand the relationship between CD4+ T-cell subsets and HIV-1 latency, we examined subset-defining markers and virus induction following multiple rounds of T-cell activation (Fig. 1B). Cells were activated with the mitogen phytohemagglutinin (PHA) and irradiated allogeneic peripheral PBMCs for 24 hours and then co-cultured with MOLT-4/CCR5 cells to expand virus released following induction of latent proviruses (Fig. 1B). Cultures were split every 9 days at which time half of each culture was restimulated while the remaining cells were cultured without further stimulation for 21 days. Supernatant p24 was measured 21 days after each stimulation to detect outgrowth.
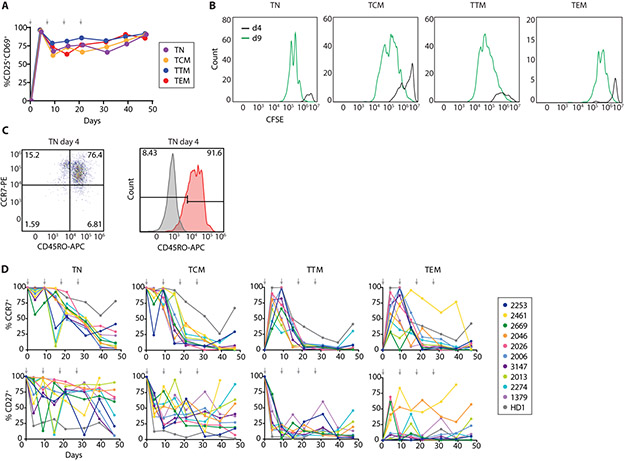
Following the initial stimulation, >70% of cells retained an activated phenotype throughout the culture, as determined by CD25 and CD69 expression (Fig. 3A). CFSE dilution in each subset after initial activation demonstrated that all cells proliferated (Fig. 3B). Phenotypic markers of CD4+ T-cell subsets assessed every 5-7 days showed notable changes. TN gained CD45RO expression by 4 days after initial activation (Fig. 3C). CCR7 expression on TN and TCM remained high after initial activation, then decreased as cells approached a TEM phenotype by day 21 (Fig. 3D). Interestingly, TTM and TEM strongly upregulated CCR7 expression after initial activation from a CCR7− state. CD27 expression after T-cell activation showed a more complex pattern that varied between individuals and subsets (Fig. 3D). In general, CCR7 and CD27 expression demonstrated changes consistent with memory subset differentiation from naïve to effector phenotype with repeated stimulations (Fig. S1), as expected based on previous studies of cells from uninfected individuals (47, 57). The dramatic changes in subset-defining markers upon T-cell activation suggest that studies of subsets are best performed on purified resting CD4+ T-cells as done here.
Fig. 3. Changes in activation state, proliferative status, and phenotypic marker expression following repeated activation of CD4+ T-cell subsets.
(A) Activation status of CD4+ T-cells throughout the MSVOA averaged across 4 representative individuals as percent of cells expressing CD25 and/or CD69. (B) CFSE dilution after first stimulation. Cells from each subset were stained with CFSE and activated on day 0, then analyzed by flow cytometry at days 4 and 9. (C) CD45RO expression on TN from representative individual. 91.6% of TN became CD45RO+ by day 4 after initial stimulation (Fig. S1A). (D) Percent of cells from each subset culture expressing CCR7 and CD27 at the indicated timepoints. Arrows denote stimulation timepoints.
Inducibility of provirus expression is not dependent on memory subset
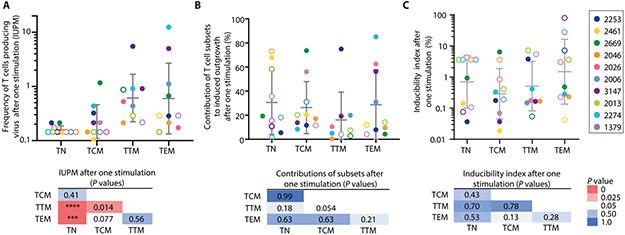
To test the hypothesis that proviral inducibility varies with memory subset, we examined viral outgrowth from each subset after each round of T-cell activation by measuring supernatant p24 21 days after each stimulation (Fig. 1B). This period is sufficient to allow outgrowth of virus released from individual latently infected cells following induction (8-10). Thus, we can determine how many rounds of T-cell activation are required to induce latent proviruses. We first determined the frequency of cells in each subset that gave rise to replication-competent virus after one round of mitogen stimulation, expressed as infectious units per million cells (IUPM) based on the number of initially plated cells. No outgrowth was induced from TN in 7 of 10 individuals (Fig. 4A), consistent with the lower intact provirus copies determined by IPDA (Fig. 2B). The highest frequencies were observed in TTM and TEM, but there was substantial overlap with frequencies in TCM (Fig. 4A). Following normalization based on subset frequency among PBMCs, we observed similar contributions of each subset to the total pool of replication-competent viruses induced after one stimulation (Fig. 4B).
Fig. 4. Viral outgrowth and inducibility of CD4+ T-cell subsets after one stimulation.
(A) Frequency of cells giving rise to viral outgrowth after one stimulation. Results expressed as IUPM. Open circles denote samples from which no outgrowth was observed; IUPM values for these represent maximum likelihood estimates based on the total number of wells plated for that subset (97). Geometric means ± SD are shown. (B) Contribution of subsets to total pool of replication-competent proviruses induced after one stimulation, calculated using frequencies of each subset in peripheral blood (Fig. 2C). Means ± SD are shown. (C) Percentage of intact proviruses that gave rise to replication-competent virus after one stimulation. Inducibility index was calculated by dividing IUPM values by ICPM values. For samples with no outgrowth or intact proviruses detected, estimates based on the number of input cells and LOD for IPDA are used in the calculation of inducibility (open circles, see Methods). TTM from participant 2461 were not cultured due to insufficient number of cells. Geometric means ± SD are shown. Two-tailed P-values were calculated using Mann-Whitney U-tests (****P <0.0001, ***P <0.001).
To compare inducibility in different subsets after one stimulation, we calculated an inducibility index, the ratio of IUPM determined by viral outgrowth to the intact copies per million cells (ICPM) determined by the IPDA. We found substantial person-to-person variation in inducibility and no significant differences between subsets (P>0.13). Importantly, for TTM and TEM, on average <1% of intact proviruses were induced by one stimulation (Fig. 4C), although this ratio widely varied between individuals. This value is similar to that reported for unfractionated resting CD4+ T-cells (75).
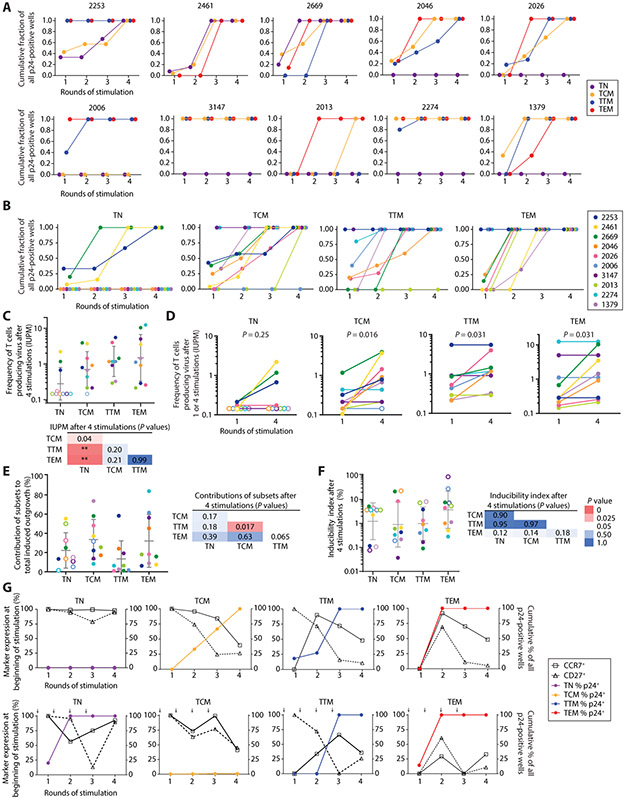
Because differentiation caused by repetitive stimulation (Fig. 3D, Fig. S1B) might alter inducibility, we evaluated viral outgrowth 21 days after each stimulation. Because most outgrowth occurs in MOLT-4/CCR5 cells added to each culture, we could assess changes in inducibility as cells differentiate without the confounding effect of changes in the ability of cells to propagate the infection. Additional viral outgrowth was observed after multiple stimulations for most individuals (Fig. 5A). For participant 3147, all outgrowth from each subset occurred after the first stimulation. In 7 of 10 individuals, there was no outgrowth from TN cultures even after 4 stimulations. For other subsets, there were no clear patterns in the number of stimulations required for outgrowth (Fig. 5B). For example, TTM cells required multiple stimulations for outgrowth in some individuals but only one stimulation for maximum outgrowth in others, and this trend was seen in other subsets (Fig. 5B). IUPM values based on the number of input cells at the beginning of the culture and the cumulative fraction of positive wells after four stimulations are shown in Fig. 5C. Frequencies were higher for memory cells than for TN but did not differ significantly among memory subsets (P>0.21). IUPM values determined after one stimulation or four stimulations are compared in Fig. 5D. To determine the relative contributions of each subset to the total pool of replication-competent viruses induced after four stimulations, IUPM values after 4 stimulations were normalized based on the frequency of subsets present in peripheral blood. There was substantial person-to-person variation and wide overlap between subsets (Fig. 5E). We observed no significant differences between subsets in inducibility after 4 stimulations (P>0.12; Fig. 5F). Outgrowth observed after multiple stimulations may have resulted from virus production by progeny cells expanded in vitro.
Fig. 5. Viral outgrowth and inducibility for T-cell subsets after multiple stimulations.
(A) Cumulative fraction of all positive wells that had positive p24 values 21 days after the indicated stimulation. Data are graphed separately for each individual. TTM were not analyzed for participant 2461 due to insufficient number of cells. (B) Cumulative fraction of all positive wells that had positive p24 values 21 days after the indicated stimulation. Data are graphed separately for each subset. (C) Frequency of cells giving rise to infectious virus in the MSVOA based on initial number of cells seeded. Geometric means ± SD are shown. P-values were calculated from Mann-Whitney U-tests (** P < 0.01). (D) Frequency of cells giving rise to infectious virus after one stimulation or after all 4 stimulations. Results are expressed as IUPM based on initial number of cells seeded and number of wells turned p24+. P-values between IUPM values after the indicated numbers of stimulations were calculated using Wilcoxon matched-pairs signed rank tests. (E) Contribution of each subset to total pool of replication-competent proviruses calculated from IUPM values (Fig. 5C) and the frequency of subsets present in peripheral blood (Fig. 2C). Means ± SD are shown. (F) Inducibility index calculated from ratio of MSVOA IUPM to ICPM from each subset (geometric means ± SD shown). On average, only 1.7% of intact proviruses measured across all subsets were induced to replicate in culture. Mann-Whitney tests showed no significant differences in inducibility indices across subsets (P>0.12). (G) Phenotype of cultured cells at the time of each stimulation (left axis) overlaid with outgrowth results from that stimulation (right axis) from representative individuals (top: 2026, bottom: 2669).
To understand the lack of a relationship between conventional CD4+ memory T-cell subsets and the distribution and inducibility of replication-competent proviruses, we examined the phenotypes of cells at the time of the stimulation that led to viral outgrowth. Data for representative individuals are shown in Fig. 5G. For some TN and TCM cultures, viral outgrowth was detected even when the phenotype at time of stimulation did not indicate differentiation to TEM (Fig. 5G, Fig. S1A). This suggests that CD4+ T-cells need not have an effector phenotype to produce virus (Fig. 5G, Fig. S1B).
Only a small fraction of intact proviruses is induced to replicate despite multiple stimulations
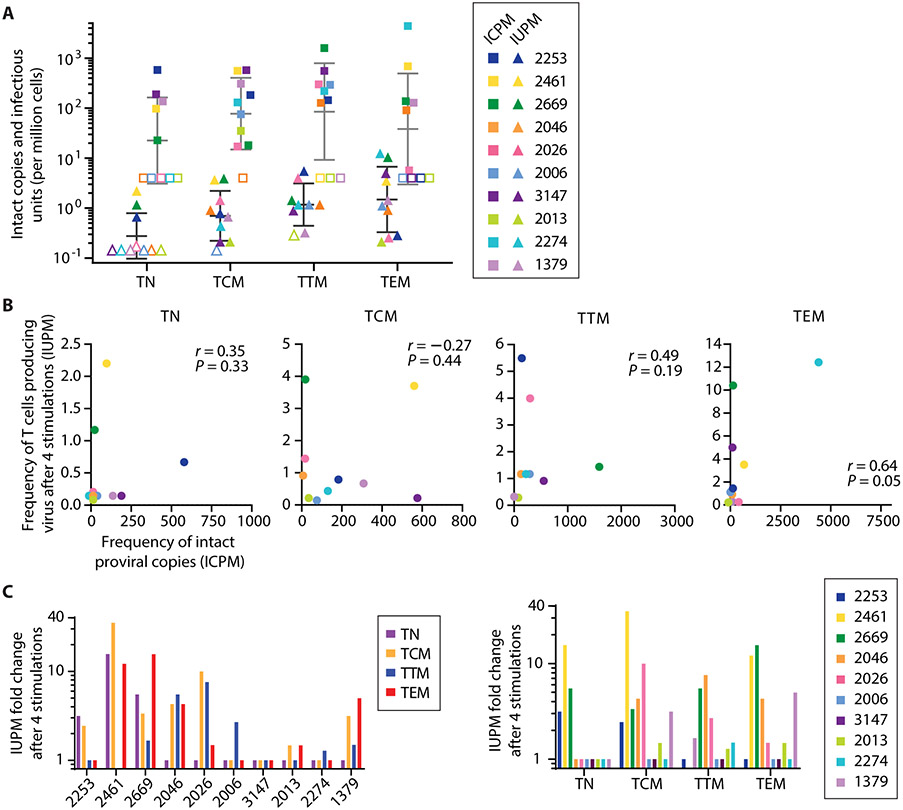
Our results suggest that only a small fraction of intact proviruses present in each subset can be induced to replicate by multiple sequential in vitro stimulations. The results of the MSVOA and the IPDA are compared in Fig. 6A. The overall fraction of intact proviruses that were induced was low, averaging 1.7% across all subsets (Fig. 5F). There was no strong correlation between IUPM and ICPM (Fig. 6B). Additional stimulations caused a 30-fold increase in IUPM in TCM cells from one individual, but no increase in other individuals (Fig. 6C). In general, inducibility varied by donor rather than by subset. The variability in numbers of stimulations required for outgrowth for the different subsets suggests that inducibility is not mainly dependent on subset.
Fig. 6. Inducibility of viruses from different subsets.
(A) Comparison of frequencies of cells with intact proviruses as measured by IPDA (squares, gray bars) and frequencies of cells giving rise to viral outgrowth as measured by MSVOA (triangles, black bars). Bars indicate geometric mean ± SD. (B) Spearman’s rank correlations between IPDA and MSVOA values within each subset. Rho and p-values are shown. (C) Fold-change in IUPM by individual and by subset (n = 10 for TN, TCM, TEM; n = 9 for TTM).
Replication-competent viruses with identical env V3-V4 sequences are distributed across different subsets
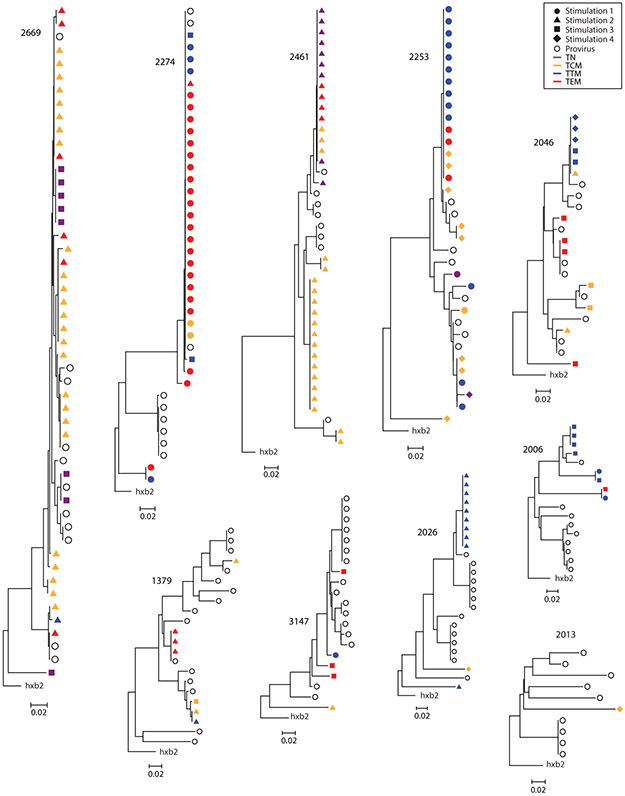
Recent studies suggest that in treated individuals, the population of CD4+ T-cells carrying replication-competent proviruses is dominated by large cellular clones with identical proviruses (13-16). To explore the distribution of proviruses with identical sequences among CD4+ T-cell subsets, we sequenced the highly variable V3-V4 region of the env gene from viral RNA from the supernatants of p24+ wells (Fig. 7). Of 175 independent isolates of replication-competent virus, 142 (81.1%) had a matching env sequence present in the same blood sample. We also obtained 109 proviral V3-V4 sequences from unfractionated resting CD4+ T-cells from these individuals. Of these, 56 (51.3%) had a matching proviral env sequence in the same blood sample, which could reflect in vivo clonal expansion given the clonal prediction score of 96 (14, 83). However, clonality cannot be definitively established without integration site analysis. Importantly, identical sequences were often obtained from different subsets after different numbers of stimulations. For example, participant 2274 had a large potential clone identified as identical sequences from TCM, TTM, and TEM that were induced after different numbers of stimulations. In cases where the identical sequences reflect proliferation of a single infected cell, these findings suggest that members of the clone can display differential inducibility and reside in different subsets.
Fig. 7. Neighbor-joining env trees of replication-competent viral isolates from the MSVOA.
Sequences of V3-V4 region of env from outgrowth viruses at 4 timepoints (solid symbols) and proviral sequences (open circles) from unfractionated resting CD4+ T-cells.
DISCUSSION
It has been unclear whether the LR is contained mainly in a particular subset of CD4+ T-cells (29, 38-44). This would allow more specific targeting of reservoir cells and provide insight into viral persistence. Therefore, we examined the distribution and inducibility of intact, replication-competent proviruses in resting CD4+ T-cells belonging to the TN, TCM, TTM, and TEM subsets. In 5 of 10 individuals, we detected no intact proviral DNA in TNs by IPDA and no viral outgrowth after 4 stimulations (Table S2). These results are consistent with previous studies (4, 80, 82) and some models for LR formation (61). However, for memory subsets, there was no significant difference in the frequency of cells with intact proviruses. Because TCM were the most abundant, they made the largest contribution to the total pool of intact proviruses, but there was dramatic person-to-person variability in the contribution of T-cell subsets, which is striking given the homogeneity of our cohort.
We also considered whether there was differential inducibility of replication-competent proviruses in different memory subsets. Previous characterization of the epigenetic landscapes and cytokine responsiveness suggested that TEM would be easiest to induce, followed by TTM, TCM, then TN (38, 50, 51, 58, 76-78). To test this hypothesis, we subjected sorted cells from each subset to 4 consecutive rounds of global T-cell activation. We found no correlation between subsets and the number of stimulations needed for outgrowth. In addition, we observed virions with identical env sequences in different culture wells after different numbers of stimulations and from different subsets. If sequence identity reflects clonal expansion, then our results suggest that infected cells can proliferate and differentiate into different memory subsets in vivo without succumbing to viral cytopathic effects or immune clearance. Another study has provided evidence of clonal proviruses with identical integration sites distributed across subsets (83). To better understand inducibility, we developed an index defined as the frequency of cells that give rise to viral outgrowth after one or more stimulations divided by the initial number of cells with intact proviruses. Average inducibility was 0.74% after one round of activation, similar to previously reported results for unfractionated CD4+ T-cells (75). On average, only 1.7% of intact proviruses were induced after 4 rounds of activation. Low inducibility has also been observed using assays for intracellular HIV-1 RNA (84-86), p24 protein (87), or cell-free HIV-1 RNA (85, 86, 88, 89). For example, Cillo et al reported that only 1.5% of resting CD4+ T-cells with HIV-1 DNA could be induced to produce virions following stimulation with anti-CD3/CD28 (88).
The alarmingly high fraction (98.3%) of intact proviruses that are not induced even after 4 stimulations demands further investigation to determine whether these proviruses contribute to the LR, residual viremia, and viral rebound. Several biological mechanisms might explain this observation. Stochastic processes may govern whether an intact provirus is induced following T-cell activation (90). T-cell exhaustion following repeated in vitro stimulation may limit induction (14). Some proviruses may be permanently silenced by epigenetic mechanisms (91, 92), and others may be integrated in genomic locations that affect inducibility (93). Induction of some intact proviruses may be suppressed by transcriptional interference (94-96), or may require signals other than those provided by mitogen stimulation. If a large proportion of these intact non-induced proviruses are replication-competent and ultimately inducible in vivo, then they represent an enormous hurdle to eradicating the reservoir.
Our study has several limitations. Participants were mainly Caucasian males over 50 who initiated therapy with low CD4 counts. The induction of latent proviruses and the replication of viruses released following latency reversal may be different in vivo. For example, outgrowth may be limited by the in vitro culture system used. However, viral outgrowth measured using MOLT-4/CCR5 cells is equivalent to that obtained using the standard donor CD4+ T lymphoblasts (9). The use of a relatively insensitive ELISA to score outgrowth ensures that our results reflect the induction of proviruses capable of robust in vitro replication. Some non-induced proviruses detected by the IPDA may have small defects affecting viral fitness. By near full genome sequencing, ~70% of proviruses called intact by the IPDA had no obvious defects at the primary sequence level, while the remaining 30% had small defects (75). CD4+ T-cells that were activated in vivo and cells that express as CD25+ such as Tregs were not analyzed. We focused on resting CD4+ T-cells because their transcriptional environment is non-permissive for viral gene expression (61) and they have a longer half-life than activated CD4+ T-cells (65). While virus can be found in activated T-cells, the stable reservoir resides mainly in resting cells. An additional reason to focus on resting CD4+ T-cells is that CD4+ T-cell subsets cannot be reliably defined without considering activation status. We show here that expression of markers used to define subsets changes dramatically upon T-cell activation (Fig. 3D), thus recent in vivo activation could have caused confounding changes in expression of canonical markers.
One approach to HIV-1 cure involves identifying markers that would allow selectively targeting latently infected cells. Our results demonstrate that latent replication-competent proviruses are widely distributed among the memory subsets with no consistent differences in inducibility.
MATERIALS AND METHODS
Study Design
This study was designed to determine differences in inducibility of latent HIV-1 across resting memory CD4+ T-cell subsets. We obtained leukapheresis samples from 10 HIV-1-infected individuals on suppressive ART, isolated resting CD4+ T-cells, and sorted TN, TCM, TTM, and TEM based on markers previously described (45-47). Frequencies of intact proviral DNA were assessed using the IPDA. Subsets were then cultured in the MSVOA to assess differential inducibility. Viral outgrowth was measured by ELISA for p24 antigen. Expression of subset-defining markers was analyzed throughout the culture. Viral RNA from culture supernatants and proviral DNA from sorted resting cells were sequenced to identify clones.
Leukopaks were obtained from 10 HIV-1-infected individuals enrolled in the UCSF SCOPE cohort. Inclusion criteria were initiation of ART during chronic infection, viral suppression for >6 months, HIV RNA <40 copies/ml, and CD4+ count >350 cells/μl. This study was approved by the Institutional Review Boards at Johns Hopkins University and the University of California San Francisco. Written informed consent was obtained from all participants.
Resting CD4+ T-cell isolation and sorting strategy
PBMCs were isolated from leukapheresis samples by Ficoll density centrifugation. CD4+ cells were isolated from PBMCs using the EasySep™ Human CD4+ T-cell Isolation Kit (Stemcell Technologies). Resting CD4+ cells were then isolated by negative depletion of cells expressing CD69, CD25, or HLA-DR (CD69 MicroBead Kit II, CD25 MicroBeads, Anti-HLA-DR MicroBeads; Miltenyi Biotec).
Cells were stained with CD27-BV421, CD3-BV510, CCR7-PE, CD45RO-APC, CD4-PECy7 (Biolegend) and sorted on a MoFlo Legacy (Beckman Coulter). Viability was determined using a propidium iodide stain. Cells were sorted from gated live singlet CD3+CD4+ lymphocytes based on the following expression patterns: (TN)CD45RO−/CCR7+/CD27+, (TCM)CD45RO+/CCR7+/CD27+, (TTM)CD45RO+/CCR7−/CD27+, (TEM)CD45RO+/CCR7−/CD27−.
Intact Proviral DNA Assay (IPDA)
The IPDA was performed on sorted resting subset cells as described (75). Primers/probes for the RPP30 gene were used to calculate shearing index and determine cell input. ICPM values were corrected for the fraction of intact proviruses quantified by IPDA (0.32) that are not replication-competent (75).
Flow cytometry
Cells were stained with Live/Dead Fixable Violet Dead Cell Stain Kit (Invitrogen), CD3-BV510, CCR7-PE, CD45RO-APC, CD4-PECy7, and CD27-BV785 (Biolegend). Activation levels were determined using CD25-APCCy7, CD69-APCCy7, and HLA-DR-APCCy7 (Biolegend). CFSE (Invitrogen) staining was done separately to prevent spillover. Stained cells were analyzed on the iQue Screener Plus (Intellicyt) at 4, 9, 14, and 21 days. Data was analyzed using FlowJo.
Multiple Stimulation Viral Outgrowth Assay (MSVOA)
The MSVOA was performed as described (14). Each sorted subset was cultured in separate transwell plates. The irradiated PBMCs were stained with CellTrace Violet (Invitrogen) to distinguish participant cells from feeders in downstream flow cytometry analysis. Infection frequencies were calculated as previously described (IUPMStats) (97).
Viral RNA sequencing
Viral RNA was isolated from culture supernatants saved after day 21. cDNA was synthesized using the SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific) and gene-specific primer ES8 (5′-CACTTCTCCAATTGTCCC TCA-3′). The V3-V4 region of env was amplified using primers ES7 (5′-CTGTTAAATGGCAGTCTAGC-3′), ES8, and Platinum SuperFi DNA Polymerase (Invitrogen). PCR products were run on a 1% agarose gel, extracted (Qiagen QIAquick Gel Extraction Kit), then Sanger sequenced (Genewiz) using sequencing primers Nesty8 (5′-CATACATTGCTTTTCCTACT-3′) and DLoop (5′-GTCTAGCAGAAGAAGAGG-3′). Sequences were analyzed in BioEdit, aligned by ClustalW, and trimmed to equal lengths. Neighbor-joining trees were generated using the maximum composite likelihood method in MEGA6. Proviral sequences were obtained from the same nested PCR reactions done on a limiting dilution of template DNA from resting subset cells.
Statistical analysis
All statistical analyses were conducted using GraphPad Prism. Data were analyzed as lognormal distributions based on D’Agostino-Pearson tests. Comparisons between subsets were analyzed using Mann-Whitney two-tailed U-tests. Correlations were assessed using Spearman rank tests. P-values <0.05 were considered significant.
Supplementary Material
Fig. S1. Representative flow cytometry plots of subset phenotypes after stimulation.
Fig. S2. MSVOA and IPDA results excluding datapoints below limit of detection.
Fig. S3. Illustration of surface marker differences on resting vs activated CD4+ subsets
Table S1. Participant characteristics.
Table S2. Frequencies of proviral copies, infection frequencies, and assay input cell numbers for samples below limits of detection.
Table S3. Frequencies of proviral copies, infection frequencies, and inducibility indices for all participants’ subsets.
Acknowledgements:
We thank Jun Lai, Subul Beg, and Mithra Kumar for their technical assistance, and Eshan Patel for assistance with statistical analyses.
Funding: This work was supported by the NIH Martin Delaney I4C (UM1 AI126603), Beat-HIV (UM1 AI126620) and DARE (UM1 AI12661) Collaboratories, by the Johns Hopkins Center for AIDS Research (P30AI094189), and by the Howard Hughes Medical Institute and the Bill and Melinda Gates Foundation (OPP1115715).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. The env sequences discussed in this publication have been deposited in GenBank under accession numbers MN441770 - MN442053.
REFERENCES
- 1.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF, Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells, Nat. Med 9, 727–728 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF, Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy, Nat. Med 5, 512–517 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF, In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency, Nat. Med 1, 1284–1290 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF, Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection, Nature 387, 183–188 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS, Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy, Proc. Natl. Acad. Sci. U. S. A 94, 13193–13197 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong JK, Hezareh M, Gunthard HF, V Havlir D, Ignacio CC, Spina CA, Richman DD, Recovery of replication-competent HIV despite prolonged suppression of plasma viremia, Science 278, 1291–1295 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF, Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy, Science 278, 1295–1300 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Siliciano JD, Siliciano RF, Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals, Methods Mol. Biol 304, 3–15 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Laird GM, Eisele EE, Rabi SA, Lai J, Chioma S, Blankson JN, Siliciano JD, Siliciano RF, Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay, PLoS Pathog. 9, e1003398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laird GM, Rosenbloom DIS, Lai J, Siliciano RF, Siliciano JD, in Methods in molecular biology (Clifton, N.J.), (Department of Medicine, Johns Hopkins University School of Medicine, 733 N. Broadway, Baltimore, MD, 21205, USA.; Department of Biomedical Informatics, Columbia University Medical Center, New York, NY, 10032, USA.; Department of Medicine, Johns (TRUNCATED, 2016), vol. 1354, pp. 239–253. [Google Scholar]
- 11.Crooks AM, Bateson R, Cope AB, Dahl NP, Griggs MK, Kuruc JD, Gay CL, Eron JJ, Margolis DM, Bosch RJ, Archin NM, Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies, J. Infect. Dis , 1–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strain MC, Günthard HF, V Havlir D, Ignacio CC, Smith DM, Leigh-Brown AJ, Macaranas TR, Lam RY, Daly OA, Fischer M, Opravil M, Levine H, Bacheler L, Spina CA, Richman DD, Wong JK, Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence., Proc. Natl. Acad. Sci. U. S. A 100, 4819–24 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenzi JCC, Cohen YZ, Cohn LB, Kreider EF, Barton JP, Learn GH, Oliveira T, Lavine CL, Horwitz JA, Settler A, Jankovic M, Seaman MS, Chakraborty AK, Hahn BH, Caskey M, Nussenzweig MC, Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA, Proc. Natl. Acad. Sci 113, E7908–E7916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosmane NN, Kwon KJ, Bruner KM, Capoferri AA, Beg S, Rosenbloom DISS, Keele BF, Ho Y-C, Siliciano JD, Siliciano RF, Proliferation of latently infected CD4 + T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics, J. Exp. Med 214, 959–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bui JK, Sobolewski MD, Keele BF, Spindler J, Musick A, Wiegand A, Luke BT, Shao W, Hughes SH, Coffin JM, Kearney MF, Mellors JW, Ross SR, Ed. Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir, PLOS Pathog. 13, e1006283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, Wells D, Su L, Luke BT, Halvas EK, Besson G, Penrose KJ, Yang Z, Kwan RW, Van Waes C, Uldrick T, Citrin DE, Kovacs J, Polis MA, Rehm CA, Gorelick R, Piatak M, Keele BF, Kearney MF, Coffin JM, Hughes SH, Mellors JW, Maldarelli F, Clonally expanded CD4 + T cells can produce infectious HIV-1 in vivo, Proc. Natl. Acad. Sci 113, 1883–1888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandergeeten C, Fromentin R, DaFonseca S, Lawani MB, Sereti I, Lederman MM, Ramgopal M, Routy JP, Sekaly RP, Chomont N, Interleukin-7 promotes HIV persistence during antiretroviral therapy, Blood 121, 4321–4329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katlama C, Lambert-Niclot S, Assoumou L, Papagno L, Lecardonnel F, Zoorob R, Tambussi G, Clotet B, Youle M, Achenbach CJ, Murphy RL, Calvez V, Costagliola D, Autran B, E.−01 study team, Treatment intensification followed by interleukin-7 reactivates HIV without reducing total HIV DNA: a randomized trial, AIDS 30, 221–230 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Wagner TA, McLaughlin S, Garg K, Cheung CYK, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM, HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection, Science 345, 570–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris a L., Mellors JW, Kearney MF, Coffin JM, Hughes SH, HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells., Science (80-. ). 345, 179–83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohn LB, Silva IT, Oliveira TY, Rosales RA, Parrish EH, Learn GH, Hahn BH, Czartoski JL, McElrath MJ, Lehmann C, Klein F, Caskey M, Walker BD, Siliciano JD, Siliciano RF, Jankovic M, Nussenzweig MC, HIV-1 integration landscape during latent and active infection, Cell 160, 420–432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM, Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults, Immunity 38, 373–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Surh CD, Sprent J, Homeostasis of naive and memory T cells, Immunity 29, 848–862 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Maeurer MJ, Lotze MT, Interleukin-7 (IL-7) knockout mice. Implications for lymphopoiesis and organ-specific immunity, Int. Rev. Immunol 16, 309–322 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Purton JF, Tan JT, Rubinstein MP, Kim DM, Sprent J, Surh CD, Antiviral CD4+ memory T cells are IL-15 dependent, J. Exp. Med 204, 951–961 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scripture-Adams DD, Brooks DG, Korin YD, Zack JA, Interleukin-7 induces expression of latent human immunodeficiency virus type 1 with minimal effects on T-cell phenotype, J. Virol 76, 13077–13082 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang FX, Xu Y, Sullivan J, Souder E, Argyris EG, Acheampong EA, Fisher J, Sierra M, Thomson MM, Najera R, Frank I, Kulkosky J, Pomerantz RJ, Nunnari G, IL-7 is a potent and proviral strain-specific inducer of latent HIV-1 cellular reservoirs of infected individuals on virally suppressive HAART, J. Clin. Invest 115, 128–137 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, Siliciano RF, Replication-Competent Noninduced Proviruses in the Latent Reservoir Increase Barrier to HIV-1 Cure, Cell 155, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fromentin R, Bakeman W, Lawani MB, Khoury G, Hartogensis W, DaFonseca S, Killian M, Epling L, Hoh R, Sinclair E, Hecht FM, Bacchetti P, Deeks SG, Lewin SR, Sékaly R-P, Chomont N, CD4+ T Cells Expressing PD-1, TIGIT and LAG-3 Contribute to HIV Persistence during ART., PLoS Pathog. 12, e1005761 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Descours B, Petitjean G, López-Zaragoza JL, Bruel T, Raffel R, Psomas C, Reynes J, Lacabaratz C, Levy Y, Schwartz O, Lelievre JD, Benkirane M, CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses, Nature 543, 564–567 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Hogan LE, Vasquez J, Hobbs KS, Hanhauser E, Aguilar-Rodriguez B, Hussien R, Thanh C, Gibson EA, Carvidi AB, Smith LCB, Khan S, Trapecar M, Sanjabi S, Somsouk M, Stoddart CA, Kuritzkes DR, Deeks SG, Henrich TJ, Douek DC, Ed. Increased HIV-1 transcriptional activity and infectious burden in peripheral blood and gut-associated CD4+ T cells expressing CD30, PLOS Pathog. 14, e1006856 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osuna CE, Lim S-Y, Kublin JL, Apps R, Chen E, Mota TM, Huang S-H, Ren Y, Bachtel ND, Tsibris AM, Ackerman ME, Jones RB, Nixon DF, Whitney JB, Evidence that CD32a does not mark the HIV-1 latent reservoir, Nature 561, E20–E28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pérez L, Anderson J, Chipman J, Thorkelson A, Chun T-W, Moir S, Haase AT, Douek DC, Schacker TW, Boritz EA, Conflicting evidence for HIV enrichment in CD32+ CD4 T cells, Nature 561, E9–E16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertagnolli LN, White JA, Simonetti FR, Beg SA, Lai J, Tomescu C, Murray AJ, Antar AAR, Zhang H, Margolick JB, Hoh R, Deeks SG, Tebas P, Montaner LJ, Siliciano RF, Laird GM, Siliciano JD, The role of CD32 during HIV-1 infection, Nature 561, E17–E19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Badia R, Ballana E, Castellví M, García-Vidal E, Pujantell M, Clotet B, Prado JG, Puig J, Martínez MA, Riveira-Muñoz E, Esté JA, CD32 expression is associated to T-cell activation and is not a marker of the HIV-1 reservoir, Nat. Commun 9, 2739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin GE, Pace M, Thornhill JP, Phetsouphanh C, Meyerowitz J, Gossez M, Brown H, Olejniczak N, Lwanga J, Ramjee G, Kaleebu P, Porter K, Willberg CB, Klenerman P, Nwokolo N, Fox J, Fidler S, Frater J, CD32-Expressing CD4 T Cells Are Phenotypically Diverse and Can Contain Proviral HIV DNA, Front. Immunol 9, 928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdel-Mohsen M, Kuri-Cervantes L, Grau-Exposito J, Spivak AM, Nell RA, Tomescu C, Vadrevu SK, Giron LB, Serra-Peinado C, Genescà M, Castellví J, Wu G, Del Rio Estrada PM, González-Navarro M, Lynn K, King CT, Vemula S, Cox K, Wan Y, Li Q, Mounzer K, Kostman J, Frank I, Paiardini M, Hazuda D, Reyes-Terán G, Richman D, Howell B, Tebas P, Martinez-Picado J, Planelles V, Buzon MJ, Betts MR, Montaner LJ, CD32 is expressed on cells with transcriptionally active HIV but does not enrich for HIV DNA in resting T cells, Sci. Transl. Med 10, 6759 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sékaly RP, Sekaly RP, HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation, Nat. Med 15, 893–900 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S, Identification of Genetically Intact HIV-1 Proviruses in Specific CD4+T Cells from Effectively Treated Participants, Cell Rep. 21, 813–822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soriano-sarabia N, Bateson RE, Dahl NP, Crooks AM, Kuruc JD, Margolis DM, Archin NM, Quantitation of Replication-Competent HIV-1 in Populations of Resting CD4+ T cells, J. Virol 88, 14070–14077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, Martin-Gayo E, Leng J, Henrich TJ, Li JZ, Pereyra F, Zurakowski R, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M, HIV-1 persistence in CD4+ T cells with stem cell–like properties, Nat. Med 20, 139–142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaafoura S, de Goër de Herve MG, Hernandez-Vargas EA, Hendel-Chavez H, Abdoh M, Mateo MC, Krzysiek R, Merad M, Seng R, Tardieu M, Delfraissy JF, Goujard C, Taoufik Y, ARTICLE Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4 þ memory T Cells, Nat. Commun 5, 5407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phetsouphanh C, Xu Y, Bailey M, Pett S, Zaunders J, Seddiki N, Kelleher AD, Ratios of effector to central memory antigen-specific CD4 + T cells vary with antigen exposure in HIV+ patients, Immunol. Cell Biol 92, 384–388 (2014). [DOI] [PubMed] [Google Scholar]
- 44.von Stockenstrom S, Odevall L, Lee E, Sinclair E, Bacchetti P, Killian M, Epling L, Shao W, Hoh R, Ho T, Faria NR, Lemey P, Albert J, Hunt P, Loeb L, Pilcher C, Poole L, Hatano H, Somsouk M, Douek D, Boritz E, Deeks SG, Hecht FM, Palmer S, Longitudinal Genetic Characterization Reveals That Cell Proliferation Maintains a Persistent HIV Type 1 DNA Pool During Effective HIV Therapy., J. Infect. Dis 212, 596–607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A, Two subsets of memory T lymphocytes with distinct homing potentials and effector functions, Nature 401, 708–712 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Sallusto F, Geginat J, Lanzavecchia A, Central memory and effector memory T cell subsets: function, generation, and maintenance., Annu. Rev. Immunol 22, 745–63 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Fritsch RD, Shen X, Sims GP, Hathcock KS, Hodes RJ, Lipsky PE, Stepwise differentiation of CD4 memory T cells defined by expression of CCR7 and CD27., J. Immunol 175, 6489–97 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Farber DL, a Yudanin N, Restifo NP, Human memory T cells: generation, compartmentalization and homeostasis., Nat. Rev. Immunol 14, 24–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahnke YD, Brodie TM, Sallusto F, Roederer M, Lugli E, The who’s who of T-cell differentiation: Human memory T-cell subsets, Eur. J. Immunol 43, 2797–2809 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Geginat J, Sallusto F, Lanzavecchia A, Cytokine-driven proliferation and differentiation of human naive, central memory, and effector memory CD4(+) T cells., J. Exp. Med 194, 1711–9 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geginat J, Campagnaro S, Sallusto F, in Lymphocyte Activation and Immune Regulation IX. Advances in Experimental Medicine and Biology, vol 512, (Springer, Boston, MA, 2011), pp. 107–112. [DOI] [PubMed] [Google Scholar]
- 52.Beverley PC, Functional analysis of human T cell subsets defined by CD45 isoform expression., Semin. Immunol 4, 35–41 (1992). [PubMed] [Google Scholar]
- 53.Pinto L, Loss of CD45RA and gain of CD45RO after in vitro activation of lymphocytes from HIV-infected patients, Immunology 73, 147–150 (1991). [PMC free article] [PubMed] [Google Scholar]
- 54.Kristensson K, Borrebaeck CAK, Carlsson R, Human CD4+ T cells expressing CD45RA acquire the lymphokine gene expression of CD45RO+ T-helper cells after activation in vitro, Immunology 76, 103–109 (1992). [PMC free article] [PubMed] [Google Scholar]
- 55.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A, Two subsets of memory T lymphocytes with distinct homing potentials and effector functions, Nature 401, 708–712 (1999). [DOI] [PubMed] [Google Scholar]
- 56.Michie CA, McLean A, Alcock C, Beverley PCL, Lifespan of human lymphocyte subsets defined by CD45 isoforms., Nature 360, 264–265 (1992). [DOI] [PubMed] [Google Scholar]
- 57.Okada R, Kondo T, Matsuki F, Takata H, Takiguchi M, Phenotypic classification of human CD4+ T cell subsets and their differentiation, Int. Immunol 20, 1189–1199 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Riou C, Yassine-Diab B, Van grevenynghe J, Somogyi R, Greller LD, Gagnon D, Gimmig S, Wilkinson P, Shi Y, Cameron MJ, Campos-Gonzalez R, Balderas RS, Kelvin D, Sekaly R-PP, Haddad EK, Convergence of TCR and cytokine signaling leads to FOXO3a phosphorylation and drives the survival of CD4+ central memory T cells, J. Exp. Med 204, 79–91 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Macallan DC, Wallace D, Zhang Y, De Lara C, Worth AT, Ghattas H, Griffin GE, Beverley PCL, Tough DF, Rapid turnover of effector-memory CD4(+) T cells in healthy humans., J. Exp. Med 200, 255–60 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pierson T, Hoffman TL, Blankson J, Finzi D, Chadwick K, Margolick JB, Buck C, Siliciano JD, Doms RW, Siliciano RF, Characterization of chemokine receptor utilization of viruses in the latent reservoir for human immunodeficiency virus type 1, J. Virol 74, 7824–7833 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shan L, Deng K, Gao H, Xing S, Capoferri AA, Durand CM, Rabi SA, Laird GM, Kim M, Hosmane NN, Yang H-C, Zhang H, Margolick JB, Li L, Cai W, Ke R, Flavell RA, Siliciano JD, Siliciano RF, Transcriptional Reprogramming during Effector-to-Memory Transition Renders CD4+ T Cells Permissive for Latent HIV-1 Infection., Immunity 47, 766–775.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, Kuo H-H, Hua S, Chen H-R, Ouyang Z, Reddy K, Dong K, Ndung’u T, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M, Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells, J. Clin. Invest 127, 2689–2696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gosselin A, Monteiro P, Chomont N, Diaz-Griffero F, Said EA, Fonseca S, Wacleche V, El-Far M, Boulassel M-R, Routy J-P, Sekaly R-P, Ancuta P, Peripheral Blood CCR4+CCR6+ and CXCR3+CCR6+ CD4+ T Cells Are Highly Permissive to HIV-1 Infection, J. Immunol 184, 1604–1616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G, Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production, J. Exp. Med 210, 143–156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murray AJ, Kwon KJ, Farber DL, Siliciano RF, The Latent Reservoir for HIV-1: How Immunologic Memory and Clonal Expansion Contribute to HIV-1 Persistence, J. Immunol 197, 407–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bosque A, Famiglietti M, Weyrich AS, Goulston C, Planelles V, Emerman M, Ed. Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells, PLoS Pathog. 7, e1002288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Imamichi H, Natarajan V, Adelsberger JW, Rehm CA, Lempicki RA, Das B, Hazen A, Imamichi T, Lane HC, Lifespan of effector memory CD4+ T cells determined by replication-incompetent integrated HIV-1 provirus, AIDS 28, 1091–1099 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Younes SA, Yassine-Diab B, Dumont AR, Boulassel MR, Grossman Z, Routy JP, Sekaly RP, HIV-1 viremia prevents the establishment of interleukin 2-producing HIV-specific memory CD4+ T cells endowed with proliferative capacity., J. Exp. Med 198, 1909–22 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hiener B, Horsburgh BA, Eden J-S, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S, Identification of Genetically Intact HIV-1 Proviruses in Specific CD4 + T Cells from Effectively Treated Participants, Cell Rep. 21, 813–822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl S. a., Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O’Doherty U, Palmer S, Deeks SG, Siliciano JD, Comparative Analysis of Measures of Viral Reservoirs in HIV-1 Eradication Studies, PLoS Pathog. 9, e1003174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Desire N, Dehee A, Schneider V, Jacomet C, Goujon C, Girard P-M, Rozenbaum W, Nicolas J-C, Quantification of Human Immunodeficiency Virus Type 1 Proviral Load by a TaqMan Real-Time PCR Assay, J. Clin. Microbiol 39, 1303–1310 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao Y, Yu M, Miller JW, Chen M, Bremer EG, Kabat W, Yogev R, Quantification of human immunodeficiency virus type 1 proviral DNA by using TaqMan technology., J. Clin. Microbiol 40, 675–8 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT, Dewar RL, Metcalf JA, Hammer S, Mellors JW, Coffin JM, Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis., J. Clin. Microbiol 43, 406–13 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Josefsson L, Palmer S, Faria NR, Lemey P, Casazza J, Ambrozak D, Kearney M, Shao W, Kottilil S, Sneller M, Mellors J, Coffin JM, Maldarelli F, Luban J, Ed. Single Cell Analysis of Lymph Node Tissue from HIV-1 Infected Patients Reveals that the Majority of CD4+ T-cells Contain One HIV-1 DNA Molecule, PLoS Pathog. 9, e1003432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, Bertagnolli LN, Capoferri AA, Kufera JT, Timmons A, Nobles C, Gregg J, Wada N, Ho Y-C, Zhang H, Margolick JB, Blankson JN, Deeks SG, Bushman FD, Siliciano JD, Laird GM, Siliciano RF, A quantitative approach for measuring the reservoir of latent HIV-1 proviruses, Nature 566, 120–125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barski A, Cuddapah S, Kartashov AV, Liu C, Imamichi H, Yang W, Peng W, Lane HC, Zhao K, Rapid Recall Ability of Memory T cells is Encoded in their Epigenome, Sci. Rep (2017), doi: 10.1038/srep39785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weng N, Araki Y, Subedi K, The molecular basis of the memory T cell response: differential gene expression and its epigenetic regulation, Nat. Rev. Immunol (2012), doi: 10.1038/nri3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Durek P, Nordström K, Gasparoni G, Salhab A, Kressler C, de Almeida M, Bassler K, Ulas T, Schmidt F, Xiong J, Glažar P, Klironomos F, Sinha A, Kinkley S, Yang X, Arrigoni L, Amirabad AD, Ardakani FB, Feuerbach L, Gorka O, Ebert P, Müller F, Li N, Frischbutter S, Schlickeiser S, Cendon C, Fröhler S, Felder B, Gasparoni N, Imbusch CD, Hutter B, Zipprich G, Tauchmann Y, Reinke S, Wassilew G, Hoffmann U, Richter AS, Sieverling L, Chang HD, Syrbe U, Kalus U, Eils J, Brors B, Manke T, Ruland J, Lengauer T, Rajewsky N, Chen W, Dong J, Sawitzki B, Chung HR, Rosenstiel P, Schulz MH, Schultze JL, Radbruch A, Walter J, Hamann A, Polansky JK, Epigenomic Profiling of Human CD4+T Cells Supports a Linear Differentiation Model and Highlights Molecular Regulators of Memory Development, Immunity 45, 1148–1161 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, Alexe G, Nagar S, McCausland MM, Gupta S, Tata P, Haining WN, McElrath MJ, Zhang D, Hu B, Greenleaf WJ, Goronzy JJ, Mulligan MJ, Hellerstein M, Ahmed R, Origin and differentiation of human memory CD8 T cells after vaccination, Nature 552, 362–367 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA, T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis, J. Virol 78, 1160–1168 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho Y-C, Richman DD, Deeks SG, Siliciano JD, Siliciano RF, Defective proviruses rapidly accumulate during acute HIV-1 infection, Nat. Med 22, 1043–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pierson T, Hoffman TL, Blankson J, Finzi D, Chadwick K, Margolick JB, Buck C, Siliciano JD, Doms RW, Siliciano RF, Characterization of Chemokine Receptor Utilization of Viruses in the Latent Reservoir for Human Immunodeficiency Virus Type 1, J. Virol 74, 7824 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boritz EA, Darko S, Swaszek L, Wolf G, Wells D, Wu X, Henry AR, Laboune F, Hu J, Ambrozak D, Hughes MS, Hoh R, Casazza JP, Vostal A, Bunis D, Nganou-Makamdop K, Lee JS, Migueles SA, Koup RA, Connors M, Moir S, Schacker T, Maldarelli F, Hughes SH, Deeks SG, Douek DC, Multiple Origins of Virus Persistence during Natural Control of HIV Infection, Cell 166, 1004–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Procopio FA, Fromentin R, Kulpa DA, Brehm JH, Bebin A-G, Strain MC, Richman DD, O’Doherty U, Palmer S, Hecht FM, Hoh R, Barnard RJO, Miller MD, Hazuda DJ, Deeks SG, Sékaly R-P, Chomont N, A Novel Assay to Measure the Magnitude of the Inducible Viral Reservoir in HIV-infected Individuals., EBioMedicine 2, 874–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Massanella M, Yek C, Lada SM, Nakazawa M, Shefa N, Huang K, Richman DD, Improved assays to measure and characterize the inducible HIV reservoir, EBioMedicine 36, 113–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Plantin J, Massanella M, Chomont N, Inducible HIV RNA transcription assays to measure HIV persistence: pros and cons of a compromise, Retrovirology 15, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pardons M, Baxter AE, Massanella M, Pagliuzza A, Fromentin R, Dufour C, Leyre L, Routy J-P, Kaufmann DE, Chomont N, Swanstrom R, Ed. Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection, PLoS Pathog. 15, e1007619 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cillo AR, Sobolewski MD, Bosch RJ, Fyne E, Piatak M, Coffin JM, Mellors JW, Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy, Proc. Natl. Acad. Sci 111, 7078–7083 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cillo AR, Hong F, Tsai A, Irrinki A, Kaur J, Sloan DD, Follen M, Geleziunas R, Cihlar T, Win SS, Murry JP, Mellors JW, Blood biomarkers of expressed and inducible HIV-1, AIDS (2018), doi: 10.1097/QAD.0000000000001748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rouzine IM, Razooky BS, Weinberger LS, Stochastic variability in HIV affects viral eradication, , doi: 10.1073/pnas.1413362111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, Tyagi M, Karn J, Epigenetic Silencing of Human Immunodeficiency Virus (HIV) Transcription by Formation of Restrictive Chromatin Structures at the Viral Long Terminal Repeat Drives the Progressive Entry of HIV into Latency, J. Virol 82, 12291–12303 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tyagi M, Pearson RJ, Karn J, Establishment of HIV Latency in Primary CD4+ Cells Is due to Epigenetic Transcriptional Silencing and P-TEFb Restriction, J. Virol 84, 6425–6437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jordan A, Defechereux P, Verdin E, The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation, EMBO J. 20, 1726 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Han Y, Lin YB, An W, Xu J, Yang H-C, O’Connell K, Dordai D, Boeke JD, Siliciano JD, Siliciano RF, Orientation-Dependent Regulation of Integrated HIV-1 Expression by Host Gene Transcriptional Readthrough, Cell Host Microbe 4, 134–146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lenasi T, Contreras X, Peterlin BM, Transcriptional Interference Antagonizes Proviral Gene Expression to Promote HIV Latency, Cell Host Microbe 4, 123–133 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gallastegui E, Millan-Zambrano G, Terme J-M, Chavez S, Jordan A, Chromatin Reassembly Factors Are Involved in Transcriptional Interference Promoting HIV Latency, J. Virol 85, 3187–3202 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rosenbloom DIS, Elliott O, Hill AL, Henrich TJ, Siliciano JM, Siliciano RF, Designing and Interpreting Limiting Dilution Assays: General Principles and Applications to the Latent Reservoir for Human Immunodeficiency Virus-1., Open forum Infect. Dis 2, 018911 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Representative flow cytometry plots of subset phenotypes after stimulation.
Fig. S2. MSVOA and IPDA results excluding datapoints below limit of detection.
Fig. S3. Illustration of surface marker differences on resting vs activated CD4+ subsets
Table S1. Participant characteristics.
Table S2. Frequencies of proviral copies, infection frequencies, and assay input cell numbers for samples below limits of detection.
Table S3. Frequencies of proviral copies, infection frequencies, and inducibility indices for all participants’ subsets.